Abstract
The recently determined crystal structure of a chimeric Kv1.2-Kv2.1 Kv channel at 2.4 Å resolution motivated this molecular-dynamics simulation study of the chimeric channel and its mutants embedded in a DPPC membrane. For the channel protein, we used two types of C-terminus: E+ and Eo. E+ contains, and Eo lacks, the EGEE residue quartet located distal to the S6 helix. For both E+ and Eo, the following trend was observed: When S4 helices were restrained at the same position as in the x-ray structure (S4high), the S6 gate remained open for 12 ns. The results were similar when the S4 helices were pulled downward 7 Å (S4low). However, S4middle (or S4low) facilitated the S6 gate-narrowing for the following mutated channels (shown in order of increasing effect): 1), E395W; 2), E395W-F401A-F402A; and 3), E395W-F401A-F402A-V478W. The amino acid numbering system is that used for the Shaker channel. Even though all four subunits were set at S4low, S6 gate-narrowing was often brought about by movements of only two opposing S6 helices toward the central axis of the pore, resulting in a twofold symmetry-like structure. A free-energy profile analysis over the ion conduction pathway shows that the two opposing S6 helices whose peptide backbones are ∼10.4 Å distant from each other lead to an energetic barrier of ∼25 kJ/mol. S6 movement was coupled with translocation of the S4–S5 linker toward the central axis of the same subunit, and the coupling was mediated by salt bridges formed between the inner (intracellular side) end of S4 and that of S6. Simulations in which S4 of only one subunit was pulled down to S4low showed that a weak intersubunit coordination is present for S5 movement, whereas the coupling between the S4–S5 linker and S6 is largely an intrasubunit one. In general, whereas subunit-based behavior appears to be dominant and to permit heteromeric conformations of the pore domain, direct intersubunit coupling of S5 or S6 is weak. Therefore, the “concerted transition” of the pore domain that has been predicted based on electrophysiological analyses is likely to be mediated mainly by the dual effects of S4 and the S4–S5 linker; these segments of one subunit can interact with both S5 of the same subunit and that of the adjacent subunit.
Introduction
Voltage-gated ion channels open and close in response to changes in transmembrane potential due to the motion of the voltage sensor domains, controlling the flow of ions through the membrane (1). The voltage-gated potassium (Kv) channels are tetramers, with each subunit containing six-transmembrane segments, termed S1–S6. The voltage sensor domain of Kv channels consists of segments containing four transmembrane helices S1–S4 (2–5). A single pore domain is formed by the S5–S6 regions from the four subunits. S6 segments have been shown to form the intracellular gate (S6 gate). For the Shaker channel, a tight closure against ions is enabled by the intracellular gate (6,7).
The movement and conformational change of segments S1–S4 within the voltage-sensing domain in response to membrane depolarization is coupled to the S6 gate of the pore domain (S5–S6) to open and close the channel (8,9). A number of experimental analyses have been done to elucidate the closed-state structure, and several different models for the closed state have been proposed (4,9–13). In general, current arguments point to a model that falls somewhere between the helical screw model and the transporter model (9,10). Most recently, it was shown that I241C (of S1) and I287C (of S2) can form disulphide and metal bridges with R362C, implying a vertical movement of ∼6.5 Å of the residue upon the open/closed transition of the Shaker channel (14).
The kinetics of voltage gating of Kv channels has been studied by electrophysiological approaches. In the model proposed by Hodgkin and Huxley (15), four independent and identical K channel subunits convert individually between a resting state (R) and an activated state (A), and four subunits in the A state make an open channel (O). Based on gating-current measurements (16) and other analyses, Zagotta et al. (17) introduced a model with two sequential voltage-dependent conformational changes per subunit.
Such models suggest that voltage-dependent gating involves two sequential conformational changes. First is the independent motion of each voltage sensor domain, which transfers most of the gating charge between the resting state and an activated-not-open state (9,18–22). Second is the concerted opening transition of the voltage sensor domain and the pore domain, comprising the transition from the activated-not-open-state to the open state that opens the intracellular gate (7,9,17,22–24). Prompted by recent developments in understanding the K channel structure, researchers have proposed the allosteric model (25,26), where different conformations of the voltage sensor lead to differences in the open-close equilibrium of the pore domain (22,25). For further understanding, it is essential to know the atomic details of the mechanism by which movement of S4 is coupled to the movements of S6.
Here we address two questions: First, to what extent is the movement due to within-subunit (as opposed to between-subunits) coupling between S4 and S5 (and S6) important for voltage gating? A recent study of two tandemly connected subunits carrying distinct types of mutations showed the independence of individual subunits from other subunits (27). The analysis showed that the S4/S6 intersubunit interactions play a relatively minor role. Second, is the sequential transmission of force from S4 to the S4–S5 linker, then to S5, and finally to S6 important, or does the direct coupling between S4 and S6 and/or that between the S4–S5 linker and S6 play a more dominant role? Of note, several studies on the human ether-a-go-go-related (HERG) K+ channel, hyperpolarization-activated cyclic nucleotide-modulated (HCN) channel, and Shaker have suggested direct coupling between the S4–S5 linker and residues in the C-terminal part of S6 (25,27–30).
Recent improvements in molecular-dynamics (MD) simulations have allowed increasingly complex systems, including ion channels, to be studied (31–35). MD simulations have been used to analyze Kv1.2 channel dynamics within a lipid bilayer (36,37). For the previous simulation analysis of Shaker K+ channel (35), due to the limited structural information at the time, a model based on the KvAP channel was used, yet the applied electric field induced upward movement of S5 as well as precession movements of S5 and S6.
The structure of a Kv1.2-Kv2.1 chimeric channel crystallized in a membrane environment was recently determined at 2.4 Å resolution (10). The chimeric channel, which comprises the S3b–S4 segment of Kv2.1 and the S1–S3a and S5–S6 segments of Kv1.2, has been shown to function as a Shaker-type Kv channel (10) (see Fig. S1 in the Supporting Material). Here we report our analyses of the chimeric Kv channel and its mutated derivatives. The results delineate the subunit-based movements of the S4–S5 linker and S6, as well as the intrasubunit coupling between them. S5 movements exhibit a slight degree of concertedness between subunits.
Materials and Methods
The Gromacs 3.3.1 program was used for the MD simulations. For DPPC, the parameters modified by Tieleman and Berendsen (38) were used. For the protein, the GROMOS96 parameter set was used. For the 1,4 interactions each set was used, whereas for the nonbonded Lennard-Jones parameters between lipid and protein molecules, the geometric mean was used as the combination rule. For water, the SPC model was used (39).
