SUMMARY
MutS homologs (MSHs) are critical components of the eukaryotic mismatch repair machinery. In addition to repairing mismatched DNA, mismatch repair enzymes are known in higher eukaryotes to directly signal cell cycle arrest and apoptosis in response to DNA damaging agents. Accordingly, mammalian cells lacking certain MSHs are resistant to chemotherapeutic drugs. Interestingly, we have discovered that the disruption of TgMSH-1, an MSH in the pathogenic parasite, T. gondii, confers drug resistance. Through a genetic selection for T. gondii mutants resistant to the antiparasitic drug monensin, we have isolated a strain that is resistant not only to monensin but also to salinomycin and the alkylating agent, methylnitrosourea. We have shown that this phenotype is due to the disruption of TgMSH-1 as the multi-drug resistance phenotype is complemented by a wild-type copy of TgMSH-1 and is recapitulated by a directed disruption of this gene in a wild-type strain. We have also shown that, unlike previously described MSHs involved in signaling, TgMSH-1 localizes to the parasite mitochondrion. These results provide the first example of a mitochondrial MutS Homolog that is involved in drug sensitivity and implicate the induction of mitochondrial stress as a mode of action of the widely used drug, monensin.
Keywords: Toxoplasma gondii, monensin, MutS, MSH, resistance, mitochondrion
INTRODUCTION
The Apicomplexa comprise a medically and economically important phylum of obligate intracellular parasites, including Plasmodium falciparum, the causative agent of malaria, Eimeria, which causes chicken coccidiosis, Cryptosporidium, which causes gastrointestinal disease, and Toxoplasma gondii, which can be pathogenic in immunocompromised hosts and the developing fetus. Drug therapies that effectively treat infection with certain apicomplexan parasites, such as Cryptosporidium are currently lacking (Haberkorn, 1996), while therapies directed against other parasites are quickly becoming ineffective due to the emergence of drug-resistant parasite strains. This is best demonstrated by the fact that chloroquine resistant Plasmodium strains have been described in almost every country in Africa, and parasites resistant to alternative antibiotics including atovaquone and pyrimethamine-sulfadiazine are also increasing in frequency (Hyde, 2002).
Due to the constant development and proliferation of drug resistant parasite strains, there is increasing pressure to identify new drugs that are effective against apicomplexan parasites, and to prevent the development of strains that are resistant to existing drugs. Thus, a thorough understanding of the targets of anti-parasitic drugs, and the mechanisms behind resistance is needed. Such information helps facilitate the discovery of new drugs that more efficiently target parasite pathways and the design of combination therapies that act synergistically, thereby allowing more effective treatment and slowing drug resistance.
One drug that has proven very effective against certain apicomplexan parasites is the coccidiostat, monensin. Monensin is a polyether monocarboxylic acid ionophore produced by Streptomyces cinnamonensis that catalyzes the exchange of Na+ for H+ across biological membranes (Pressman, 1976). Soon after its initial isolation in 1967 (Agtarap et al., 1967) monensin’s activity against gram positive bacteria and Eimeria parasites was discovered (Haney & Hoehn, 1967, Shumard & Callender, 1967). Monensin was first used commercially in 1971 for the prevention of chicken coccidiosis caused by Eimeria parasites and has also been used to reduce abortions caused by T. gondii in sheep (Buxton et al., 1987, Buxton et al., 1988). Unfortunately, monensin-resistant Eimeria maxima parasites were reported within three years of the drug’s introduction (Jeffers, 1974), and by 1987, a field isolate of Eimeria tenella with a 20-fold reduced susceptibility to monensin had been described (Augustine et al., 1987). Nonetheless, monensin and other ionophorous antibiotics continue to be among the most widely used anticoccidial drugs in the poultry industry (Chapman, 2001).
Previous investigators have shown that treatment of extracellular Eimeria tenella sporozoites with monensin increases the parasite Na+-K+ ATPase activity, causes lactate accumulation, and decreases the intracellular ATP concentration (Smith & Galloway, 1983). These authors suggest that following treatment with monensin, the Na+-K+ ATPase works to restore proper intraparasitic ion concentration, but that energy depletion and pump saturation allow a net influx of Na+ and Cl− into the parasite, resulting in osmotic swelling and non-specific parasite death. This assertion is supported by studies employing electron microscopy, which have shown that T. gondii tissue cysts and Eimeria tenella sporozoites appear swollen and have a distorted membrane structure following treatment with monensin (Couzinet et al., 2000, Smith & Strout, 1980).
Although some authors have suggested that monensin resistance in Eimeria is a result of decreased drug uptake caused by changes in membrane fluidity (Augustine et al., 1987), monensin’s mode of action and the genetic basis of drug resistance remain unknown. This is due, in part, to the fact that most studies involving monensin have utilized Eimeria parasites, which cannot be easily cultured in vitro, and which lack methods for effective genetic and molecular manipulation. However, monensin is also effective against T. gondii, both in vivo and in vitro (Buxton et al., 1988, Ricketts & Pfefferkorn, 1993). In contrast to Eimeria, T. gondii can be easily manipulated genetically and has become a model system for the study of apicomplexan parasites (Kim & Weiss, 2004). Thus, T. gondii presents a good model for studying monensin’s mode of action and the mechanisms underlying resistance. Indeed, studies employing chemical mutagenesis in T. gondii have suggested that the in vitro development of drug resistance in T. gondii is comparable to that of Eimeria in the field (Ricketts & Pfefferkorn, 1993).
Here we report the isolation and characterization of a T. gondii mutant that is resistant to monensin and several other drugs. We have shown that the drug-resistant phenotype is caused by a disruption of TgMSH-1, a homolog of the DNA repair enzyme, MutS. Given that TgMSH-1 localizes to the mitochondrion, and that mammalian MutS Homologs are known to signal cell death in response to genotoxic drugs (Jiricny, 2006), we hypothesize that monensin induces parasite stress and propose a role for TgMSH-1 in detecting and responding to mitochondrial stress induced by monensin.
RESULTS
Isolation of a Toxoplasma gondii monensin-resistant mutant
To gain a better understanding of the mechanism by which monensin kills coccidian parasites, we aimed to identify genes that when mutated, confer resistance to the drug. Accordingly, we designed and performed a genetic selection for monensin-resistant T. gondii parasites. Briefly, parasites from two insertionally mutagenized populations and a non-mutagenized control (see Materials and Methods) were allowed to invade host cells and intracellular parasites were incubated for 4–6 days in the presence of 1.0 ng/ml monensin, which kills 100% of wild-type parasites (Fig. 1A). Following this treatment, the media was changed to eliminate monensin and to allow surviving parasites to propagate. As expected, no surviving parasites were recovered from the non-mutagenized population. The selection was repeated with the surviving mutant parasites and after a total of three rounds of selection, parasites were cloned by limiting dilution.
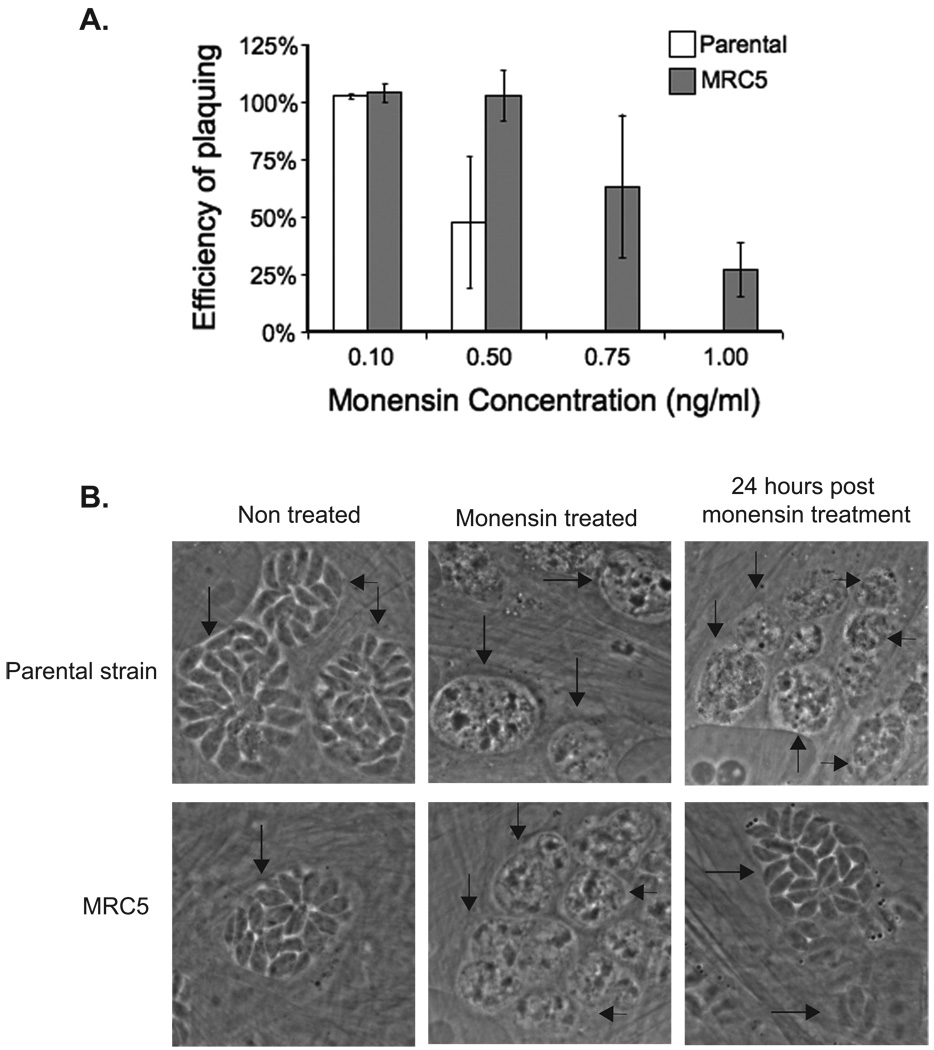
Figure 1.
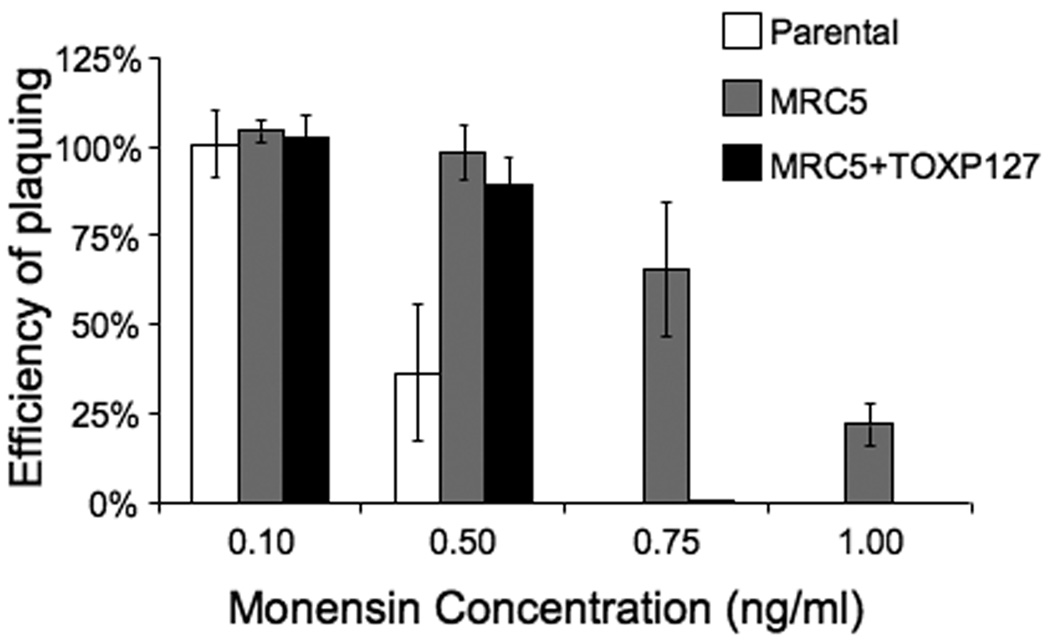
Effect of monensin on parental strain and MRC5 parasites. A. Intracellular parasites were allowed to form plaques in the media containing monensin or solvent control. The efficiency of plaquing represents the number of plaques formed in the presence of monensin divided by the number of plaques formed in the presence of the solvent. Data bars represent the average of three independent experiments, and error bars denote the standard deviation. B. Control parasites of the parental strain (top panels) and MRC5 (bottom panels) that were either not treated with monensin, treated with 2 ng/ml monensin for 24 hours, and parasites that were treated with 2 ng/ml monensin for 24 hours followed by a 24 hour recovery period in the absence of monensin are shown. Arrows point at the vacuoles as confirmed by the presence of GFP signal (data not shown).
