Abstract
Cytokinesis requires coordination of actomyosin ring (AMR) contraction with rearrangements of the plasma membrane and extracellular matrix. In Saccharomyces cerevisiae, new membrane, the chitin synthase Chs2 (which forms the primary septum [PS]), and the protein Inn1 are all delivered to the division site upon mitotic exit even when the AMR is absent. Inn1 is essential for PS formation but not for Chs2 localization. The Inn1 C-terminal region is necessary for localization, and distinct PXXP motifs in this region mediate functionally important interactions with SH3 domains in the cytokinesis proteins Hof1 (an F-BAR protein) and Cyk3 (whose overexpression can restore PS formation in inn1Δ cells). The Inn1 N terminus resembles C2 domains but does not appear to bind phospholipids; nonetheless, when overexpressed or fused to Hof1, it can provide Inn1 function even in the absence of the AMR. Thus, Inn1 and Cyk3 appear to cooperate in activating Chs2 for PS formation, which allows coordination of AMR contraction with ingression of the cleavage furrow.
Introduction
Cytokinesis in animal and fungal cells involves actomyosin ring (AMR) contraction and targeted plasma membrane and ECM rearrangements, which appear to be interdependent processes (Balasubramanian et al., 2004; Strickland and Burgess, 2004). Many components of the AMR and many proteins involved in targeted membrane trafficking have been identified, most of which are conserved from yeast to humans (Balasubramanian et al., 2004; Echard et al., 2004; Skop et al., 2004). The major question at present is how these components interact to form the efficient and high fidelity molecular machines that drive cytokinesis in a spatially and temporally coordinated fashion.
Targeted membrane trafficking presumably increases membrane surface area in the cleavage furrow and also delivers specific molecules (whose precise nature may differ between cell types) that are required for cytokinesis. In the budding yeast Saccharomyces cerevisiae, one important function of targeted membrane trafficking is delivery of the chitin synthase Chs2 (Chuang and Schekman, 1996; VerPlank and Li, 2005), which is chiefly responsible for assembly of the primary septum (PS; Shaw et al., 1991). The PS is a thin chitin-rich layer of cell wall that forms centripetally at the mother–bud neck during AMR contraction; once PS formation is complete, secondary septa (SS) are laid down on both sides of the PS. Deletion of MYO1, which encodes the sole type-II myosin in S. cerevisiae, eliminates the AMR but is not lethal in most strain backgrounds. However, myo1Δ cells are typically delayed in cytokinesis and/or cell separation (Rodriguez and Paterson, 1990; Bi et al., 1998), and transmission EM (TEM) has shown that although both PS and SS can form, they are frequently misoriented and/or disorganized in structure (Schmidt et al., 2002; unpublished data). Thus, the AMR and its contraction appear to guide membrane trafficking such that cleavage furrow and PS formation are properly oriented and organized (Vallen et al., 2000; Bi, 2001). In contrast, deletion of CHS2 completely blocks PS formation and results in abortive AMR contraction, suggesting that the PS may stabilize the contracting ring or the associated plasma membrane (Bi, 2001; Schmidt et al., 2002; VerPlank and Li, 2005).
The viability of myo1Δ cells indicates that AMR-independent mechanisms, presumably involving septum formation, can sustain cytokinesis in yeast (Bi et al., 1998). The proteins Iqg1, Cyk3, Hof1, and Mlc1 appear to play important roles in the AMR-independent pathway. Iqg1 is the sole IQGAP protein in S. cerevisiae and is essential for AMR formation (Epp and Chant, 1997; Lippincott and Li, 1998; Shannon and Li, 1999), but the near lethality of an iqg1Δ mutation can be suppressed by overexpression of Cyk3 without restoration of the AMR (Korinek et al., 2000). In addition, the growth defect of a myo1Δ mutant can be suppressed by overexpression of either Iqg1 or Cyk3 (Ko et al., 2007). Cyk3 contains SH3 and putative transglutaminase (Makarova et al., 1999) domains, whereas Hof1 contains SH3 and F-BAR (putative membrane interaction domain; Heath and Insall, 2008) domains. Deletion of either CYK3 or HOF1 has no effect on AMR assembly, but either deletion causes severe synthetic growth defects in combination with myo1Δ (Korinek et al., 2000; Vallen et al., 2000). In addition, hof1Δ and cyk3Δ are synthetically lethal (or nearly so) with each other. Mlc1 is a light chain for Myo1 and for the type V myosin Myo2 as well as for Iqg1, whose localization to the neck it appears to mediate (Stevens and Davis, 1998; Boyne et al., 2000; Shannon and Li, 2000; Luo et al., 2004).
Collectively, the aforementioned observations have led to the hypotheses that Hof1 and Cyk3 play distinct roles in septum formation downstream of Iqg1/Mlc1 (Bi, 2001; Luo et al., 2004; unpublished data) and that yeast cells can tolerate either loss of the AMR (myo1Δ) or a partial defect in septum formation (hof1Δ or cyk3Δ) but not both. To identify other genes involved in the AMR-dependent and -independent pathways of cytokinesis, we performed a screen for mutations that are synthetically lethal in combination with a hof1Δ mutation. We identified a variety of previously known cytokinesis genes and a previously uncharacterized gene, ORF YNL152W, which has also recently been studied (and named INN1) by Sanchez-Diaz et al. (2008). In this study, we report our functional analyses of the role of Inn1 in cytokinesis, which suggest that Inn1 interacts with Hof1 and Cyk3 to promote PS formation in coordination with AMR contraction. Our conclusions differ radically from those reached by Sanchez-Diaz et al. (2008).
Results
Identification of INN1 in a hof1Δ synthetic lethal screen
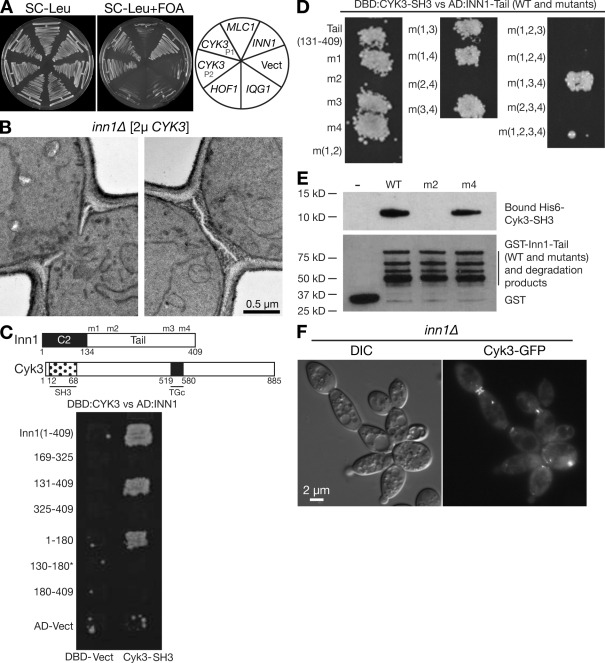
A hof1Δ mutation is not lethal by itself but is lethal in combination with several other mutations affecting cytokinesis proteins (see Introduction). To identify additional cytokinesis proteins, we used a colony-sectoring assay (Bender and Pringle, 1991) to screen systematically for ethyl methanesulphonate–induced mutations that were synthetically lethal with hof1Δ. From ∼33,000 colonies screened, we found 38 such mutations, which defined at least 13 genes (Table S1), 11 of which encode proteins already known to be involved in cytokinesis. These proteins are in four general groups: septins and proteins that regulate septin function (Cdc12, Gin4, Elm1, and Bni5), proteins involved in the function of the AMR (Myo1 and Bni1), proteins that appear to regulate both the AMR and some aspects of membrane and/or cell wall deposition (Mlc1 and Iqg1), and proteins that regulate septal cell wall assembly and/or cell separation (Chs2, Cyk3, and Psa1). The synthetic lethality of hof1Δ with mutations in MYO1, BNI1, CYK3, and BNI5 was shown previously (Korinek et al., 2000; Vallen et al., 2000; Lee et al., 2002). It should be noted that we recovered point mutations in several essential (or nearly so) genes, which would have been missed in a genome-wide synthetic array analysis using the viable deletion strains (Tong et al., 2001).
The twelfth gene identified was YNL152W/INN1, uncharacterized at the time but subsequently studied also by Sanchez-Diaz et al. (2008). INN1 is predicted to encode a protein of 409 amino acids with a possible C2 domain at its N terminus (Sanchez-Diaz et al., 2008) and multiple PXXP motifs in its C-terminal region (Fig. S1). The roles of these domains are discussed in the following sections. Inn1 has unambiguous homologues in a variety of other fungi; homologues outside the fungi are less clear and may be limited to the putative C2 domains.
Essential role of Inn1 in PS formation
Tetrad analysis of an INN1/inn1Δ heterozygous diploid on YPD-rich medium suggested that INN1 is an essential gene (Sanchez-Diaz et al., 2008; unpublished data), but we found that inn1Δ cells could grow vegetatively, although slowly (Fig. 1 A). Similar observations have been made with other cytokinesis mutants (Bulawa and Osmond, 1990; unpublished data). The inn1Δ cells formed extensive cell clusters with abnormal-looking septal regions (Fig. 1 B, left); decoration of the plasma membrane with GFP-Ras2 (Fig. 1 B, right) revealed that cytokinesis (cytoplasmic separation) was complete in some of these septal regions (neck 1) but not in others (neck 2). These data suggest that Inn1 plays a role in membrane invagination, septum synthesis, and/or cell separation.
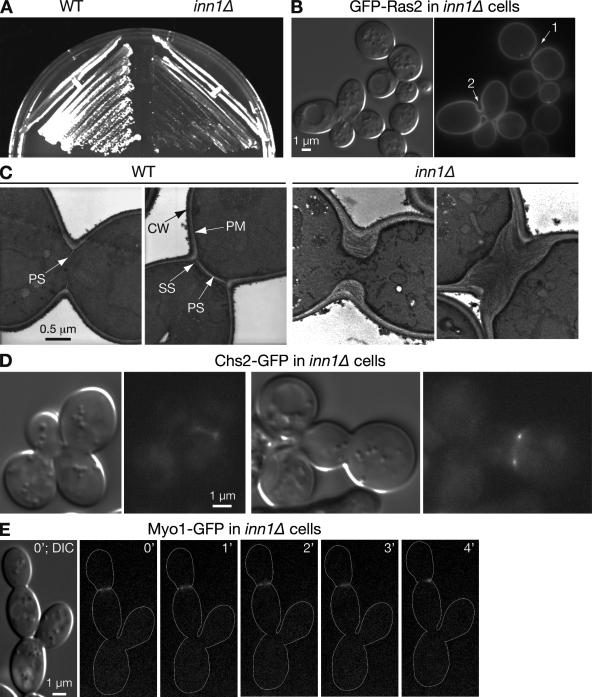
Figure 1.
Dependence of PS formation on Inn1. (A) Slow growth of inn1Δ cells. Wild-type (WT; YEF473A) and inn1Δ (YEF5216) cells were streaked on an SC plate and incubated at 25°C for 3 d. (B) Abnormal but complete cytokinesis in inn1Δ cells. YEF5216 cells carrying plasmid pRS315-GFP-RAS2 were grown to exponential phase in SC-Leu liquid medium at 23°C and imaged by DIC and fluorescence microscopy. 1 and 2 indicate bud necks. (C) Absence of PS formation in inn1Δ cells. Strains YEF473A and YEF5216 were grown to exponential phase in SC medium at 24°C and examined by TEM. CW, cell wall; PM, plasma membrane. (D) Localization of Chs2 to the neck in inn1Δ cells. Strain LY1373 (inn1Δ CHS2-GFP [pUG36-INN1]) was transferred from an SC plate to an SC+FOA plate, incubated overnight at 25°C to select for loss of the URA3-marked INN1 plasmid, and examined by fluorescence microscopy. (E) Abnormal contraction of the AMR in inn1Δ cells. inn1Δ MYO1-GFP cells (YEF5291) were observed by time-lapse microscopy. Cell bodies are outlined in the GFP panels.
To explore these possibilities, we used TEM. In wild-type cells, a thin, chitinous PS forms first and is sandwiched by layers of SS (see Introduction; Fig. 1 C, left). In contrast, in 50 inn1Δ cells scored, no sign of a PS could be seen; instead, the necks filled with SS-like material (Fig. 1 C, right). Similar results were obtained when temperature-sensitive inn1 mutants (created by PCR mutagenesis) were incubated at restrictive temperature (unpublished data). Because digestion of the PS normally leads to cell separation (Yeong, 2005), the absence of the PS in inn1 mutant cells presumably accounts for the delay in cell separation and resultant formation of cell clusters. The lack of PS formation might mean that Inn1 is required for recruitment of Chs2 to the mother–bud neck. However, the localization of GFP-tagged Chs2 to the vicinity of the neck (Fig. 1 D) was similar to that seen in wild type (Chuang and Schekman, 1996; VerPlank and Li, 2005; Zhang et al., 2006). Thus, Inn1 presumably controls PS formation by controlling the activation and/or precise localization of Chs2.