Our choice of the Kv1.2-Kv2.1 chimera (10) is based on the relatively high resolution of the crystal structure. The chimera channel exhibits a voltage dependence of macroscopic current similar to the Kv1.2 channel (10). For the initial coordinates we used PDB ID code 2R9R, which is considered to be the open-state structure. For mutants such as E395W, we used SwissPDBViewer (http://www.us.expasy.org/spdbv) for substituting the side chain. The N-terminus was truncated before the F, which is 16 residues upstream of the S1 helix (Fig. S1). We used two types of C-terminus: E+ and Eo. E+ has the EGEE residue quartet, which is just downstream of the S6 helix, at its end. For Eo, the C-terminus is truncated before the EGEE. EGEE is aligned with D490QEE493 of Shaker (Fig. S1). A previous x-ray structural study was not able to determine the structure of the EGEE segment (10). During our preliminary 2 ns simulations in which EGEE was initially modeled as an α-helix extending from S6, the EGEE segments unfolded and its conformation varied among subunits. Therefore, a conformation randomly sampled at the end of the 2 ns simulations was used for the initial structure of EGEE. All N- and C-termini were capped with an acetyl and amino group, respectively, and thus had no net charge. (The conditions and procedure used to set up the simulation system are described in Section 1 of the Supporting Material.)
In many models proposed for the Shaker channel, it is accepted that the S1 and S2 segments do not move extensively upon gating (9,13,44). To reduce between-subunit difference due to drift, the z-positions of the Cα of S1 and S2 were restrained. For the S1 and S2 helices, the dihedral angles (ϕ and ψ) were harmonically restrained with the coefficient of 125.6 kJ/mol(rad)2 during all production runs. To move S4 to S4high, S4middle, or S4low (see Results), we used the steered-dynamics procedure. We pulled the Cα of R2, R3, R4, and K5 downward without restraining the x- and y-coordinates of the S4 atoms, whereas the atoms of the pore domain were restrained (as Fig. S1 shows, Q of Kv2.1 corresponds to R1 of Kv1.2). During productive runs, positional restraint of S4 at S4high, S4middle, or S4low was accomplished by restraining the z-coordinates of the Cαs of the gating charge carrying R and Ks (i.e., R365, R368, R371, and K374). The two K+ ions were placed at the S1S3 configuration and harmonically restrained (for the K+ configuration, see Shrivastava and Sansom (45)). After the energy minimization was completed, an equilibration run was carried out for 2 ns to restrain the peptide Cα, followed by a free productive run. (We acknowledge that we have not evaluated the potential biases on the results caused by the artificial restraints on S1, S2, and S4; much longer equilibration and production runs should be necessary for these helices and lipid molecules to undergo full relaxation after the forced movement of S4). We analyzed the movement of the peptide using Gromacs utilities and our own programs.
In their kinetics model, Zagotta et al. (17) proposed a small degree of voltage dependency in the final concerted transition. To mimic the resting membrane potential, we applied an inward electric field of 0.05 V/nm for all the simulations listed in Table 1. Each charged atom “feels” a local field that is the sum of the externally introduced field and the field due to the charges of the neighboring atoms (34,46).
Table 1.
Simulation summary
| Simulation | Mutant∗ | S4 Position | Pore radius (Å) ± SD† |
|---|---|---|---|
| 1 | wt-chimera-Eo | S4high | 4.36 ± 0.26 |
| 2 | wt-chimera-Eo | S4middle | 3.86 ± 0.32. |
| 3 | wt-chimera-Eo | S4low | 3.92 ± 0.23 |
| 4 | E395W-Eo | S4high | 3.37 ± 0.17 |
| 5 | E395W-Eo | S4middle | 2.14 ± 0.20 |
| 6 | E395W-Eo | S4low | 3.66 ± 0.26 |
| 7 | E395W-F401A-F402A-Eo | S4high | 3.09 ± 0.23 |
| 8 | E395W-F401A-F402A-Eo | S4middle | 3.06 ± 0.22 |
| 9 | E395W-F401A-F402A-Eo | S4low | 1.69 ± 0.41 |
| 10 | E395W-F401A-F402A-V478W-Eo | S4high | 1.91 ± 0.37 |
| 11 | E395W-F401A-F402A-V478W-Eo | S4middle | 1.44 ± 0.13 |
| 12 | E395W-F401A-F402A-V478W-Eo | S4low | 1.44 ± 0.10 |
| 13 | wt-chimera-E+ | S4high | 3.98 ± 0.30 |
| 14 | wt-chimera-E+ | S4middle | 4.05 ± 0.28 |
| 15 | wt-chimera-E+ | S4low | 3.41 ± 0.33 |
| 16 | E395W-E+ | S4high | 2.87 ± 0.26 |
| 17 | E395W-E+ | S4middle | 1.94 ± 0.33 |
| 18 | E395W-E+ | S4low | 2.83 ± 0.28 |
| 19 | E395W-F401A-F402A-E+ | S4high | 2.44 ± 0.21 |
| 20 | E395W-F401A-F402A-E+ | S4middle | 2.70 ± 0.23 |
| 21 | E395W-F401A-F402A-E+ | S4low | 1.66 ± 0.31 |
| 22 | E395W-F401A-F402A-V478W-E+ | S4high | 1.59 ± 0.23 |
| 23 | E395W-F401A-F402A-V478W-E+ | S4middle | 1.62 ± 0.29 |
| 24 | E395W-F401A-F402A-V478W-E+ | S4low | 1.55 ± 0.15 |
The amino acid number denotes the corresponding amino acid of Shaker. The wt-chimera was used as the template for all of the mutants. For all simulations, an inward (resting) electric field of 0.05 V/nm was applied.
The mean and SD calculated from the 10 frames covering the final 1 ns.
For pore radius analyses, we used our own source code based on the HOLE algorithm developed by Smart et al. (47), with the modification that the simulated annealing-based search of the center of the sphere to probe the pore was repeated five times to reduce the chance of being trapped in the local minima. Umbrella sampling was performed with the Gromacs suite (for details, see the legend for Fig. S4). The data were analyzed using the weighted histogram analysis method (48).
Results
Mutations facilitate narrowing of the S6 gate in silico
For our simulations, we chose the recently studied chimeric Kv1.2 channel (10). In the chimeric channel, the helix-turn-helix motif comprising the S3b and S4 helices was replaced by the corresponding segment from rat Kv2.1 channel (Fig. S1).
One problem in approaches like ours is that it has not been precisely determined how long it takes for the pore domain to close after membrane repolarization brings about the S4 translocation. Some perturbations may facilitate the transition in MD simulations, and thus can be helpful for such studies. Many mutants that perturb the G-V relationship of the Shaker K+ channel in favor of the closed state are known (19,21,22,49). To be safe, we included mutants that harbor multiple mutations that, when combined, cause a strong bias toward the closed state (Table 1). The mutations were introduced onto the original Kv1.2-Kv2.1 chimera (wt-chimera). Mutation E395W (E327W of Kv1.2) shifts the G-V relationship in a positive direction to +210.9 mV (22). Similarly, F401A and F402A shifts the G-V curve by >80 mV (22,49). We also included a triple mutant, E395W-F401A-F402A, and a quadruple mutant, E395W-F401A-F402A-V478W. V478 (V410 of Kv1.2) has been proposed to create a tight closure in the closed state (6,9). V478W significantly shifts the closed-to-open equilibrium toward the closed state, causing the phenotype to become nonconducting (50). Note that E395W-F401A-F402A modifies the G-V relationship, but only slightly perturbs the Q-V relationship, reflecting the voltage dependency of S4 translocation. The C-terminus was also varied. For sim1–12, the C-terminus was truncated just after T of HRETEGEE (which we refer to as Eo), whereas for sim13–24 the C-terminus contained the four extra residues EGEE (E+). This HRETEGEE corresponds to H486RETDGEE493 of Shaker (Fig. S1). We did not include the C-terminus region located downstream of EGEE, although they play an important role in modulating the dynamics of S6 (51).
