Abstract
Bacterial biofilms are the most prevalent mode of bacterial growth in nature. Adhesive and viscoelastic properties of bacteria play important roles at different stages of biofilm development. Following irreversible attachment of bacterial cells onto a surface, a biofilm can grow in which its matrix viscoelasticity helps to maintain structural integrity, determine stress resistance, and control ease of dispersion. In this study, a novel application of force spectroscopy was developed to characterize the surface adhesion and viscoelasticity of bacterial cells in biofilms. By performing microbead force spectroscopy with a closed-loop atomic force microscope, we accurately quantified these properties over a defined contact area. Using the model gram-negative bacterium Pseudomonas aeruginosa, we observed that the adhesive and viscoelastic properties of an isogenic lipopolysaccharide mutant wapR biofilm were significantly different from those measured for the wild-type strain PAO1 biofilm. Moreover, biofilm maturation in either strain also led to prominent changes in adhesion and viscoelasticity. To minimize variability in force measurements resulting from experimental parameter changes, we developed standardized conditions for microbead force spectroscopy to enable meaningful comparison of data obtained in different experiments. Force plots measured under standard conditions showed that the adhesive pressures of PAO1 and wapR early biofilms were 34 ± 15 Pa and 332 ± 47 Pa, respectively, whereas those of PAO1 and wapR mature biofilms were 19 ± 7 Pa and 80 ± 22 Pa, respectively. Fitting of creep data to a Voigt Standard Linear Solid viscoelasticity model revealed that the instantaneous and delayed elastic moduli in P. aeruginosa were drastically reduced by lipopolysaccharide deficiency and biofilm maturation, whereas viscosity was decreased only for biofilm maturation. In conclusion, we have introduced a direct biophysical method for simultaneously quantifying adhesion and viscoelasticity in bacterial biofilms under native conditions. This method could prove valuable for elucidating the contribution of genetic backgrounds, growth conditions, and environmental stresses to microbial community physiology.
Introduction
Biofilms are sessile microbial communities growing on surfaces and encased in matrices composed of extracellular polymers (1). Because of their persistence and ubiquity, bacterial biofilms have particularly profound impact on human health, the environment, and industrial systems. Bacterial biofilms have been shown to adhere strongly to interfaces and to behave as viscoelastic materials (2). During the initial stage of biofilm formation (designated as “early biofilm”), the adhesive properties of a collection of bacterial cells cause irreversible attachment to a colonizable surface. Once a biofilm is established and matures (designated as “mature biofilm”), exopolymeric substances are produced, and the viscoelastic properties of the resultant matrix determine its structural integrity, resistance to stresses, and ease of dispersion. Since the biofilm forming ability of a bacterium has often been linked to persistence and virulence, a thorough understanding of how adhesion and viscoelasticity modulate biofilm establishment may be important for the proper design of control strategies.
Atomic force microscopy (AFM) was originally developed by Binnig et al. in 1986 (3) and has evolved into a sophisticated technique for high-resolution imaging of surfaces and for measurement of their mechanical properties. Because of the ability to examine nonconductive surfaces under native conditions in air or in fluids, AFM quickly distinguishes itself as the most powerful and versatile scanning probe technique available for characterizing soft materials and biological samples. In microbiological research, AFM has routinely been employed to image properly-immobilized microbial samples immersed in liquids at nanometre resolution (4). AFM has also been used for elucidating the physical properties of microbial cells through force measurements with piconewton sensitivity (4,5). Since samples were often examined in their native state, the requirement for sample preparation is minimal, thus greatly reducing the potential for artifacts. On a slightly larger scale, AFM can be used to obtain high-resolution images and mechanical properties of microbial biofilms. For instance, Oh et al., using AFM, observed that Escherichia coli biofilms developed faster in a low nutrient medium and became more adhesive to a cantilever tip as they matured while their elastic properties varied across cell surfaces (5).
Bacterial adhesion in the context of biofilm formation has previously been examined using biophysical techniques such as optical tweezers (OT), total internal reflection fluorescence (TIRF) microscopy, surface plasma resonance (SPR) and quartz crystal microbalance (QCM) (6,7), but none of these methods rival the flexibility and sensitivity of AFM for directly probing nanoscale interaction forces. A number of quantitative studies of bacterial adhesion by AFM have been reviewed and summarized (Table S1 in the Supporting Material). Methods have varied from using regular tips interacting with cells (6), to tips coated with cells interacting with different surfaces (7–11), to modified tips interacting with cells (12,13), to colloid probes interacting with cells (14). To date, however, no AFM methodology has combined the flexibility of tips coated with cells for examining bacterial interaction with multiple surfaces and the quantifiable contact areas afforded by the use of spherical probes. Additionally, although adhesive measurements clearly depend on experimental conditions such as loading pressure, retraction speed and contact time, no attempts have yet been made to standardize these conditions used in force spectroscopy for comparison of data obtained in different experiments.
The matrix viscoelasticity of a mature biofilm determines its resistance to stress and its eventual dispersal mechanism. Like other viscoelastic materials, biofilms exhibit a mixture of elastic and viscous properties (15). A number of quantitative viscoelastic studies of intact biofilms using different methods have been reviewed and summarized (Table S2). Some studies examined shear stresses with flow cells combined with light microscopy (2,16–19) or rotational and oscillatory rheometry (20–24), whereas others looked at compressional stresses using film rheometry (25) or microindentation (26). Recently, the use of nanoindentation to measure the viscoelastic response of polymeric substances at the microscale had been validated by Lu et al., who compared nanoindentation data to results obtained using conventional techniques (27). In a nanoindentation experiment, viscoelastic materials have a time-dependent stress-strain relation that manifests as a creep response involving an increase in depth (strain) at a constant applied load (stress). This phenomenon of creep can be modeled using conventional linear spring and dashpot elements to obtain the viscoelastic properties of the material (28). Although the use of AFM in phase imaging or force modulation mode for qualitative differentiation of bacterial viscoelastic properties had been attempted for a number of years (29), the adaptation of AFM force spectroscopy for quantitative analysis of bacterial viscoelasticity by nanoindentation is a recent innovation (30) that has yet to be applied to studying bacterial biofilms.
In this study, we report a novel application of AFM force spectroscopy, coined “microbead force spectroscopy” (MBFS), for measuring the adhesive and viscoelastic properties of bacterial biofilms quantitatively and reproducibly. In MBFS, a 50-μm diameter glass bead attached to a tipless AFM cantilever is coated with a bacterial biofilm and brought into brief contact with a clean glass surface using a closed-loop AFM instrument to accurately gather force versus distance data over time (Fig. 1). This approach essentially combines the defined contact geometry of a spherical probe (14,31–33) with the sample flexibility obtained by using an AFM tip coated with cells (7–11). Adhesive properties of biofilm cells were readily deduced from the force versus separation plots during the retraction, whereas their viscoelastic properties could be determined from the indentation versus time plots during the hold period on the interacting surface (34). Because of its ease of manipulation, its natural ability to grow as biofilms, its genetic tractability, and the availability of genome sequences, Pseudomonas aeruginosa is the model gram-negative biofilm organism of choice in the microbiological literature. We therefore aimed to accurately measure the adhesion between bacterial cells in biofilms and a glass surface, and to quantify biofilm viscoelasticity for comparative analysis. Our study shows that P. aeruginosa wild-type strain PAO1 biofilms possess very different adhesive and viscoelastic properties when compared with those of its isogenic mutant strain wapR, which has defective expression of lipopolysaccharides (LPS) on its cell envelope (35,36). We also discovered that maturation of biofilms resulted in changes in these mechanical properties. Since MBFS is highly amenable to the standardization of conditions for data acquisition, routine comparison of measurements performed on different samples is possible. Additionally, the ability to remove and reattach microbead probes for repeated usages can lead to much time and cost savings.
Figure 1.
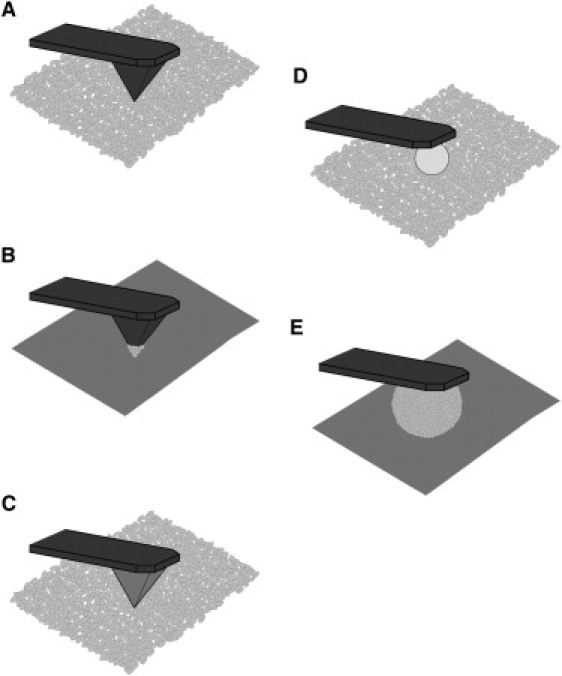
A comparison of microbead force spectroscopy to other tip-surface configurations. (A) Native tip on cells. (B) Cell probe on surfaces. (C) Modified tip on cells. (D) Colloid probe on cells. (E) Microbead biofilm probe on surfaces.