Assembly and contraction of the AMR occurred in inn1Δ cells. However, the actin rings were generally less tight and stained more faintly than those in wild-type cells (Fig. S2), and the Myo1-GFP rings invariably (n = 7) appeared to detach from part of the plasma membrane within 3–4 min after the initiation of contraction, resulting in an asymmetrically localized dot at one side of the neck (Fig. 1 E) in contrast to the symmetrical AMR contraction that occurs over 6–8 min in wild-type cells under these conditions (Bi et al., 1998; Vallen et al., 2000). This behavior is similar to that of the AMR in chs2Δ cells (Bi, 2001; Schmidt et al., 2002; VerPlank and Li, 2005), which is consistent with the hypothesis that Inn1 plays an essential role in PS formation.
Mitotic exit network (MEN)–dependent, AMR-independent localization of Inn1 to the division site
Analysis of Inn1 levels using cells that had been synchronized in G1 indicated that Inn1 is present at an approximately constant level throughout the cell cycle (unpublished data). However, time-lapse analysis showed that Inn1-GFP did not localize to the neck until the septin hourglass split into two cortical rings (Fig. 2 A and Video 1), an event that is under the control of the MEN (Lippincott et al., 2001). Once a ring of Inn1-GFP was visible at the neck, it began to contract almost immediately. Contraction from a full-sized ring to a dot took ∼8 min (n = 9), as did the centripetal synthesis of the septum (Fig. 2 A, differential interference contrast [DIC] images). Immediately after contraction, Inn1-GFP disappeared from the neck.
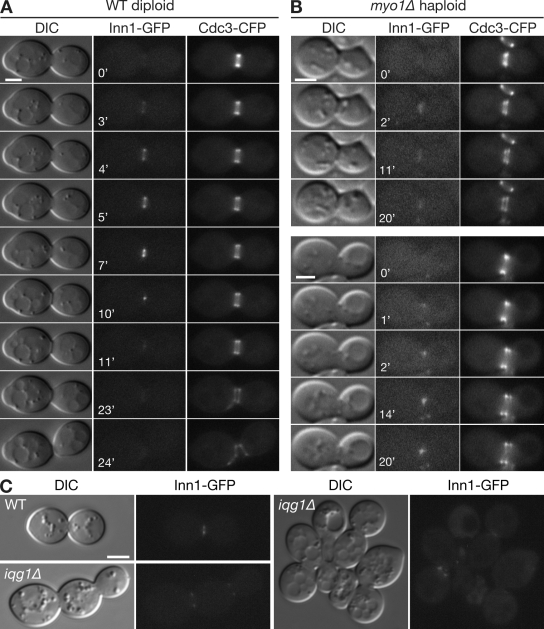
Figure 2.
Localization of Inn1 to the bud neck at mitotic exit in wild-type and AMR-deficient cells. (A and B) Strains LY1302 (INN1-GFP; A) and YEF5293 (myo1Δ INN1-GFP; B) were transformed with plasmid YCp111-CDC3-CFP and observed by time-lapse microscopy (Videos 1–4). (C) Wild-type (WT; RNY2395) and iqg1Δ (RNY2393) cells expressing Inn1-GFP and containing plasmid YCp50-IQG1 were grown overnight at 25°C on an SC+FOA plate to eliminate the plasmid, were scraped from the plate, and imaged by DIC and fluorescence microscopy. Bars, 2 µm.
These data suggest that the localization of Inn1 is regulated posttranslationally and might occur in response to activation of the MEN in which a Polo kinase (Cdc5) and a GTPase-controlled kinase cascade (Cdc15, Dbf2, and Dbf20) lead to activation of the protein phosphatase Cdc14 (Stegmeier and Amon, 2004). The MEN controls mitotic exit (by down-regulating Cdk/mitotic cyclins) and cytokinesis in a largely independent manner whose mechanisms remain obscure (Balasubramanian et al., 2004). The MEN is not required for AMR assembly but is required for its contraction and for septum formation (Vallen et al., 2000; Lippincott et al., 2001; Hwa Lim et al., 2003). To ask whether Inn1 localization depends on the MEN, we examined various temperature-sensitive mutants. As expected, Inn1-GFP localized to the neck in large-budded cells of all MEN mutants at permissive temperature (Fig. 3 A, top), although the percentage of cells in which localized Inn1-GFP could be seen was less than in wild-type cells (Fig. 3 B). In contrast, at restrictive temperature, Inn1-GFP failed to accumulate at the necks of large-budded cells in all MEN mutants (Fig. 3, A [bottom] and B), suggesting that Inn1 localization to the bud neck is directly or indirectly regulated by the MEN.
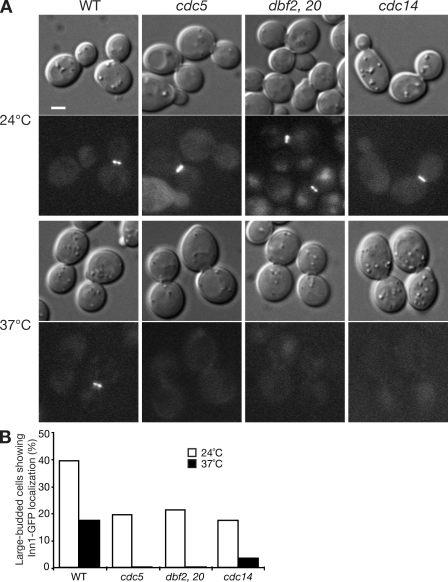
Figure 3.
Dependence of Inn1 localization on the MEN. (A and B) Wild-type (WT; LY1313), cdc5 (LY1357), dbf2 dbf20 (LY1355), and cdc14 (LY1360) cells expressing Inn1-GFP were grown to exponential phase in YM-P–rich medium at 24°C and shifted to 37°C for 3.5 (LY1360) or 2.5 h (the other strains) before imaging (A) and scoring large-budded cells (B) for Inn1-GFP localization. In B, the numbers of cells scored were as follows: 24°C, 62 for wild type and 102–131 for the mutants; 37°C, 78 for wild type and 177–199 for the mutants. The experiments were repeated three times with similar results. Bar, 2 µm.
The contraction of the Inn1-GFP ring was almost identical to that of the Myo1-GFP (Bi et al., 1998) and Iqg1-GFP (Shannon and Li, 1999) rings, suggesting that Inn1 might be associated with the AMR. Indeed, Sanchez-Diaz et al. (2008) reported that Inn1 failed to localize in either Myo1- or Iqg1-depleted cells. In contrast, we found that Inn1 localized to the neck at the normal time in myo1Δ cells (Fig. 2 B; Table I; and Videos 2–4). However, the appearance and behavior of the Inn1-GFP signal were abnormal; it usually appeared either as a faint band that never displayed a clear contraction (in 10 of the 19 cells observed by time-lapse analysis; Fig. 2 B, top; and Video 3) or as one or two relatively bright dots that moved asymmetrically across the bud neck (in the other nine cells; Fig. 2 B, bottom; and Video 4). Similarly, in random fields of cells, 16% of myo1Δ cells with split septin rings displayed an asymmetric line or dot of Inn1-GFP at the neck, whereas this was rarely seen in control cells (Table I). This behavior might reflect the asymmetric PS formation that occurs in some myo1Δ cells (unpublished data). We also observed Inn1-GFP localization to the neck in iqg1Δ cells (Fig. 2 C), although the signal was generally weaker than in wild type. Collectively, our results indicate that the normal contraction of the Inn1 ring depends on the AMR, but the initial localization of Inn1 does not. This suggests that Inn1 is not a true component of the AMR but rather part of a functional complex that associates and cooperates with it.
Table I.
Localization of Inn1-GFP in wild-type and cytokinesis mutant strains
| Strain | Percentage of cells with the indicated localization pattern | ||||
| Faint or no signal | Symmetric line or two dotsa | One dot (center)b | Asymmetric dot or lineb | Otherc | |
| myo1Δ [YCp50-MYO1] | 62d | 27 | 11 | 0 | 0 |
| myo1Δ | 42 | 22 | 11 | 16 | 9 |
| Wild type | 68d | 20 | 11 | 2 | 0 |
| hof1Δ | 56 | 15 | 12 | 17 | 0 |
| cyk3Δ | 35 | 54 | 9 | 2 | 0 |
| hof1Δ cyk3Δe | 54 | 24 | 12 | 10 | 0 |
After transformation of each strain with plasmid YCp111-CDC3-CFP and growth to exponential phase in SC-Leu or SC-Leu-Ura liquid medium at 24°C, cells with split septin rings were scored in strains LY1364 (myo1Δ INN1-GFP [YCp50-MYO1]; n = 81), YEF5293 (myo1Δ INN1-GFP; n = 95), LY1314 (INN1-GFP; n = 66), LY1328 (hof1Δ INN1-GFP; n = 94), LY1321 (cyk3Δ INN1-GFP; n = 117), and LY1325 (hof1Δ cyk3Δ INN1-GFP; n = 50). Strains LY1328 and LY1325 were first cured of their URA3 HOF1 plasmids by growth on an FOA plate. The patterns of Inn1 localization were assessed by 3D microscopy as described in Materials and methods (Videos 1–7).
Both types of images presumably represent views of a more or less normal ring of Inn1-GFP.
If an Inn1-GFP dot was positioned within one third of the diameter of the Cdc3-CFP ring from either side, it was scored as asymmetric; if within the middle one third of the neck, it was scored as one dot (center). Asymmetric lines presumably represent asymmetrically contracting rings.
Other indicates a variety of asymmetric patterns, including asymmetries along the mother–bud axis (presumably related to the misdirected membrane invagination that occurs in many myo1Δ cells; unpublished data).
The higher number of cells with faint or no signal in wild-type strains, in comparison with myo1Δ and cyk3Δ strains, presumably reflects the more efficient completion of cytokinesis and corresponding rapid disappearance of the Inn1-GFP signal in wild-type cells.
Like several other mutants (see Introduction and Results), hof1Δ cyk3Δ strains appear to be inviable on rich medium but can be cultured on SC medium.
Inn1–Hof1 interaction and its role in the symmetric localization of Inn1 at the neck
The C-terminal region of Inn1 contains eight PXXP motifs, which represent generic binding sites for SH3 domains (Feller et al., 1994) and might interact with the Hof1 SH3 domain (Ito et al., 2001; Tong et al., 2002). Inn1 was also among the Hof1-binding proteins identified by mass spectrometry (unpublished data). To examine possible Inn1–Hof1 interaction during the cell cycle, we used a coimmunoprecipitation assay. We observed that Inn1 interacted strongly with Hof1 throughout the 90 min after release from an MEN block (Fig. 4 A), suggesting that Inn1 forms a tight complex with Hof1 before, during, and after cytokinesis.
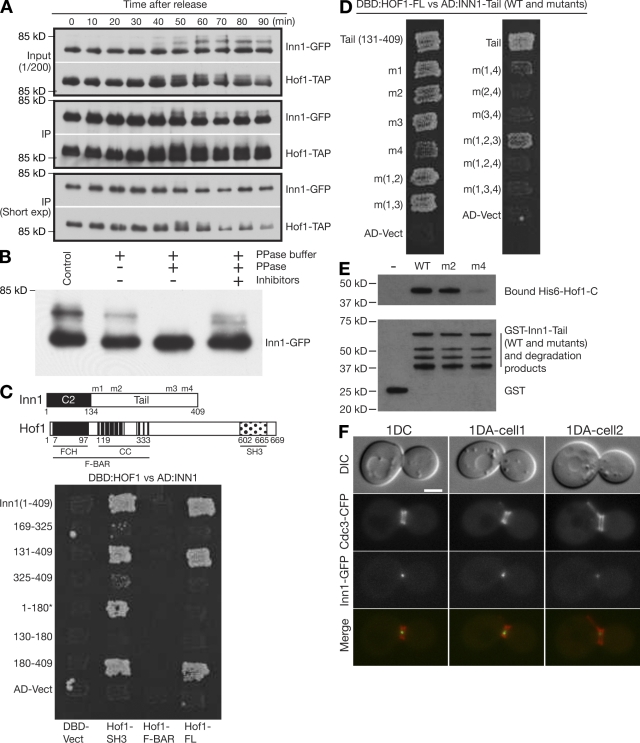
Figure 4.