For each mutant, three 12-ns simulations, each with S4 positioned differently (i.e., S4high, S4middle, and S4low), were performed (Table 1). For S4high, the z-position of the center of mass (COM) of the Cαs of the S4 helix was restrained to the reported position. To prepare S4middle and S4low, the S4 helix was moved by steered-dynamics procedures and restrained at 3 and 7 Å, respectively, downward from S4high (Fig. 1 B; also see Materials and Methods). The choice of 7 Å is greater than the 6.5 Å derived by recent experiments (14) and 6.7 Å by our simulation study (52). However, the S4 position in the activated state for the chimera channel, which has been crystallized in a lipid environment, is likely to be ∼2.4 Å higher than in the Kv1.2 crystal structure (53).
Figure 1.
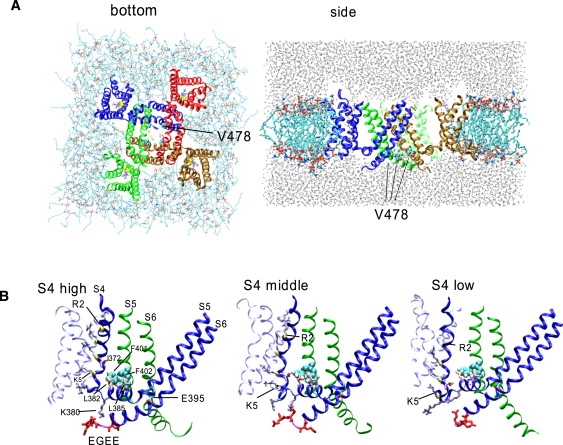
The simulation box. (A detailed version of this part of the legend is given in Section 4 of the Supporting Material.) (A) The view from the bottom (left) and side (right) of the initial configuration for the wt-chimera channel with S4high settings is shown. (B) The initial conformation for wt-chimera-S4high, -S4middle, and -S4low. Only subunit 1 (blue) is shown, along with S5 and S6 of the adjacent subunit 2 (green).
For the trajectories, inspection showed that all well-defined helical segments remain conserved (data not shown). Conformational changes during simulation were mostly accounted for by rigid-body movements of the S4–S5 linker, S5, and S6. Of importance, the pore radius measured at the height of the Cα of V478 exhibited a decrease over the trajectory in many simulations (Fig. 2). For both wt-chimera-Eo and -E+, no significant narrowing of the S6 gate occurred for all three S4 positions. For E395W-Eo-S4middle, the S6 gate narrowed quickly (∼3 ns), resulting in a radius of ∼0.2 nm. Neither E395W-Eo-S4low nor E395W-Eo-S4high showed clear narrowing, yet the radius reached 0.35 nm, which is narrower than that of the wt-chimera (∼0.4 nm). It should be noted that although S4middle exhibited a more prominent narrowing than S4low in this particular set of simulations, S6 behavior appears to be fairly stochastic and unpredictable, and therefore more simulation runs (perhaps >10) may be necessary to figure out which is more effective. Similarly, for E395W-E+, S4middle and S4low simulations showed a mild degree of narrowing, although S4middle exhibited fluctuation in the pore radius. Inspection showed that the S6 helices of two opposing subunits moved toward and then away from each other. For E395W-F401A-F402A (i.e., the triple mutant)-Eo, S4high and S4middle resulted in a slight narrowing of the S6 gate (∼0.3 nm), whereas S4low resulted in a further narrowing of ∼0.15 nm. For the triple mutant-E+, all simulations exhibited a slight narrowing to ∼0.25 nm. For both Eo and E+, the quadruple mutant exhibited a significant narrowing of the conduction pathway that occurred quickly (∼300 ps∼1 ns) for all three S4 positions.
Figure 2.
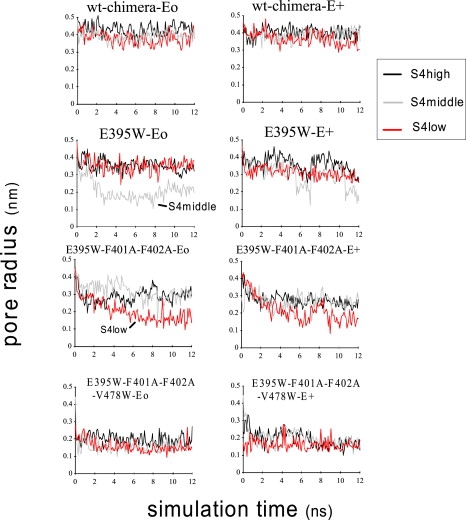
The pore radius over the simulation trajectory measured at the height (the average for four subunits) of the Cα in V478.
The pore radius profile at the end of the simulation (12 ns) is shown along with the graphical image of S6 at 12 ns (Fig. 3). For wt-chimera-Eo and -E+, the profile was largely similar for S4high, S4middle, and S4low. For E395W-Eo-S4middle, narrowing at the segment PVPVIV (i.e., P473–V478) is evident (gray line) and, intriguingly, the central cavity is also narrowed; for a segment located at −1 to −0.5 nm from the bilayer center, the radius decreased to ∼0.2–0.3 nm (Fig. 3 A). For the triple mutant-Eo, the S4low simulation showed narrowing (to ∼0.2 nm) of the PVPVIV segment and also the upstream segment located at −1 to −0.5 nm from the bilayer center. For the triple mutant-Eo-S4high and -S4middle, a modest amount of narrowing was observed. On the other hand, a comparison between the triple mutant-Eo-S4high and the quadruple mutant-Eo-S4high showed that although the central cavity narrowed to a similar degree, narrowing at the PVPVIV segment was very strong for the quadruple mutant. This difference was also observed for the S4middle simulation and the corresponding E+ simulations. Thus, for the quadruple mutant, narrowing was relatively limited to the short segment containing V478W.
Figure 3.
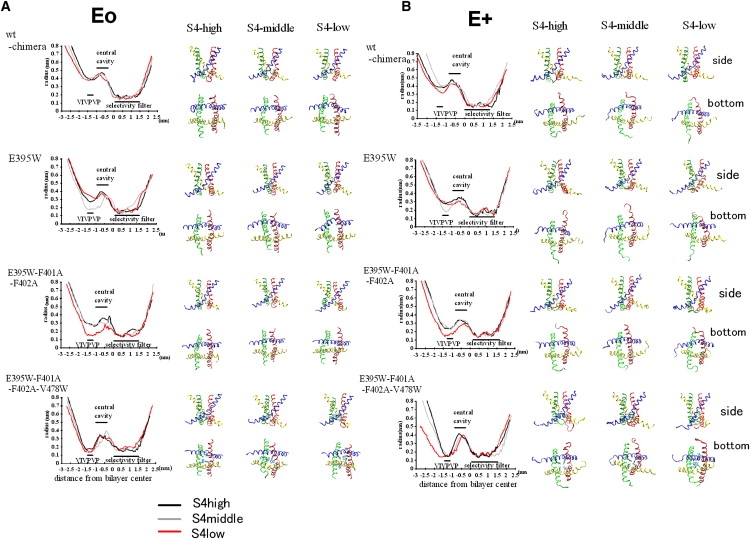
Conformation of the intracellular S6 gate at the end of the simulations. (A) Results for the Eo simulation series (sim1–12 in Table 1). Left: Analysis of the pore radius profile along the z axis for the structure at 12 ns. The same procedure as in the pore radius profile analysis (Fig. 2) was used. Right: Graphical image of four S6 helices viewed from the side (upper) and the intracellular side (lower) at the end of each simulation. Subunits 1–4 are colored blue, green, yellow, and red, respectively. (B) Results for the E+ simulations (sim13–24 in Table 1), shown in the same manner as in A.