Materials and Methods
Bacterial strains and growth
P. aeruginosa wild-type strain PAO1 and mutant strain wapR were used in this study. PAO1 is a serotype O5 strain according to the International Antigen Typing Scheme (IATS) and its genome has been sequenced and annotated (35). Strain wapR is an isogenic mutant of PAO1 with the rhamnosyltransferase gene, wapR (PA5000), disrupted by a gentamicin-resistance cassette, and the disrupted gene was incorporated into the chromosome by allelic replacement to generate a nonpolar knockout (36). The phenotype of the wapR mutant shows a truncation of the LPS core oligosaccharide and the absence of O-antigen. P. aeruginosa cells were grown overnight (16 h) in Trypticase Soy Broth (TSB) (Becton, Dickinson and Company, Sparks, MD) at 37°C with shaking at 125 rpm. Bacterial cells were harvested by centrifugation at 2300 × g for 5 min, and the pellets were resuspended in sterile deionized water and centrifuged. After a second wash in deionized water, 10-fold dilutions were made, and each diluted cell suspension was measured for its optical density at 600 nm (OD600). Finally, the washed cells were adjusted to an OD600 of 2.0 for use in force spectroscopy.
Calibration of AFM cantilevers for force spectroscopy
Force measurements were performed using an MFP-3D atomic force microscope (Asylum Research, Santa Barbara, CA). This instrument was controlled using the MFP-3D software version 070111+217 provided by the manufacturer and operating within the Igor Pro 6.02A software environment (Wavemetrics, Lake Oswego, OR). For absolute and simultaneous quantitation of adhesion and viscoelasticity in bacterial biofilms, rectangular tipless silicon cantilevers CSC12/Tipless/No Al Type E (Mikromasch USA, San Jose, CA), with manufacturer's quoted resonance frequencies of 10 kHz (range: 7–14 kHz) and force constants of 0.03 N/m (range: 0.01–0.08 N/m) were used. We used the thermal method of Hutter and Bechhoefer (37) to obtain an accurate value of the spring constant for each individual cantilever. Cantilevers with a calibrated spring constant outside of the 0.015–0.060 N/m range (i.e., from half to twice the quoted force constant), or with a resonant frequency outside of 8–12 kHz, were rejected from use.
Preparation of early biofilms on glass beads attached to cantilevers
To prepare early biofilms for MBFS, microsized glass beads with diameters of 50 μm (Polysciences, Inc., Warrington, PA) were attached to the distal ends of the cantilevers with two-component epoxy glue using a micromanipulator and dried at room temperature overnight (Fig. 2). Beaded cantilevers were then coated with 0.01% poly-d-lysine and allowed to dry for 10 min. Finally, overnight bacterial cultures grown, washed and adjusted as detailed above were applied to the poly-d-lysine-coated beads three times to ensure complete bead coverage with bacteria before immediate use in MBFS measurements (Fig. 2 A).
Figure 2.
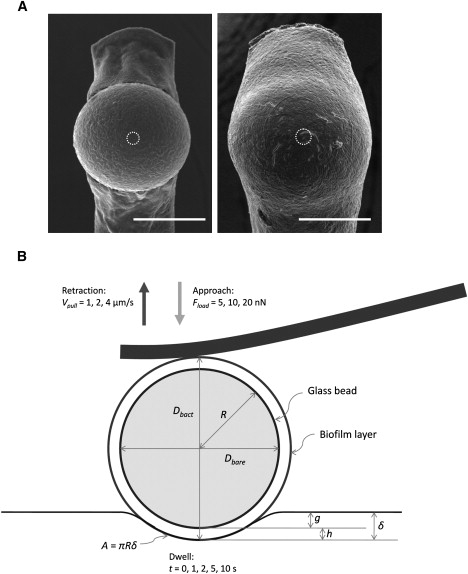
Bacterial biofilm-coated glass beads used in microbead force spectroscopy. (A) Scanning electron micrographs of early biofilm-(left) and mature biofilm-(right) coated glass beads that were attached to cantilevers for microbead force spectroscopy, taken after adhesion and viscoelasticity measurements. Typical area of contact is indicated by the white dashed circle at the center of each image. Average thicknesses of early and mature biofilms were ∼0.5 μm and 3 μm, respectively (Scale bars: 30 μm). (B) Schematic diagram illustrating microbead contact geometry during the hold period of force spectroscopy.
Growth of mature biofilms on beaded cantilevers
To prepare mature biofilms for MBFS, a biofilm drip reactor system was devised to culture bacteria on the surfaces of glass beads attached to cantilevers. The setup consisted of a 6-L media reservoir flask containing sterile 10-fold diluted TSB (dTSB) with four stretches of silicone tubing (VWRBrand Select Silicone, size: 0.062″ inner diameter × 0.125″ outer diameter × 0.032″ wall thickness; VWR International, Mississauga, ON, Canada) leading out of it and threaded through a peristaltic pump (Minipuls 2; Gilson, Inc., Middleton, WI) into four 1-L Erlenmeyer flasks. An aliquot of the microsized glass beads were first precoated with 0.01% poly-d-lysine by rinsing in the solution and drying in air on a Petri dish. The beads were then attached to the same tipless silicon cantilevers with epoxy glue and allowed to cure overnight as previously done. Subsequently, each cantilever-bead assembly was suspended from silicone tubing at the mouth of a collection flask, slightly below the drilled stopper through which the tubing from the reservoir was fed. Beaded cantilevers were connected to the ends of the silicone tubings, bead-side down, via adaptors consisting of precut 200-μL polystyrene micropipette tips.
Before we initiated operation of the drip reactor system to culture biofilms, tubings and cantilevers were sterilized by flowing 75% ethanol from a 1-L sterilizing flask through the system at 1 mL/min overnight. The source of flow was then switched to bacterial medium by disconnecting the tubing from the sterilizing flask and reconnecting it to the media reservoir flask aseptically. Following the switch, dTSB was allowed to flow for 30 min at the same rate to push out all of the ethanol. Finally, the cantilever beads were inoculated with P. aeruginosa by briefly submerging the chip end three times in overnight bacterial cultures that were grown, washed and adjusted as described earlier. Flow of dTSB was immediately resumed at 0.1 mL/min to wash off bacterial cells not attaching to the polylysine-coated bead. The biofilms were allowed to grow for 72 h before they were harvested and rinsed in sterile deionized water before immediate use in MBFS measurements (Fig. 2 A).
Light microscopy of coated glass beads attached to cantilevers
To ensure proper coverage of glass beads prior to MBFS experiments, bright field images of coated beads were captured in air immediately after bacterial application using an Eclipse TE2000-E inverted microscope (Nikon Canada Inc., Mississauga, ON, Canada), situated directly under the MFP-3D atomic force microscope. Light micrographs were also used to estimate bead radius for calculating surface area of contact (Fig. 2 B).
Microbead force spectroscopy: experimental regimes for force curve collection
After loading the cantilever onto the head of the MFP-3D atomic force microscope and centering the infrared laser spot behind the bead, the cantilever assembly was immersed in 200 μL of sterile deionized water on a precleaned glass slide. The microbead was lowered gradually to approach the glass surface, and force-separation plots (i.e., force plots) were gathered under two regimens. In variable-push experiments, sets of 10 force plots each were collected at 5, 10, and 20 nN loading force (Fload, i.e., “trigger point”) with zero contact time (t, i.e., “dwell time”) and a constant approach (Vpush) and retract velocity (Vpull) at 2 μm/s. This series was then repeated four more times with contact time t varied to 1, 2, 5, and 10 s. In variable-pull experiments, 10 force-distance curves each were collected at retraction velocities Vpull of 1, 2, and 4 μm/s with zero contact time and a constant approach velocity Vpush of 2 μm/s and a loading force of 10 nN. This series was then repeated four more times with the contact time t varied to 1, 2, 5 and 10 s. Under either experimental regime, a total of 150 force plots were collected per experiment. Three independent experiments were performed for each biofilm sample, as well as for the control (in which bare glass beads were pressed onto glass surfaces), under each regime. Blinded experiments were carried out in which bacterial strains were assigned numbers unknown to the experimenter, and strain-number correlations were revealed after all experiments were completed.
Cleaning and regeneration of cantilever tips
Routine cleaning of cantilevers to regenerate used tips for MBFS was performed using the following protocol. Chemical reagents were obtained from Fisher Scientific (Fair Lawn, NJ). Briefly, Piranha solution (70% sulfuric acid, 9% hydrogen peroxide) was prepared using the method of Lo et al. by slowly adding 3 mL 30% hydrogen peroxide to 7 mL concentrated sulfuric acid in a clean 20-mL capped glass vial (38). Each cantilever chip was immersed in 1 mL of this solution in a separate capped glass vial for 18 h, then rinsed by submersion in sterile deionized water in a glass Petri dish for 5 min, and finally rinsed briefly in anhydrous ethanol and air dried on Grade 1 filter paper (Whatman plc, Kent, UK). The cleaned cantilever tips were examined under optical microscopy to ensure the attached glass beads had become detached from the cantilever after such treatment, and that the cantilever bodies were devoid of organic debris. Finally, the regenerated cantilevers were recalibrated and new glass beads reattached as described previously.
Force plot analysis for adhesive properties
Under precisely controlled conditions for MBFS as detailed above, the adhesive properties of bacterial biofilms can be readily deduced from force plots and creep curves (Fig. 3). For routine data analysis, it is convenient to distinguish three classes of experimental quantities: controlled, measured and calculated quantities. Controlled quantities included the loading force Fload, the approach and retraction velocities Vpush and Vpull, as well as the contact time t. Measured quantities included the adhesive force Fad, the instantaneous indentation g, and the creep indentation h. Calculated quantities included the total indentation δ, the contact radius a, the contact area A, the loading pressure Pload, the adhesive pressure Pad, and the adhesive efficiency €ad.
Figure 3.