In order to confirm the monensin-resistant phenotype, all established clones (11 clones from mutant population #1 and 12 clones from population #2) were tested for the ability to form plaques in the presence of media containing either monensin or an equivalent concentration of solvent. For these assays, monensin was present in the media for the duration of the experiment, as opposed to being removed after 4–6 days of incubation, as was done during the selection. In this manner we can identify those clones that have the strongest monensin-resistant phenotypes (i.e. the clones that can develop and divide normally in the presence of drug) without physically disturbing the cultures, which can interfere with plaque formation and quantification. Of the clones tested, monensin-resistant clone C5 (MRC5) had the highest efficiency of plaquing in the presence of the drug, and was therefore chosen for further analysis.
MRC5’s efficiency of plaquing in the presence of monensin was quantified and compared to the parental strain. While no plaques were detected when parasites of the parental strain were grown in the presence of either 0.75 ng/ml or 1.0 ng/ml monensin, mutant strain MRC5 exhibited an efficiency of plaquing of 64% in 0.75 ng/ml monensin and 27% in 1.0 ng/ml monensin (Fig. 1A). While the mutants were selected for resistance to 1.0 ng/ml monensin we do not see 100% survival at this concentration. A possible reason is that, as stated above, during the selection, the drug was removed after 4–6 days, which allows a surviving parasite to form a plaque more quickly, while in the quantification assay, monensin was left in the culture medium for the duration of the assay (8–15 days). As an additional control, we carried out the resistance assay using a parasite clone that had incorporated the insertional vector into its genome, but was not selected for resistance to monensin. This control strain had a monensin-sensitive phenotype similar to that of the parental strain (data not shown). Therefore, the monensin-resistant phenotype of MRC5 is not a consequence of the expression of the selectable marker present in the mutagenic vector.
To better understand the nature of the MRC5’s monensin-resistant phenotype, we used microscopy to examine the morphological changes that occur in MRC5 and parental stain parasites in response to monensin. For this purpose, parasites were allowed to invade host cells, and then 24 hours after invasion, the culture medium was changed to include 2 ng/ml monensin. The parasites were then incubated in the presence of drug for an additional 24 hours. Following this incubation period, the parasites were examined by microscopy. As shown in Figure 1B, both parental strain and MRC5 parasites lose their elongated morphology following treatment with 2 ng/ml monensin, and vacuoles containing these monensin-treated parasites appear to be filled with debris. However, when the parasites are allowed to recover in the absence of monensin for 24 hours, MRC5 parasites regain their normal elongated morphology, while parental parasites do not recover. These results indicate that monensin is affecting both the parental strain and MRC5 parasites, but in MRC5, the effects of monensin do not result in parasite death.
MRC5 is also resistant to the coccidiostat, salinomycin
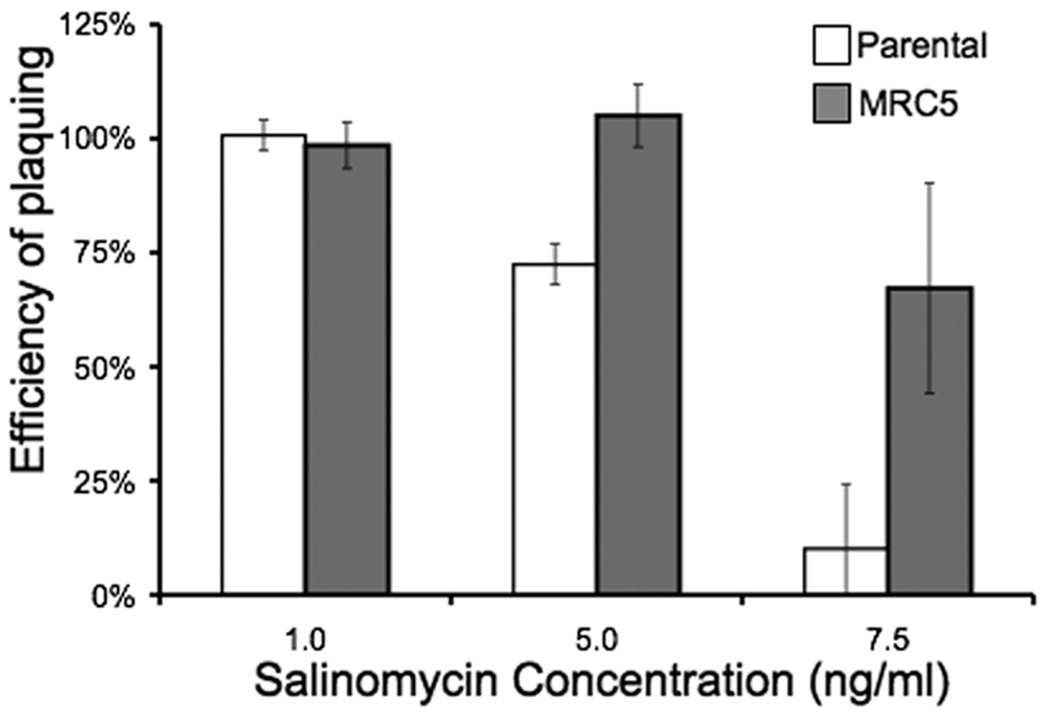
It has been reported that some monensin-resistant Eimeria parasite isolates can exhibit cross-resistance to other coccidiostats including maduramicin, salinomycin, nicarbazin, halofuginone, toltrazuril, and diclazuril (Stephen et al., 1997). We were therefore interested in determining whether the mutation that confers resistance to monensin in MRC5 also confers resistance to other anticoccidial drugs. Accordingly, we tested MRC5 for resistance to salinomycin, a H+/K+ ionophore and determined that MRC5 parasites exhibit cross-resistance to this drug (Fig. 2). While 67% of MRC5 parasites formed plaques in the presence of 7.5 ng/ml salinomycin, only 10% of the parental strain parasites formed plaques at this drug concentration. These results indicate that mutations present in MRC5 confer resistance to multiple anticoccidial drugs.
Figure 2.
Effect of salinomycin on parental strain and MRC5 parasites. Intracellular parasites were allowed to form plaques in media containing salinomycin or an equivalent concentration of solvent. The efficiency of plaquing represents the number of plaques formed in the presence of salinomycin divided by the number of plaques formed in the presence of solvent. Data bars represent the average of three independent experiments, and error bars denote the standard deviation.
The mutagenic insertion in MRC5 falls within a region of homology to a DNA repair enzyme
As a first step to characterize MRC5 at the molecular level, we determined the number of mutagenic insertions that are present in this mutant clone. For this purpose, Southern blot analysis was performed on EcoRI digested genomic DNA from the mutant and the parental strain (Fig. 3A) using a probe that consists of a 635 bp region of the hypoxanthine/xanthine guanine phosphoribosyl transferase (HPT) cDNA, which is the selectable marker in the mutagenic insertion. In DNA from both the parental and MRC5 parasites, a fragment of approximately 16 kb in length can be observed, which corresponds to the remnants of the endogenous HPT gene that remains in the parental RHΔhpt strain (Donald et al., 1996) (Fig. 3A). In addition, probing the MRC5 DNA with HPT revealed only one additional band of approximately 7 kb in length (Fig. 3A) suggesting that MRC5 contains a single insertion. The band representing the insertion is more intense than the band corresponding to the endogenous locus. This is most likely due to the fact that only a portion of the probe corresponds to the remaining region of the endogenous gene while the entire probe hybridizes to the inserted plasmid.
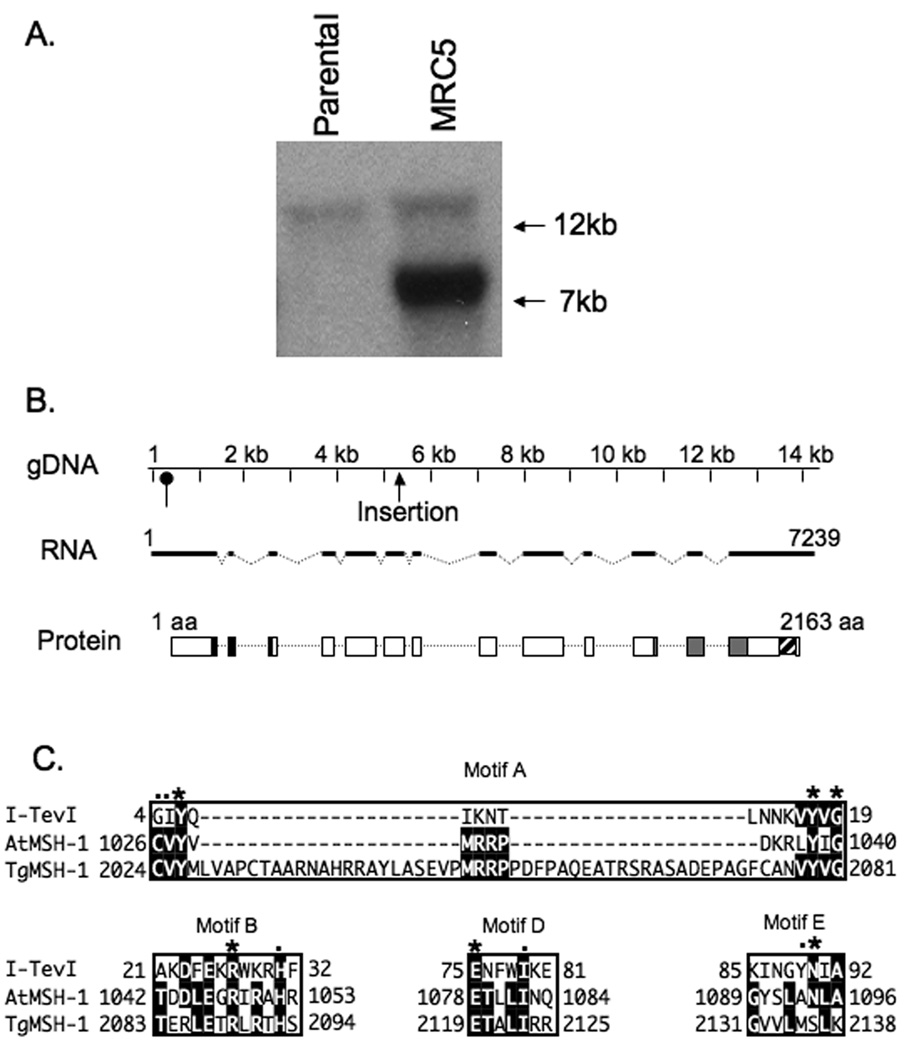
Figure 3.