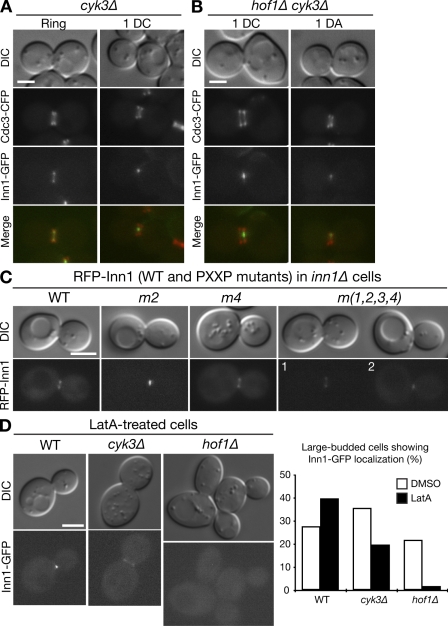
Inn1–Hof1 interaction and its role in the symmetric localization of Inn1 at the neck. (A) Coimmunoprecipitation and cell cycle–dependent modification of Inn1 and Hof1. Strain MOY157 (INN1-GFP HOF-TAP cdc15-2) was grown to exponential phase in YM-P medium at 24°C, shifted to 37°C for 2.5 h to synchronize cells at mitotic exit using the cdc15-2 block, released to permissive temperature by rapidly cooling to 24°C, and sampled at intervals. Hof1-TAP was precipitated from protein extracts, and samples of the extracts (input) and precipitates (IP) were analyzed by SDS-PAGE and immunoblotting. In a control in which no TAP-tagged protein was present, no Inn1-GFP was detected in the precipitate (not depicted). (B) Phosphorylation of Inn1. Strain MOY215 (INN1-GFP cdc15-2) was synchronized as in A and sampled 45 min after release. Inn1-GFP was immunoprecipitated and subjected to phosphatase treatments as indicated. (C) Two-hybrid analysis of Inn1–Hof1 interaction. The diagram shows the domain structures of Inn1 (m1-m4 are the mutations introduced into the PXXP motifs; see Results; Fig. S1) and Hof1. FCH, FER/CIP4 homology; CC, coiled coil; SH3, Src homology 3. Various Inn1 fragments carried on the AD (AD-Vect) were tested pairwise for interaction with full-length Hof1 (Hof1-FL), Hof1 amino acids 576–669 (Hof1-SH3), and Hof1 amino acids 1–340 (Hof1–F-BAR) carried on the DBD vector (DBD-Vect). Asterisk indicates that Inn1(1–180) interacted with Hof1-SH3 for unknown reasons. (D) Role of Inn1 amino acids 377–383 (PXXPPXP) in the Inn1–Hof1 interaction. Two-hybrid analysis was conducted using full-length Hof1-DBD and Inn1(131–409)-AD. The Inn1 sequence was wild type (tail[131–409]) or carried mutations m1, m2, m3, and/or m4 individually or in combinations. (E) Direct binding of Inn1 to Hof1 and its mediation by Inn1 amino acids 377–383 (PKLPPLP). Purified GST-Inn1-tail (amino acids 131–409; wild type [WT] or carrying mutation m2 or m4) and His6-Hof1-C (amino acids 341–669) were tested for binding in vitro as described in Materials and methods. (F) Asymmetric localization of Inn1 at the neck in hof1Δ cells. Strain LY1328 (INN1-GFP hof1Δ [pRS316-HOF1]) was transformed with plasmid YCp111-CDC3-CFP and incubated on an FOA plate to eliminate the HOF1 plasmid. Cells from a population growing exponentially in SC-Leu medium at 24°C were examined by 3D microscopy (see Materials and methods). 1DC, one central dot (as typically observed in wild-type cells); 1DA, one asymmetric dot (as often observed in hof1Δ cells; Table I; and Videos 5 and 6). Bar, 2 µm.
Like Hof1 (Fig. 4 A; Vallen et al., 2000; Blondel et al., 2005; Corbett et al., 2006), Inn1 undergoes cell cycle–regulated modification, as indicated by the multiple retarded forms of Inn1 seen by SDS-PAGE (Fig. 4 A). The modification appears to be phosphorylation, as phosphatase treatment reduced all high molecular weight forms of Inn1 to a single band (Fig. 4 B). The modified forms of both Inn1 and Hof1 first appear at ∼40 min after release from the MEN block, which corresponds closely to the time at which AMR contraction and PS formation occur under these conditions, as judged from parallel time course analyses (unpublished data).
To define the interacting regions of Inn1 and Hof1, we used two-hybrid analysis. As shown in Fig. 4 C, full-length Hof1 interacted with the Inn1 C terminus (residues 180–409) but not with the N terminus (residues 1–180). Any Hof1 fragment lacking the C-terminal SH3 domain failed to interact with any region of Inn1, whereas the isolated Hof1 SH3 domain was sufficient for binding to the Inn1 C terminus (and also weakly or perhaps artifactually to the Inn1 N terminus; Fig. 4 C; unpublished data). When P to A mutations (m1-m4; Fig. 4 C and Fig. S1) were introduced into the Inn1 PXXP motifs, mutations m1-m3, alone or in combination, had no detectable effect on binding to Hof1, but mutation m4 dramatically reduced binding particularly when combined with m2 or m3 (Fig. 4 D). These data suggest that the PKLPPLP motif at Inn1 amino acids 377–383 is primarily responsible for interaction with the Hof1 SH3 domain, although there may also be some interaction with the PIPPLP (amino acids 160–165) and PPLPPIP (amino acids 329–325) motifs.
To determine whether Inn1 interacts directly with Hof1, we used a pull-down assay using bacterially expressed GST-tagged Inn1 C terminus (wild type or mutant) and His6-tagged Hof1 C terminus. His6-Hof1 bound strongly to both wild-type and m2 mutant GST-Inn1 in comparison with the negative control, GST alone (Fig. 4 E). In contrast, the m4 mutation nearly eliminated the interaction between Inn1 and Hof1. These results support the conclusion from two-hybrid analysis that Hof1 binds to Inn1 primarily via the Inn1 PKLPPLP motif.
Hof1 localizes to the neck much earlier in the cell cycle than does Inn1 (Vallen et al., 2000), suggesting that Inn1 localization might depend on Hof1. We found that Inn1-GFP localized to the neck with essentially normal timing in hof1Δ cells; 44% of cells with split septin rings had detectable signal compared with 33% in wild type (Fig. 4 F and Table I). However, although Inn1-GFP localization was almost always (∼95%) symmetric in wild-type cells, it was asymmetric in 39% of the hof1Δ cells with detectable signal (Fig. 4 F; Table I; and Videos 5 and 6). Thus, Hof1 appears to be required for the initiation or maintenance of symmetric Inn1 localization at the neck.
Functional and physical interactions between Inn1 and Cyk3
To explore further the interactions among the proteins important for AMR-independent cytokinesis, we asked whether overexpression of Iqg1, Hof1, Cyk3, or Mlc1 could suppress the effects of an inn1Δ mutation. We found that Cyk3, but not the other proteins, could partially suppress the growth (Fig. 5 A) and cytokinesis defects of inn1Δ cells. The cluster index for inn1Δ cells (indicative of a cytokinesis and/or cell separation defect; see Materials and methods) was reduced from 67 to 44% by a CYK3 plasmid. Remarkably, this suppression involved the formation of almost normal-looking PS in many cells (38% of the 50 cells examined; Fig. 5 B).
Figure 5.
Functional and physical interactions between Inn1 and Cyk3. (A) Suppression of inn1Δ growth defect by overexpression of Cyk3. Strain LY1310 (inn1Δ [pUG36-INN1]) was transformed with a vector control (Vect; YEplac181) or with LEU2-marked high copy plasmids carrying IQG1 (YEp181-IQG1), HOF1 (pTSV30A-HOF1), CYK3 (P1, pBK132; P2, pBK133), MLC1 (pBK65), or INN1 (pGP564-INN1). Transformants were streaked on SC-Leu and SC-Leu+FOA plates and incubated at 25°C for 3 d to ask whether any of the candidate plasmids could replace the URA3-marked pUG36-INN1. (B) Restoration of PS formation in inn1Δ cells by overexpression of Cyk3. Strain LY1310 (inn1Δ [pUG36-INN1]) was transformed with pRS425-CYK3, incubated on an SC-Leu+FOA plate at 24°C for 3 d to eliminate plasmid pUG36-INN1, grown to exponential phase in SC-Leu medium at 24°C, and examined by TEM. (C–E) Interaction of the SH3 domain of Cyk3 with the PIPPLP motif (amino acids 159–165) of Inn1 as determined by two-hybrid analysis (C and D) and in vitro protein-binding assays (E). Experiments were performed as described for Fig. 4 (C–E) using a Cyk3 SH3 domain fragment (amino acids 1–70) instead of Hof1. In C, the diagram shows the motifs of Cyk3 (SH3 and TGc [putative transglutaminase domain]). Asterisk indicates that Inn1(130–180) failed to interact with Cyk3-SH3 for unknown reasons. (F) Localization of Cyk3 in inn1Δ cells. Strain YEF5216 (inn1Δ) was transformed with plasmid pRS315GW-CYK3-2GFP, grown overnight on an SC-Leu plate at 25°C, and imaged by DIC and fluorescence microscopy.
We next tested for physical interaction between Inn1 and Cyk3. Two-hybrid and in vitro protein-binding analyses like those used to characterize the Inn1–Hof1 interaction (see previous section) indicated that Inn1 and Cyk3 interact directly and that this interaction is mediated by the SH3 domain of Cyk3 and the PIPPLP motif (amino acids 159–165) of Inn1 (Fig. 5, C–E). Cyk3-GFP could localize to the neck in inn1Δ cells (Fig. 5 F), although its localization was somewhat less well ordered than the tight band observed in wild-type cells (Korinek et al., 2000).
One possible interpretation of these data is that Cyk3 localizes to the neck independently of Inn1 but is activated by Inn1 for a role in promoting PS formation; on this model, overexpression of Cyk3 would partially bypass the activation requirement. Alternatively, Inn1 and Cyk3 might act in parallel to promote PS formation.
Dependence of Inn1 localization on both Hof1 and the AMR
Inn1 could also localize to the neck in cyk3Δ cells, and, unlike hof1Δ cells, nearly all cyk3Δ cells with detectable Inn1-GFP signal at the neck showed a symmetric pattern like that in wild type (Fig. 6 A and Table I). The fraction of cyk3Δ cells with split septin rings that showed localized Inn1-GFP was greater than that in wild type (Table I), presumably reflecting the increased duration of cytokinesis (accompanied by persistent Inn1-GFP at the neck; Video 7) that results from the delayed PS formation in cyk3Δ cells (unpublished data). Although Inn1 interacts physically with both Hof1 and Cyk3 (see previous sections), these interactions do not appear sufficient to account for the neck localization of Inn1 because Inn1 could localize to the neck both in hof1Δ cyk3Δ cells (Fig. 6 B and Table I) and when the PXXP motifs involved in the interactions were mutated (Fig. 6 C).
Figure 6.
Mechanisms of Inn1 bud neck localization. (A and B) Strains LY1321 (INN1-GFP cyk3Δ; A) and LY1325 (INN1-GFP cyk3Δ hof1Δ [pRS316-HOF1]; B) were transformed with plasmid YCp111-CDC3-CFP, and the LY1325 transformants were incubated on an FOA plate to eliminate the HOF1 plasmid. Cells were examined as described in Fig. 4 F. 1DC, one central dot; 1DA, one asymmetric dot (Table I and Video 7). (C) Localization of Inn1 lacking its Hof1- and Cyk3-binding sites. Strain LY1310 (inn1Δ [pUG36-INN1]) was transformed with HIS3-marked plasmids carrying RFP-tagged wild-type (WT) or mutant INN1 alleles. After growth on an SC-His+FOA plate at 25°C to eliminate pUG36-INN1, DIC and fluorescence images were captured. (D) Loss of Inn1 localization in latA-treated hof1Δ cells. Wild-type (LY1324), cyk3Δ (LY1321), and hof1Δ (LY1328 after eliminating plasmid pRS316-HOF1 by growth on an FOA plate) strains were grown to exponential phase in YM-P medium at 25°C. Portions of each culture were treated with latA for 20 min, and cells were imaged by DIC and fluorescence microscopy. Images of representative latA-treated cells (left) and percentages of large-budded cells with localized Inn1-GFP (right) are shown. The experiments were performed twice with similar results. Bars, 2 µm.
Because Inn1-GFP localized weakly and/or asymmetrically to the neck in both AMR-deficient (myo1Δ and iqg1Δ) and hof1Δ mutants (see previous sections), it seemed possible that the AMR and Hof1 might act in concert to localize Inn1 during cytokinesis. Because myo1Δ and hof1Δ are synthetically lethal (Vallen et al., 2000), we could not examine Inn1 localization in the double mutant. Thus, we instead examined Inn1 localization in wild-type, hof1Δ, and cyk3Δ cells that had been treated with latrunculin A (latA), which disrupts all filamentous actin structures, including the actin ring (Ayscough et al., 1997). Inn1-GFP localized efficiently to the neck in latA-treated wild-type and cyk3Δ cells but not in latA-treated hof1Δ cells (Fig. 6 D), which is consistent with the hypothesis that Hof1 and the AMR cooperate in Inn1 localization.