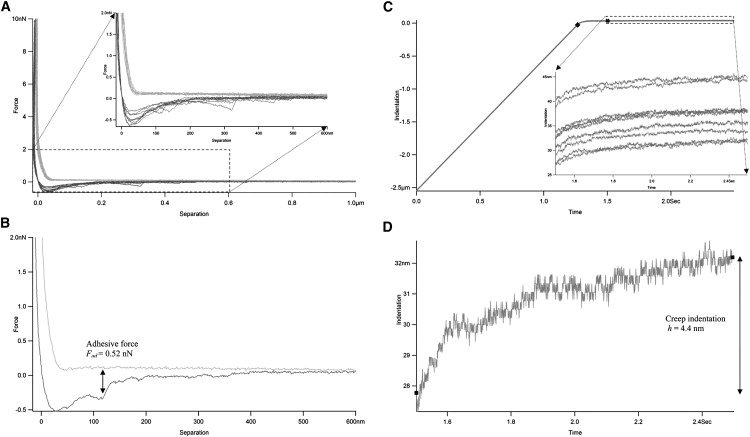
Determination of biofilm adhesion and creep indentation by microbead force spectroscopy. (A) Representative force-separation plots (force plots) during approaches (light lines) and retractions (dark lines) in MBFS experiments. Inset shows more details of the expanded interaction region. Negative separation resulted from sample indentation. (B) Measurement of adhesive force from a sample force plot. (C) Representative indentation-time curves (creep curves) during dwell in MBFS experiments. Inset shows more details of the expanded creep region. Negative indentation indicated the ramp region before sample contact. (♦) Beginning of contact and instantaneous indentation; (#) End of instantaneous indentation and beginning of delayed or creep indentation. (D) Measurement of creep indentation from a sample creep curve. (■) Start and end of the creep curve.
Representative force plots in MBFS consist of approach and retraction curves (Fig. 3 A). The adhesive force Fad is measured as the difference in force experienced by the cantilever between the base level of the retraction curve and its lowest point on a force plot (Fig. 3 B). Representative creep curves in MBFS during the dwell period (i.e., the contact time after approach but before retraction) show that creep begins where instantaneous indentation ends, and that it increases over time (Fig. 3 C). The creep indentation h is taken as the difference in indentation experienced by the sample between the beginning and the end of the dwell period (Fig. 3 D).
To estimate the thickness y of the bacterial layer and the surface area A of contact, the diameter of each attached glass bead before (Dbare) and after (Dbact) sample application were measured from optical micrographs taken at 600× magnification with the bead edge in focus. To obtain better estimates of bead dimensions, measurements were made along two perpendicular directions and averaged. Half of the difference between Dbare and Dbact was taken as thickness y of the bacterial layer on the bead surface, while half of Dbact gave an estimate of the coated bead radius R. An estimate of the actual contact area can be derived from Hertz's theory of spherical contact (39), which relates the indentation δ to the contact radius a and the radius of the spherical indenter R as:
| (1) |
Therefore, the actual contact area A at maximum load can be calculated as:
| (2) |
bearing in mind that contact area A and indentation δ are both functions of time. The indentation δ is calculated by adding its instantaneous component g, equivalent to the cantilever deflection x at the end of the approach curve, to its delayed component h, or the creep indentation:
| (3) |
where k is the cantilever spring constant. The loading pressure Pload and adhesive pressure Pad are related to the loading force Fload and adhesive force Fad, respectively, by the contact area A:
| (4) |
| (5) |
Finally, the adhesive efficiency €ad can be defined as the ratio between Fad and Fload:
| (6) |
By establishing standardized conditions, the adhesive properties derived from different MBFS experiments can be meaningfully compared. The standard loading force Fload, contact time t and approach and retraction velocities Vpush and Vpull (SFTV) used in our experiments were as follows: Fload = 10 nN, t = 1 s, Vpush = Vpull = 2 μm/s. The optimal distance from the surface at the start of approach and end of retraction (i.e., “ramp distance”) was found to be 3 μm. For each experimental regime (i.e., variable-push or variable-pull), measured and calculated quantities from three independent experiments (3 × 150 = 450 force plots in total) were averaged and graphed to show trends for each sample. As for data derived under SFTV conditions, results from six independent experiments (60 force plots in total) were averaged and tabulated for comparison between the four bacterial biofilm samples.
Creep curve fitting for viscoelastic properties
Viscoelastic properties of bacterial biofilms can be extracted from the fitting of MBFS experimental creep data to an equation derived from theoretical models. In these mechanical analogs, springs and dashpots represent elastic and viscous properties of the systems, respectively. One particularly useful model for viscoelastic materials is the Voigt version of the Standard Linear Solid (SLS) model (40), which consists of a spring in series with a spring-dashpot Voigt element in parallel (Fig. 4 A). In this three-element model, the compression distance d as a function of time can be calculated as:
| (7) |
where F is the loading force, k1 and k2 are stiffness of the springs representing elasticity, and λ is the damping coefficient of the dashpot representing viscosity. However, since the biofilm-glass interaction in our experiment occurs over a finite area, not at a single point, we can improve our model by rewriting this equation in terms of strain, stress and modulus. The analogous equation describing strain ɛ as a function of time is:
| (8) |
where σ is the stress, E1 and E2 are the instantaneous and delayed elastic moduli, respectively, and η is the viscosity in the model. Since stress σ (loading force over a unit area; unit: N/m2 or Pa) and strain ɛ (change in length divided by the original length; unitless) are related by modulus E or compliance D, i.e.,
| (9) |
Figure 4.
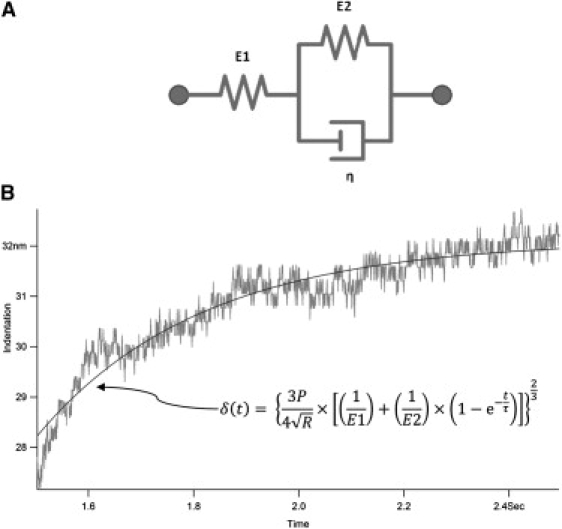
Determination of biofilm viscoelasticity by microbead force spectroscopy. (A) The three-element Voigt Standard Linear Solid model for viscoelastic materials. (B) Fitting of creep plots to the Standard Linear Solid model to derive viscoelastic properties. The model is described by Eq. 12. E1: instantaneous elasticity. E2: delayed elasticity. η: viscosity.
Eq. 8 can be rewritten as:
| (10) |
where D is the creep compliance and E is the combined elastic modulus of the system. According to the Hertzian theory of contact between a sphere and a plane (41), indentation δ is related to the elastic modulus E, the loading force P and the radius of the spherical indenter R by:
| (11) |
Substituting Eq. 10 into Eq. 11 gives:
| (12) |
where τ is the retardation time, the time at which ∼63% of creep has occurred, and is related to E2 and η by:
| (13) |
In a general sense, the retardation time marks the initial interval in which rapid creep occurs after a constant stress is applied in a creep test, just as relaxation time τ′ delineates the initial period of rapid relaxation as a constant strain is maintained in a stress-relaxation experiment. The latter can be calculated from elastic moduli and viscosity as:
| (14) |
For meaningful comparison between the results of different experiments, only creep curves gathered under SFTV conditions established above were used for fitting. Before fitting, creep curves were shifted so that δ = 0 coincided with the start of contact and t = 0 coincided with the start of creep. Using a nonlinear regression algorithm within the Igor Pro environment, creep curves were fitted to Eq. 12 with P = Fload set to 10 nN and R held at 25 or 28 μm (for early and mature biofilms, respectively) to derive E1, E2, and τ (Fig. 4 B). Parameters derived from fitting 10 curves each in three independent experiments (30 creep curves in total) were averaged and tabulated for comparison between the four P. aeruginosa biofilm samples.
Scanning electron microscopy
Scanning electron microscopy was performed to examine used MBFS beaded cantilevers with early or mature biofilm coatings. Briefly, used cantilevers were coated with gold for 2 min using an Emitech K550 sputter coater (Emitech, Ashford, Kent, UK) and imaged with a Hitachi S-570 scanning electron microscope (Hitachi, Tokyo, Japan). Images were captured digitally using the Quartz PCI imaging software (Quartz Imaging Corp., Vancouver, BC, Canada).
Results
Quantitation of adhesive properties of P. aeruginosa cells by MBFS
Adhesive force and total indentation measurements were performed on early and mature biofilms of P. aeruginosa PAO1 wild-type strain and the wapR mutant. From these results, histograms of adhesive force distribution were plotted (Fig. 5), and both adhesive pressure and adhesive efficiency were calculated (Table 1). The frequency distribution of P. aeruginosa adhesive forces to glass under SFTV condition revealed that early biofilms of strain PAO1 exhibited adhesive forces of 0 to 3 nN (average Fad = 0.66 ± 0.27 nN), while those of the LPS defective mutant wapR adhered with forces that were approximately an order of magnitude larger, ranging from 2 to 13 nN (average Fad = 6.9 ± 1.2 nN) (Fig. 5, A and B). When biofilms were allowed to mature for 3 days, however, both strains became less adherent to glass. The adhesive forces of PAO1 mature biofilms dropped slightly to 0 to 2 nN (average Fad = 0.56 ± 0.30 nN), whereas those of the wapR mature biofilms decreased significantly to a range of 0 to 3 nN (average Fad = 1.4 ± 0.2 nN) (Fig. 5, C and D).
Figure 5.