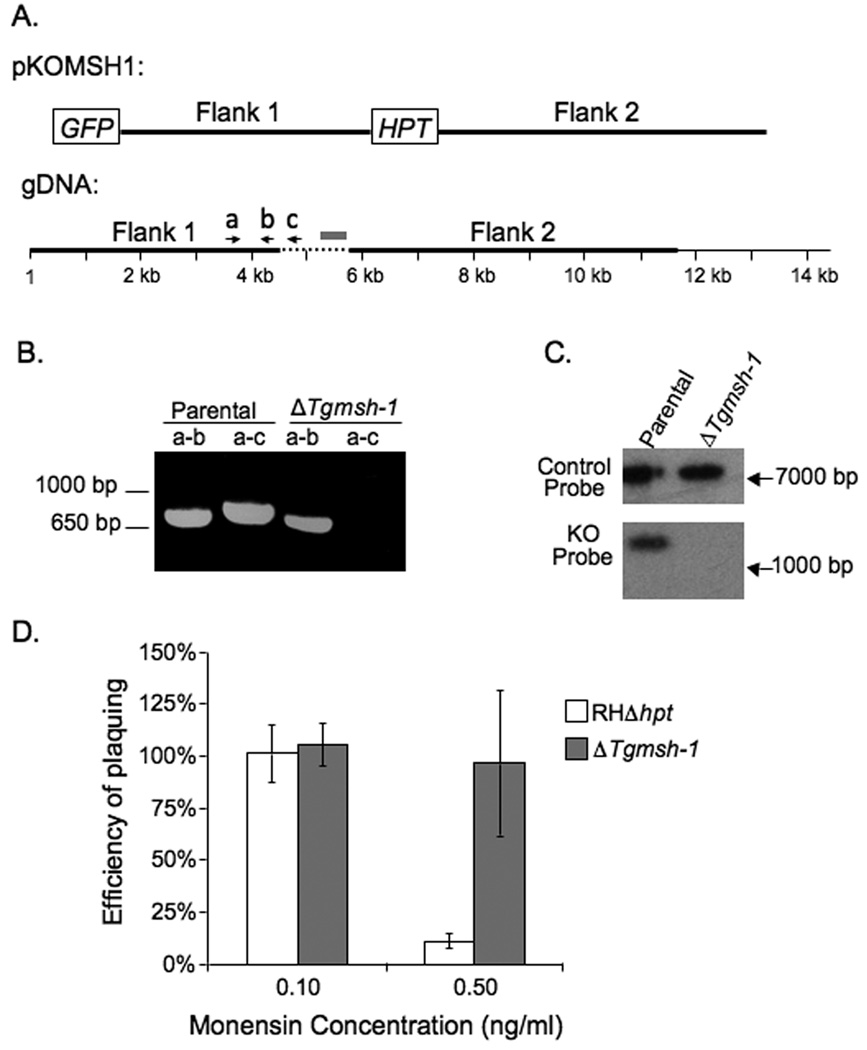
Identification of insertion site and characterization TgMSH-1. A. EcoRI digested genomic DNA from the parental strain and MRC5 was analyzed by Southern Blot using HPT cDNA as a probe. The 16 Kb band corresponds to the remnants of the endogenous hpt gene that was present in the parental strain, while the 7 Kb band corresponds to hpt present in the mutagenic insertion. B. The distribution of exons (dark lines), introns (broken lines) and coding sequences (white boxes) along the genomic region encoding TgMSH-1 are shown. Base 1 in the gDNA and RNA sequence represents the first putative transcription start site, as determined by 5’ RACE. The position of an alternative transcription start detected by 5’ RACE is indicated with an oval arrow. The location of the mutagenic insertion within the genomic DNA is also indicated. Conceptual translation of TgMSH-1 reveals sequences with homology to MutS domain I (black boxes in protein diagram, amino acids 290 to 393), sequences with homology to MutS domain V (grey boxes, amino acids 1521 to 1799), and a region with homology to the GIY-YIG endonuclease domain (striped box, amino acids 2024 to 2138). C. Alignment of the predicted TgMSH-1 GIY-YIG endonuclease domain with GIY-YIG motifs found in the T4 phage homing endonuclease, I-TevI, and in the Arabidopsis thaliana MSH1 protein. Sequences were aligned using the MegAlign program (DNA Star). Asterisks denote residues that are more than 80% conserved among GIY-YIG family members, while dots denote residues that are more than 90% conserved (Van Roey et al., 2002). The numbers adjacent to the protein sequences denote the location of the first and last aligned residue within the corresponding protein. The accession number for I-TevI is P13299 and for AtMSH-1 is NP_189075.
To determine the specific region of the genome disrupted by the insertional vector in MRC5, we performed a molecular cloning approach to recover the integrated plasmid along with flanking genomic DNA fragments (Arrizabalaga et al., 2004), (Materials and Methods). Fragments flanking both ends of the insertion were recovered in this manner and then sequenced. Bioinformatic analysis of these sequences allowed us to determine that the insertion was located between bases 1,216,140 and 1,216,141 of chromosome XII (http://ToxoDB.org). This integration site and the orientation of the insertion predicts the presence of a 7.7 kb HPT containing band when MRC5 DNA is digested with EcoRI, which is consistent with our Southern blot results (Fig. 3A). Importantly, in three independent attempts, only this insertion site was detected, emphasizing that MRC5 is likely to have only one insertion.
Sequence analysis of the region disrupted by the insertion revealed the presence of a predicted gene encoding conserved N-terminal and C-terminal domains found in the MutS family of mismatch repair enzymes. MutS proteins in prokaryotes and MutS homologs (MSH) in eukaryotic cells are part of the mismatch repair (MMR) pathway, which is responsible for correcting errors that occur during DNA replication. Within this pathway, MutS is responsible for the initial recognition of the mismatch and recruitment of other MMR enzymes that enact the actual repair (Kunkel & Erie, 2005). Interestingly, mutations in certain MSHs are known to confer resistance to various drugs in cell systems including yeast (Drotschmann et al., 2004) and human cancer cells (Aebi et al., 1997, Umar et al., 1997, Wu et al., 2004). Thus, drug resistance as a consequence of an MSH disruption in T. gondii might be consistent with these previous observations.
The disrupted gene encodes an atypical MutS homolog
To further characterize this undescribed T. gondii gene and to confirm that the insertion was within it, we cloned and sequenced the entire putative gene using PCR approaches and wild-type parasite cDNA (see material and methods for details). We have named this gene and its predicted product MutS Homolog-1 (TgMSH-1) since it is the first protein in the MutS family to be characterized in T. gondii, and because analysis of the genome reveals the presence of at least two additional proteins with homology to MutS. We have deposited the TgMSH-1 cDNA sequence on GenBank (accession number FJ167393). The TgMSH-1 gene is encompassed within a genomic region of approximately 14.4 kilobases and contains 13 exons and 12 introns (Fig. 3B). Analysis of the gene’s sequence confirms that the insertion is located within the sixth exon of TgMSH-1 (Fig. 3B), with the selectable marker oriented in the opposite direction as the TgMSH-1 open reading frame. 5’ and 3’ RACE analysis revealed the presence of two putative transcripts that are 7,239 bases and 7,025 bases in length respectively. These transcripts differ only in the length of the 5’ untranslated regions and are likely the result of two alternative transcription start sites.
Conceptual translation of the coding sequence reveals that TgMSH-1 is a 2,163 amino acid protein with a predicted molecular weight of approximately 240 kDa (Fig. S1). Bioinformatic analysis of this protein using BLAST indicates that TgMSH-1 is most homologous to predicted MSHs in photosynthetic organisms, including the green algae, Ostreococcus tauri, the tomato (Lycopersicon esculentum), corn (Zea mays), rice (Oryza sativa), and to the MSH-1 protein in Arabidopsis thaliana, with which TgMSH-1 shares a 27% sequence identity (Fig. S2).
Typical MutS proteins contain 5 structural domains of which domains I and IV interact with DNA and domain V contains an ATPase and dimerization interface (Kunkel & Erie, 2005, Obmolova et al., 2000). Domains II and III of MutS form a connector domain and the core domain, respectively (Lamers et al., 2000, Obmolova et al., 2000). Interestingly, TgMSH-1 contains amino acid sequences with homology to MutS domains I and V, but lacks sequences homologous to domains II, III, and IV (Fig. 3B). While this is an unusual arrangement for MutS homologs, the MSH-1 protein in Arabidopsis thaliana (AtMSH-1) shares a similar domain organization with TgMSH-1 at the sequence level (Abdelnoor et al., 2003). AtMSH-1, which localizes to the mitochondria, is thought to play a role in the suppression of recombination or amplification of mitochondrial DNA molecules (Abdelnoor et al., 2003).
Unlike typical MSH proteins, AtMSH-1 also encodes a C-terminal GIY-YIG endonuclease domain that is necessary for protein function (Abdelnoor et al., 2006). GIY-YIG endonucleases include either four or five conserved sequence motifs that serve structural roles and which contain catalytic residues (Kowalski et al., 1999). Of the 5 conserved motifs present in GIY-YIG family members, TgMSH-1 contains regions of identity to motifs A, B, D and E (Fig. 3C). We were unable to find sequences with strong homology to motif C, but numerous other GIY-YIG family members lack region (Kowalski et al., 1999). Two residues that are completely conserved among GIY-YIG family members, including a catalytic arginine residue in motif B, and a divalent cation-binding glutamate residue in motif D, are present in TgMSH-1. In addition, motif A of TgMSH-1 contains the two conserved tyrosine residues (Y2026 and Y2079) as well as a glycine residue (G2081) that are invariant among GIY-YIG family members. Interestingly, TgMSH-1 contains a 42 amino acid insertion within motif A. Thus, the homology of TgMSH-1 to the plant MSH-1 extends beyond the DNA binding domain typical of MutS homologs.
Disruption of TgMSH-1 confers resistance to the alkylating agent, methylnitrosourea
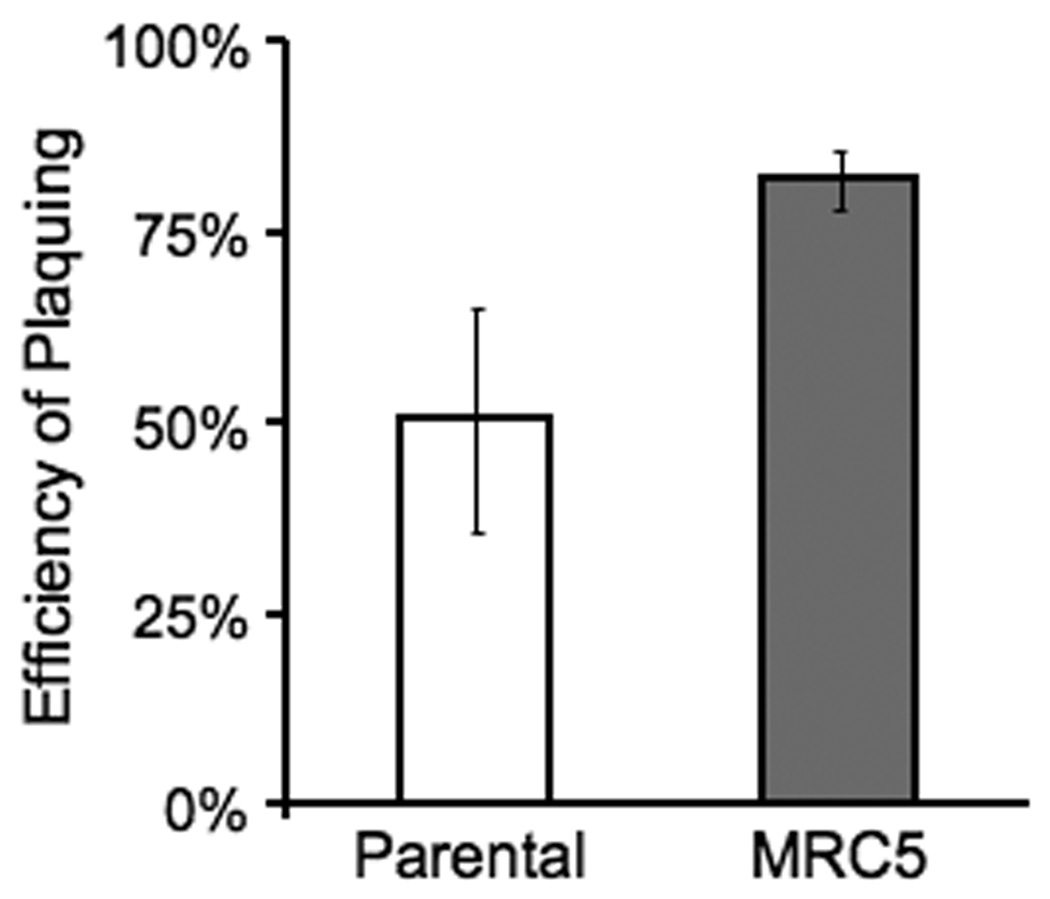
It is well established that mutations in mammalian mismatch repair enzymes confer resistance to certain DNA damaging drugs, including the alkylating agent methylnitrosourea (MNU). Accordingly, we were interested in determining whether mutations in TgMSH-1 confer resistance to this drug. In order to assess the role of TgMSH-1 in MNU induced parasite death, intracellular parental strain and MRC5 parasites were allowed to form plaques in media containing either 1.0 mM MNU or an equivalent concentration of the solvent, and the efficiency of plaquing in the presence of MNU was determined for each strain. As shown in figure 4, the disruption of TgMSH-1 results in an increased ability to form plaques in the presence of 1.0 mM MNU, since 82% of MRC5 parasites form plaques in this drug concentration, while only 51% of the parental strain parasites form plaques under the same conditions.