Distinct roles of Inn1 domains in localization and the activation of PS formation
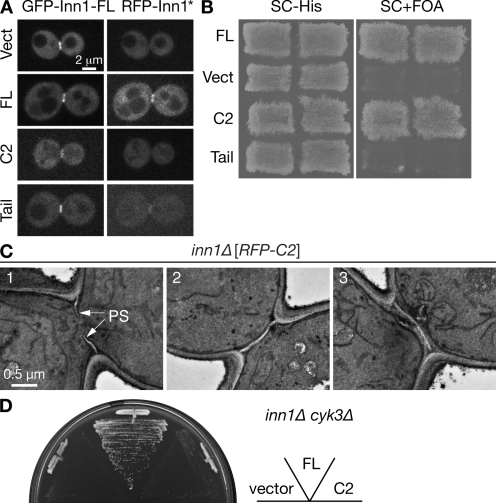
To further analyze the functions of the Inn1 N-terminal and C-terminal regions, appropriate fragments were tagged with RFP at their N termini, expressed from a methionine-regulatable promoter, and assessed for their abilities to localize and provide Inn1 function. Consistent with its binding to Hof1 and Cyk3, the C-terminal region was able to localize to the bud neck in telophase in either the presence (Fig. 7 A) or absence (unpublished data) of full-length Inn1. However, the RFP signal was less intense than with the full-length protein (Fig. 7 A), and no contraction was seen in the absence of full-length protein (unpublished data). Despite its localization to the neck, the C-terminal fragment was unable to rescue the growth of an inn1Δ mutant (Fig. 7 B).
Figure 7.
Structure function analysis of Inn1. (A) Role of the Inn1 C-terminal region in neck localization. Strain LY1310 (inn1Δ [pUG36-INN1]) was transformed with the pUG34mCherry vector (Vect) or its derivatives containing sequences encoding full-length (FL) INN1, the putative C2 domain (amino acids 1–140), or the C-terminal tail (amino acids 130–409). Transformants were incubated on an SC-His-Ura plate for 2 d at 25°C, scraped off, and imaged by spinning-disk confocal microscopy for GFP-Inn1-FL and RFP-Inn1 derivatives (asterisk). (B) Critical role of the putative C2 domain in Inn1 function. The transformants described in A were patched onto SC-His and SC+FOA (to select against pUG36-INN1) plates and incubated at 25°C for 3 d to assess the ability of the INN1 fragments to provide Inn1 function. (C) Restoration of PS formation in inn1Δ cells by the putative C2 domain. Strain YEF5202 (inn1Δ [pUG34mCherry-INN1-C2]), obtained as described in B, was grown to exponential phase in SC-His medium at 24°C and examined by TEM. (D) Cooperative function of Cyk3 and the putative C2 domain of Inn1. Strain MWY1171 (inn1Δ cyk3Δ [pUG36-INN1]) was transformed with the plasmids described in A. The transformants were streaked on an SC-His-Met+FOA plate and incubated for 4 d at 24°C.
In striking contrast, the RFP-tagged N-terminal fragment showed no detectable localization to the neck and appeared to be cytosolic, as judged by confocal microscopy (Fig. 7 A), but it could nonetheless rescue the growth (Fig. 7 B) and PS formation (Fig. 7 C) defects of an inn1Δ mutant. Most of the inn1Δ cells expressing the N-terminal fragment formed either a seemingly normal PS (Fig. 7 C, cells 1 and 2) or a seemingly normal PS with additional PS-like structures (cell 3); some cells formed an asymmetrically localized PS sandwiched by SS (unpublished data). The ability of the N-terminal fragment to provide Inn1 function appears to depend on its overexpression because a single chromosomal copy under the normal INN1 promoter was not sufficient for colony formation (Fig. S3 A), whereas the same construct rescued the growth of inn1Δ cells when overexpressed from a GAL promoter (Fig. S3 B); presumably, overexpression produces a sufficient concentration of the fragment at its site of action despite its inability to localize efficiently to this site. The ability of the N-terminal fragment to provide Inn1 function also appears to depend on Cyk3, as the overexpressed N-terminal fragment was unable to rescue the growth of an inn1Δ cyk3Δ double mutant (Fig. 7 D).
Collectively, these results suggest that the Inn1 N-terminal domain collaborates with Cyk3 to provide the activity necessary for PS formation and cytokinesis, whereas the C-terminal domain is responsible for targeting Inn1 to its site of action.
Apparent lack of phospholipid binding by the putative C2 domain of Inn1
Sanchez-Diaz et al. (2008) proposed that Inn1 might help to physically link the AMR to the plasma membrane based in part on the resemblance of the Inn1 N-terminal region to C2 domains, which are typically involved in calcium-dependent lipid binding (Rizo and Sudhof, 1998; Cho and Stahelin, 2006). However, C2 domains have also been implicated in protein–protein interactions (Benes et al., 2005; Lu et al., 2006; Dai et al., 2007), and the Inn1 N-terminal domain does not appear to possess aspartates in positions corresponding to those critical for Ca2+ binding in the C2 domains of rat synaptotagmin-I (Shao et al., 1996) and the yeast Tcb proteins (Schulz and Creutz, 2004). Moreover, in lipid overlay assays, we could not detect significant lipid binding by Inn1 in either the presence or absence of Ca2+ (Fig. S4 A), although Ca2+-dependent phospholipid binding was observed with a positive control (Fig. S4 B).
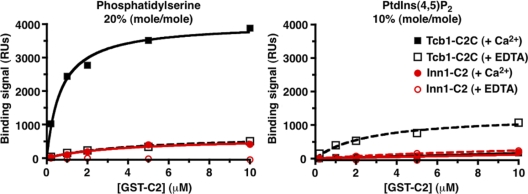
To analyze possible phospholipid binding in a membrane environment and in a more quantitative manner, we also used the surface plasmon resonance (SPR) approach (Narayan and Lemmon, 2006). As shown in Fig. 8, the Inn1 N-terminal region showed no significant binding to surfaces containing 20% (mol/mol) PtdSer or 10% (mol/mol) PtdIns(4,5)P2 in a dioleoylphosphatidylcholine background in the presence or absence of Ca2+. In contrast, the positive control Tcb1 C2C showed robust binding to PtdSer in the presence of Ca2+ (KD = 0.95 ± 0.57 µM) but did not bind significantly to PtdIns(4,5)P2 (a low level of binding was observed in the absence of Ca2+). Because phospholipid binding by the Inn1 N-terminal region was barely above background even when 10 mM protein was applied, the KD for binding is likely to exceed 100 µM. Based on other experiments with GST fusion proteins (which are known to dimerize; Klein et al., 1998), the monomeric Inn1 N-terminal region presumably binds phospholipids with a KD in the 1-mM range or weaker. No binding of either the Tcb1 C2 domain or Inn1 N-terminal region was detected by SPR for PtdIns3P, PtdIns4P, or PtdIns(3,5)P2 surfaces regardless of Ca2+ levels (unpublished data).
Figure 8.
Lack of detectable phospholipid binding by the putative C2 domain of Inn1. Bacterially expressed GST-Inn1(1–134) and the positive control GST-Tcb1-C2C (the third C2 domain [amino acids 979–1,186] in the tricalbin Tcb1; Schulz and Creutz, 2004) were tested by SPR for binding of phosphatidylserine and Ptdlns(4,5)P2. RU, response unit.
In summary, the apparently cytosolic localization of the Inn1 N-terminal fragment (see previous section), the apparent lack of amino acids critical for Ca2+-dependent lipid binding, and the biochemical data all suggest that the Inn1 N-terminal region is not a lipid-binding domain.
Function of Inn1 in AMR-independent cytokinesis
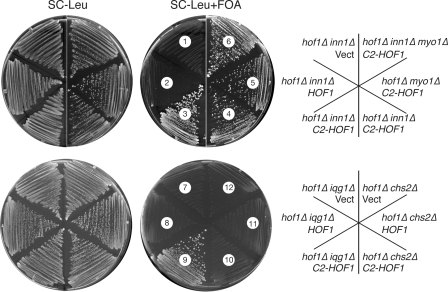
In the model of Sanchez-Diaz et al. (2008), Inn1 couples plasma membrane ingression to contraction of the AMR. However, we found that an Inn1(1–134)–Hof1 fusion similar to that described by Sanchez-Diaz et al. (2008) could not only provide Hof1 function (see Introduction; Fig. 9, sector 5; note that the hof1Δ myo1Δ double mutant is essentially inviable) and Inn1 function (Fig. 9, compare sectors 3 and 4 with sectors 1 and 2), but it could do so in the absence of Myo1 and thus of an AMR (Fig. 9, sector 6). Moreover, the fusion protein could also suppress an iqg1Δ mutation (Fig. 9, compare sector 9 with sectors 7 and 8) even though Iqg1 is essential for AMR formation (see Introduction). In striking contrast, the Inn1(1–134)–Hof1 fusion protein showed no detectable suppression of a chs2Δ mutation (Fig. 9, compare sector 10 with sectors 11 and 12), which is consistent with the other evidence that the primary function of Inn1 is to stimulate synthesis of the PS by Chs2 (see Discussion). Because Iqg1 is also essential for PS formation (unpublished data), the data suggest that Inn1 functions downstream of Iqg1 but upstream of Chs2 in PS formation. It should also be noted that the Inn1(1–134)–Hof1 fusion protein could provide Inn1 function even when expressed from low copy vectors (Fig. 9), whereas the free Inn1 N terminus required overexpression to do so (Fig. S3). This difference presumably reflects the ability of the Hof1 portion of the fusion protein to target the Inn1 N-terminus to the neck (Sanchez-Diaz et al., 2008), thus increasing its effective concentration at that site.
Figure 9.
Evidence that Inn1 functions downstream of Iqg1 and upstream of Chs2 in AMR-independent cytokinesis. Strains MWY1145 (hof1Δ inn1Δ [pUG36-INN1]; sectors 1–3), MWY764 (hof1Δ iqg1Δ [pRS316GW-IQG1]; sectors 7–9), or RNY2225 (hof1Δ chs2Δ [pJC328]; sectors 10–12) were transformed with pRS315GW, pRS315GW-NotI-HOF1, or pRS315GW-C2-HOF1. The resulting transformants and strains MOY632 (hof1Δ innΔ [pUG36-INN1][pRS315GW-C2-HOF1]; sector 4), MOY630 (hof1Δ myo1Δ [pUG36-INN1][pRS315GW-C2-HOF1]; sector 5), and MOY634 (hof1Δ inn1Δ myo1Δ [pUG36-INN1][pRS315GW-C2-HOF1]; sector 6) were streaked on SC-Leu and SC-Leu+FOA plates and incubated at 24°C for 3 d. Vect, vector.
Discussion
In most if not all animal and fungal cells, the contractile AMR is important for efficient cytokinesis. However, it is also clear that a variety of cell types, including yeast, Dictyostelium discoideum amoebae (DeLozanne and Spudich, 1987; Neujahr et al., 1997; Hibi et al., 2004), and at least some kinds of mammalian cells (Kanada et al., 2008) can undergo cell cycle–regulated division at appropriate sites in the absence of AMR function when grown under appropriate conditions. These observations focus attention on the processes of membrane deformation, membrane addition and compositional specialization, and ECM (e.g., cell wall) formation that normally work in close concert with AMR contraction but can also form a cleavage furrow even when the AMR is absent or nonfunctional (Finger and White, 2002; Mizuguchi et al., 2003; Strickland and Burgess, 2004; Albertson et al., 2005; Szafer-Glusman et al., 2008). They also suggest that animal and fungal cytokinesis may have more in common mechanistically with plant cytokinesis than has traditionally been thought (Hales et al., 1999; Otegui et al., 2005).
In yeast, Iqg1, Mlc1, Hof1, and Cyk3 have all been implicated in the AMR-independent processes of cytokinesis (see Introduction). In this study, we have identified Inn1 as another critical contributor to these processes. Specifically, we have shown that Inn1 interacts directly with Hof1 and Cyk3, plays an essential role in PS formation, and can function in cytokinesis independently of the AMR, as summarized in Fig. 10. Our study has some overlap with an independent study of Inn1 (Sanchez-Diaz et al., 2008) but reaches a very different conclusion about the role of Inn1 in cytokinesis.
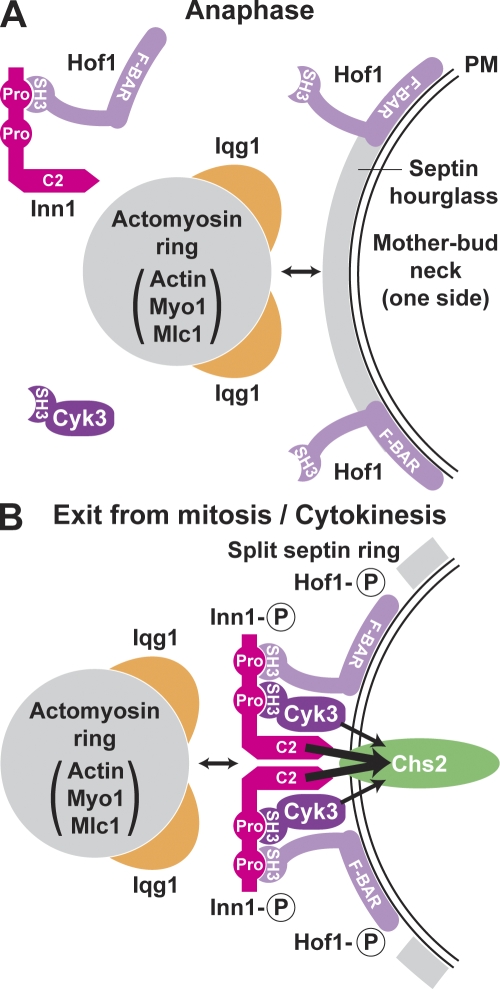
Figure 10.