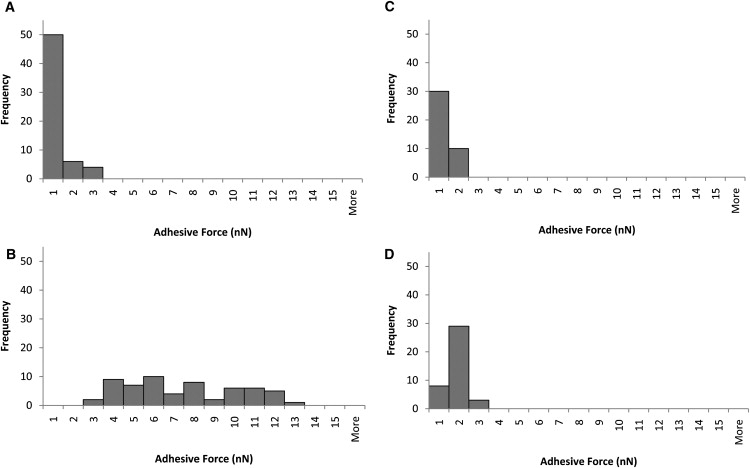
Frequency distributions of Pseudomonas aeruginosa adhesive forces to glass under standard conditions. (A) PAO1 early biofilm. (B) wapR early biofilm. (C) PAO1 mature biofilm. (D) wapR mature biofilm. Histograms show data derived from six independent experiments per early biofilm sample and four independent experiments per mature biofilm sample. Each experiment consists of 10 replicate force plots at standard loading force (10 nN), contact time (1 s), and ramp velocity (2 μm/s) (SFTV).
Table 1.
Summary of bacterial adhesive properties measured by MBFS under SFTV
| Sample | PAO1 early biofilm | wapR early biofilm | PAO1 mature biofilm | wapR mature biofilm |
|---|---|---|---|---|
| Adhesive force Fad (nN) | 0.66 ± 0.27 | 6.9 ± 1.2 | 0.56 ± 0.30 | 1.4 ± 0.2 |
| Fold difference | 1 | 10.49 | 0.85 | 2.17 |
| Adhesive pressure Pad (Pa) | 34 ± 15 | 332 ± 47 | 19 ± 7 | 80 ± 22 |
| Fold difference | 1 | 9.64 | 0.56 | 2.34 |
| Adhesive efficiency €ad | 6.6% | 69% | 5.6% | 14% |
Means and standard errors of data shown from six independent experiments for early biofilms and from four independent experiments for mature biofilms. Each experiment consists of 10 replicate force plots under standard load force (10 nN), contact time (1 s), and ramp velocity (2 μm/s). Because loading forces under SFTV were fixed at 10 nN, adhesive efficiencies calculated were directly proportional to adhesive forces.
Adhesive pressure and adhesive efficiency are measures of adhesion that take into account differences in contact area and loading force, respectively. When the average adhesive pressures of the four samples were compared (Table 1), it is apparent that the trends were similar to those observed for adhesive forces. The average adhesive pressure of PAO1 biofilms was reduced from 34 ± 15 Pa to 19 ± 7 Pa during maturation, whereas the Pad of the wapR biofilms dropped about fourfold from 332 ± 47 Pa down to 80 ± 22 Pa in the same period. Correspondingly, adhesive efficiency dropped from 6.6% to 5.6% for PAO1 biofilms and from 69% to 14% for wapR biofilms within 3 days of maturation.
Effect of loading force, contact time and retraction velocity on the measurement of adhesive properties in bacterial biofilms
As already alluded to in the previous section, quantitation of adhesive properties by MBFS or any other method depends on experimental conditions such as loading force, contact time and ramp velocity. We therefore examined how the measured adhesion in terms of adhesive force, pressure and efficiency were influenced by these factors. The dependence of P. aeruginosa adhesion to glass on loading force and retraction velocity can be illustrated by data from variable-push (Fig. 6) and variable-pull experiments (Fig. 7), respectively, while, its dependence on contact time was demonstrated by the data from both kinds of experiments (Fig. 8). Inverse relations are also illustrated by plots of the inverse of the measured quantity against the controlled quantity (see Fig. 6, B and C, insets). The equation and R-squared value in the upper right corner of each graph were those for the linear best fit line through each of the wapR mutant early biofilm data set, since this sample showed the clearest trends among all of the samples. The adhesive force Fad was moderately correlated to loading force (slope of Fad versus Fload = 0.16; Fig. 6 A), almost unrelated to retraction velocity (slope of Fad versus Vpull = 0.0006 nN·s/μm; Fig. 7 A), and strongly proportional to contact times (slope of Fad versus t = 0.80 nN/s; Fig. 8 A). The adhesive pressure Pad, on the other hand, showed an inverse correlation with loading force (slope of 1/Pad versus Fload = 0.00020 Pa−1nN−1; Fig. 6 B), but was weakly tied to the retraction velocity (slope of Pad versus Vpull = −2.26 Pa·s/μm; Fig. 7 B), and strongly dependent on contact time (slope of Pad versus t = 52.87 Pa/s; Fig. 8 B). The adhesive efficiency €ad was inversely proportional to loading force (slope of 1/€ad versus Fload = 0.082 nN−1; Fig. 6 C), weakly linked to retraction velocity (slope of €ad vesus Vpull = 5.965 × 10−5 s/μm; Fig. 7 C), but directly proportional to contact time (slope of €ad versus t = 0.098 s−1; Fig. 8 C). In general, contact time was the most important factor in determining biofilm adhesion, followed by loading force, with retraction velocity not having significant influence. Moreover, the total indentation and the contact area are both directly proportional to the loading force (slope of δ versus Fload = 34.63 nm/nN, slope of A versus Fload = 3 × 10−12 m2/nN; Fig. 9, A and B). For further details regarding how different experimental conditions affected measurements, additional data have been graphed and included in the Supporting Material.
Figure 6.
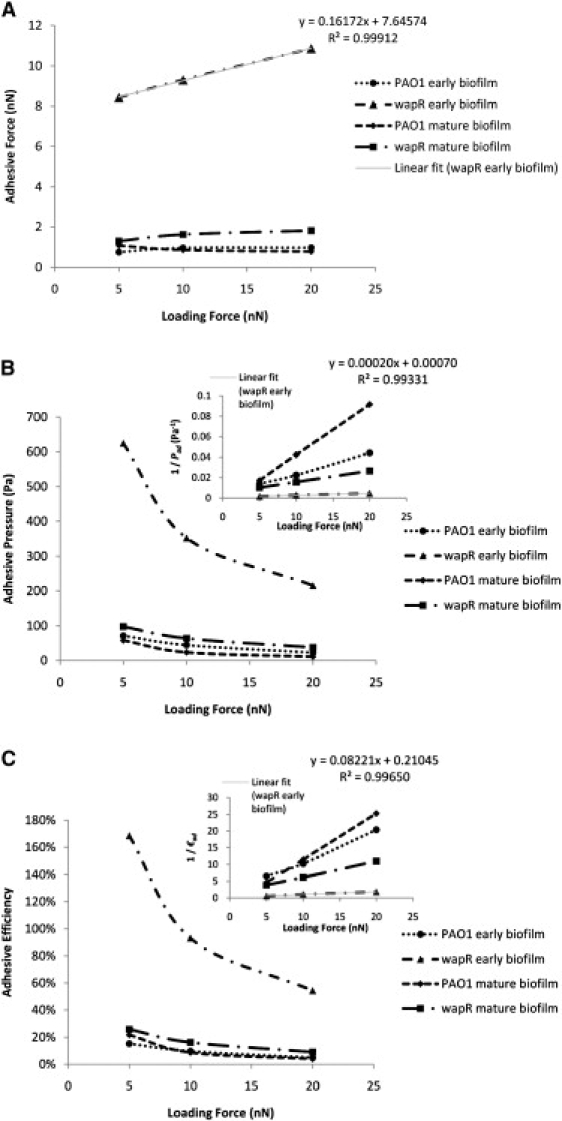
Loading force dependence of Pseudomonas aeruginosa adhesion to glass—variable-push experiments. (A) Adhesive force. (B) Adhesive pressure. (C) Adhesive efficiency. Means and trends of data are shown from three independent experiments per early biofilm sample and two independent experiments per mature biofilm sample. Each experiment consists of three sets of 10 replicate force plots, one set for each loading force setting, with contact time held at 1 s and ramp velocity at 2 μm/s.
Figure 7.
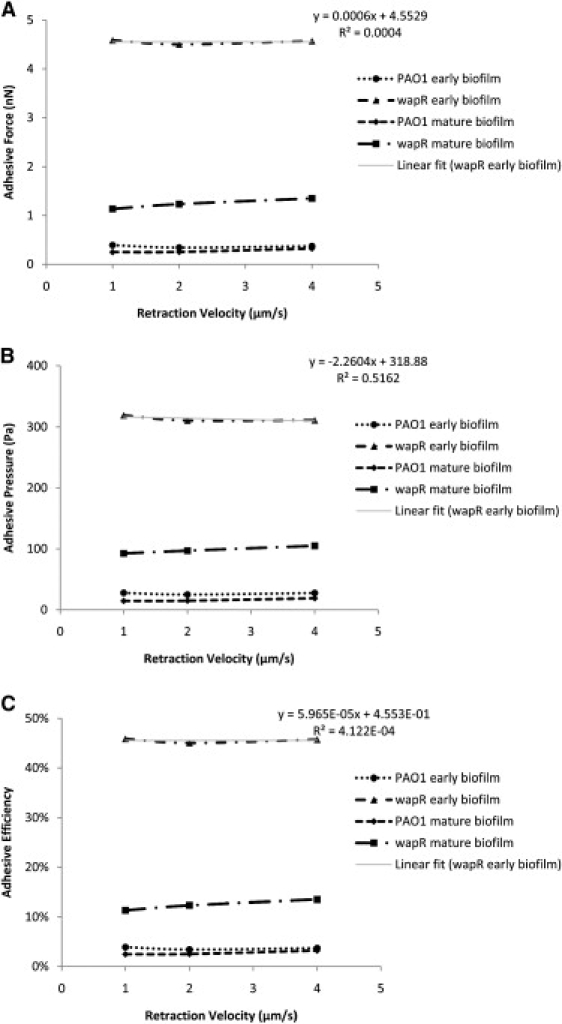
Retraction velocity dependence of Pseudomonas aeruginosa adhesion to glass—variable-pull experiments. (A) Adhesive force. (B) Adhesive pressure. (C) Adhesive efficiency. Means and trends of data are shown from three independent experiments per early biofilm sample and two independent experiments per mature biofilm sample. Each experiment consists of three sets of 10 replicate force plots, one set for each retraction velocity setting, with contact time set at 1 s and loading force at 10 nN.