Figure 4.
Effect of MNU on TgMSH-1 deficient parasites. Intracellular MRC5 and parental strain parasites were allowed to form plaques in the presence of 1.0 mM MNU or an equivalent concentration of solvent control. The efficiency of plaquing for each strain was determined by dividing the number of plaques formed in the presence of MNU by the number of plaques formed in the presence of solvent. Data bars represent the average of three independent experiments, and the error bars denote the standard deviation.
Complementation of MRC5 with functional copy of TgMSH-1 restores a monensin sensitivity
While the simplest explanation for the phenotype observed in MRC5 is that the lack of TgMSH-1 results in multi-drug resistance, other possibilities must be considered. First, the methods used to develop random insertional mutants can at times result in sporadic mutations in genes other than the one disrupted by the insertion and, while not detected by our tests, there is always a possibility that multiple insertions are present within a mutant. Second, given that MRC5 has an insertion within a gene that could be responsible for maintaining the fidelity of DNA replication, it is possible that MRC5’s drug resistant phenotype is a result of secondary mutations that accumulated within the parasite during selection, and not a direct consequence of lacking TgMSH-1. In order to exclude the possibility that secondary mutations are responsible for conferring drug resistance in strains lacking TgMSH-1, we transformed MRC5 with a cosmid, TOXP127, which contains a full copy of the TgMSH-1 gene as well as the T. gondii DHFR-TS pyrimethamine resistant marker. An MRC5 clone that carries this cosmid and expresses TgMSH-1 protein (see below) was tested for resistance to monensin and as anticipated, incorporation of the cosmid almost completely eliminates MRC5’s ability to form plaques in the presence of 0.75 ng/ml monensin (Fig. 5). While less than 1% of MRC5 parasites that incorporated the cosmid were capable of forming plaques in the presence of 0.75 ng/ml monensin, 66% of MRC5 parasites were capable of forming plaques under the same conditions. Reintroduction of TgMSH-1 did not completely complement the monensin-resistant phenotype, however, since the complemented strain shows increased resistance as compared to the parental strain when grown in the presence of 0.5 ng/ml of monensin. This could be due to a decrease in TgMSH-1 expression in the complemented strain. Nevertheless, the restoration of sensitivity to 0.75 ng/ml and 1.0 ng/ml monensin by the addition of a wild-type copy of the gene provides strong evidence that the disruption of TgMSH-1, and not another mutation, is responsible for conferring a monensin-resistant phenotype.
Figure 5.
Complementation of MRC5 parasites with wild type copy of TgMSH-1. Intracellular parental strain, MRC5, and TgMSH-1 complemented parasites (C5+TOXP127) were allowed to form plaques in media containing monensin or an equivalent concentration of solvent. The efficiency of plaquing represents the number of plaques formed in the presence of monensin divided by the number of plaques formed in the presence of the solvent. Data bars represent the average of three independent experiments and error bars denote the standard deviation.
The monensin-resistant phenotype is recapitulated with a directed knockout of TgMSH-1
To further confirm that the disruption of TgMSH-1 is directly responsible for the phenotypes seen, we performed a directed knocked-out of TgMSH-1 in a wild-type strain using homologous recombination. For this purpose we made a construct, pKOMSH1, which consists of the selectable marker HPT, flanked by genomic regions of TgMSH-1 and a copy of the green fluorescent protein (GFP) gene, which can be used as a negative selectable marker (Fig. 6A). Double homologous recombination between pKOMSH1 and the TgMSH-1 locus results in the loss of GFP and the replacement of 1201 bp of TgMSH-1 gDNA with HPT. In total, we established 145 GFP negative, HPT expressing clones and 3 GFP positive, HPT expressing clones from 8 different populations. Using a PCR approach (see Materials and Methods) we determined that only one of these clones, which we have named RHΔmsh-1 has a deletion in TgMSH-1 (Fig. 6B). Interestingly, this clone expresses GFP, which indicates that it either has a second insertion or a single crossover insertion in the TgMSH-1 locus. To unequivocally show that RHΔmsh-1 has the expected deletion, Southern analysis of XbaI digested gDNA from this strain was performed using a 474 bp segment of TgMSH-1 that should be absent from the RHΔmsh-1 strain, as a probe (Fig. 6C). As expected, Southern analysis indicated that parental strain parasites contained the TgMSH-1 DNA segment, while the knockout strain lacked the corresponding region of DNA. As a positive control to confirm the presence of DNA on the membrane, the blot was also probed with a 600 bp sequence of DNA, which corresponds to a genomic segment on chromosome IX (Fig. 6C).
Figure 6.
Establishment and phenotypic analysis of directed TgMSH-1 knockout strain. A. Schematic diagram of the knockout construct, pKOMSH1 and the genomic location of the flanking regions (dark lines). Base 1 of the gDNA corresponds the TgMSH-1 transcription start site, as determined by 5’ RACE, and the dashed line corresponds to the genomic region of TgMSH-1 that is replaced by the selectable marker HPT following double homologous recombination between the knockout construct and TgMSH-1. The grey box represents the region of DNA used for the Southern probe (see Fig. 6C). B. A putative TgMSH-1 knockout clone, RHΔmsh-1 was identified in a PCR based screen. Genomic DNA from the parental strain (RHΔhpt) and from RHΔmsh-1 was used for PCR with primers a and b or primers a and c (primers are shown as arrows in A). C. Southern blot analysis of XbaI digested DNA from the RHΔhpt and RHΔmsh-1 parasite strains. Upper panel: Membrane was probed with a 600 bp region of chromosome IX. Lower panel: Membrane was probed with a 474 bp region of TgMSH-1 (grey box in 6A). D. The effect of monensin on RHΔmsh-1 parasites. Intracellular RHΔhpt and RHΔmsh-1 parasites were allowed to form plaques in the media containing monensin or solvent control. The efficiency of plaquing represents the number of plaques formed in the presence of monensin divided by the number of plaques formed in the presence of the solvent. Data bars represent the average of three independent experiments, and error bars denote the standard deviation.
After confirming that RHΔmsh-1 has a disruption in the TgMSH-1 locus, parasites from this strain were tested for sensitivity to monensin (Fig. 6D). RHΔmsh-1 parasites showed reduced sensitivity to monensin as compared to the parental strain used for the knockout. When grown in the presence of 0.5 ng/ml monensin, 97% of intracellular RHΔmsh-1 parasites were capable of forming plaques, while only 11% of RHΔhpt parasites, which are more sensitive to monensin than the parental strain that was used for the selection (Fig 1 and Fig 6), were capable of forming plaques at the same concentration. In addition, RHΔmsh-1 parasites exhibit an increased efficiency of plaquing when grown in media containing salinomycin or MNU, as compared to the parental strain just as observed with MRC5 (Fig. S3). Given that the directed disruption of TgMSH-1 recapitulates all the phenotypes seen with the original mutant without any selection for drug resistance, the phenotypes observed can be ascribed directly to the lack of TgMSH-1 and not to the accumulation of downstream mutations caused by a lack of DNA damage control.
TgMSH-1 is localized to the mitochondrion of the parasite
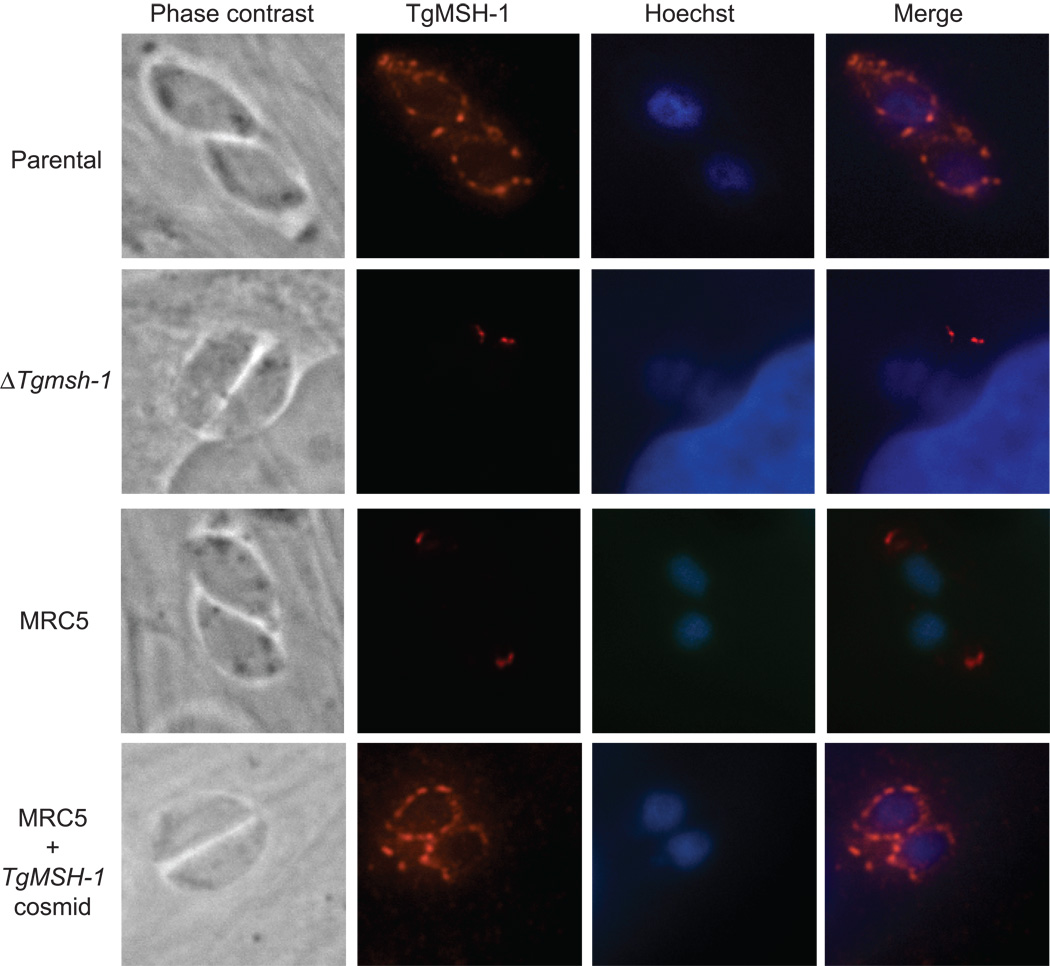
To identify the subcellular localization of TgMSH-1, we made mouse polyclonal antibodies against a bacterially expressed fragment of the protein (amino acids 1752–2095), which were used in an indirect immunofluorescence assay (IFA) to visualize the location of TgMSH-1 within the parasite. The IFA revealed that TgMSH-1 is not located in the nucleus of wild-type parasites, as would be expected for most DNA repair proteins, but rather, the protein appears to form a punctate circular pattern around the nucleus (Fig. 7). As anticipated, this signal is present in wild-type parasites and in MRC5 parasites that have been complemented with TgMSH-1, yet absent in MRC5 and RHΔmsh-1 parasites (Fig. 7), confirming that the pattern seen is specific to the antigen used. The faint fluorescent signal seen at the apical end of all parasite strains is due to background staining as it is also seen at a very low level with pre-immune sera (data not shown).