A model for the assembly and function of Inn1 in cytokinesis. (A and B) Circled P symbols indicate phosphorylation of the proteins. PM, plasma membrane; Pro, PXXP motif. See Discussion for details.
Assembly of cytokinesis proteins at the mother–bud neck
Myo1 forms a ring at the presumptive bud site in late G1 (Bi et al., 1998; Lippincott and Li, 1998). This process is septin dependent, and after bud emergence, the Myo1 ring lies near the center of the hourglass-shaped septin assembly. The mechanisms by which Myo1 associates with the septins and/or the plasma membrane remain obscure. Later in the cell cycle, other cytokinesis proteins are recruited to the neck. By anaphase (Fig. 10 A), Mlc1 has joined Myo1 and has also helped to recruit Iqg1 to the neck (Shannon and Li, 2000; Luo et al., 2004). Actin recruitment occurs just before mitotic exit and depends on Myo1, Mlc1, and Iqg1 (Bi et al., 1998; Shannon and Li, 1999; Korinek et al., 2000; Yoshida et al., 2006) but not on Inn1 (Fig. S2; Sanchez-Diaz et al., 2008), which is not yet localized to the neck (Fig. 2 A and Fig. 3). At this stage, Hof1 forms a double ring at the neck; its recruitment depends on the septins but not on other known proteins (Vallen et al., 2000). Some Hof1 also appears to be present in complexes with the as yet unlocalized Inn1 (Fig. 4 A).
As cells enter cytokinesis, multiple events occur that depend directly or indirectly on the MEN (Fig. 10 B). The septin ring splits (Kim et al., 1991; Lippincott et al., 2001) and defines a domain to which other proteins are confined (Dobbelaere and Barral, 2004). Chs2 is recruited to the neck (Chuang and Schekman, 1996; Zhang et al., 2006), an event that depends on the septins and the secretory apparatus (VerPlank and Li, 2005) but not on the other proteins discussed in this study (Fig. 1 D; unpublished data). Cyk3 is also recruited to the neck (Korinek et al., 2000); this recruitment is less efficient (or the recruited Cyk3 is less well organized) in the absence of either Hof1 or Inn1 (Fig. 5 F; unpublished data). Cyk3 presumably is bound to Inn1 at this time (Fig. 5, C–E), although it is not yet known whether this binding also occurs earlier in the cell cycle. Hof1 reorganizes into a single ring, an event that is correlated with its MEN-dependent phosphorylation (Fig. 4, A and B; Vallen et al., 2000; Corbett et al., 2006). Inn1 is recruited to the neck, an event that depends on its C-terminal region but not on its N-terminal region (Fig. 7 A; Sanchez-Diaz et al., 2008) or the presence of Cyk3 (Fig. 6 A and Table I). Inn1 localization also occurs in myo1Δ, iqg1Δ, and hof1Δ cells (Fig. 2, B and C; Fig. 4 F; and Table I), as well as when interactions with Hof1 and Cyk3 are disrupted by mutation of the Inn1 PXXP motifs (Fig. 6 C). However, Inn1 localization appears weak and/or asymmetric in each case and was abolished when hof1Δ cells (but not wild-type or cyk3Δ cells) were treated with latA (Fig. 6 D), suggesting that Inn1 localization depends jointly on Hof1 and the AMR.
Like Hof1, Inn1 undergoes MEN-dependent phosphorylation (Fig. 4 B), and it seems likely that the rearranged protein localizations and associations that occur at this time depend at least in part on these phosphorylations. Because the MEN component Dbf2 is also targeted to the neck upon actin ring assembly and is required for the phosphorylation and/or localization of both Hof1 and Inn1 (Fig. 3; Vallen et al., 2000; Corbett et al., 2006), Inn1, Hof1, or both may be direct substrates of this protein kinase.
Functions of the assembled proteins during cytokinesis
Once the cytokinesis apparatus is fully assembled, contraction of the AMR, membrane ingression, and PS synthesis all normally begin almost immediately. AMR contraction has long been presumed to involve motor activity of Myo1 upon actin filaments. This view has been challenged by the findings that the Myo1 tail (lacking the motor domain) assembles at the neck and supports efficient cytokinesis (Lord et al., 2005) and that even some myo1Δ cells form nearly normal-looking cleavage furrows and PSs (unpublished data). However, a role for Myo1-actin force generation is supported by the observations that in inn1Δ (Fig. 1 E) and chs2Δ (VerPlank et al., 2005) mutants, the AMR can apparently continue to contract after it has pulled away from the plasma membrane over much of its circumference.
These observations also suggest one possible role for Inn1, namely that it might help to physically tether the AMR to the membrane during contraction (Sanchez-Diaz et al., 2008). However, such a role appears to be ruled out by the following arguments. First, the myosin ring (later AMR) can associate with the cell cortex in inn1Δ cells (Fig. 1 E) and in wild-type cells does so long before Inn1 has localized to the neck. Second, the plasma membrane can ingress without force production by the AMR (see previous paragraph). Third, the putative C2 domain of Inn1 does not appear to bind phospholipids (Fig. 8 and Fig. S4), and indeed, its sequence is quite different from C2 domains that are known to bind lipids (Cho and Stahelin, 2006). Fourth, although the Inn1-binding partner Hof1 might possibly help to tether the AMR to the membrane via the presumed interaction of its F-BAR domain with the membrane (Fig. 10 B), there is no good evidence for a role of Inn1 in linking Hof1 to the AMR. The contraction of the Inn1 ring (Fig. 2 A; Sanchez-Diaz et al., 2008) would be seen with any protein that is associated with the leading edge of the cleavage furrow, and although Sanchez-Diaz et al. (2008) detected weak binding of Inn1 to Iqg1, Iqg1 is a multifunctional protein that is involved in AMR-independent processes as well as in AMR formation (see Introduction). More compellingly, an N-terminal fragment of Inn1 can provide Inn1 function when overexpressed (Fig. 7, B and C; and Fig. S3) despite its inability to bind Hof1 (Fig. 4, C–E) or concentrate at the neck (Fig. 7 A). Fifth, a fusion of Inn1(1–134) to Hof1 can provide Inn1 function not only in otherwise wild-type cells (Sanchez-Diaz et al., 2008) but also in myo1Δ and iqg1Δ cells (Fig. 9), showing that Inn1 function does not depend on the AMR. Finally, the formation of reasonably well-oriented PSs in inn1Δ cells overexpressing either Cyk3 (Fig. 5 B) or an Inn1 fragment that cannot concentrate at the division site (Fig. 7 C) shows that the AMR can direct furrow ingression without an Inn1-dependent link to the membrane.
Thus, we favor a different model in which the role of Inn1 is to cooperate with Cyk3 in the activation of Chs2 for PS formation (Fig. 10 B). This model is supported by (a) the absence of PS formation in inn1Δ cells (Fig. 1 C) and its delay in cyk3Δ cells (unpublished data), (b) the restoration of PS formation in inn1Δ cells overexpressing either Cyk3 (Fig. 5 B) or an N-terminal fragment of Inn1 (Fig. 7 C), (c) the observation that the Inn1 N-terminal fragment (whose function is presumably inefficient because of its inability to localize) can only provide Inn1 function when Cyk3 is present (Fig. 7 D), and (d) the inability of the Inn1(1–134)–Hof1 fusion to suppress the growth defect of a chs2Δ mutant (Fig. 9). Moreover, the behavior of the AMR in inn1Δ cells (Fig. 1 E) appears very similar to that in chs2Δ cells (VerPlank et al., 2005); thus, in the absence of PS formation, the membrane apparently cannot ingress rapidly enough to keep pace with AMR contraction, resulting in detachment of the AMR from the membrane and/or its disassembly. Because the Inn1(1–134)–Hof1 fusion rescues an iqg1Δ but not a chs2Δ mutant (Fig. 9), Inn1 presumably functions downstream of Iqg1 but upstream of Chs2 in the PS formation pathway, as also seems likely for Cyk3 (Korinek et al., 2000; unpublished data). Because the PS formation defects of iqg1Δ and inn1Δ mutants are more severe than that of a cyk3Δ mutant, the simplest model is that Inn1 and Cyk3 function in parallel to activate Chs2 by mechanisms that remain to be determined. The MEN-regulated localization of Inn1 and Cyk3 to the division site presumably allows proper coordination of PS formation and furrow ingression with AMR contraction. It will be interesting to explore the interplay between AMR contraction and ECM synthesis during cytokinesis in other types of cells.
Materials and methods
Strains, growth conditions, and genetic methods
Yeast strains are described in Table II. Standard culture media and genetic techniques were used (Guthrie and Fink, 1991); where noted, cells were grown in YM-P, a rich, buffered liquid medium (Lillie and Pringle, 1980). To select for the loss of URA3-containing plasmids, 1 mg/ml 5-fluoroorotic acid (FOA; Research Products International) was added to media. To depolymerize filamentous actin (Ayscough et al., 1997), latA (Wako Chemicals USA, Inc.) was dissolved in DMSO as a 20-mM stock solution and added to media at a final concentration of 200 µM; an identical concentration of DMSO alone was added to control cultures. Oligonucleotide primers were purchased from Integrated DNA Technologies.
Table II.