Figure 8.
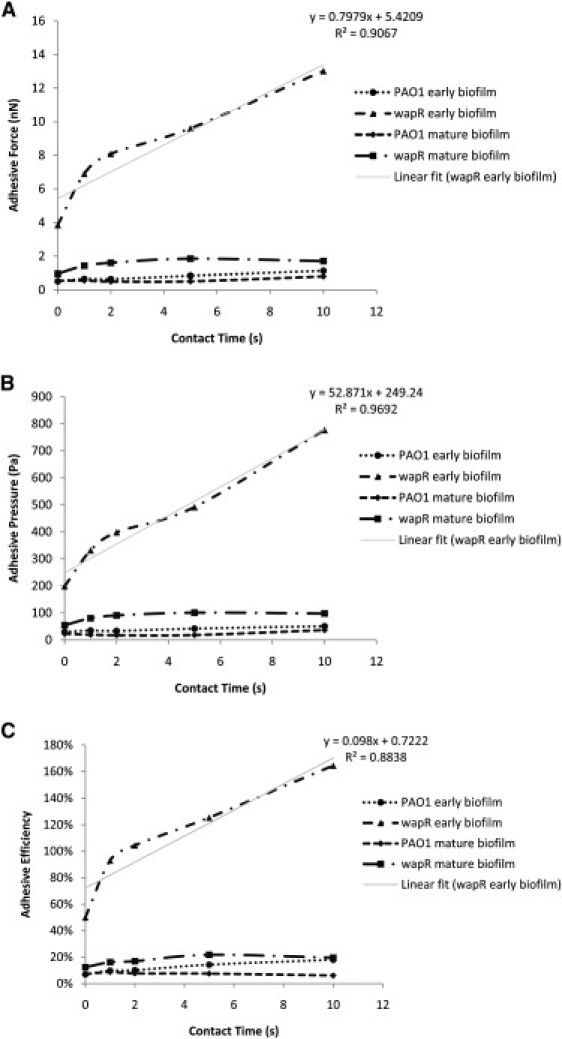
Contact time dependence of Pseudomonas aeruginosa adhesion to glass—variable-push and pull experiments. (A) Adhesive force. (B) Adhesive pressure. (C) Adhesive efficiency. Means and trends of data are shown from six independent experiments per early biofilm sample and four independent experiments per mature biofilm sample. Each experiment consists of five sets of 10 replicate force plots, one set for each contact time setting, with loading force set at 10 nN and retraction velocity at 2 μm/s.
Figure 9.
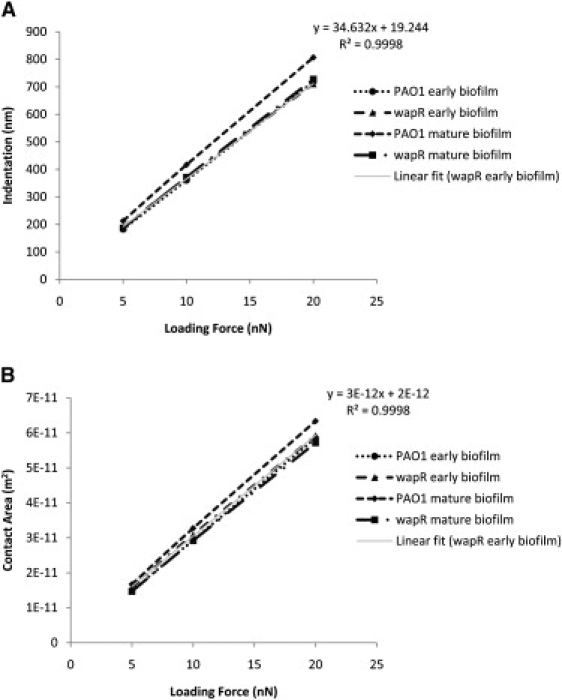
Loading force dependence of Pseudomonas aeruginosa indentation and contact area on glass—variable-push experiments. (A) Indentation. (B) Contact area. Means and trends of data are shown from three independent experiments for each early biofilm sample or from two independent experiments for each mature biofilm sample. Each experiment consists of three sets of 10 replicate force plots, one set for each loading force setting, with contact time held at 1 s and ramp velocity at 2 μm/s.
Nanoindentation measurement by force spectroscopy
A comparison of average instantaneous and creep indentations observed in nanoindentation experiments with P. aeruginosa biofilm samples was performed (Fig. 10). When examining the instantaneous indentation data, both PAO1 and wapR biofilms exhibited higher instantaneous indentation after maturation (PAO1: 50 ± 17 nm versus 109 ± 3 nm; wapR: 43 ± 30 nm versus 249 ± 84 nm). The effect of LPS deficiency on instantaneous indentation, however, was not immediately apparent. Whereas early biofilms of both PAO1 and wapR had comparable instantaneous indentation, mature biofilms of wapR showed ∼2.3 times higher indentation depth than their PAO1 counterparts. In contrast, maturation of biofilms had different effects on the creep indentation of P. aeruginosa biofilms of PAO1 and wapR. Although PAO1 biofilms experienced a six-fold increase in creep indentation during maturation (4.2 ± 0.3 nm versus 26 ± 22 nm), the wapR mutant biofilms showed negligible changes (13 ± 11 nm versus 13 ± 3 nm). On the other hand, LPS deficiency in P. aeruginosa seemed to have opposing effects on the creep indentation of biofilms depending on their growth stage: early biofilms with a lack of LPS O-antigens experienced a three-fold increase in creep compared to their wild-type counterparts, but such a defect in mature biofilms resulted in a two-fold reduction in creep.
Figure 10.
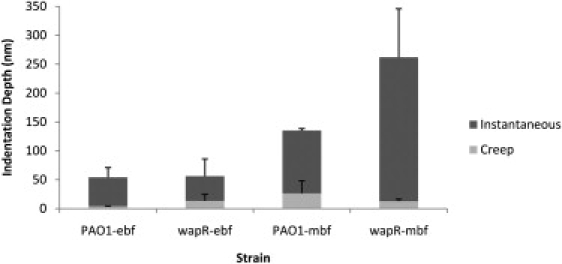
Instantaneous and creep indentations of Pseudomonas aeruginosa biofilms at SFTV—variable-push and -pull experiments. Means and standard errors of data are shown from two independent experiments per sample. Each experiment consists of 10 replicate force plots under standard loading force (10 nN), contact time (1 s), and ramp velocity (2 μm/s). Indentation data were extracted from the resulting creep curves. PAO1-ebf: PAO1 early biofilm; wapR-ebf: wapR early biofilm; PAO1-mbf: PAO1 mature biofilm; wapR-mbf: wapR mature biofilm.
Measurement of bacterial viscoelastic properties by MBFS
Nonlinear regression fitting of MBFS data to the three-element Voigt SLS model was performed to derive quantitative bacterial viscoelastic parameters (Table 2). A comparison of the viscoelasticity measurements revealed that LPS deficiency and biofilm maturation exerted the same drastic effect in reducing both instantaneous and delayed elastic moduli of P. aeruginosa biofilms. For instance, when compared to the instantaneous elastic modulus E1 of PAO1 early biofilms (1.7 × 105 ± 8 × 104 Pa), those of wapR early biofilms, PAO1 mature biofilms and wapR mature biofilms were reduced by a factor of 2.7, 4.8 and 11.6, respectively. Similarly, the delayed elastic moduli E2 for these three samples were reduced by a factor of 1.8, 2.1 and 5.3, respectively, when compared to that measured for early biofilms of PAO1 (1.1 × 106 ± 1 × 105 Pa). On the other hand, the impact of biofilm maturation on viscosity η was much more pronounced than that of LPS deficiency, with the corresponding reductions being factors of 1.1, 2.3 and 3.4, respectively, relative to the viscosity of early PAO1 biofilms (2.3 × 105 ± 1.1 × 105 Pa·s). Finally, the retardation time τ, measured at 0.20 ± 0.08 s for PAO1 early biofilms, was found to increase as a result of LPS deficiency (1.6-fold increase in early biofilms and 1.7-fold increase in mature biofilms) but remained more or less constant despite biofilm maturation in either strain PAO1 or wapR.
Table 2.