Figure 7.
Intracellular localization of TgMSH-1. Parental strain, RHΔmsh-1, MRC5, and MRC5+TOXP127 parasites were stained with mouse antiserum generated against amino acids 1752–2095 of TgMSH-1, and the signal was visualized with an alexa fluor-594 conjugated goat anti-mouse antibody. Phase contrast, Hoechst stain, and anti-TgMSH-1 images are shown for each vacuole.
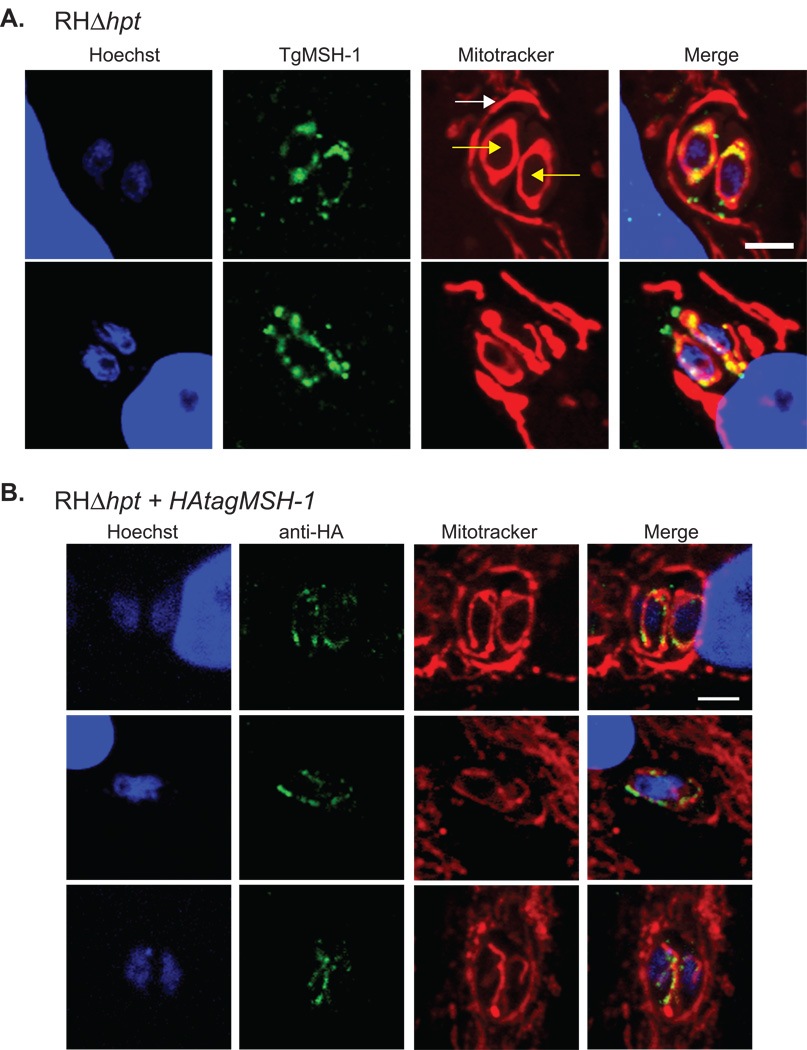
Since the peri-nuclear staining pattern exhibited by TgMSH-1 is somewhat reminiscent of mitochondrial localization (Melo et al., 2000, Pino et al., 2007), and because TgMSH-1 is expected to be located within a DNA-containing organelle, we also performed the IFA using parasites that were labeled with MitoTracker (Fig. 8A). MitoTracker stains the host cell mitochondria, which tightly associate with the parasitophorous vacuole membrane (Sinai & Joiner, 2001), as well as the single mitochondrion of each parasite. Dual labeling of the mitochondria and TgMSH-1 in all parasites analyzed confirmed that the TgMSH-1 signal coincides with MitoTracker labeled parasite mitochondria.
Figure 8.
Mitochondrial localization of TgMSH-1. The mitochondria of intracellular wild-type (RHΔhpt) parasites and of the HFF host cells were stained with 300 nM MitoTracker Red CMX-ROS and the nuclei were stained with Hoechst Stain. A. TgMSH-1 was labeled with serum generated against amino acids 1752–2095 of TgMSH-1 and was visualized with an alexa fluor-488 conjugated goat anti-mouse antibody. Yellow arrows point at the 2 intracellular parasites and the white arrow indicates the host mitochondria surrounding the parasitophorous vacuole, which contains the parasites. Signal from each fluorophore and a merged image is shown. Scale bar, 4 microns. B. HA-tagged TgMSH-1 was visualized in parasites expressing the tagged protein using an anti-HA primary antibody in combination with an alexa fluor-488 conjugated goat anti-rabbit secondary antibody. Scale bar, 4 microns.
Because the punctate staining pattern demonstrated by TgMSH-1 is not completely typical of mitochondrial proteins, and because background staining was observed in the apical end of both wild-type and msh-1 mutant parasites, additional tests were necessary to definitively show that TgMSH-1 is specifically localized to the parasite mitochondrion. In order to verify this localization, we used double homologous recombination to introduce an HA tag into the endogenous TgMSH-1 gene of wild-type parasites, just upstream of the stop codon. As expected, visualization of the HA-tagged protein by immunofluorescence in MitoTracker labeled parasites demonstrates that TgMSH-1-HA forms punctate foci within the parasite mitochondrion (Fig. 8B). Taken together, these dual labeling experiments unequivocally demonstrate that TgMSH-1 is localized to the parasite mitochondrion. This localization of TgMSH-1 is consistent with the mitochondrial localization of the Arabidopsis AtMSH-1 protein (Abdelnoor et al., 2003). However, TgMSH-1 appears to be concentrated within discrete subdomains of the mitochondria. While additional tests would be required to understand the mechanisms underlying the formation of this punctate pattern, it is possible that since TgMSH-1 contains putative DNA binding domains, these foci could represent the sites of the parasite mitochondrial nucleoids.
Given that TgMSH-1 localizes to the parasite mitochondrion, we examined its predicted protein sequence to determine whether it contains a mitochondrial localization signal at its N-terminus. If translation of TgMSH-1 begins at the first in-frame AUG, computer algorithms (TargetP, MitoProt II, MitoPred) do not predict mitochondrial localization (Fig. S4). However, if translation initiates at the second, third, or fourth in-frame AUG, a mitochondrial localization signal is predicted. Therefore, our localization data suggests that translation of TgMSH-1 does not necessarily begin at the first in-frame AUG, but rather could initiate at the second third, or fourth in-frame AUG.
DISCUSSION
Using a forward genetic approach, we have shown that the disruption of TgMSH-1, a protein with homology to the mismatch repair protein MutS, confers multi-drug resistance in the apicomplexan parasite, T. gondii. A connection between the disruption of proteins in the mismatch repair (MMR) pathway and drug resistance has been established in many other cell types but never in a protozoan parasite. In several mammalian cell lines, mutations in MutS Homologs are known to directly confer drug resistance, since, just as we have shown for TgMSH-1, complementation with a wild-type copy of the MSH gene restores drug sensitivity. For example, HEC59 human endometrial adenocarcinoma cells containing mutations in both MSH2 alleles are resistant to the drugs MNNG, 6-thioguanine, cisplatin, carboplatin, and etoposide, and this drug resistant phenotype is complemented by the addition of a chromosome containing a functional MSH2 gene (Aebi et al., 1997). Furthermore, it has been shown that these DNA damaging drugs exert their cytotoxic effects by inducing a G2 cell cycle arrest and apoptosis, and that these effects are dependent on functional MMR (O'Brien & Brown, 2006). Thus, the drug resistant phenotype of MMR deficient cells appears to be a consequence of the cell’s inability to detect the DNA damage and undergo cell cycle arrest and programmed cell death, which would be the normal response to severe genotoxic insult.
Two models have been proposed to explain the mechanism behind mismatch repair-dependent cell cycle arrest and apoptosis, and given that both models are supported by ample evidence, it is likely that both proposed mechanisms contribute to the mismatch repair dependent response to DNA damaging drugs. The first model proposes that mismatch repair enzymes play an indirect role in DNA damage signaling by catalyzing repeated futile cycles of repair (Karran, 2001). In contrast, the “direct signaling model” proposes that in addition to repairing replication errors, MMR enzymes are also responsible for directly signaling cell cycle arrest and apoptosis in response to stress. A direct role for MMR in signaling is supported by the fact that over expression of MSH2 and the mismatch repair enzyme MLH1, results in the induction of apoptosis (Hickman & Samson, 1999, Salsbury et al., 2006). Furthermore, direct signaling by MMR is supported by the analysis of cell lines with mutations in the ATP binding sites of MSH2 and MSH6 (Lin et al., 2004, Yang et al., 2004, Salsbury et al., 2006). Such mutations result in cells which are defective in the repair of mismatched DNA, but which exhibit a normal sensitivity and apoptotic response to DNA damaging drugs. Thus, MSH proteins can be themselves signaling molecules that translate DNA damage to cell death.
At present we do not know the mechanism by which the disruption of TgMSH-1 leads to monensin resistance in T. gondii. However, given the known role of MutS homologs in signaling cell death, it is plausible that monensin is causing a stress within the parasite that induces TgMSH-1 to signal cell death. While we have shown that the disruption of TgMSH-1 confers resistance to MNU, a drug that is known to directly alkylate DNA, it is unlikely that monensin and salinomycin exert their antiparasitic effects by directly damaging DNA, since these drugs are known to be selective ionophores that affect the cellular pH and osmolarity. Nevertheless, monensin may indirectly damage DNA in Toxoplasma, since it reportedly affects the DNA integrity in rat testicular cells (Singh et al., 2006) and can induce apoptosis in the leukemic cell line HL-60 (Zhu & Loh, 1995) and cell cycle arrest and apoptosis in various cancer cell lines (Park et al., 2003, Park et al., 2002). Unfortunately, the role of MMR in monensin-induced apoptotic death has not been analyzed in any of these cell lines.
One striking difference between the MSH-1 dependent drug sensitivity in T. gondii and the MSH dependent drug sensitivities in other systems is the localization of the respective MSH proteins. All previously described MSH proteins that are involved in signaling cell cycle arrest and apoptosis localize to the nucleus, while TgMSH-1 localizes to the parasite mitochondrion. In our experiments, we never detected TgMSH-1 in the parasite nucleus, and its localization was unaffected by monensin treatment or the cell cycle status of the parasite (data not shown). Thus, TgMSH-1 is unlikely to be involved in T. gondii nuclear DNA repair. The localization of TgMSH-1 is consistent with the mitochondrial localization of its close homolog, the MSH-1 protein of Arabidopsis thaliana (Abdelnoor et al., 2003). While the mitochondria of most organisms, including humans, are thought to be devoid of MutS homologs, mitochondrial MutS homologs have been described in several systems including yeast and plants (Abdelnoor et al., 2003, Chi & Kolodner, 1994, Mookerjee et al., 2005), where they appear to play a role in DNA stability (Chi & Kolodner, 1994, Abdelnoor et al., 2003). Whether the T. gondii msh-1 mutants that we have generated accumulate mutations or rearrangements in the mitochondrial DNA is unknown. Our msh-1 mutants grow easily in tissue culture, however they form smaller plaques than their wild-type counterparts, indicating that the TgMSH-1 mutation may retard the growth rate or invasion frequency of the parasite (data not shown). This slow plaquing phenotype is probably not due to the irreversible damage of mitochondrial DNA, however, since complementation of TgMSH-1 mutant parasites with a functional TgMSH-1 gene allows the parasites to form plaques at the same rate as wild-type parasites (data not shown).