Yeast strains used in this study
| Strain | Genotype | Source |
| YEF473 | a/α his3/his3 leu2/leu2 lys2/lys2 trp1/trp1 ura3/ura3 | Bi and Pringle, 1996 |
| YEF473A | a his3 leu2 lys2 trp1 ura3 | Bi and Pringle, 1996 |
| YEF473B | α his3 leu2 lys2 trp1 ura3 | Bi and Pringle, 1996 |
| Y860 | α his3-11,15 leu2-3,112 trp1-1 ade2-1 can1-100 ura3-1∷URA3:lexAop-ADE2 | C. Boone |
| Y1026 | a his3-11,15 leu2-3,112 trp1-1 ade2-1 can1-100 ura3-1∷URA3:lexAop-lacZ | C. Boone |
| MOY157 | As YEF473B except INN1-GFP:TRP1 HOF1-TAP:His3MX6 cdc15-2 | This studya |
| MOY215 | As YEF473B except INN1-GFP:TRP1 cdc15-2 | This studya |
| MOY609 | As YEF473 except hof1Δ∷TRP1/hof1Δ∷TRP1 INN1/inn1Δ∷kanMX6/ MYO1/myo1Δ∷kanMX6 [pUG36-INN1][pRS315GW-C2-HOF1] | This study |
| MOY630 | As YEF473B except hof1Δ∷TRP1 myo1Δ∷kanMX6 [pUG36-INN1][pRS315GW-C2-HOF1] | Segregant from MOY609 |
| MOY632 | As YEF473B except hof1Δ∷TRP1 innΔ∷kanMX6 [pUG36-INN1][pRS315GW-C2-HOF1] | Segregant from MOY609 |
| MOY634 | As YEF473B except hof1Δ∷TRP1 inn1Δ∷kanMX6 myo1Δ∷kanMX6 [pUG36-INN1] [pRS315GW-C2-HOF1] | Segregant from MOY609 |
| MWY764 | As YEF473A except hof1Δ∷TRP1 iqg1Δ∷His3MX6 [pRS316GW-IQG1] | This study |
| MWY1145 | As YEF473A except hof1Δ∷TRP1 inn1Δ∷kanMX6 [pUG36-INN1] | This study |
| MWY1171 | As YEF473B except cyk3Δ∷kanMX6 inn1Δ∷kanMX6 [pUG36-INN1] | This study |
| RNY2225 | As YEF473A except hof1Δ∷TRP1 chs2Δ∷kanMX6 [pJC328] | This study |
| RNY2393 | As YEF473A except iqg1Δ∷His3MX6 INN1-GFP:kanMX6 [YCp50-IQG1] | This study |
| RNY2395 | As YEF473A except INN1-GFP:kanMX6 [YCp50-IQG1] | This study |
| RNY2494 | As YEF473 except INN1/pINN1-inn1(1–134)-GFP:His3MX6 | This study |
| RNY2498 | As YEF473 except INN1/TRP1:pGAL1-GFP-inn1(1–134):His3MX6 | This study |
| RNY2499 | As YEF473 except INN1/TRP1:pGAL1-GFP-INN1 | This study |
| LY1065 | α hof1Δ∷kanMX6 ade2 ade3 his3 leu2 trp1 ura3 [pTSV30A-HOF1] | This studyb |
| LY1067 | a hof1Δ∷kanMX6 ade2 ade3 leu2 lys2 ura3 [pTSV31A-HOF1] | This studyb |
| LY1302 | As YEF473 except INN1-GFP:kanMX6/INN1-GFP:kanMX6 | This study |
| LY1310 | As YEF473A except inn1Δ∷kanMX6 [pUG36-INN1] | This study |
| LY1313 | As YEF473A except INN1-GFP:kanMX6 | This study |
| LY1314 | As YEF473B except INN1-GFP:kanMX6 | This study |
| LY1321 | As YEF473A except INN1-GFP:kanMX6 cyk3Δ∷His3MX6 | This study |
| LY1324 | As YEF473A except INN1-GFP:kanMX6 | This study |
| LY1325 | As YEF473A except INN1-GFP:kanMX6 cyk3Δ∷His3MX6 hof1Δ∷kanMX6 [pRS316-HOF1] | This study |
| LY1328 | As YEF473A except INN1-GFP:kanMX6 hof1Δ∷kanMX6 [pRS316-HOF1] | This study |
| LY1355 | a dbf2-1 dbf20Δ∷TRP1 INN1-GFP:kanMX6 ade1 leu2 trp1 ura3 | This studyc |
| LY1357 | a cdc5ts∷URA3 INN1-GFP:kanMX6 leu2 trp1 ura3 | This studyc |
| LY1360 | a cdc14 INN1-GFP:kanMX6 can1 his7 leu2 ura3 | This studyc |
| LY1364 | As YEF473A except myo1Δ∷His3MX6 INN1-GFP:kanMX6 [YCp50-MYO1] | This study |
| LY1373 | As YEF473A except inn1Δ∷kanMX6 CHS2-GFP:kanMX6 [pUG36-INN1] | This study |
| YEF1951 | As YEF473A except hof1Δ∷kanMX6 | Vallen et al., 2000 |
| YEF5202 | As YEF473A except inn1Δ∷kanMX6 [pUG34mCherry-INN1-C2] | This study |
| YEF5216 | As YEF473A except inn1Δ∷kanMX6 | This studyd |
| YEF5291 | As YEF473A except inn1Δ∷kanMX6 MYO1-GFP:His3MX6 | This study |
| YEF5293 | As YEF473A except myo1Δ∷His3MX6 INN1-GFP:kanMX6 | This study |
Bolded a′s and α′s indicate mating types of haploid yeast cells. Genes were deleted (the entire coding region in each case) or tagged at their C termini using the PCR method (Baudin et al., 1993). Template plasmids were as described previously by Longtine et al. (1998) except for pFA6a-TAP-His3MX6 (provided by P. Walter, University of California, San Francisco, San Francisco, CA) and pFA6a-link-mCherry-His3MX6 (see Materials and methods). In some cases, genomic DNA from previously transformed strains was used as a template to generate transformation fragments with longer flanking regions. Other steps in strain constructions were conventional genetic crosses and plasmid transformations.
cdc15-2 was derived from strain DLY3034 (provided by D. Lew, Duke University, Durham, NC) and backcrossed more than seven times into the YEF473 background.
Derived from strains CDV38 and CDV39 (provided by C. De Virgilio, University of Fribourg, Fribourg, Switzerland).
Strains J230-2D (provided by L. Johnston, National Institute for Medical Research, London, England, UK), KKY021 (provided by L. Johnston), and 4078-14-3a (provided by L. Hartwell, Fred Hutchinson Cancer Research Center, Seattle, WA) were transformed with a PCR-generated INN1-GFP:kanMX6 cassette.
Generated by growing LY1310 on SC+FOA medium.
Plasmids
Plasmids are listed in Table III and/or described here. A genomic DNA library in the low copy vector YCp50LEU2 was supplied by F. Spencer and P. Hieter (University of British Columbia, Vancouver, British Columbia, Canada; Bi and Pringle, 1996). Plasmid YCp50LEU2-HOF1, carrying full-length HOF1, was isolated from this library by complementing the temperature-sensitive growth of a hof1Δ strain (YEF1951). Plasmids pTSV30A-HOF1 and pTSV31A-HOF1 were constructed by first subcloning an ∼6.3-kb BamHI fragment containing HOF1 from YCp50LEU2-HOF1 into the BamHI sites of pTSV30A (2µ, LEU2, and ADE3) and pTSV31A (2µ, URA3, and ADE3); in each case, an ∼2.9-kb XbaI fragment (one site in the insert DNA and the other in the vector) was then deleted to remove the neighboring gene ARP9 to avoid possible complications during the synthetic lethal screen.
Table III.
Plasmids used in this study
| Plasmid | Description | Reference or source |
| YEplac181 | 2µ, LEU2 | Gietz and Sugino, 1988 |
| pRS315GW | CEN, LEU2 | Pringle laboratory |
| pRS425 | 2µ, LEU2 | Christianson et al., 1992 |
| pRS315-GFP-RAS2 | CEN, LEU2, GFP-RAS2 | Luo et al., 2004 |
| YCp111-CDC3-CFP | CEN, LEU2, CDC3-CFP | Pringle laboratory |
| YCp50-MYO1 | CEN, URA3, MYO1 | S. Browna |
| pBK65 | 2µ, LEU2, MLC1 | J. Chantb |
| pJC328 | CEN, URA3, CHS2-MYC | Chuang and Schekman, 1996 |
| pRS316GW-IQG1 | CEN, URA3, IQG1 | Pringle laboratory |
| YCp50-IQG1 (=p1868) | CEN, URA3, IQG1 | Korinek et al., 2000 |
| YEp181-IQG1 | 2µ, LEU2, IQG1 | Ko et al., 2007 |
| pBK132 | 2µ, LEU2, CYK3 | Korinek et al., 2000 |
| pBK133 | 2µ, LEU2, CYK3 | Korinek et al., 2000 |
| pRS425-CYK3 | 2µ, LEU2, CYK3 | Ko et al., 2007 |
| pRS315GW-CYK3-2GFP | CEN, LEU2, CYK3-2GFP | Pringle laboratory |
| pRS315GW-NotI-HOF1 | CEN, LEU2, HOF1 | This study |
| pRS316-HOF1 | CEN, URA3, HOF1 | Vallen et al., 2000 |
| YCp50LEU2-HOF1 | CEN, LEU2, HOF1 | This study |
| pTSV30A-HOF1 | 2µ, LEU2, ADE3, HOF1 | This study |
| pTSV31A-HOF1 | 2µ, URA3, ADE3, HOF1 | This study |
| YCp50LEU2-INN1-17C | CEN, LEU2, INN1 | This study |
| pGP564-INN1 | 2µ, LEU2, INN1 | F. Lucac |
| pUG34mCherry | CEN, HIS3, pMET25-mCherry | This study |
| pUG34mCherry-INN1d | CEN, HIS3, pMET25-mCherry-INN1 | This study |
| pUG36-INN1 | CEN, URA3, pMET25-yEGFP-INN1 | This study |
| pRS315GW-C2-HOF1 | CEN, LEU2, C2-HOF1 | This study |
CEN indicates low copy number plasmids; 2µ indicates high copy number plasmids.
University of Michigan, Ann Arbor, MI.
Harvard University, Cambridge, MA.
University of Pennsylvania, Philadelphia, PA.
Related plasmids contain the wild-type INN1 N terminus (amino acids 1–140) or C terminus (amino acids 130–409) or full-length INN1 into which mutations m1-m4 (Fig. S1) had been introduced singly or in combinations (see Materials and methods).
Plasmid YCp50LEU2-INN1-17C, carrying the full-length ORF YNL152W/INN1 and flanking DNA, was isolated from the YCp50-LEU2 library by rescuing the sectoring ability of mutant 5033 from the synthetic lethal screen (see the following section). Mutant 5033 showed a temperature-sensitive growth defect even in the presence of the HOF1 plasmid. To recover the mutant inn1-5033 allele by gap repair, mutant 5033 was transformed with a PvuII-digested plasmid (derived in several steps from YCp50LEU2-INN1-17C) in which the INN1 ORF had been replaced by a PvuII site. After selection for a Leu+ phenotype, a plasmid was isolated and shown to confer Ts growth to strain LY1310 in the absence of plasmid pUG36-INN1. Sequencing of this plasmid revealed a single mutation in the INN1 ORF (Fig. S1).
To generate plasmid pUG34mCherry, the mCherry RFP ORF without its stop codon was PCR amplified from pKT355 (or pFA6a-link-mCherry-His3MX6), provided by K. Thorn (University of California, San Francisco, San Francisco, CA), and gap repaired into XbaI-digested pUG34 (provided by J. Hedgemann, Heinrich-Heine-Universität, Düsseldorf, Germany) to replace the yEGFP allele in pUG34 (confirmed by sequencing). Plasmids pUG36-INN1 and pUG34mCherry-INN1 were constructed by gap repairing the PCR-amplified INN1 ORF into EcoRI-digested pUG36 (provided by J. Hedgemann) or pUG34mCherry, generating N-terminally tagged GFP-INN1 and RFP-INN1 fusions that are under MET25 promoter control. pUG34mCherry-INN1-C2 and pUG34mCherry-INN1-tail were made similarly and contain INN1 codons 1–140 and 130–409, respectively. pUG34mCherry-INN1 was subjected to site-directed mutagenesis using the QuickChange Site-directed Mutagenesis kit (Agilent Technologies) to generate plasmids containing PXXP motif mutations (m1 to m4, either individually or in different combinations; see Results; Fig. S1).
To generate plasmid pRS315GW-C2-HOF1, the HOF1 gene (−1,000 to +2,510 bp relative to the start codon) was amplified by PCR from yeast genomic DNA and cloned into the pCR8/GW/TOPO vector (Invitrogen). A NotI site was introduced at the position immediately downstream of the HOF1 start codon by site-directed mutagenesis using the QuickChange Site-directed Mutagenesis kit, creating plasmid pCR8/GW-NotI-HOF1. A DNA fragment encoding the putative C2 domain of Inn1 (amino acids 1–134) flanked by two NotI sites was amplified by PCR, digested with NotI, and cloned into the NotI site of pCR8/GW-NotI-HOF1. The resulting plasmid was subjected to Gateway recombination (Invitrogen) into pRS315-attR, yielding pRS315GW-C2-HOF1.
The parent vectors for two-hybrid analyses were the DNA-binding domain (DBD) plasmid pEG202 (2µ and HIS3) and the activation domain (AD) plasmid pJG4-5 (2µ and TRP1; Gyuris et al., 1993). pEG202-HOF1-SH3 (residues 576–669) was supplied by C. Boone (University of Toronto, Toronto, Ontario, Canada). Other two-hybrid plasmids were constructed by PCR amplifying and cloning full-length INN1, HOF1, CYK3, and fragments of these genes (Figs. 4 and 5) into plasmids pEG202 and pJG4-5. In addition, pJG4-5-INN1-tail (residues 131–409) was subjected to site-directed mutagenesis to generate plasmids containing PXXP motif mutations (Fig. S1). The structures of these plasmids were confirmed by sequencing.
Plasmids for lipid binding and in vitro protein interaction assays were constructed as follows. DNA fragments encoding Inn1 amino acids 1–134 and Tcb1 amino acids 979–1,186 (the third C2 domain in Tcb1) were PCR amplified, digested with BamHI and XhoI (sites included in the primers), and cloned into BamHI–XhoI-digested pGSTag3vM (Narayan and Lemmon, 2006) to create plasmids encoding GST fusion proteins. DNA fragments encoding HOF1 amino acids 341–669 and CYK3 amino acids 1–70 were PCR amplified, digested with BamHI and SalI (sites included in the primers), and cloned into BamHI–SalI-digested pCOLADuet-1 (EMD) to create plasmids encoding His6-tagged proteins. An ∼840-bp BamHI–XhoI fragment encoding the wild-type or PXXP mutant derivatives of INN1 amino acids 131–409 was subcloned from wild-type or mutant pJG4-5-INN1-tail into the corresponding sites of pGEX-5X-1 (GE Healthcare) to create plasmids encoding GST fusion proteins.
Identification of synthetic lethal mutations
To screen for mutations synthetically lethal with hof1Δ, we used a hof1Δ ade2 ade3 leu2 ura3 strain harboring a high copy HOF1 ADE3 URA3 plasmid (strain LY1067). After mutagenesis with ethyl methanesulphonate to ∼50% viability, cells were grown overnight at 23°C to allow the expression of mutant phenotypes, plated, and screened for an inability to lose the HOF1 plasmid. Colonies lacking white sectors (indicating an inability to lose ADE3) were screened for sensitivity to FOA (indicating an inability to lose URA3) and then for recovery of growth on FOA after transformation with a HOF1 LEU2 plasmid (YCp50LEU2-HOF1) but not with a similar plasmid lacking HOF1, indicating that growth depended on HOF1 and not on some other feature of the plasmid.