Summary of bacterial viscoelastic parameters measured by MBFS under SFTV
| Sample | PAO1 early biofilm | wapR early biofilm | PAO1 mature biofilm | wapR mature biofilm |
|---|---|---|---|---|
| Instantaneous elastic modulus E1 (Pa) | 1.7 × 105 ± 8 × 104 | 6.2 × 104 ± 1.9 × 104 | 3.51 × 104 ± 1 × 102 | 1.5 × 104 ± 7 × 103 |
| Fold difference | 1 | 0.37 | 0.21 | 0.086 |
| Delayed elastic modulus E2 (Pa) | 1.1 × 106 ± 1 × 105 | 6.1 × 105 ± 4.6 × 105 | 5.1 × 105 ± 4.3 × 105 | 2.1 × 105 ± 5 × 104 |
| Fold difference | 1 | 0.56 | 0.47 | 0.19 |
| Viscosity η (Pa·s) | 2.3 × 105 ± 1.1 × 105 | 2.1 × 105 ± 1.7 × 105 | 1.0 × 105 ± 9 × 104 | 6.7 × 104 ± 2.5 × 104 |
| Fold difference | 1 | 0.92 | 0.43 | 0.29 |
| Retardation time τ (s) | 0.20 ± 0.08 | 0.33 ± 0.03 | 0.18 ± 0.02 | 0.32 ± 0.04 |
| Fold difference | 1 | 1.60 | 0.89 | 1.55 |
Means and standard errors of data shown from two independent experiments for each sample. Data were derived from nonlinear regression fit of creep curves using the enhanced Voigt Standard Linear Solid equation (see Materials and Methods). Each experiment consists of 10 replicate force plots under standard load force (10 nN), contact time (1 s), and ramp velocity (2 μm/s) to derive 10 creep curves for data extraction. Fold difference shown is relative to PAO1 early biofilm.
Discussion
In this study, we described a powerful biophysical method which allows direct and simultaneous measurement of the adhesive and viscoelastic properties of bacterial biofilms. Although single-cell force spectroscopy may be useful for elucidating cellular mechanisms, the application of microbead force spectroscopy for analyzing a collection of cells is much preferred for the study of multicellular systems like biofilms. Using MBFS, we were able to quantitatively characterize bacterial biofilm adhesion and viscoelasticity, and to determine the effects of LPS deficiency and biofilm maturation on these parameters. Furthermore, we found that different experimental variables affected our measurements to various extents. Therefore, experimental conditions were carefully standardized to allow for meaningful comparison of adhesive and viscoelastic parameters observed among different samples. Finally, we developed a tip cleaning and regeneration protocol that permitted the reuse of AFM cantilevers for MBFS, thereby resulting in substantial time and cost savings.
The MBFS technique was successfully applied to the quantitation of adhesive properties displayed by P. aeruginosa PAO1 and wapR biofilms to a glass surface. Although direct comparison of our results with literature values may not be easily interpreted because of differences in instrumentation and experimental conditions, the adhesive forces that we measured (Table 1 and Fig. 5) did fall into the same nanonewton range as measurements from similar studies (Table S1). In a report by Sheng et al., AFM probes coated with Pseudomonas sp. cells were found to bind to various metals with adhesive forces of 0.5-5.6 nN (11), while Abu-Lail et al. observed that P. aeruginosa cells adhered to glass colloid probes with forces in the range of 1.4-3.9 nN (14). These adhesive forces measured are comparable to those that we measured between the biofilms of P. aeruginosa and glass using MBFS, which ranged from 0.56 to 6.9 nN. To take into account the effect of contact area, we calculated the corresponding adhesive pressures under standard conditions (SFTV) and discovered that these ranged from 19 to 332 Pa. The extent to which biofilms adhered to glass relative to the applied force was quantified by adhesive efficiencies, which ranged from 5.6% to 69% and were proportional to adhesive forces under SFTV because loading forces were fixed at 10 nN. Our observations that cells with truncated LPS resulted in higher adhesion were consistent with results from macro-scale assays by Lindhout et al. indicating that P. aeruginosa strains with defective LPS O-antigens adhered much more strongly to hydrophilic surfaces like glass and thereby became less motile (42). In contrast, biofilm maturation led to a reduction in adhesion to glass, a phenomenon not previously reported quantitatively for P. aeruginosa, and could be a result of the accumulation of extracellular polymers leading to higher surface heterogeneity after an extended period of growth. Increase in the physical roughness of microcolony surface could contribute to reduced adhesion, as evidenced by the work of Butt et al. (34) and that of Santos and coworkers (43). Moreover, Bruinsma et al. showed that the capacity of P. aeruginosa cells to adhere decreased with damages to bacterial cell surfaces (44), which are more likely to occur as biofilms mature.
Many previous comparative adhesion studies using force spectroscopy appeared to have arrived at inconclusive results because the effects of contact area and loading force were not taken into account. For instance, attempts by Burks et al. to correlate bacterial adhesion data in macroscopic column tests and nanoscale AFM experiments were not successful (45). Another AFM study by Atabek et al. investigating the effect of LPS and exopolymers on adhesion of P. aeruginosa cells to silicon did not reveal significant differences in adhesion forces between a smooth strain and a rough strain, although the adhesion events were shown to have occurred over shorter distances for the latter (46). In contrast, we were able to use a spherical geometry and obtained calculable contact areas in MBFS with a defined set of interaction conditions. As a result, the adhesion data collected were highly reproducible. Knowing the contact area and loading force, our investigation of adhesive pressures Pad and adhesive efficiency €ad allowed a more thorough interpretation of adhesion data beyond the limited perspective provided by adhesive force alone. Furthermore, it has become theoretically possible for us to calculate the average adhesive force exerted by a single cell on the glass surface by dividing our measured adhesive force by the number of cells counted within the contact area in SEM images. For the interaction of early biofilms with glass under SFTV, the measured adhesive forces of 0.66 nN for PAO1 and 6.9 nN for wapR can be translated to 51 pN per PAO1 cell and 530 pN per wapR cell, respectively. This is based on the observation that 13 cells on average were visible within the calculated contact area of 2.2 × 10−11 m2 (Fig. 2 A). Similar calculations for mature biofilms were not performed since attributing forces to individual cells could be problematic with the abundance of exopolymers present.
In MBFS experiments, loading forces, contact times and retraction velocities were varied to evaluate how these variables might affect adhesion as suggested by previous work (47,48). Dwell times were increased incrementally from 0 to 1, 2, 5 and 10 s to minimize total accumulative contact time, as adhesive properties could change for prolonged contact duration. When experiments were performed in the reverse order with dwell times decreasing incrementally from 10 to 0 s, similar adhesive forces were observed (data not shown), although such an order was not preferred since force curves collected later in the sequence could become less reliable because of increased accumulative contact time. The loading forces in the variable-push experiments (5, 10 and 20 nN) and the retraction velocities in the variable-pull experiments (1, 2 and 4 μm/s) were likewise increased incrementally over time as a result of expected increases in resultant adhesive forces. In these sequences, a sudden decay of adhesive force could signal bacterial detachment from the bead or cantilever failure, thus allowing the exclusion of invalid data. Since trends in adhesion were similar for different samples but most prominent for the wapR early biofilms, we focused on this particular sample and calculated equations of the linear best fit lines (see Figs. 6 to 9). Taking into account all of these data, we observed that the contact time was the single most important factor affecting adhesive measurements, as it was shown to be strongly and consistently proportional to all three measures of adhesion in variable-push and variable-pull experiments, although a “kink” exists at the start of each plot (Fig. 8). The apparent inflection point, which delineates the boundary between the initial jump in adhesion from zero to finite contact time and the subsequent linearity with increasing contact time, probably arises as a result of the difference between kinetic and static friction, because friction is known to contribute to adhesion (49). The coefficient of kinetic friction involved at zero contact time is much lower in magnitude than the coefficient of static friction at finite contact time. As a result, adhesion becomes nonlinear with regard to contact time near the plot origin. The loading force, in contrast, was only weakly proportional to adhesive force, but inversely related to both adhesive pressure and adhesive efficiency (Fig. 6). This observation revealed that both the contact area and the loading force increase faster than the resultant adhesive force. Surprisingly, the retraction velocity was found to have negligible effect on adhesion (Fig. 7). Perhaps a larger effect could be observed at higher velocities, although the resultant viscous drag on the glass bead would lead to incorrect adhesion measurements. Incidentally, the loading pressure was found to be nearly constant for each microbead, regardless of different loading forces or contact times (data not shown), since a higher loading force also resulted in a larger contact area.
Although quantitative measurements of bacterial adhesion and viscoelasticity could be carried out simultaneously using MBFS, the associated data analyses were quite distinct. Whereas adhesive data were determined by analyzing force-separation plots, viscoelastic data were derived from analyzing indentation-time curves. The common link existing between these two kinds of analyses is the need to quantify indentation depths accurately when pressing the biofilm-covered probe against the sample. On the one hand, the total indentation must be determined to derive the contact area for the adhesive pressure calculation. On the other hand, instantaneous indentation must be found to give the y-intercept of the creep indentation function for fitting to the Voigt SLS model to derive the viscoelastic parameters. Simply comparing the indentation depths of P. aeruginosa biofilms could also tell us about their compliance. We found that the total indentation values obtained were affected differently by LPS deficiency and biofilm maturation. In general, LPS deficiency had little effect on the total indentation of early biofilms, but increased that of mature biofilms by about a factor of 2. As the biofilms matured, the total indentation increased by more than a factor of 2.5 in PAO1 and by more than a factor of 5 in wapR. Whereas the increased thickness in mature biofilms was expected to increase their compliance, the defect in LPS appeared also to weaken the overall mechanical structure of the bacterial cells, making them more compressible.