Intriguingly, for simulations that exhibit narrowing of the S6 gate (e.g., for E395W-Eo-S4middle, the triple mutant-Eo-S4low and E395W-E+-S4middle), motion of the four subunits was not equivalent (Fig. 3). For example, in the cases of E395W-Eo-S4middle and the triple mutant-Eo-S4low, the S6 segment of only two opposing subunits moved toward the central axis, resulting in a conformation somewhat like a twofold symmetrical structure. This gave us a chance to examine whether S5 (and the S4–S5 linker) exhibits strong coordination with S6 in the same subunit or in the adjacent subunit.
Gate-closing movement of S6 is associated with translocation of the S4–S5 linker of the same subunit
The translocation of various segments of the channel protein over the simulation time was analyzed. Strikingly, the gate-narrowing translocation of V478 is associated with the S4–S5 linker of the same subunit. For example, for E395W-Eo-S4middle, the overall feature of the four-subunit data of toward-central-axis movement of V478 Cα (Fig. 4 A, second column) resembles that of the COM of the S4–S5 linker (Fig. 4 B). Such a resemblance is evident for several other simulations (Fig. 4, A and B). For the quadruple mutant-Eo, the resemblance is not obvious, but this can be explained by the relatively local effect of W on the gate-narrowing movement of S6. In general, the horizontal movements of the S4–S5 linker and S6 are tightly coordinated and exhibit a subunit-based behavior.
Figure 4.
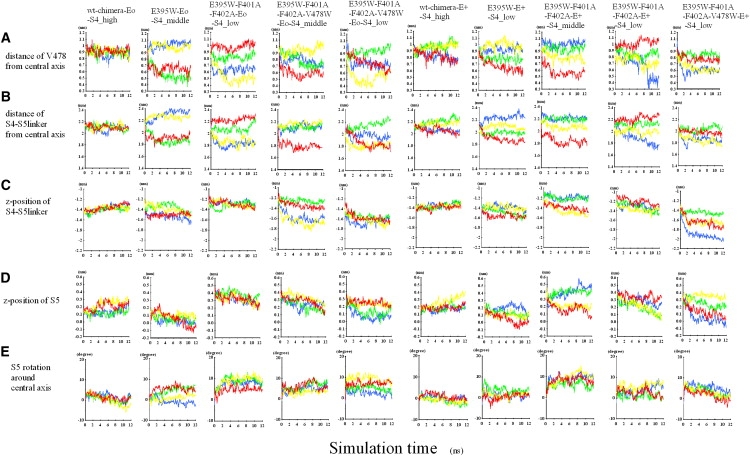
Time course of the location of the S4–S5 linker, S5, and S6. Four subunits are shown with the same coloring scheme as in the graphical images of Fig. 3. Only the results of more notable simulations are shown. (A) The distance of V478 from the central axis of the pore domain. (B) The distance of the COM of S4–S5 linker from the central axis. (C) The z-position (i.e., vertical position) of the COM of the S4–S5 linker. (D) The z-position of the COM of S5. (E) Rotational angle (in degrees) around the central axis. A positive value indicates clockwise rotation when viewed from the intracellular side.
The translocation of the S4–S5 linker toward the central axis of the channel is more or less accompanied by a downward translocation (Fig. 4, B and C). This implies an oblique direction of the linker translocation, but because there is a substantial difference between Fig. 4, B and C, the direction of movement of the linker is not the same among subunits. As Fig. S2 shows, upon the gate-narrowing movement of S6, the typical translocation of the S4–S5 linker is clockwise (when viewed from the bottom) and also directed somewhat downward and toward the central axis. The distance of S5 from the central axis remained largely unchanged during the S6 gate narrowing (data not shown). For S4middle and S4low, S5 exhibited a trend of downward translocation (Fig. 4 D) as well as precession clockwise when viewed from the bottom (Fig. 4 E).
Fig. 4, A and D appear to be different, suggesting that the vertical motion of S5 is not tightly coupled to the gate-narrowing movement of S6 in the same subunit. This and inspection of Fig. 4 D may reflect a tendency of S5 toward concerted movement among subunits. However, we cannot rule out the possibility that the vertical movement of S5 may simply reflect its propensity to follow that of S4, because S4 of four subunits is held at the same position in our case.
To address this issue, we performed simulations in which S4 of only one subunit was held at S4low. The left column of Fig. 5 shows a representative simulation, in which S4 of the blue subunit was held at the S4low position. The S4–S5 linker and S6 can move in a subunit-based manner (Fig. 5, A–C). Intriguingly, S5 of the same subunit exhibits precession, whereas S5 of the other subunits also exhibits precession as well as downward movement (Fig. 5, D and E). Therefore, S5 movements are concerted: the motion of one subunit influences the movement of adjacent subunits. However, inspection did not show any obvious structural feature that could allow S5–S5 coupling or S6–S6 coupling among subunits (details not shown). Of note, S5 of the green subunit, which interacts with S4 of the blue subunit, underwent the downward motion more quickly than the other subunits (Fig. 5 D, left column). Moreover, when we pulled down the S5 of only one subunit and held S4 at S4high, it did not lead to downward motion of the adjacent subunit S5 (Fig. 5, right column). These results suggest that the direct S5–S5 coupling between subunits is not strong. It is possible that the concerted movement of S5 is mediated by the dual effect of S4 and the S4–S5 linker, which have inter- and intrasubunit interactions with S5 (see Discussion).
Figure 5.
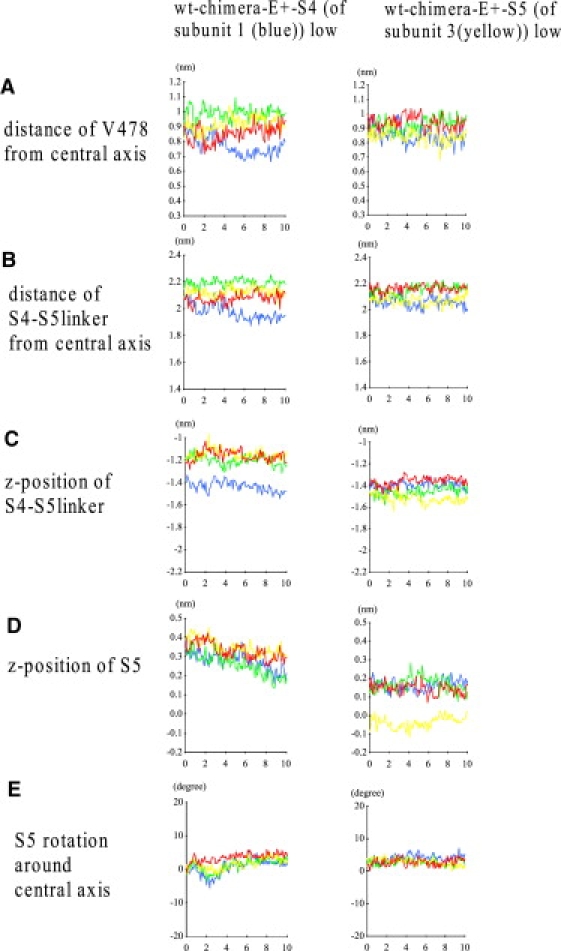
Effect of perturbation of one subunit on the location of the S4–S5 linker, S5, and S6. Shown are the results for the simulation in which only S4 of the blue subunit (subunit 1, left column) or S5 of the yellow subunit (subunit 3, right column) was pulled down. (A detailed version of the legend is given in Section 5 of the Supporting Material.)