To identify the genes defined by the synthetic lethal mutations, each mutant was crossed to strain LY1065, and appropriate segregants were mated and tested for complementation as judged by the ability to grow without plasmid-borne HOF1. Similar tests asked whether the new mutations could complement mutations in genes previously known to be synthetically lethal with hof1Δ. We also tested for the ability of low copy plasmids carrying known cytokinesis genes to rescue the mutants and/or analyzed the genes on plasmids obtained by rescuing the mutants using a YCp50LEU2-based genomic library (Bi and Pringle, 1996). Collectively, these tests showed that the mutations fell into 13–18 genes (see Results; Table S1).
Light and EM
The DIC and fluorescence microscopy images in Fig. 1 (B, D, and E), Fig. 2 C, Fig. 5 F, Fig. 6 (C and D), and Fig. S2 were acquired and processed using a computer-controlled microscope (Eclipse 800; Nikon), a Plan Apo 60×/1.40 NA oil immersion objective (Nikon), a high resolution charge-coupled device (CCD) camera (C4742-95; Hamamatsu Photonics), Image-Pro Plus software (Media Cybernetics), and Photoshop (CS3; Adobe). Time-lapse microscopy was performed as described previously by Vallen et al. (2000). Time-lapse experiments throughout this study were performed at 24°C. Actin rings and DNA were stained with Alexa Fluor 568 phalloidin (Invitrogen) and bisbenzimide (Sigma-Aldrich) as described previously by Bi et al. (1998).
The images in Fig. 7 A were acquired using IPLab software (BD) and a spinning-disk confocal microscope system comprising a scanner (CSU 10; Yokogawa), a microscope (IX 71; Olympus), a Plan S-Apo 100×/1.4 NA oil immersion objective (Olympus), and an ImagEM back-thinned EM CCD camera (C9100-13; Hamamatsu Photonics); components were integrated by BioVision Technologies. Diode lasers for excitation (488 nm for GFP; 561 nm for RFP) were housed in a launch constructed by Spectral Applied Research.
Other DIC and fluorescence images were acquired using a microscope (Eclipse 600-FN; Nikon), an Apo 100×/1.40 NA oil immersion objective (Nikon), a cooled CCD camera (ORCA-2; Hamamatsu Photonics), and MetaMorph software (version 5.0 or 7.0; MDS Analytical Technologies). Image contrast was enhanced using the MetaMorph and/or Photoshop software. GFP signal was observed using a triple-band filter set except in experiments involving GFP/CFP double staining in which YFP and CFP filter sets were used. To assess the asymmetry of Inn1 localization, DIC and Cdc3-CFP images were captured in the mid-cell focal plane, and a z series of 11 steps (0.2 µm) was captured for Inn1-GFP. The maximum projection images created from the z stacks using MetaMorph were analyzed for the Inn1 distribution patterns. Time-lapse microscopy was performed essentially as described previously by Salmon et al. (1998). To determine cluster indices (number of clusters with ≥3 connected cell bodies divided by this number plus the numbers of unbudded [one cell body] and budded [two cell bodies] cells), 400 cells plus clusters were scored for strain LY1310 transformed with either pRS425 or pRS425-CYK3, cured of plasmid pUG36-INN1 by growth on SC-Leu+FOA medium, and grown to exponential phase in SC-Leu medium.
For EM, cells were fixed with glutaraldehyde and potassium permanganate, embedded in LR white resin, and stained with uranyl acetate and lead citrate (details are available upon request). Images were obtained and processed using an electron microscope (JEM1230; JEOL), a cooled CCD camera (967; Gatan), Digital Micrograph software (Gatan), and Photoshop.
Lipid-binding assays
The lipid overlay and SPR assays were performed as described previously by Narayan and Lemmon (2006).
Coimmunoprecipitation and phosphatase treatment
Samples of cells from a synchronized culture (Fig. 4) were collected by centrifugation and frozen immediately in liquid nitrogen. Protein extracts were prepared using glass beads in NP-40 buffer (6 mM Na2HPO4, 4 mM NaH2PO4, 1% NONIDET P-40, 150 mM NaCl, and 2 mM EDTA) supplemented with 1 mM DTT, 50 mM NaF, 0.1 mM Na3VO4, and a complete protease inhibitor cocktail (Roche) and centrifuged at 2,000 g for 10 min. To precipitate Hof1–tandem affinity purification (TAP), 15 mg of each extract were incubated with 15 µl Dynabeads pan-mouse IgG (Invitrogen) for 1 h at 4°C, washed three times with NP-40 buffer, and eluted with SDS sample buffer. Samples were analyzed by SDS-PAGE (7.5% gel) and Western blotting using a mouse anti-GFP antibody (Roche) and an HRP-conjugated rabbit anti–mouse antibody (MP Biomedicals) to detect Inn1-GFP and peroxidase antiperoxidase soluble complex (Sigma-Aldrich) to detect Hof1-TAP.
For the phosphatase treatment experiment, 10 mg protein extract (prepared as described in the previous paragraph) was incubated for 1 h at 4°C with 4 µg mouse anti-GFP antibody (Roche) bound to 40 µl protein G–Sepharose. The beads were washed three times with NP-40 buffer and separated into four aliquots. As a control, SDS sample buffer was added to one aliquot. The other aliquots were washed twice with lambda protein phosphatase buffer (New England Biolabs, Inc.) and incubated for 30 min at 30°C in 30 µl of the same buffer with or without lambda protein phosphatase and phosphatase inhibitors (50 mM NaF and 1 mM Na3VO4). Reactions were terminated by adding 10 µl of 4× SDS sample buffer, and samples were analyzed by SDS-PAGE and Western blotting using anti-GFP and HRP-conjugated antibodies.
Two-hybrid interactions
Strain Y1026 carrying various DBD plasmids (see previous section) was mated to strain Y860 carrying various AD plasmids. Diploids were selected on SC-His-Trp plates, replica plated to SC-His-Trp-Ade plates containing 1% raffinose plus 2% galactose (to induce production of the fusion proteins), and incubated at 30°C for ≥4 d to detect interactions.
In vitro protein-binding assays
To purify His6-tagged proteins, Escherichia coli strain BL21 (Invitrogen) was transformed with pCOLADuet-based plasmids (see previous section), grown to exponential phase at 37°C, and induced with 1 mM IPTG for 3 h at 23°C. Cells were washed twice with double-distilled water, frozen at −20°C, thawed in Ni-NTA lysis buffer (300 mM NaCl, 20 mM Tris-HCl, pH 8.0, 20 mM imidazole, 10 mM β-mercaptoethanol, and 0.1% NP-40) containing a cocktail of protease inhibitors, sonicated seven times, placed on ice for 30–60 min, and centrifuged at 15,000 rpm for 20 min. The supernatant was mixed with Ni-NTA beads that had been freshly washed with Ni-NTA lysis buffer. After rocking for 1 h at 4°C, the beads were collected by centrifugation, washed three times with Ni-NTA buffer, and eluted five times with elution buffer (PBS containing 5 mM EDTA, 5 mM DTT, and 0.1% NP-40).
To purify GST-tagged proteins, E. coli BL21 was transformed with pGEX-5X–based plasmids (see previous section). Protein extracts were prepared essentially as described for the His6-tagged proteins except that the lysis buffer was PBS containing 5 mM EDTA, 5 mM DTT, and 0.1% NP-40. The 15,000-rpm supernatant was mixed with prewashed glutathione beads and rocked for 1 h at 4°C. The beads were collected by centrifugation, washed three times with lysis buffer, and resuspended in lysis buffer.
To test for protein binding in vitro, ∼3 µg His6-tagged protein was mixed with ∼5–7 µg GST (as negative control) or GST-tagged protein that was still bound to the glutathione beads (400 µl total volume) and rocked for 1 h at 4°C. The beads were washed five times with the GST fusion lysis buffer (see previous paragraph) and resuspended in 50 µl SDS sample buffer, and proteins were analyzed by SDS-PAGE (12% gel) and Western blotting using mouse monoclonal anti–penta-His (QIAGEN) and anti-GST (Covance) primary antibodies and an HRP-conjugated rabbit anti–mouse IgG secondary antibody (Jackson ImmunoResearch Laboratories). The anti-His signal was detected using the Immobilon Western chemiluminescent HRP substrate (Millipore), and the blot was incubated with the Restore blot-stripping buffer (Thermo Fisher Scientific) for 15 min at 37°C before reprobing with the anti-GST antibody, which was then detected by ECL (GE Healthcare).
Online supplemental materials
Table S1 shows genes identified by screening for synthetic lethality with hof1Δ. Fig. S1 shows sequence features of Inn1. Fig. S2 shows staining of actin rings and DNA in wild-type and inn1Δ cells. Fig. S3 shows dosage-dependent suppression of the inn1Δ growth defect by the putative C2 domain of Inn1. Fig. S4 shows lipid overlay assay of possible phospholipid binding by the putative C2 domain of Inn1. Time-lapse videos of Inn1-GFP and Cdc3-CFP in a wild-type cell (Video 1), a myo1Δ cell transformed with YCp50-MYO1 (Video 2), myo1Δ cells (Videos 3 and 4), hof1Δ cells (Videos 5 and 6), and cyk3Δ cells (Video 7). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200903125/DC1.
Acknowledgments
We thank M. Wang, J.H. Hedgemann, K. Thorn, J. Chant, F. Luca, A. Gitler, P. Walter, D. Lew, C. De Virgilio, L. Johnston, R.H. Valdivia, and L. Hartwell for strains and plasmids, J. Mulholland, J. Perrino, C. Adams, and A. Chien for assistance with EM and mass spectrometry, and many people in the Vallen (K. Bass, H. Bhattarai, S. Boyle, F. Castro, M.J. Hussain, N. Negrey, C. Palmer, S. Prow, L. Thé, M. Yang, and P. Yang) and Bi (M. Iwase, J. Luo, D. Lysko, and P. Martin) laboratories for assistance in the analysis of the genes defined by the hof1Δ synthetic lethal mutations.
This work was supported by Swarthmore College, by Howard Hughes Medical Institute grants to Swarthmore College (52002999, 52002654, and 52005202), by the National Institutes of Health (grants R15 GM065883-01 to E.A. Vallen, GM59216 to E. Bi, GM56846 to M.A. Lemmon, and GM31006 to J.R. Pringle), by a postdoctoral fellowship from the Uehara Memorial Foundation (to M. Onishi), by a predoctoral fellowship from the Great Rivers Affiliate of the American Heart Association (grant 0715300U to K. Moravcevic), and by the American Cancer Society (grant RSG-02-039-01-CSM to E. Bi).
Footnotes
Abbreviations used in this paper: AD, activation domain; AMR, actomyosin ring; CCD, charge-coupled device; DBD, DNA-binding domain; DIC, differential interference contrast; FOA, 5-fluoroorotic acid; latA, latrunculin A; MEN, mitotic exit network; PS, primary septum; SPR, surface plasmon resonance; SS, secondary septum; TAP, tandem affinity purification; TEM, transmission EM.