By extending the contact time during the MBFS experiments, the viscoelastic properties of P. aeruginosa PAO1 and wapR biofilms could be directly measured during their compression by a stiff surface such as glass. Previous quantitative studies of biofilm rheology by other researchers were performed with various microscopy or rheometry techniques and examined responses to shear stresses or compressional stresses. To the best of our knowledge, this study is the first report of the quantitation of biofilm viscoelasticity by force spectroscopy and thus represents a novel application in AFM. A comparison of viscoelastic data from published reports revealed that the values for moduli and viscosity obtained were quite wide ranging (Table S2). Focusing on the compressional studies, Korstgens et al. found an apparent elastic modulus of 6.5 kPa and a yield strength (stress at the point of failure) of 0.99 kPa for P. aeruginosa biofilms using a film rheometer, whereas Cense et al. measured storage moduli of 0.73-8.56 kPa, loss moduli of 5.03–10.4 kPa, and viscosities of 256–2140 Pa·s for S. mutans biofilms using microindentation combined with confocal microscopy. These values were much lower than ours for E1, E2 and η in P. aeruginosa biofilms, which were calculated to be in the ranges of 15-170 kPa, 210–1100 kPa, and 67-230 kPa·s, respectively (Table 2). Such differences in measured quantitative viscoelastic data may not be surprising, since it is probable that factors such as the growth conditions, the type and age of the samples, the length scale and duration of the measurements, the type of theoretical model used, and the type and magnitude of applied stresses all have profound effects on the viscoelastic parameters derived, making comparative analyses of these figures extremely difficult. Despite problems associated with interstudy comparisons, intrastudy comparisons of viscoelastic parameters can still give us useful insights within the context of the particular study.
In this study, we compared the differential responses of bacterial biofilms to compression arising from LPS deficiency and biofilm maturation. Our results showed that biofilms exhibited reductions in instantaneous and delayed elastic moduli and viscosity (i.e., became less stiff) as a result of either LPS deficiency or biofilm maturation, although the latter had a stronger influence than the former. Interestingly, retardation time τ was found to be only slightly influenced by the status of LPS O-antigens but not by the stage of biofilm maturation. This small variation in τ during creep (range: 0.18–0.33 s) is analogous to the finding by Shaw et al. that elastic relaxation times τ′ of different biofilms during shear stress-relaxation were more or less constant (∼1100 s) (22). According to the interpretation by Klapper et al., the elastic relaxation time demarcates disturbances that produce a reversible elastic response and those that produce an irreversible viscous deformation (2). Our calculated retardation times could be easily converted to relaxation times (see Eq. 14), whereas Young's modulus E could be theoretically related to shear modulus G by the relation E = 3G (50) (i.e., viscoelastic materials conceptually respond three times faster to compression than to shear). However, quantitative comparison of the time scales from our experiments with those found by Shaw and co-workers is not meaningful because of the dissimilar experimental configurations, the different stages of biofilm growth, and the fact that we were concerned only with delayed indentation as opposed to total shear (22).
In recent years, mathematical models have become powerful tools in the structural and functional studies of biofilms. For instance, a model of biofilm matrix constructed by Cogan and Keener indicated that biomass redistribution would occur through swelling of exopolymers, leading to typical tower and mushroom cluster formation (51). In another study by Towler et al., a two-dimensional fluid-biofilm interaction model showed that softer biofilms would be more susceptible to lift forces, and that hydrodynamic stress could induce detachment most effectively during the initial relaxation time (52). Although the use of mathematical models may be insightful, the interpretation of quantitative data derived from them must be done with caution. Since the calculations of viscoelastic parameters in our case were based on a model consisting of simplified mechanical analogs with springs and dashpots, these parameters merely gave a mathematical representation of how bacterial biofilms might behave. The Voigt SLS model cannot be regarded as an exact solution of the system, and may not entirely reflect the actual physical mechanisms of deformation, which are likely to be much more complex. Despite its potentially limited mechanistic significance, the model is able to provide a quantitative description of differences in the viscoelastic properties that exist between different samples and under different experimental conditions.
In addition to the technical innovation in quantitatively measuring bacterial biofilm adhesive and viscoelastic properties, an added advantage of the MBFS technique is its potential for AFM cantilevers to be reused. Since only the glass bead was in contact with bacteria, bacteria induced corrosion of the cantilevers could be avoided. The cantilevers could thus be reused numerous times with proper cleaning after usage. We therefore sought a method for cleaning our cantilevers while dislodging the used glass beads so that new beads could be reattached for further experimentation. Using the method described herein, we found that the cleaning of cantilever tips with Piranha solution to be highly satisfactory for their regeneration for repeated usage. Furthermore, recalibration of the cleaned cantilevers before new bead reattachment showed little changes in their resonant frequencies and spring constants, indicating that their mechanical properties were retained after the cleaning. The ability to reuse cantilevers thus improves productivity and provides substantial cost savings, since AFM cantilevers are microfabricated and not trivial consumable items. In our hands, reusable cantilevers deployed in MBFS under standard experimental conditions allowed the consistent and reproducible derivation of quantitative adhesive and viscoelastic data that could be compared across different independent experiments.
In conclusion, we have developed a novel application of force spectroscopy to quantify bacterial biofilm adhesion forces and viscoelastic parameters quantitatively and reproducibly. Experimental conditions that affect data acquisition have been standardized and robust data analysis protocols have been established to allow for consistent comparison of acquired data across experiments. The method was validated for the absolute and simultaneous quantitation of biofilm adhesion and viscoelasticity in P. aeruginosa, and for discerning changes in these measurements associated with LPS deficiency and biofilm maturation. In the future, MBFS may be applied to evaluate the effects that other genetic, growth and environmental factors have on adhesion and viscoelasticity, physical aspects that are crucial to the establishment and persistence of bacterial biofilms.
Acknowledgments
The authors thank Dr. Ahmed Touhami (Polymer Surface and Interface Group, University of Guelph) and Dr. Colin Grant (Radford Research Group, University of Leeds) for their thoughtful comments; Dr. Alexandra Smith for assistance with scanning electron microscopy; and Dr. Jason Bemus (Asylum Research) for technical support on the use of the AFM software.
This work was supported by AFMNet funding and Discovery Grants to T.J.B. and J.R.D. from the Natural Sciences and Engineering Council of Canada, and an operating grant to J.S.L. from the Canadian Cystic Fibrosis Foundation. T.J.B. was a Canada Research Chair in the Structure of Microorganisms, J.R.D. is a Canada Research Chair in Soft Matter Physics, and J.S.L. holds a Canada Research Chair in Cystic Fibrosis and Microbial Glycobiology.
Footnotes
This work is dedicated to the memory of our colleague T.J.B.; P.C.L. is honored to be T.J.B.'s last graduate student and eternally thankful for his guidance.
Supporting Material
References
- 1.Costerton J.W., Stewart P.S., Greenberg E.P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 2.Klapper I., Rupp C.J., Cargo R., Purvedorj B., Stoodley P. Viscoelastic fluid description of bacterial biofilm material properties. Biotechnol. Bioeng. 2002;80:289–296. doi: 10.1002/bit.10376. [DOI] [PubMed] [Google Scholar]
- 3.Binnig G., Quate C.F., Gerber C. Atomic force microscope. Phys. Rev. Lett. 1986;56:930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- 4.Nunez M.E., Martin M.O., Chan P.H., Duong L.K., Sindhurakar A.R. Atomic force microscopy of bacterial communities. Methods Enzymol. 2005;397:256–268. doi: 10.1016/S0076-6879(05)97015-8. [DOI] [PubMed] [Google Scholar]
- 5.Oh Y.J., Jo W., Yang Y., Park S. Influence of culture conditions on Escherichia coli O157:H7 biofilm formation by atomic force microscopy. Ultramicroscopy. 2007;107:869–874. doi: 10.1016/j.ultramic.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 6.Fang H.H., Chan K.Y., Xu L.C. Quantification of bacterial adhesion forces using atomic force microscopy (AFM) J. Microbiol. Methods. 2000;40:89–97. doi: 10.1016/s0167-7012(99)00137-2. [DOI] [PubMed] [Google Scholar]
- 7.Bowen W.R., Fenton A.S., Lovitt R.W., Wright C.J. The measurement of Bacillus mycoides spore adhesion using atomic force microscopy, simple counting methods, and a spinning disk technique. Biotechnol. Bioeng. 2002;79:170–179. doi: 10.1002/bit.10321. [DOI] [PubMed] [Google Scholar]
- 8.Emerson R.J.T., Camesano T.A. Nanoscale investigation of pathogenic microbial adhesion to a biomaterial. Appl. Environ. Microbiol. 2004;70:6012–6022. doi: 10.1128/AEM.70.10.6012-6022.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao T., Tang H., Liang X., Wang A., Auner G.W. Nanoscale investigation on adhesion of E. coli to surface modified silicone using atomic force microscopy. Biotechnol. Bioeng. 2006;94:167–176. doi: 10.1002/bit.20841. [DOI] [PubMed] [Google Scholar]