Horizontal movement of the voltage sensor domains was also observed. Fig. S3 A shows the COM positions of the S1–S4 helices viewed from the extracellular side. Intriguingly, for the S4low simulations, each voltage-sensor domain is slightly rotated counter-clockwise. This is in accordance with a recent model (9), although the extent of translocation is much less in our studies, possibly because of the short simulation time. For reference, Fig. S3 B shows the COM positions of the open- and closed-state models reported previously (9). Pathak et al. (9) predicted that S4 approaches the N-terminus segment of S1 upon transition to the closed state, creating a sharp (i.e., concave) bend at the joint between S4 and the S4–S5 linker (red line of Fig. S3 B). Intriguingly, although the movement of S4 toward S1 was weak, it was observed in all S4low simulations (Fig. S3 A). More extended simulations are necessary to examine whether the sharp bend predicted in the model is indeed created.
Umbrella sampling analysis of S6 gate closure
To examine the effect of gate narrowing on the K+ ion passage, we carried out free-energy analyses based on the umbrella sampling procedure. As shown in Fig. 6, we chose three conformations from those presented in Fig. 3. The curve for E395W-Eo-S4high shows that two opposing S6 helices located ∼5.5 Å distant from each other (based on the pore radius measurement) can act as a barrier of ∼25 kJ/mol. For this conformation, the distance between the two opposing S6 helices measured by the Cα–Cα distance exhibited the minimum value (∼10.4 Å) at P475, which is the second P of the PVP motif. The E395W-Eo-S4middle shows that even two opposing S6 helices located ∼4 Å distant from each other lead to two alternative results depending on the number of water molecules that remain in the pore (Fig. 6 B). Here, the distance between the two opposing S6 helices measured by the Cα–Cα distance again exhibited the minimum value (∼5.6 Å) at P475. Although we rehydrated the pore before performing the umbrella sampling, some water molecules occasionally moved out during the sampling. For this structure, we repeated the umbrella sampling six times and found that, whereas the central cavity always contained a reproducible number of water molecules (∼15), the number of water molecules that remained at the height of A471 and L472 varied among trials, exhibiting somewhat of an all-or-nothing feature (the number of water molecules for the six trials was 1, 2, 2, 10, 11, and 13, respectively). That is, for three of the six trials, water near A471 and L472 was almost depleted, whereas for the remaining three trials, this portion of the pore remained hydrated. As Fig. 6 B shows, the water-depleted cases exhibited a higher energetic barrier compared with the case with the hydrated pore. Of note, even for the latter cases, the barrier was as high as ∼50 kJ/mol. This level of energy barrier was not observed for the conformation taken from the wt-chimera-Eo-S4high, which has a ∼0.4 nm pore radius (Fig. 6 C). These findings suggest that the physiologically relevant S6 gate closure can be realized not only by the tight binding of the four V478 side chains, but also by the two opposing S6 helices approaching one another to within ∼5.5 Å, which is likely to create an energetic barrier of ∼25 kJ/mol.
Figure 6.
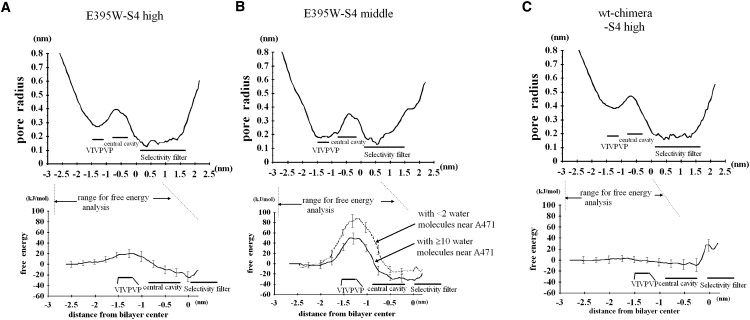
Free-energy profile for representative structures obtained from our simulations. Pore radius profile (top) and the corresponding free energy (bottom) are shown. (A) E395W-Eo-S4high. (B) E395W-Eo-S4middle. (C) wt-chimera-Eo-S4high. (A) The average and SD from three sets of the umbrella sampling procedure are shown. (B) Six trials were carried out and divided into two groups depending on the degree of the hydration (i.e., the number of the water molecules that remained at the height of A471 or L472) as described in the text. For all cases, the free energy at the position in the bulk water (0.8 nm below the Cα of V478) was set to zero.
Internal motions of S6
Previous studies have modeled or simulated the internal motions of S6 conformation upon the open-/closed-state transition (9,35,54). We tried various procedures and found it useful to examine the change in distance of the S6 residues from the central axis relative to the initial structure (Fig. S4). (See Section 2 of the Supporting Material for further comments.)
Discussion
We performed simulations on the Kv1.2-Kv2.1 chimera and its mutated analogs. Despite the relatively short simulation time (∼12 ns), S6 gate-narrowing was observed for mutated channel simulations carried out with S4 placed at lower positions than in the reported x-ray structure. It is experimentally difficult to measure the time lag, after the downward movement of S4, required for the pore domain to undergo conformational change toward the closed state. Therefore, the finding that MD simulation can reproduce the effects of at least some mutations is important.
Many mutants that modify the kinetics of the voltage gating of Shaker, perturbing the G-V relationship in favor of the closed state, are known (19,21,22,49). It has been shown that there are two clusters (external and internal) of residues critical for gating kinetics (22). The external cluster consists of V408, L409, A413, I457, and V458, and affects the dynamics of the voltage sensor. On the other hand, the mutations E395W, L398W, F401A, F402A, I405W, L472W, and S479W, which alter residues in internal clusters, have pronounced effects on the concerted opening transition. For example, E395W slightly shifts the Q-V curve while strongly altering the G-V curve (+210.9 mV), indicating a shift in favor of the closed state. F401A and F402A show a −8.9 mV and −3.8 mV shift of the Q-V curve, respectively, and both shift the G-V curve by >+80 mV.
Inspection suggested two possible mechanisms for the E395W effect. First, the direct or indirect (mediated by lipid headgroups) interaction between E395 of one subunit and S4 of the adjacent subunit may provide an attractive force, keeping S5 away from the central axis. (Note that polar headgroups and water are more abundant outside than inside the S6 helices.) The E395W mutation appears to weaken this attractive force, causing S5 to be closer to the central axis of the channel. Second, the side chain of the introduced W tends to be located between S5 and S6 of the same subunit, pushing S6 toward the channel axis (Fig. S5).