The fact that TgMSH-1 most closely resembles mitochondrial MutS Homologs in algae and plants is consistent with the geno-wide similarities between Toxoplasma and plants that have been observed. Indeed, Toxoplasma harbors a relict plastid (McFadden et al., 1996), and like plants, this parasite is sensitive to a number of herbicides including clodinafop (Zuther et al., 1999) and glyphosate (Roberts et al., 1998). In addition, numerous pathways within the parasite have plant-like characteristics. For example, Toxoplasma reportedly synthesizes the plant hormone, abscisic acid, which controls the release of intracellular calcium stores within the parasite in a pathway mirroring that of plants (Nagamune et al., 2008). Toxoplasma also encodes two enolases that contain amino acid insertions that are typically found in plant enolases (Dzierszinski et al., 2001). Previous investigators have hypothesized that these plant-like genes were acquired by the Apicomplexa by horizontal gene transfer from the algal plastid precursor to the nuclear genome (Soldati, 1999). Therefore, TgMSH-1 could be yet another example of a gene whose origins in the apicomplexa trace back to its ancient endosymbiont.
Studies involving the A. thaliana MSH-1 protein have shown the mutation of this gene results in the amplification of a rearranged portion of mitochondrial DNA. Accordingly, previous investigators have suggested that AtMSH-1 is involved in the suppression of replication or recombination of mitochondrial DNA molecules (Abdelnoor et al., 2003). Recombination of the plant mitochondrial genome is a common occurrence, and it has been suggested that mitochondrial genome replication in some plants is recombination-dependent (Backert et al., 1997). While little is known about mitochondrial DNA replication in Toxoplasma, previous studies in Plasmodium have shown that complex recombination products are present in the parasite’s mitochondrial DNA, and that these recombination products appear to be present only in replicating mitochondria (Preiser et al., 1996). Consequently, it has been suggested that the parasites’ mitochondrial DNA is replicated in a recombination dependent manner as it is in plants (Wilson & Williamson, 1997). Interestingly, Plasmodium has a protein with high homology to TgMSH-1 and the plant MSHs.
Like Toxoplasma, the closely related coccidian parasites Eimeria tenella and Neospora caninum, encode orthologs of TgMSH-1 (www.genedb.org, http://www.sanger.ac.uk/cgi-bin/blast/submitblast/n_caninum, www.genedb.org). The observation that all of these parasites are sensitive to monensin (Lindsay et al., 1994, Ricketts & Pfefferkorn, 1993), and that they all encode MSH-1 orthologs opens up the possibility that monensin may act in an MSH-1 dependent pathway in each of these parasites. Sequencing of MSH-1 in monensin-resistant Eimeria isolates could be helpful in determining a possible role for this protein in the high sensitivity to monensin observed in most coccidian parasites.
Interestingly, we were unable to identify a TgMSH-1 ortholog in coccidians of the genus, Cryptosporidium (www.cryptodb.org). This absence of a TgMSH-1 ortholog in Cryptosporidium spp. is intriguing given that unlike other apicomplexan parasites, which possess mitochondria and mitochondrial genomes, Cryptosporidium spp. possess only a mitochondrion-like organelle, which appears to completely lack its own genome (Lindsay et al., 1994, Ricketts & Pfefferkorn, 1993). The observation that MSH-1 appears to be present only in parasites that have mitochondrial DNA also supports the notion that MSH-1 is involved in signaling, replication, or recombination involving the parasite mitochondrial genome.
TgMSH-1 is the first mitochondrial MutS homolog to be linked to any type of drug resistance. Nevertheless, given the important role mitochondria play in stress signaling in higher eukaryotes, it is not surprising that these enzymes might play a dual role in repair and signaling cell death, just like their nuclear homologs do. As is the case in many higher eukaryotes, the mitochondrion is central to the stress signaling response in T. gondii. In its intermediate hosts, which can be any warm blooded animal, T. gondii converts between two stages: the rapidly dividing tachyzoite, which predominates in the acute phase of the infection, and the slower growing bradyzoite, which predominates in the chronic phase (Dubey, 1998). While the exact cues that trigger bradyzoite development in vivo are unknown, conditions that stress the parasite can induce bradyzoite differentiation in vitro (Soete et al., 1994, Narasimhan et al., 2008). Accordingly, treatment of tachyzoites with mitochondrial inhibitors including atovaquone, rotenone, antimycin A, myxothiazol, and CCCP have all been shown to induce the expression of bradyzoite-specific genes, while treatment of parasites with drugs such as clindamycin and sulfadiazine, which do not target the parasite mitochondrion, failed to induce bradyzoite gene expression (Tomavo & Boothroyd, 1995). The particular signaling mechanisms by which disruption of mitochondrial function is translated to conversion to the bradyzoite stage is not known. Since the parental strain used to generate the msh-1 mutants does not easily convert to the bradyzoite stage in vitro, we have been unable to unequivocally show that these mutants are defective in bradyzoite development, which would implicate TgMSH-1 in stage conversion. Nevertheless, we have been able to observe that treating wild-type parasites of the Prugniaud strain with monensin induces the selective expression of a GFP marker regulated by a bradyzoite specific promoter (data not shown).
The results reported here suggest that monensin is directly or indirectly affecting the mitochondria. Monensin could act directly on mitochondria by catalyzing the exchange of H+ for Na+ or K+ across the mitochondrial membrane, or alternatively, it could act indirectly on mitochondria by affecting another cellular target, which, in turn causes mitochondrial stress. Indeed, a role for monensin in affecting the mitochondrion is consistent with previous studies in other systems. For example, condensed and vacuolized muscle mitochondria were frequently observed in a previous study utilizing histological sections taken from monensin-poisoned animals, while the morphology of other organelles appeared normal (Mollenhauer et al., 1990). In another study, a monensin concentration of 0.1 µM (approximately 70x the maximum concentration used in our study) affected the morphology of mitochondria in cultured L929 fibroblasts (Souza et al., 2005). It is important to note that the effects of monensin on mitochondria have only been described in vertebrate cells, and therefore, this phenomenon does not necessarily occur in apicomplexan parasites. Nonetheless, our results suggesting that monensin is affecting mitochondrial function are consistent with the reported effects of this drug.
Our data and previous studies of MutS homologs are compatible with the hypothesis that monensin induces a mitochondrial stress that is detected by TgMSH-1, and that parasite death can result either from TgMSH-1 directing the parasite to undergo apoptosis, or from aberrant processing of the stress by TgMSH-1. Although an apoptotic pathway has not been clearly established in T. gondii, previous investigators have reported that T. gondii parasites undergo an apoptotic-like cell death in response to the nitric oxide donor, sodium nitroprusside, and cell death resembling apoptosis has also been described in Plasmodium (Hurd et al., 2006, Peng et al., 2003). According to this idea, the RHΔmsh-1 parasites still experience monensin-induced stress, but since the parasites lack functional TgMSH-1, they do not respond to the drug-induced stress, and consequently, the parasites survive. Consistent with this hypothesis, we have observed that both TgMSH-1 mutants and wild-type strains change their morphology after being treated with high concentrations of monensin, but only the wild-type strains die from this drug treatment (Fig. 1B).
The results presented here strongly suggest that monensin directly or indirectly affects the parasite mitochondrion, and that at least one parasite protein, TgMSH-1, contributes to monensin-induced parasite death. The role of nuclear MSH proteins in drug sensitivity has been well established. Our work now suggests that mitochondrial MSH enzymes might also play a similar role in some organisms. The elucidation of the nature of monensin-induced stress and the other factors involved in TgMSH-1 signaling will provide further insights into monensin’s mode of action, and may prove useful in determining why coccidian parasites are so susceptible to the drug.
EXPERIMENTAL PROCEDURES
Parasite and host cell maintenance
For the experiments described here we used parasites of the RH strain lacking a functional hpt gene (Donald et al., 1996), RHΔhpt, as well as a modified line of that strain, RHΔhpt+GFP+βGal, that expresses the green fluorescent protein (GFP) and β-Galactosidase (βGal), (Fruth & Arrizabalaga, 2007). Parasites were maintained by passage through human foreskin fibroblasts (HFFs) in a humidified incubator at 37°C with 5% CO2. The normal culture medium was Dulbecco’s Modified Eagle Medium (DMEM, Gibco), supplemented with 10% fetal bovine serum (Atlanta Biologicals), 2 mM L-glutamine, 100 units penicillin per ml and 100 µg streptomycin per ml. A 10 µg/ml monensin A sodium salt (Sigma) stock solution and a 40 µg/ml salinomycin (Sigma) stock solution were prepared using absolute ethanol as a solvent. A 1.5 M methylnitrosourea (MNU) solution was prepared by dissolving 77% MNU (Sigma) in DMSO.
Insertional mutagenesis and selection for monensin resistance
To generate insertional mutants, two populations of RHΔhpt+GFP+βGal parasites were electroporated according to established methods (Soldati & Boothroyd, 1993) with 30 µg of NotI linearized pHANA vector (Arrizabalaga et al., 2004), which contains a functional copy of the HPT gene for selection in the parasite and an ampicillin resistance gene, which aids in the rescue of the mutagenic plasmid and in the identification of the insertion site. Twenty-four hours following electroporation, parasites were treated with culture medium containing 50 µg/ml mycophenolic acid and 50 µg/ml xanthine in order to select for parasites that had incorporated pHANA into the genome. Monensin-resistant mutants were obtained by exposing intracellular parasites from the two mutant populations to culture medium containing 1.0 ng/ml monensin, which we determined kills 100% of parental parasites. Parasites were treated with monensin for 4–6 days, and were then allowed to recover in the absence of the drug. The surviving parasites were mechanically lysed from the fibroblast monolayer with syringe and a 27.5 G needle, then transferred to a fresh HFF monolayer, and the monensin selection was repeated 24 hours later. After a total of three rounds of selection, individual parasite clones were established from each population by limiting dilution. To screen for clones with the greatest monensin-resistant phenotype, eleven clones from population 1, twelve clones from population 2, and the parental strain were passed in duplicate onto HFFs grown in 24 well tissue culture plates containing culture media. The following day, the media in the tissue culture plates was changed to either normal culture medium containing 0.75 ng/ml monensin or to normal culture medium with an equivalent amount of ethanol as a solvent control, and parasites were allowed to form plaques. The wells were observed microscopically 4–8 days later in order to determine which clones had the desired phenotype of growing in the presence of monensin.
Quantification of monensin, salinomycin, and MNU resistance
Monensin-resistant phenotypes were quantified by assessing each strain’s ability to form plaques in the presence of monensin. Four hundred parasites of each strain were added to each of six wells in 12 well tissue culture plates containing HFF monolayers grown in normal culture medium. After 24 hrs, the media was changed to normal culture medium with monensin in three of the wells or normal culture medium containing ethanol in the remaining three wells. This assay was conducted with four concentrations of monensin (0.1 ng/ml, 0.5 ng/ml, 0.75 ng/ml and 1.0 ng/ml) and equivalent amounts of ethanol (which did not exceed 0.01%). Parasites were incubated under these conditions until plaques formed, after which point cultures were fixed with 100% methanol, stained with crystal violet, and the plaques were counted using a stereomicroscope. The percent survival (i.e. efficiency of plaquing) was determined by dividing the number of plaques formed in the presence of monensin by the number of plaques formed in media containing an equivalent concentration of ethanol, and was expressed as a percentage. The salinomycin and MNU resistant phenotypes were quantified as described above using normal media containing either 1.0 ng/ml, 5.0 ng/ml, 7.5 ng/ml or 10.0 ng/ml of salinomycin, with controls containing equivalent amounts of ethanol, or media with 1.0 mM MNU, with controls containing an equivalent volume of DMSO.
Plasmid rescue of MRC5
Genomic DNA was isolated from parasites according to the TELT DNA isolation protocol (Medina-Acosta & Cross, 1993). 1 µg genomic DNA was digested with AgeI, EcoRI or BamHI in order to obtain the flanking DNA from both ends of the insertion. The restriction digestions were heat inactivated and 0.5 µg digested DNA was ligated at a DNA concentration of 1.25 ng/µl to favor intramolecular ligation. Ligated DNA was phenol: chloroform extracted and subsequently transformed into chemically competent DH5α E. coli. Plasmids were isolated from ampicillin-resistant bacteria and were analyzed by restriction digestion and then sequenced to confirm the presence and identity of flanking genomic sequences. Plasmids from the EcoRI and BamHI digestions were sequenced with the M13 Forward Primer (Invitrogen), and plasmids from the AgeI digestion were sequenced with a primer that recognizes a site within pHANA that is near the presumed junction site, (5’-CGCTCGGTCGTTCGGCTGCG-3).