- 10.Touhami A., Jericho M.H., Boyd J.M., Beveridge T.J. Nanoscale characterization and determination of adhesion forces of Pseudomonas aeruginosa pili by using atomic force microscopy. J. Bacteriol. 2006;188:370–377. doi: 10.1128/JB.188.2.370-377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheng X., Ting Y.P., Pehkonen S.O. Force measurements of bacterial adhesion on metals using a cell probe atomic force microscope. J. Colloid Interface Sci. 2007;310:661–669. doi: 10.1016/j.jcis.2007.01.084. [DOI] [PubMed] [Google Scholar]
- 12.Alsteens D., Dague E., Rouxhet P.G., Baulard A.R., Dufrene Y.F. Direct measurement of hydrophobic forces on cell surfaces using AFM. Langmuir. 2007;23:11977–11979. doi: 10.1021/la702765c. [DOI] [PubMed] [Google Scholar]
- 13.Busscher H.J., van de Belt-Gritter B., Dijkstra R.J.B., Norde W., Petersen F.C. Intermolecular forces and enthalpies in the adhesion of Streptococcus mutans and an antigen I/II-deficient mutant to laminin films. J. Bacteriol. 2007;189:2988–2995. doi: 10.1128/JB.01731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abu-Lail L.I., Liu Y., Atabek A., Camesano T.A. Quantifying the adhesion and interaction forces between Pseudomonas aeruginosa and natural organic matter. Environ. Sci. Technol. 2007;41:8031–8037. doi: 10.1021/es071047o. [DOI] [PubMed] [Google Scholar]
- 15.Lakes R.S. CRC Press; Madison, WI: 1999. Viscoelastic Solids. [Google Scholar]
- 16.Stoodley P., Lewandowski Z., Boyle J.D., Lappin-Scott H.M. Structural deformation of bacterial biofilms caused by short-term fluctuations in fluid shear: an in situ investigation of biofilm rheology. Biotechnol. Bioeng. 1999;65:83–92. [PubMed] [Google Scholar]
- 17.Stoodley P., Cargo R., Rupp C.J., Wilson S., Klapper I. Biofilm material properties as related to shear-induced deformation and detachment phenomena. J. Ind. Microbiol. Biotechnol. 2002;29:361–367. doi: 10.1038/sj.jim.7000282. [DOI] [PubMed] [Google Scholar]
- 18.Dunsmore B.C., Jacobsen A., Hall-Stoodley L., Bass C.J., Lappin-Scott H.M. The influence of fluid shear on the structure and material properties of sulphate-reducing bacterial biofilms. J. Ind. Microbiol. Biotechnol. 2002;29:347–353. doi: 10.1038/sj.jim.7000302. [DOI] [PubMed] [Google Scholar]
- 19.Rupp C.J., Fux C.A., Stoodley P. Viscoelasticity of Staphylococcus aureus biofilms in response to fluid shear allows resistance to detachment and facilitates rolling migration. Appl. Environ. Microbiol. 2005;71:2175–2178. doi: 10.1128/AEM.71.4.2175-2178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Towler B.W., Rupp C.J., Cunningham A.B., Stoodley P. Viscoelastic properties of a mixed culture biofilm from rheometer creep analysis. Biofouling. 2003;19:279–285. doi: 10.1080/0892701031000152470. [DOI] [PubMed] [Google Scholar]
- 21.Vinogradov A.M., Winston M., Rupp C.J., Stoodley P. Rheology of biofilms formed from the dental plaque pathogen Streptococcus mutans. Biofilms. 2004;1:49–56. [Google Scholar]
- 22.Shaw T., Winston M., Rupp C.J., Klapper I., Stoodley P. Commonality of elastic relaxation times in biofilms. Phys. Rev. Lett. 2004;93:098102. doi: 10.1103/PhysRevLett.93.098102. [DOI] [PubMed] [Google Scholar]
- 23.Lahaye E., Aubry T., Fleury V., Sire O. Does water activity rule P. mirabilis periodic swarming? II. Viscoelasticity and water balance during swarming. Biomacromolecules. 2007;8:1228–1235. doi: 10.1021/bm070115w. [DOI] [PubMed] [Google Scholar]
- 24.Houari A., Picard J., Habarou H., Galas L., Vaudry H. Rheology of biofilms formed at the surface of NF membranes in a drinking water production unit. Biofouling. 2008;24:235–240. doi: 10.1080/08927010802023764. [DOI] [PubMed] [Google Scholar]
- 25.Korstgens V., Flemming H.C., Wingender J., Borchard W. Uniaxial compression measurement device for investigation of the mechanical stability of biofilms. J. Microbiol. Methods. 2001;46:9–17. doi: 10.1016/s0167-7012(01)00248-2. [DOI] [PubMed] [Google Scholar]
- 26.Cense A.W., Peeters E.A., Gottenbos B., Baaijens F.P., Nuijs A.M. Mechanical properties and failure of Streptococcus mutans biofilms, studied using a microindentation device. J. Microbiol. Methods. 2006;67:463–472. doi: 10.1016/j.mimet.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 27.Lu H., Wang B., Ma J., Huang G., Viswanathan H. Measurement of creep compliance of solid polymers by nanoindentation. Mech. Time-Depend. Mater. 2003;7:189–207. [Google Scholar]
- 28.Fischer-Cripps A.C. A simple phenomenological approach to nanoindentation creep. Mater. Sci. Eng. 2004;385:74–82. [Google Scholar]
- 29.Kolari M., Schmidt U., Kuismanen E., Salkinoja-Salonen M.S. Firm but slippery attachment of Deinococcus geothermalis. J. Bacteriol. 2002;184:2473–2480. doi: 10.1128/JB.184.9.2473-2480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vadillo-Rodriguez V., Beveridge T.J., Dutcher J.R. Surface viscoelasticity of individual gram-negative bacterial cells measured using atomic force microscopy. J. Bacteriol. 2008;190:4225–4232. doi: 10.1128/JB.00132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowen W.R., Hilal N., Lovitt R.W., Wright C.J. Direct measurement of interactions between adsorbed protein layers using an atomic force microscope. J. Colloid Interface Sci. 1998;197:348–352. doi: 10.1006/jcis.1997.5247. [DOI] [PubMed] [Google Scholar]
- 32.Cho J.M., Sigmund W.M. Direct surface force measurement in water using a nanosize colloidal probe technique. J. Colloid Interface Sci. 2002;245:405–407. doi: 10.1006/jcis.2001.8075. [DOI] [PubMed] [Google Scholar]
- 33.Berg I.C., Rutland M.W., Arnebrant T. Lubricating properties of the initial salivary pellicle–an AFM study. Biofouling. 2003;19:365–369. doi: 10.1080/08927010310001618571. [DOI] [PubMed] [Google Scholar]
- 34.Butt H.J., Cappella B., Kappl M. Force measurements with the atomic force microscope: technique, interpretation and applications. Surf. Sci. Rep. 2005;59:1–152. [Google Scholar]
- 35.Hancock R.E., Carey A.M. Outer membrane of Pseudomonas aeruginosa: heat- 2-mercaptoethanol-modifiable proteins. J. Bacteriol. 1979;140:902–910. doi: 10.1128/jb.140.3.902-910.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poon K.K., Westman E.L., Vinogradov E., Jin S., Lam J.S. Functional characterization of MigA and WapR: putative rhamnosyltransferases involved in outer core oligosaccharide biosynthesis of Pseudomonas aeruginosa. J. Bacteriol. 2008;190:1857–1865. doi: 10.1128/JB.01546-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hutter J.L., Bechhoefer J. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 1993;64:1868–1873. [Google Scholar]
- 38.Lo Y.S., Huefner N.D., Chan W.S., Dryden P., Hagenhoff B. Organic and inorganic contamination on commercial AFM cantilevers. Langmuir. 1999;15:6522–6526. [Google Scholar]
- 39.Johnson K.L. Cambridge University Press; Cambridge: 1985. Contact Mechanics. [Google Scholar]
- 40.Findley W.N., Lai J.S., Onaran K. Dover Publications; New York: 1989. Creep and Relaxation of Nonlinear Viscoelastic Materials: With an Introduction to Linear Viscoelasticity. [Google Scholar]
- 41.Fischer-Cripps A.C. Nanoindentation. In: Ling F.F., Hart W.H., editors. Mechanical Engineering Series. Springer-Verlag; New York: 2004. pp. 39–68. [Google Scholar]
- 42.Lindhout T. University of Guelph; Guelph, Ontario, Canada: 2007. Investigation of the effect of lipopolysaccharide truncation on motility in Pseudomonas aeruginosa PAO1. MSc thesis. [Google Scholar]
- 43.Santos R.P., Arruda T.T., Carvalho C.B., Carneiro V.A., Braga L.Q. Correlation between Enterococcus faecalis biofilms development stage and quantitative surface roughness using atomic force microscopy. Microsc. Microanal. 2008;14:150–158. doi: 10.1017/S1431927608080227. [DOI] [PubMed] [Google Scholar]
- 44.Bruinsma G.M., Rustema-Abbing M., van der Mei H.C., Busscher H.J. Effects of cell surface damage on surface properties and adhesion of Pseudomonas aeruginosa. J. Microbiol. Methods. 2001;45:95–101. doi: 10.1016/s0167-7012(01)00238-x. [DOI] [PubMed] [Google Scholar]
- 45.Burks G.A., Velegol S.B., Paramonova E., Lindenmuth B.E., Feick J.D. Macroscopic and nanoscale measurements of the adhesion of bacteria with varying outer layer surface composition. Langmuir. 2003;19:2366–2371. [Google Scholar]
- 46.Atabek A., Camesano T.A. Atomic force microscopy study of the effect of lipopolysaccharides and extracellular polymers on adhesion of Pseudomonas aeruginosa. J. Bacteriol. 2007;189:8503–8509. doi: 10.1128/JB.00769-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cross S.E., Kreth J., Zhu L., Sullivan R., Shi W. Nanomechanical properties of glucans and associated cell-surface adhesion of Streptococcus mutans probed by atomic force microscopy under in situ conditions. Microbiology. 2007;153:3124–3132. doi: 10.1099/mic.0.2007/007625-0. [DOI] [PubMed] [Google Scholar]
- 48.Bremmell K.E., Evans A., Prestidge C.A. Deformation and nano-rheology of red blood cells: an AFM investigation. Colloids Surf. B Biointerfaces. 2006;50:43–48. doi: 10.1016/j.colsurfb.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 49.Yoshizawa H., Chen Y.L., Israelachvili J. Fundamental mechanisms of interfacial friction. 1. relation between adhesion and friction. J. Phys. Chem. 1993;97:4128–4140. [Google Scholar]
- 50.Aklonis J.J., MacKnight W.J., Shen M. Wiley-Interscience; New York: 1972. Introduction to Polymer Viscoelasticity. [Google Scholar]
- 51.Cogan N.G., Keener J.P. The role of the biofilm matrix in structural development. Math. Med. Biol. 2004;21:147–166. doi: 10.1093/imammb/21.2.147. [DOI] [PubMed] [Google Scholar]
- 52.Towler B.W., Cunningham A., Stoodley P., McKittrick L. A model of fluid-biofilm interaction using a Burger material law. Biotechnol. Bioeng. 2007;96:259–271. doi: 10.1002/bit.21098. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