F401A and F402A and the ILT mutant (V369I, I372L, and S376T (of S4)) cause a similar degree of perturbation in favor of the closed state (21,22,55). Because F401A and F402A are located near or facing V369 and I372 of the adjacent subunit S4 (Fig. 1), it seems possible that F401A-F402A and V369I-I372L exert an effect by a common mechanism, namely, weakening hydrophobic interactions between S5 and the adjacent subunit S4 (and the S4–S5 linker), as noted in the legend for Fig. S5. However, our simulations are clearly insufficient for a quantitative evaluation.
The V478 in Shaker (V410 in Kv1.2) faces the inside of the pore and has been implicated in hydrophobic seal formation (6). It has been proposed that V478W causes a profound shift of the closed-to-open equilibrium toward the closed state, rather than creating a local steric hindrance to ion conduction (50,56). In our cases, the quadruple mutants showed a strong trend toward the S6 gate narrowing. This is not caused solely by the bulkiness of W; when V478 is replaced with W without changing the peptide backbone structure, the pore radius is smaller by only ∼0.13 nm (e.g., when the pore radius was 0.48 nm for the wt-chimera, it was 0.35 nm for the V478W mutant; details not shown). For both Eo and E+, the position of S5 is similar between the triple mutant-S4middle and the quadruple mutant-S4middle (not shown). It appears that S6 is not strongly attached to S5 of the same subunit, and therefore the substitution of V478 with W appears to be sufficient for promoting the toward-the-center movement of S6. On the other hand, despite the strong attractive force of W, the quadruple mutant simulations ended with a >0.1 nm pore radius, with two W side chains opposite from one another being at least 2 Å apart. At present, we cannot rule out the possibility of simulation artifacts, but it is tempting to envision the closed state corresponding to a pore with a small radius and effectively reduced conduction, and not to conformations with a tight contact between hydrophobic side chains.
Direct interaction between the S4–S5 linker and the C-terminus of S6
Although S5 showed a weak degree of concertedness between subunits, the movement of the S4–S5 linker and S6 within the same subunit is largely independent from those of other subunits (Fig. 4). Several studies on HERG, HCN, and Shaker have suggested direct coupling between the S4–S5 linker and residues in the COOH-terminal part of S6 (25,27–30). In the case of HCN, a mutational study suggested that Arg of the S4–S5 linker directly interacts with Asp of the C-linker, which is immediately downstream of S6 (30). A specific pairing of the S4–S5 linker with the C-terminal portion of S6 was indicated in a study where the voltage-sensing domain of Shaker was transplanted onto the KcsA (25). In addition, F484C and several other mutants of Shaker shift the Q-V curve negatively, suggesting coupling between S4 gating-charge movement and aromatic residues at the S6 tail (29).
In most of the E+ simulations, E residues of the EGEE segment were interacting with the S4–S5 linker (Fig. 1 B; data not shown). However, even without EGEE (i.e., Eo mutants), inspection showed that the K residue located at the inner end of S4 (K380 of Shaker) could make a salt bridge with the E488 that is located two residues upstream of the C-terminus of S6. The overall features of the sim1–12 and sim13–24 results were similar (Table 1), arguing against the idea that EGEE motif solely determines the binding energy between the K380 and S6 C-terminus. Moreover, our cumulative radial distribution function analysis showed that even for the E+ simulations, E488 is subjected to a frequent interaction with K380 (with about three out of four subunits forming the salt bridge), whereas the Es of the EGEE segment are involved in the interaction less frequently (with about one to two subunits forming a salt bridge). These findings suggest that E488 accounts for a major part of the coordination of S4–S5 the linker and S6 movements.
S5 concerted movements (downward and precession) are also likely to affect the S6 movements. However, when we pulled down the S5 of one subunit and held S4 at S4high, it did not lead to downward motion of the adjacent subunit S5 (Fig. 5, right), suggesting that the direct S5–S5 coupling between subunits is not strong.
Implications for short-lived closed states and heteromeric closed states
Several models, including allosteric models, have been proposed to link voltage-dependent activation and channel opening (57–61). In allosteric models, the probability that the pore domain will visit the open state increases with the number of activated subunits (where “activated” means that the voltage sensor domain is in the activated state; Fig. S6 A). In the complete Modod-Wyman-Changeux model shown in Fig. S6 B (63,64), heteromeric pore conformations are permitted in which some, but not all, subunits are in the open-state structure (rows H1–H3; this presentation is based on work by Chapman and VanDongen (62).
For wild-type Kv2.1, at least two short-lived subconductance levels are visited when the gate moves between the closed and fully open states (65). The subconductance levels may correspond to transient heteromeric pore conformations in which some, but not all, subunits are in the open state (62,65). It is likely that activation of one or two subunits can partially open the pore of the Kv2.1 channel (62). Subconductance levels have been reported for T442 of Shaker (66).
In our case, even though S4 in all four subunits was held at the same z-position, the S4–S5 linker and the position of S6 showed a between-subunit difference. However, we did not restrain x- and y-coordinates for S4, allowing for between-subunit differences in the S4 location. Further between-subunit differences may originate from the ambiguity in the choice of E residues near the C-terminal end of S6 for the salt bridge formation with K380. However, it is possible that much longer simulations may reduce the between-subunit difference in the S4 and S4–S5 linker position. (Implications for the short-lived closed states Cf and Ci (67) are given in Section 3 of the Supporting Material.)
Finally, we would like to consider the cooperativity between subunits (26). As we note in the legend for Fig. S6, Hill analysis should place a large weight on S4 movement, and therefore the Hill coefficient is not a good index of the concerted movement of the pore domain. Another factor that may lead to the small Hill coefficient is that the between-subunit coupling is weak and merely dependent on the dual effect of S4 and the S4–S5 linker. Fig. S6 D presents our hypothesis of the dual control of S6 by S4 translocation. S4 and the S4–S5 linker can directly control the S6 in the same subunit (Fig. S6 C). At the same time, S4 and the S4–S5 linker can control the S5 position in the same and adjacent subunit (Fig. S6 D). This enables, to some extent, concerted S5 movement, which is likely to be a factor influencing the probability of S6 translocation.
Conclusions
Our study shows that by combining the mutations and S4 positions that can be expected to facilitate the closing transition, one can study gate-narrowing conformational changes in 12 ns MD simulations. The final structure for many simulations that exhibited narrowing was often not the fourfold symmetry-like structure, but a heteromeric one in which one or two opposing S6s approached the central axis.
Coupling between the S4–S5 linker and the segment distal to S6 is largely an intrasubunit one and is mediated by several charged residues near the S4–S5 linker. The S4–S5 linker and S6 are largely independent of individual subunits. S5 movements (precessional and vertical) proceed in a relatively concerted manner, which is likely to be mediated by S4 and the S4–S5 linker.
The nonuniqueness and heteromeric conformation may be an inherent property of the closed-state structure of the pore domain. However, many more simulation studies considering various closed-state models are necessary to address this issue.
Supporting Material
Six figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(09)00847-9.