Southern Blot
For Southern analysis, 5 µg MRC5 and RHΔhpt+GFP+βgal gDNA was digested overnight with EcoRI. The resulting DNA fragments were separated by electrophoresis and the DNA was transferred to a neutral nylon membrane (Magna, Osmonics, Inc.) using established methods (Sambrook & Russell, 2001). After cross-linking the DNA to the membrane using a UV Stratalinker 1800 (Stratagene), the DNA was hybridized to a 635 bp fragment of HPT cDNA using standard methodology. This probe was obtained by PCR using pHANA as a template and the primer set 5’-gccgtattgagcccatgt-3’ and 5’-gcgagcggcgtcgctcagga-3’ and labeled with [α32P-dCTP] using the RadPrime DNA Labeling System (Invitrogen). After two 20 minute washes with 2x SSC wash solution (2x SSC, 0.1% SDS, 0.1% Na2H2P2O7), followed by two 20 minute washes with 0.2x SSC wash lution (02x SSC, 0.1% SDS, 0.1% Na2H2P2O7) the membrane was used to expose Kodak Maximum Sensitivity film.
Southern analysis of RHΔmsh-1 was conducted as described above with some modifications. 10 µg RHΔmsh-1 and RHΔhpt gDNA were digested with XbaI. Following digestion, 5 µg DNA from each reaction was loaded in duplicate into lanes on opposite sides of a 1% agarose gel and the DNA fragments were separated by electrophoresis and transferred to a membrane. One half of the membrane was probed with a 474 bp PCR fragment of TgMSH-1 gDNA generated using the primer set 5’-gtgtctccttgatgcgctg-3’ and 5’-cagggagcgtggaagcgtc-3’, (Fig. 4A). As a control, the other half of the membrane was probed with a 600 bp PCR fragment (primers 5’-tgttggctcgtgtctctccg-3’ and 5’- tgttggctcgtgtctctccg-3’), which corresponds to a predicted sodium hydrogen exchanger gene, which is present on chromosome IX (Francia and Arrizabalaga, unpublished results).
Cloning of TgMSH-1
The TgMSH-1 gene from both Prugniaud and RH strains was cloned and sequenced using a series of primer pairs designed against predicted exon sequences to expand overlapping fragments of the cDNA. The ends of the cDNA were identified using the GeneRacer Kit (Invitrogen) with RNA isolated from Prugniaud (Zenner et al., 1993). The 5’ end was reverse transcribed with the TgMSH-1 specific primer, 5’-gggtgttcagatcgggtc-3’ and the resulting cDNA was used in nested PCR reactions using the primers provided with the kit along with the gene specific primers 5’-aatcggctctgagtctggagtgt-3’ (for the primary reaction) and 5’-ggcatgtgggctgggagataca-3’ (for the nested reaction).
To identify the 3’ end of the transcript, RNA was reverse transcribed using the GeneRacer Oligo dT primer and the resulting cDNA was used as a template in nested PCR reactions using the GeneRacer 3’ primers in combination with 5’-cgcactcgtcgtcctcgcaa-3’ (for the primary reaction) and 5’-agccacgacggtacctcggtacg-3’ (for the nested reaction).
Generation of TgMSH-1 knockout strain
A knockout construct was generated using the pminiGFP.ht vector (Arrizabalaga et al., 2004), which contains the T. gondii HPT gene flanked two multiple cloning sites. To design the knock-out construct, first a PCR fragment from bases 52–4582 (Flank 1, Fig. 6A) of TgMSH-1 gDNA was digested with NotI and XbaI and cloned into pminiGFP.ht digested with the same restriction enzymes. The resulting plasmid was digested with KpnI and ligated to a PCR fragment from genomic bases 5734–11718 (Flank 2, Fig. 6A) also cut with KpnI. Since this cloning step can produce vectors with Flank 2 in either direction, we used a diagnostic digestion with HindIII to identify those with Flank 2 in the correct orientation. The primers used to expand the flanking regions were 5’-aaggaaaaaagcggccgcgttctgactctcctcgcacc-3’ and 5’-cgtccgtcttcagcgagtcc-3’ for Flank 1 and 5’-cgctactgcctccgtcgg-3’ and 5’-gagccatgtacgacgacgc-3’ for Flank 2.
The resulting vector, pKOMSH1, was linearized with XmaI and 30 µg of DNA were introduced into RHΔhpt strain parasites by standard methods (Soldati & Boothroyd, 1993). Eight independent transformations were performed and parasites were then grown in T25 flasks containing HFFs in normal culture medium for 24 hours at which point the media was changed to culture medium containing 50 µg/ml MPA and 50 µg/ml xanthine. After parasites stably expressing HPT were established, the parasites were cloned by limiting dilution. 148 clones were tested by PCR for the desired disruption using genomic DNA isolated with the DNeasy Blood and Tissue Kit (QIAGEN). The PCR reaction included a forward primer (5’-gcaggttacgcttgcggaga-3’) that anneals to a sequence in TgMSH-1 and a reverse primer (5’-cgtccgtcttcagcgagtcc-3’) that anneals to a sequence that should be absent in the knockout strain (Fig. 6A). Using this primer set, an 810 bp band is amplified from DNA of wild-type parasites, but not from parasites with a deletion in TgMSH-1. As a control, PCR was also performed using the forward primer described above and the reverse primer, 5’-ggcggcacttgacagttg-3’, which yield a 746 bp band with DNA from both parental and knock-out strains.
Complementation of resistance phenotype
An end-sequenced cosmid, TOXP127, which carries a 39.7 kb region of chromosome XII, including the entire TgMSH-1 coding sequence was identified on the Ancillary GBrowse site for T. gondii (http://roos-compbio2.bio.upenn.edu/toxo/cgi-bin/gbrowse/gbtoxo_amit/) and was obtained from David Sibley, Washington University School of Medicine. MRC5 parasites were electroporated with 100 µg PacI linearized cosmid using established methods (Soldati & Boothroyd, 1993). Twenty-four hours following electroporation, the media was changed to contain 1 µM pyrimethamine and 2.5% dialyzed FBS (rather than regular FBS) in order to select for parasites carrying the cosmid. Pyrimethamine resistant parasites were cloned by limiting dilution and a clone expressing TgMSH-1 was identified in an immunofluorescence assay using an α-TgMSH-1 antibody (see below).
Generation of TgMSH-1 antibodies
A fragment encoding amino acids 1752 through 2095 of TgMSH-1 was generated from a cloned cDNA template using Phusion High-Fidelity PCR Master Mix (Finnzymes) and the primer set, 5’-caccgggtctctcggtgatgatagg-3’ and 5’-acggctgtgagttcgcaaacg-3’. This PCR fragment was cloned into the pBAD202/D-TOPO vector (Invitrogen) in frame with vector sequences encoding an NH3-terminal his-patch thioredoxin peptide and a COOH-terminal 6x histidine tag. The resulting plasmid was transformed into TOP10 chemically competent E. coli and protein expression was induced according to the pBAD202/D-TOPO manual with 0.02% arabinose. Protein was purified under denaturing conditions with Ni-NTA agarose (Qiagen) according to manufacturer’s instructions. A female BALB/C mouse (Simonsen Laboratories) was injected with the recombinant protein fragment and 100 µL MPL+TDM adjuvant (Sigma), and six additional boosts were performed at intervals of approximately 3 weeks.
TgMSH-1 Epitope Tagging
A construct for tagging the endogenous TgMSH-1 locus was created from the parental pTKO vector (Zeiner, Reese and Boothroyd, personal communication) (Kafsack et al., 2009). Briefly, a 3.02 kb region of gDNA corresponding to sequences that immediately precede the TgMSH-1 stop codon followed by a sequence encoding a hemagglutinin (HA) tag (Fragment 1) and a 2.34 kb region of gDNA corresponding to sequences directly downstream of the TgMSH-1 stop codon (Fragment 2) were cloned into separate multiple cloning sites of the pTKO vector which flank the T. gondii HPT gene and the Gra2 3’ UTR. First, fragment 2 was generated by PCR using the primer set, 5’-ctagctagcggtaacgcgaaattcagtctgg-3’ and 5’-gggcccccctttgttccacctgtgtt-3’, which introduce NheI and ApaI sites into the amplified DNA. The resulting PCR fragment was digested with NheI and ApaI, and was cloned into the pTKO vector digested with the same enzymes. Next, Fragment 1 was generated using the forward primer, 5’-aaggaaaaaagcggccgccgggtgatgacgcttgtcgag-3’, which contains a NotI site, and the reverse primer, 5’-ggaattctcacgcgtagtccgggacgtcgtacgggtagtctctcccatcctgtgactcgg-3’, which generates an HA tag, stop codon, and EcoRI site in the amplified DNA. The resulting PCR fragment was digested with NotI and EcoRI, and was cloned into the pTKO+Fragment 2 construct, upstream of the Gra2 3’ UTR. The resulting vector, pTAGMSH-1, was linearized with ApaI, and 30 µg of the linearized DNA was transfected into RHΔhpt parasites. Following selection for the presence of the vector with mycophenolic acid, tagged TgMSH-1 was visualized by immunofluorescence in parasites that had incorporated the pTAGMSH-1 vector into the genome by double homologous recombination.
Immunofluorescence assays
Immunofluorescence assays (IFA) were performed as described previously (Arrizabalaga et al., 2004) using mouse anti-TgMSH-1 or rabbit anti-HA antibodies (Rockland Immunochemicals) in combination with Alexa fluor-594 or Alexa fluor-488 conjugated goat anti-mouse or goat anti-rabbit secondary antibodies (Molecular Probes). Slides were viewed with a 100x objective lens on a Nikon Eclipse E1000 microscope, and images were captured using a Hamamatsu C4742-95 camera. Colocalization images were generated using a 60x objective lens with an Olympus FluoView FV1000 confocal laser-scanning microscope. Mitochondria were labeled with MitoTracker Red CMXRos (Molecular Probes) at a concentration of 300 nM according to the manufacturer’s protocol.
ACKNOWLEDGEMENTS
This work was supported by an NIH grant from the NCRR Center of Biomedical Research Centers P20 RR15587 (G.A.) and the NIAID K22 program AI061293-01 (G.A.). E.G. was supported in part by the Idaho IDEA Network of Biomedical Research Excellence grant from the NIH/NCRR (P20 RR016454). We would like to thank Dr. Kurt Gustin, Dr. Peter Bradley and Dr. Mark Lavine for their careful reading of the manuscript and their scientific input into this work. We are also grateful to Dr. Gus Zeiner and Dr. John Boothroyd for kindly providing the HA tagging vector.