References
- Albertson R., Riggs B., Sullivan W. 2005. Membrane traffic: a driving force in cytokinesis.Trends Cell Biol. 15:92–101 [DOI] [PubMed] [Google Scholar]
- Ayscough K.R., Stryker J., Pokala N., Sanders M., Crews P., Drubin D.G. 1997. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor Latrunculin-A.J. Cell Biol. 137:399–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A., Ozier-Kalogeropoulos O., Denouel A., Lacroute F., Cullin C. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae.Nucleic Acids Res. 21:3329–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian M.K., Bi E., Glotzer M. 2004. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells.Curr. Biol. 14:R806–R818 [DOI] [PubMed] [Google Scholar]
- Bender A., Pringle J.R. 1991. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae.Mol. Cell. Biol. 11:1295–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benes C.H., Wu N., Elia A.E., Dharia T., Cantley L.C., Soltoff S.P. 2005. The C2 domain of PKCdelta is a phosphotyrosine binding domain.Cell. 121:271–280 [DOI] [PubMed] [Google Scholar]
- Bi E. 2001. Cytokinesis in budding yeast: the relationship between actomyosin ring function and septum formation.Cell Struct. Funct. 26:529–537 [DOI] [PubMed] [Google Scholar]
- Bi E., Pringle J.R. 1996. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae.Mol. Cell. Biol. 16:5264–5275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E., Maddox P., Lew D.J., Salmon E.D., McMillan J.N., Yeh E., Pringle J.R. 1998. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis.J. Cell Biol. 142:1301–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel M., Bach S., Bamps S., Dobbelaere J., Wiget P., Longaretti C., Barral Y., Meijer L., Peter M. 2005. Degradation of Hof1 by SCF(Grr1) is important for actomyosin contraction during cytokinesis in yeast.EMBO J. 24:1440–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyne J.R., Yosuf H.M., Bieganowski P., Brenner C., Price C. 2000. Yeast myosin light chain, Mlc1p, interacts with both IQGAP and class II myosin to effect cytokinesis.J. Cell Sci. 113:4533–4543 [DOI] [PubMed] [Google Scholar]
- Bulawa C.E., Osmond B.C. 1990. Chitin synthase I and chitin synthase II are not required for chitin synthesis in vivo in Saccharomyces cerevisiae.Proc. Natl. Acad. Sci. USA. 87:7424–7428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W., Stahelin R.V. 2006. Membrane binding and subcellular targeting of C2 domains.Biochim. Biophys. Acta. 1761:838–849 [DOI] [PubMed] [Google Scholar]
- Christianson T.W., Sikorski R.S., Dante M., Shero J.H., Hieter P. 1992. Multifunctional yeast high–copy–number shuttle vectors.Gene. 110:119–122 [DOI] [PubMed] [Google Scholar]
- Chuang J.S., Schekman R.W. 1996. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p.J. Cell Biol. 135:597–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett M., Xiong Y., Boyne J.R., Wright D.J., Munro E., Price C. 2006. IQGAP and mitotic exit network (MEN) proteins are required for cytokinesis and re-polarization of the actin cytoskeleton in the budding yeast, Saccharomyces cerevisiae.Eur. J. Cell Biol. 85:1201–1215 [DOI] [PubMed] [Google Scholar]
- Dai H., Shen N., Arac D., Rizo J. 2007. A quaternary SNARE-synaptotagmin-Ca2+-phospholipid complex in neurotransmitter release.J. Mol. Biol. 367:848–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lozanne A., Spudich J.A. 1987. Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination.Science. 236:1086–1091 [DOI] [PubMed] [Google Scholar]
- Dobbelaere J., Barral Y. 2004. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex.Science. 305:393–396 [DOI] [PubMed] [Google Scholar]
- Echard A., Hickson G.R., Foley E., O'Farrell P.H. 2004. Terminal cytokinesis events uncovered after an RNAi screen.Curr. Biol. 14:1685–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epp J.A., Chant J. 1997. An IQGAP-related protein controls actin-ring formation and cytokinesis in yeast.Curr. Biol. 7:921–929 [DOI] [PubMed] [Google Scholar]
- Feller S.M., Ren R., Hanafusa H., Baltimore D. 1994. SH2 and SH3 domains as molecular adhesives: the interactions of Crk and Abl.Trends Biochem. Sci. 19:453–458 [DOI] [PubMed] [Google Scholar]
- Finger F.P., White J.G. 2002. Fusion and fission: membrane trafficking in animal cytokinesis.Cell. 108:727–730 [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Sugino A. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites.Gene. 74:527–534 [DOI] [PubMed] [Google Scholar]
- Guthrie C., Fink G.R. 1991. Guide to yeast genetics and molecular biology.Methods Enzymol. 194:1–863 [PubMed] [Google Scholar]
- Gyuris J., Golemis E., Chertkov H., Brent R. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2.Cell. 75:791–803 [DOI] [PubMed] [Google Scholar]
- Hales K.G., Bi E., Wu J.Q., Adam J.C., Yu I.C., Pringle J.R. 1999. Cytokinesis: an emerging unified theory for eukaryotes? Curr. Opin. Cell Biol. 11:717–725 [DOI] [PubMed] [Google Scholar]
- Heath R.J., Insall R.H. 2008. F-BAR domains: multifunctional regulators of membrane curvature.J. Cell Sci. 121:1951–1954 [DOI] [PubMed] [Google Scholar]
- Hibi M., Nagasaki A., Takahashi M., Yamagishi A., Uyeda T.Q. 2004. Dictyostelium discoideum talin A is crucial for myosin II-independent and adhesion-dependent cytokinesis.J. Muscle Res. Cell Motil. 25:127–140 [DOI] [PubMed] [Google Scholar]
- Hwa Lim H., Yeong F.M., Surana U. 2003. Inactivation of mitotic kinase triggers translocation of MEN components to mother-daughter neck in yeast.Mol. Biol. Cell. 14:4734–4743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome.Proc. Natl. Acad. Sci. USA. 98:4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanada M., Nagasaki A., Uyeda T.Q. 2008. Novel functions of Ect2 in polar lamellipodia formation and polarity maintenance during “contractile ring-independent” cytokinesis in adherent cells.Mol. Biol. Cell. 19:8–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.B., Haarer B.K., Pringle J.R. 1991. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC3 gene product and the timing of events at the budding site.J. Cell Biol. 112:535–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein D.E., Lee A., Frank D.W., Marks M.S., Lemmon M.A. 1998. The pleckstrin homology domains of dynamin isoforms require oligomerization for high affinity phosphoinositide binding.J. Biol. Chem. 273:27725–27733 [DOI] [PubMed] [Google Scholar]
- Ko N., Nishihama R., Tully G.H., Ostapenko D., Solomon M.J., Morgan D.O., Pringle J.R. 2007. Identification of yeast IQGAP (Iqg1p) as an anaphase-promoting-complex substrate and its role in actomyosin-ring-independent cytokinesis.Mol. Biol. Cell. 18:5139–5153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek W.S., Bi E., Epp J.A., Wang L., Ho J., Chant J. 2000. Cyk3, a novel SH3-domain protein, affects cytokinesis in yeast.Curr. Biol. 10:947–950 [DOI] [PubMed] [Google Scholar]
- Lee P.R., Song S., Ro H.S., Park C.J., Lippincott J., Li R., Pringle J.R., De Virgilio C., Longtine M.S., Lee K.S. 2002. Bni5p, a septin-interacting protein, is required for normal septin function and cytokinesis in Saccharomyces cerevisiae.Mol. Cell. Biol. 22:6906–6920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie S.H., Pringle J.R. 1980. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation.J. Bacteriol. 143:1384–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J., Li R. 1998. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis.J. Cell Biol. 140:355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J., Shannon K.B., Shou W., Deshaies R.J., Li R. 2001. The Tem1 small GTPase controls actomyosin and septin dynamics during cytokinesis.J. Cell Sci. 114:1379–1386 [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, DeMarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae.Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Lord M., Laves E., Pollard T.D. 2005. Cytokinesis depends on the motor domains of myosin-II in fission yeast but not in budding yeast.Mol. Biol. Cell. 16:5346–5355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Machius M., Dulubova I., Dai H., Sudhof T.C., Tomchick D.R., Rizo J. 2006. Structural basis for a Munc13-1 homodimer to Munc13-1/RIM heterodimer switch.PLoS Biol. 4:e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Vallen E.A., Dravis C., Tcheperegine S.E., Drees B.L., Bi E. 2004. Identification and functional analysis of the essential and regulatory light chains of the only type II myosin Myo1p in Saccharomyces cerevisiae.J. Cell Biol. 165:843–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K.S., Aravind L., Koonin E.V. 1999. A superfamily of archaeal, bacterial, and eukaryotic proteins homologous to animal transglutaminases.Protein Sci. 8:1714–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi S., Uyama T., Kitagawa H., Nomura K.H., Dejima K., Gengyo-Ando K., Mitani S., Sugahara K., Nomura K. 2003. Chondroitin proteoglycans are involved in cell division of Caenorhabditis elegans.Nature. 423:443–448 [DOI] [PubMed] [Google Scholar]
- Narayan K., Lemmon M.A. 2006. Determining selectivity of phosphoinositide-binding domains.Methods. 39:122–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neujahr R., Heizer C., Gerisch G. 1997. Myosin II-independent processes in mitotic cells of Dictyostelium discoideum: redistribution of the nuclei, re-arrangement of the actin system and formation of the cleavage furrow.J. Cell Sci. 110:123–137 [DOI] [PubMed] [Google Scholar]
- Otegui M.S., Verbrugghe K.J., Skop A.R. 2005. Midbodies and phragmoplasts: analogous structures involved in cytokinesis.Trends Cell Biol. 15:404–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo J., Sudhof T.C. 1998. C2-domains, structure and function of a universal Ca2+-binding domain.J. Biol. Chem. 273:15879–15882 [DOI] [PubMed] [Google Scholar]
- Rodriguez J.R., Paterson B.M. 1990. Yeast myosin heavy chain mutant: maintenance of the cell type specific budding pattern and the normal deposition of chitin and cell wall components requires an intact myosin heavy chain gene.Cell Motil. Cytoskeleton. 17:301–308 [DOI] [PubMed] [Google Scholar]
- Salmon E.D., Shaw S.L., Waters J., Waterman-Storer C.M., Maddox P.S., Yeh E., Bloom K. 1998. A high-resolution multimode digital microscope system.Methods Cell Biol. 56:185–215 [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Diaz A., Marchesi V., Murray S., Jones R., Pereira G., Edmondson R., Allen T., Labib K. 2008. Inn1 couples contraction of the actomyosin ring to membrane ingression during cytokinesis in budding yeast.Nat. Cell Biol. 10:395–406 [DOI] [PubMed] [Google Scholar]
- Schmidt M., Bowers B., Varma A., Roh D.-H., Cabib E. 2002. In budding yeast, contraction of the actomyosin ring and formation of the primary septum at cytokinesis depend on each other.J. Cell Sci. 115:293–302 [DOI] [PubMed] [Google Scholar]
- Schulz T.A., Creutz C.E. 2004. The tricalbin C2 domains: lipid-binding properties of a novel, synaptotagmin-like yeast protein family.Biochemistry. 43:3987–3995 [DOI] [PubMed] [Google Scholar]
- Shannon K.B., Li R. 1999. The multiple roles of Cyk1p in the assembly and function of the actomyosin ring in budding yeast.Mol. Biol. Cell. 10:283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon K.B., Li R. 2000. A myosin light chain mediates the localization of the budding yeast IQGAP-like protein during contractile ring formation.Curr. Biol. 10:727–730 [DOI] [PubMed] [Google Scholar]
- Shao X., Davletov B.A., Sutton R.B., Sudhof T.C., Rizo J. 1996. Bipartite Ca2+-binding motif in C2 domains of synaptotagmin and protein kinase C.Science. 273:248–251 [DOI] [PubMed] [Google Scholar]
- Shaw J.A., Mol P.C., Bowers B., Silverman S.J., Valdivieso M.H., Duran A., Cabib E. 1991. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle.J. Cell Biol. 114:111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skop A.R., Liu H., Yates J., III, Meyer B.J., Heald R. 2004. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms.Science. 305:61–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F., Amon A. 2004. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation.Annu. Rev. Genet. 38:203–232 [DOI] [PubMed] [Google Scholar]
- Stevens R.C., Davis T.N. 1998. Mlc1p is a light chain for the unconventional myosin Myo2p in Saccharomyces cerevisiae.J. Cell Biol. 142:711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland L.I., Burgess D.R. 2004. Pathways for membrane trafficking during cytokinesis.Trends Cell Biol. 14:115–118 [DOI] [PubMed] [Google Scholar]
- Szafer-Glusman E., Giansanti M.G., Nishihama R., Bolival B., Pringle J.R., Gatti M., Fuller M.T. 2008. A role for very-long-chain fatty acids in furrow ingression during cytokinesis in Drosophila spermatocytes.Curr. Biol. 18:1426–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A.H., Evangelista M., Parsons A.B., Xu H., Bader G.D., Page N., Robinson M., Raghibizadeh S., Hogue C.W., Bussey H., et al. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants.Science. 294:2364–2368 [DOI] [PubMed] [Google Scholar]
- Tong A.H., Drees B., Nardelli G., Bader G.D., Brannetti B., Castagnoli L., Evangelista M., Ferracuti S., Nelson B., Paoluzi S., et al. 2002. A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules.Science. 295:321–324 [DOI] [PubMed] [Google Scholar]
- Vallen E.A., Caviston J., Bi E. 2000. Roles of Hof1p, Bni1p, Bnr1p, and Myo1p in cytokinesis in Saccharomyces cerevisiae.Mol. Biol. Cell. 11:593–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VerPlank L., Li R. 2005. Cell cycle-regulated trafficking of Chs2 controls actomyosin ring stability during cytokinesis.Mol. Biol. Cell. 16:2529–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeong F.M. 2005. Severing all ties between mother and daughter: cell separation in budding yeast.Mol. Microbiol. 55:1325–1331 [DOI] [PubMed] [Google Scholar]
- Yoshida S., Kono K., Lowery D.M., Bartolini S., Yaffe M.B., Ohya Y., Pellman D. 2006. Polo-like kinase Cdc5 controls the local activation of Rho1 to promote cytokinesis.Science. 313:108–111 [DOI] [PubMed] [Google Scholar]
- Zhang G., Kashimshetty R., Ng K.E., Tan H.B., Yeong F.M. 2006. Exit from mitosis triggers Chs2p transport from the endoplasmic reticulum to mother–daughter neck via the secretory pathway in budding yeast.J. Cell Biol. 174:207–220 [DOI] [PMC free article] [PubMed] [Google Scholar]