Supporting Material
Acknowledgments
We thank Dr. Yarov-Yarovoy for providing the coordinates of the Kv1.2 open- and resting-state models.
References
- 1.Hille B. 3rd ed. Sinauer; Sunderland, MA: 2001. Ion Channels of Excitable Membranes. 131–160. [Google Scholar]
- 2.Jan L.Y., Jan Y.N. Voltage-gated and inwardly rectifying potassium channels. J. Physiol. 1997;505:267–282. doi: 10.1111/j.1469-7793.1997.267bb.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li-Smerin Y., Swartz K.J. Gating modifier toxins reveal a conserved structural motif in voltage-gated Ca2+ and K+ channels. Proc. Natl. Acad. Sci. USA. 1998;95:8585–8589. doi: 10.1073/pnas.95.15.8585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang Y., Lee A., Chen J., Ruta V., Cadene M. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 5.Long S.B., Campbell E.B., MacKinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 6.del Camino D., Yellen G. Tight steric closure at the intracellular activation gate of a voltage-gated K+ channel. Neuron. 2001;32:649–656. doi: 10.1016/s0896-6273(01)00487-1. [DOI] [PubMed] [Google Scholar]
- 7.del Camino D., Kanevsky M., Yellen G. Status of the intracellular gate in the activated-not-open state of Shaker K+ channels. J. Gen. Physiol. 2005;126:419–428. doi: 10.1085/jgp.200509385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aggarwal S.K., MacKinnon R. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 1996;16:1169–1177. doi: 10.1016/s0896-6273(00)80143-9. [DOI] [PubMed] [Google Scholar]
- 9.Pathak M.M., Yarov-Yarovoy V., Agarwal G., Roux B., Barth P. Closing in on the resting state of the Shaker K+ channel. Neuron. 2007;56:124–140. doi: 10.1016/j.neuron.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 10.Long S.B., Tao X., Campbell E.B., MacKinnon R. Atomic structure of s voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 11.Gandhi C.S., Isacoff E.Y. Molecular models of voltage sensing. J. Gen. Physiol. 2002;120:455–463. doi: 10.1085/jgp.20028678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lecar H., Larsson H.P., Grabe M. Electrostatic model of S4 motion in voltage-gated ion channels. Biophys. J. 2003;85:2854–2864. doi: 10.1016/S0006-3495(03)74708-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chanda B., Asamoah O.K., Blunck R., Roux B., Bezanilla F. Gating charge displacement in voltage-gated ion channels involves limited transmembrane movement. Nature. 2005;436:852–856. doi: 10.1038/nature03888. [DOI] [PubMed] [Google Scholar]
- 14.Campos F.V., Chanda B., Roux B., Bezanilla F. Two atomic constraints unambiguously position the S4 segment relative to S1 and S2 segments in the closed state of Shaker K channel. Proc. Natl. Acad. Sci. USA. 2007;104:7904–7909. doi: 10.1073/pnas.0702638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hodgkin A.L., Huxley A.F. A quantitative description of membrane current and its application to conduction and excitation in nerve. J. Physiol. 1952;117:500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Armstrong C.M., Bezanilla F. Currents related to movement of the gating particles of the sodium channels. Nature. 1973;242:459–461. doi: 10.1038/242459a0. [DOI] [PubMed] [Google Scholar]
- 17.Zagotta W.N., Hoshi T., Aldrich R.W. Shaker potassium channel gating III: evaluation of kinetic models for activation. J. Gen. Physiol. 1994;103:321–362. doi: 10.1085/jgp.103.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bezanilla F., Perozo E., Stefani E. Gating of Shaker K+ channels: II. The components of gating currents and a model of channel activation. Biophys. J. 1994;66:1011–1021. doi: 10.1016/S0006-3495(94)80882-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perozo E., MacKinnon R., Bezanilla F., Stefani E. Gating currents from a non-conducting mutant reveal open-closed conformations in Shaker K+ channels. Neuron. 1993;11:353–358. doi: 10.1016/0896-6273(93)90190-3. [DOI] [PubMed] [Google Scholar]
- 20.Horn R., Ding S., Gruber H.J. Immobilizing the moving parts of voltage-gated ion channels. J. Gen. Physiol. 2000;116:461–476. doi: 10.1085/jgp.116.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ledwell J.L., Aldrich R.W. Mutations in the S4 region isolate the final voltage-dependent cooperative step in potassium channel activation. J. Gen. Physiol. 1999;113:389–414. doi: 10.1085/jgp.113.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soler-Llavina G.J., Chang T.-H., Swartz K.J. Functional interactions at the interface between voltage-sensing and pore domains in the Shaker Kv channel. Neuron. 2006;52:623–634. doi: 10.1016/j.neuron.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 23.Schoppa N.E., Sigworth F.J. Activation of Shaker potassium channels I. Characterization of voltage-dependent transitions. J. Gen. Physiol. 1998;111:271–294. doi: 10.1085/jgp.111.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mannuzzu L.M., Isacoff E.Y. Independence and cooperativity in rearrangements of a potassium channel voltage sensor revealed by single subunit fluorescence. J. Gen. Physiol. 2000;115:257–268. doi: 10.1085/jgp.115.3.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Z., Klem A.M., Ramu Y. Coupling between voltage sensors and activation gate in voltage-gated K+ channels. J. Gen. Physiol. 2002;120:663–676. doi: 10.1085/jgp.20028696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yifrach O. Hill coefficient for estimating the magnitude of cooperativity in gating transitions of voltage-dependent ion channels. Biophys. J. 2004;87:822–830. doi: 10.1529/biophysj.104.040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Labro A.J., Raes A.L., Snyders D.J. Coupling of voltage sensing to channel opening reflects intrasubunit interactions in Kv channels. J. Gen. Physiol. 2005;125:71–80. doi: 10.1085/jgp.200409194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tristani-Firouzi M., Chen J., Sanguinetti M.C. Interactions between S4–S5 linker and S6 transmembrane domain modulate gating of HERG K+ channels. J. Biol. Chem. 2002;277:18994–19000. doi: 10.1074/jbc.M200410200. [DOI] [PubMed] [Google Scholar]
- 29.Ding S., Horn R. Effect of S6 tail mutations on charge movement in Shaker potassium channels. Biophys. J. 2003;84:295–305. doi: 10.1016/S0006-3495(03)74850-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Decher N., Chen J., Sanguinetti M.C. Voltage-dependent gating of hyperpolarization-activated, cyclic nucleotide-gated pacemaker channels. J. Biol. Chem. 2004;279:13859–13865. doi: 10.1074/jbc.M313704200. [DOI] [PubMed] [Google Scholar]
- 31.Ash W.L., Zlomislic M.R., Oloo E.O., Tieleman D.P. Computer simulations of membrane proteins. Biochim. Biophys. Acta. 2004;1666:158–189. doi: 10.1016/j.bbamem.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 32.Gumbart J.Y., Wang A., Aksimentiev E., Schulten K. Molecular dynamics simulations of proteins in lipid bilayers. Curr. Opin. Struct. Biol. 2005;15:423–431. doi: 10.1016/j.sbi.2005.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kandt C., Ash W.L., Tieleman D.P. Setting up and running molecular dynamics simulations of membrane proteins. Methods. 2007;41:475–488. doi: 10.1016/j.ymeth.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 34.Sotomayor M., Vasquez V., Perozo E., Schulten K. Ion conduction through MscS as determined by electrophysiology and simulation. Biophys. J. 2007;92:886–902. doi: 10.1529/biophysj.106.095232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Treptow W., Maigret B., Chipot C., Tarek M. Coupled motions between pore and voltage-sensor domains: a model for Shaker B, a voltage-gated potassium channel. Biophys. J. 2004;87:2365–2379. doi: 10.1529/biophysj.104.039628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Treptow W., Tarek M. Environment of the gating charges in the Kv1.2 Shaker potassium channel. Biophys. J. 2006;90:L64–L66. doi: 10.1529/biophysj.106.080754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jogini V., Roux B. Dynamics of the Kv1.2 voltage-gated K+ channel in a membrane environment. Biophys. J. 2007;93:3070–3082. doi: 10.1529/biophysj.107.112540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tieleman D.P., Berendsen H.J.C. Molecular dynamics simulations of fully hydrated DPPC with different macroscopic boundary conditions and parameters. J. Chem. Phys. 1996;105:4871–4880. [Google Scholar]
- 39.Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., Hermans J. D. Reidel Publishing Co.; Dordrecht, The Netherlands: 1981. Intermolecular Forces, Interaction Models for Water in Relation to Protein Hydration. 331–342. [Google Scholar]
- 40.Nishizawa M., Nishizawa K. Molecular dynamics simulations of a stretch-activated channel inhibitor GsMTx4 with lipid membranes: two binding modes and effects of lipid structure. Biophys. J. 2007;92:4233–4243. doi: 10.1529/biophysj.106.101071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 42.Darden T., York D., Pedersen L. Particle mesh Ewald: an Nlog(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 43.Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., DiNola A., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 44.Ruta V., Chen J., MacKinnon R. Calibrated measurement of gating-charge arginine displacement in the KvAP voltage-dependent K+ channel. Cell. 2005;123:463–475. doi: 10.1016/j.cell.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 45.Shrivastava I.H., Sansom M.S.P. Simulations of ion permeation through a potassium channel: molecular dynamics of KcsA in a phospholipid bilayer. Biophys. J. 2000;78:557–570. doi: 10.1016/S0006-3495(00)76616-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tieleman D.P. The molecular basis of electroporation. BMC Biochem. 2004;5:10. doi: 10.1186/1471-2091-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smart O.S., Goodfellow J.M., Wallace B.A. The pore dimensions of gramicidin A. Biophys. J. 1993;65:2455–2460. doi: 10.1016/S0006-3495(93)81293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar S., Bouzida D., Swensen R.H., Kollman P.A., Rosenberg J.M. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J. Comput. Chem. 1992;13:1011–1021. [Google Scholar]
- 49.Kanevsky M., Aldrich R.W. Determinants of voltage-dependent gating and open-state stability in the S5 segment of Shaker potassium channels. J. Gen. Physiol. 1999;114:215–242. doi: 10.1085/jgp.114.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kitaguchi T., Sukhareva M., Swartz K.J. Stabilizing the closed S6 gate in the Shaker Kv channel through modification of a hydrophobic seal. J. Gen. Physiol. 2004;124:319–332. doi: 10.1085/jgp.200409098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Magidovich E., Orr I., Fass D., Abdu U., Yifrach O. Intrinsic disorder in the C-terminal domain of the Shaker voltage-activated K+ channel modulates its interaction with scaffold proteins. Proc. Natl. Acad. Sci. USA. 2007;104:13022–13027. doi: 10.1073/pnas.0704059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nishizawa M., Nishizawa K. Molecular dynamics simulation of Kv channel voltage sensor helix in a lipid membrane with applied electric field. Biophys. J. 2008;95:1729–1744. doi: 10.1529/biophysj.108.130658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lewis A., Jogini V., Blachowicz L., Laine M., Roux B. Atomic constrains between the voltage sensor and the pore domain in a voltage-gated K+ channel of known structure. J. Gen. Physiol. 2008;131:549–561. doi: 10.1085/jgp.200809962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bright J.N., Shrivastava I.H., Cordes F.S., Sansom M.S.P. Conformational dynamics of helix S6 from Shaker potassium channel: simulation studies. Biopolymers. 2002;64:303–313. doi: 10.1002/bip.10197. [DOI] [PubMed] [Google Scholar]
- 55.Smith-Maxwell C.J., Ledwell J.L., Aldrich R.W. Uncharged S4 residues and cooperativity in voltage-dependent potassium channel activation. J. Gen. Physiol. 1998;111:421–439. doi: 10.1085/jgp.111.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hackos D.H., Chang T.-H., Swartz K.J. Scanning the intracellular S6 activation gate in the Shaker K+ channel. J. Gen. Physiol. 2002;119:521–531. doi: 10.1085/jgp.20028569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marks T.N., Jones S.W. Calcium currents in the A7r5 smooth muscle-derived cell line. An allosteric model for calcium channel activation and dihydropyridine agonist action. J. Gen. Physiol. 1992;99:367–390. doi: 10.1085/jgp.99.3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cox D.H.J.C., Aldrich R.W. Allosteric gating of a large conductance Ca-activated K+ channel. J. Gen. Physiol. 1997;110:257–281. doi: 10.1085/jgp.110.3.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horrigan F.T., Aldrich R.W. Allosteric voltage gating of potassium channels II. Mslo channel gating charge movement in the absence of Ca(2+) J. Gen. Physiol. 1999;114:305–336. doi: 10.1085/jgp.114.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Altomare C., Bucchi A., Camatini E., Baruscotti M., Viscomi C. Integrated allosteric model of voltage gating of HCN channels. J. Gen. Physiol. 2001;117:519–532. doi: 10.1085/jgp.117.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sukhareva M., Hackos D.H., Swartz K.J. Constitutive activation of the Shaker Kv channel. J. Gen. Physiol. 2003;122:541–556. doi: 10.1085/jgp.200308905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chapman M.L., VanDongen A.M.J. K channel subconductance levels results from heteromeric pore conformations. J. Gen. Physiol. 2005;126:87–103. doi: 10.1085/jgp.200509253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Monod J., Wyman J., Changeux J.P. On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 64.Changeux J.P., Edelstein S.J. Allosteric receptors after 30 years. Neuron. 1998;21:959–980. doi: 10.1016/s0896-6273(00)80616-9. [DOI] [PubMed] [Google Scholar]
- 65.Chapman M.L., VanDongen H.M.A., VanDongen A.M.L. Activation-dependent subconductance levels in the drk1 K channel suggest a subunit basis for ion permeation and gating. Biophys. J. 1997;72:708–719. doi: 10.1016/s0006-3495(97)78707-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng J., Sigworth F.J. Selectivity changes during activation of mutant Shaker potassium channels. J. Gen. Physiol. 1997;110:101–117. doi: 10.1085/jgp.110.2.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hoshi T., Zagotta W.N., Aldrich R.W. Shaker potassium channel gating I: transition near the open state. J. Gen. Physiol. 1994;103:249–278. doi: 10.1085/jgp.103.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