ABBREVIATIONS
- HFF
human foreskin fibroblasts
- HPT
hypoxanthine-xanthine-guanine-phosphoribosyl transferase
- MSH
MutS Homolog
- MMR
mismatch repair system
- MNU
methylnitrosourea
REFERENCES
- Abdelnoor RV, Christensen AC, Mohammed S, Munoz-Castillo B, Moriyama H, Mackenzie SA. Mitochondrial genome dynamics in plants and animals: convergent gene fusions of a MutS homologue. J Mol Evol. 2006;63:165–173. doi: 10.1007/s00239-005-0226-9. [DOI] [PubMed] [Google Scholar]
- Abdelnoor RV, Yule R, Elo A, Christensen AC, Meyer-Gauen G, Mackenzie SA. Substoichiometric shifting in the plant mitochondrial genome is influenced by a gene homologous to MutS. Proc Natl Acad Sci U S A. 2003;100:5968–5973. doi: 10.1073/pnas.1037651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebi S, Fink D, Gordon R, Kim HK, Zheng H, Fink JL, Howell SB. Resistance to cytotoxic drugs in DNA mismatch repair-deficient cells. Clin Cancer Res. 1997;3:1763–1767. [PubMed] [Google Scholar]
- Agtarap A, Chamberlin JW, Pinkerton M, Steinrauf L. The structure of monensic acid, a new biologically active compound. J Am Chem Soc. 1967;89:5737–5739. doi: 10.1021/ja00998a062. [DOI] [PubMed] [Google Scholar]
- Arrizabalaga G, Ruiz F, Moreno S, Boothroyd JC. Ionophore-resistant mutant of Toxoplasma gondii reveals involvement of a sodium/hydrogen exchanger in calcium regulation. J Cell Biol. 2004;165:653–662. doi: 10.1083/jcb.200309097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustine PC, Smith CK, 2nd, Danforth HD, Ruff MD. Effect of ionophorous anticoccidials on invasion and development of Eimeria: comparison of sensitive and resistant isolates and correlation with drug uptake. Poult Sci. 1987;66:960–965. doi: 10.3382/ps.0660960. [DOI] [PubMed] [Google Scholar]
- Backert S, Nielsen BL, Borner T. The mystery of the rings: structure and replication of mitochondrial genomes from higher plants. Trends in Plant Science. 1997;2:477–483. [Google Scholar]
- Buxton D, Blewett DA, Trees AJ, McColgan C, Finlayson J. Further studies in the use of monensin in the control of experimental ovine toxoplasmosis. J Comp Pathol. 1988;98:225–236. doi: 10.1016/0021-9975(88)90021-7. [DOI] [PubMed] [Google Scholar]
- Buxton D, Donald KM, Finlayson J. Monensin and the control of experimental ovine toxoplasmosis: a systemic effect. Vet Rec. 1987;120:618–619. doi: 10.1136/vr.120.26.618. [DOI] [PubMed] [Google Scholar]
- Chapman HD. Use of anticoccidial drugs in broiler chickens in the USA: analysis for the years 1995 to 1999. Poult Sci. 2001;80:572–580. doi: 10.1093/ps/80.5.572. [DOI] [PubMed] [Google Scholar]
- Chi NW, Kolodner RD. Purification and characterization of MSH1, a yeast mitochondrial protein that binds to DNA mismatches. J Biol Chem. 1994;269:29984–29992. [PubMed] [Google Scholar]
- Couzinet S, Dubremetz JF, Buzoni-Gatel D, Jeminet G, Prensier G. In vitro activity of the polyether ionophorous antibiotic monensin against the cyst form of Toxoplasma gondii. Parasitology. 2000;121(Pt 4):359–365. doi: 10.1017/s0031182099006605. [DOI] [PubMed] [Google Scholar]
- Donald RG, Carter D, Ullman B, Roos DS. Insertional tagging, cloning, and expression of the Toxoplasma gondii hypoxanthine-xanthine-guanine phosphoribosyltransferase gene. Use as a selectable marker for stable transformation. J Biol Chem. 1996;271:14010–14019. doi: 10.1074/jbc.271.24.14010. [DOI] [PubMed] [Google Scholar]
- Drotschmann K, Topping RP, Clodfelter JE, Salsbury FR. Mutations in the nucleotide-binding domain of MutS homologs uncouple cell death from cell survival. DNA repair. 2004;3:729–742. doi: 10.1016/j.dnarep.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Dubey JP. Advances in the life cycle of Toxoplasma gondii. Int J Parasitol. 1998;28:1019–1024. doi: 10.1016/s0020-7519(98)00023-x. [DOI] [PubMed] [Google Scholar]
- Dzierszinski F, Mortuaire M, Dendouga N, Popescu O, Tomavo S. Differential expression of two plant-like enolases with distinct enzymatic and antigenic properties during stage conversion of the protozoan parasite Toxoplasma gondii. J Mol Biol. 2001;309:1017–1027. doi: 10.1006/jmbi.2001.4730. [DOI] [PubMed] [Google Scholar]
- Fruth IA, Arrizabalaga G. Toxoplasma gondii: induction of egress by the potassium ionophore nigericin. Int J Parasitol. 2007;37:1559–1567. doi: 10.1016/j.ijpara.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberkorn A. Chemotherapy of human and animal coccidioses: state and perspectives. Parasitol Res. 1996;82:193–199. doi: 10.1007/s004360050094. [DOI] [PubMed] [Google Scholar]
- Haney ME, Jr, Hoehn MM. Monensin, a new biologically active compound. I. Discovery and isolation. Antimicrob Agents Chemother (Bethesda) 1967;7:349–352. doi: 10.1128/AAC.7.3.349. [DOI] [PubMed] [Google Scholar]
- Hickman MJ, Samson LD. Role of DNA mismatch repair and p53 in signaling induction of apoptosis by alkylating agents. Proc Natl Acad Sci U S A. 1999;96:10764–10769. doi: 10.1073/pnas.96.19.10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd H, Grant KM, Arambage SC. Apoptosis-like death as a feature of malaria infection in mosquitoes. Parasitology. 2006;132(Suppl):S33–S47. doi: 10.1017/S0031182006000849. [DOI] [PubMed] [Google Scholar]
- Hyde JE. Mechanisms of resistance of Plasmodium falciparum to antimalarial drugs. Microbes Infect. 2002;4:165–174. doi: 10.1016/s1286-4579(01)01524-6. [DOI] [PubMed] [Google Scholar]
- Jeffers TK. Eimeria acervulina and E. maxima: incidence and anticoccidial drug resistance of isolants in major broiler-producing areas. Avian Dis. 1974;18:331–342. [PubMed] [Google Scholar]
- Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- Kafsack BF, Pena JD, Coppens I, Ravindran S, Boothroyd JC, Carruthers VB. Rapid membrane disruption by a perforin-like protein facilitates parasite exit from host cells. Science. 2009;323:530–533. doi: 10.1126/science.1165740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karran P. Mechanisms of tolerance to DNA damaging therapeutic drugs. Carcinogenesis. 2001;22:1931–1937. doi: 10.1093/carcin/22.12.1931. [DOI] [PubMed] [Google Scholar]
- Kim K, Weiss LM. Toxoplasma gondii: the model apicomplexan. Int J Parasitol. 2004;34:423–432. doi: 10.1016/j.ijpara.2003.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski JC, Belfort M, Stapleton MA, Holpert M, Dansereau JT, Pietrokovski S, Baxter SM, Derbyshire V. Configuration of the catalytic GIY-YIG domain of intron endonuclease I-TevI: coincidence of computational and molecular findings. Nucleic Acids Res. 1999;27:2115–2125. doi: 10.1093/nar/27.10.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de Wind N, Sixma TK. The crystal structure of DNA mismatch repair protein MutS binding to a G × T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- Lin DP, Wang Y, Scherer SJ, Clark AB, Yang K, Avdievich E, Jin B, Werling U, Parris T, Kurihara N, Umar A, Kucherlapati R, Lipkin M, Kunkel TA, Edelmann W. An Msh2 point mutation uncouples DNA mismatch repair and apoptosis. Cancer Res. 2004;64:517–522. doi: 10.1158/0008-5472.can-03-2957. [DOI] [PubMed] [Google Scholar]
- Lindsay DS, Rippey NS, Cole RA, Parsons LC, Dubey JP, Tidwell RR, Blagburn BL. Examination of the activities of 43 chemotherapeutic agents against Neospora caninum tachyzoites in cultured cells. Am J Vet Res. 1994;55:976–981. [PubMed] [Google Scholar]
- McFadden GI, Reith ME, Munholland J, Lang-Unnasch N. Plastid in human parasites. Nature. 1996;381:482. doi: 10.1038/381482a0. [DOI] [PubMed] [Google Scholar]
- Medina-Acosta E, Cross GA. Rapid isolation of DNA from trypanosomatid protozoa using a simple ‘mini-prep’ procedure. Mol Biochem Parasitol. 1993;59:327–329. doi: 10.1016/0166-6851(93)90231-l. [DOI] [PubMed] [Google Scholar]
- Melo EJ, Attias M, De Souza W. The single mitochondrion of tachyzoites of Toxoplasma gondii. J Struct Biol. 2000;130:27–33. doi: 10.1006/jsbi.2000.4228. [DOI] [PubMed] [Google Scholar]
- Mollenhauer HH, Morre DJ, Rowe LD. Alteration of intracellular traffic by monensin; mechanism, specificity and relationship to toxicity. Biochim Biophys Acta. 1990;1031:225–246. doi: 10.1016/0304-4157(90)90008-Z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mookerjee SA, Lyon HD, Sia EA. Analysis of the functional domains of the mismatch repair homologue Msh1p and its role in mitochondrial genome maintenance. Curr Genet. 2005;47:84–99. doi: 10.1007/s00294-004-0537-1. [DOI] [PubMed] [Google Scholar]
- Nagamune K, Hicks LM, Fux B, Brossier F, Chini EN, Sibley LD. Abscisic acid controls calcium-dependent egress and development in Toxoplasma gondii. Nature. 2008;451:207–210. doi: 10.1038/nature06478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan J, Joyce BR, Naguleswaran A, Smith AT, Livingston MR, Dixon SE, Coppens I, Wek RC, Sullivan WJ., Jr Translation regulation by eukaryotic initiation factor-2 kinases in the development of latent cysts in Toxoplasma gondii. J Biol Chem. 2008;283:16591–16601. doi: 10.1074/jbc.M800681200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien V, Brown R. Signalling cell cycle arrest and cell death through the MMR System. Carcinogenesis. 2006;27:682–692. doi: 10.1093/carcin/bgi298. [DOI] [PubMed] [Google Scholar]
- Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- Park WH, Kim ES, Kim BK, Lee YY. Monensin-mediated growth inhibition in NCI-H929 myeloma cells via cell cycle arrest and apoptosis. Int J Oncol. 2003;23:197–204. [PubMed] [Google Scholar]
- Park WH, Lee MS, Park K, Kim ES, Kim BK, Lee YY. Monensin-mediated growth inhibition in acute myelogenous leukemia cells via cell cycle arrest and apoptosis. Int J Cancer. 2002;101:235–242. doi: 10.1002/ijc.10592. [DOI] [PubMed] [Google Scholar]
- Peng BW, Lin J, Lin JY, Jiang MS, Zhang T. Exogenous nitric oxide induces apoptosis in Toxoplasma gondii tachyzoites via a calcium signal transduction pathway. Parasitology. 2003;126:541–550. [PubMed] [Google Scholar]
- Pino P, Foth BJ, Kwok LY, Sheiner L, Schepers R, Soldati T, Soldati-Favre D. Dual targeting of antioxidant and metabolic enzymes to the mitochondrion and the apicoplast of Toxoplasma gondii. PLoS Pathog. 2007;3:e115. doi: 10.1371/journal.ppat.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiser PR, Wilson RJ, Moore PW, McCready S, Hajibagheri MA, Blight KJ, Strath M, Williamson DH. Recombination associated with replication of malarial mitochondrial DNA. Embo J. 1996;15:684–693. [PMC free article] [PubMed] [Google Scholar]
- Pressman BC. Biological applications of ionophores. Annu Rev Biochem. 1976;45:501–530. doi: 10.1146/annurev.bi.45.070176.002441. [DOI] [PubMed] [Google Scholar]
- Ricketts AP, Pfefferkorn ER. Toxoplasma gondii: susceptibility and development of resistance to anticoccidial drugs in vitro. Antimicrob Agents Chemother. 1993;37:2358–2363. doi: 10.1128/aac.37.11.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts F, Roberts CW, Johnson JJ, Kyle DE, Krell T, Coggins JR, Coombs GH, Milhous WK, Tzipori S, Ferguson DJ, Chakrabarti D, McLeod R. Evidence for the shikimate pathway in apicomplexan parasites. Nature. 1998;393:801–805. doi: 10.1038/31723. [DOI] [PubMed] [Google Scholar]
- Salsbury FR, Jr, Clodfelter JE, Gentry MB, Hollis T, Scarpinato KD. The molecular mechanism of DNA damage recognition by MutS homologs and its consequences for cell death response. Nucleic Acids Res. 2006;34:2173–2185. doi: 10.1093/nar/gkl238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning : a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2001. p. 3 v. [Google Scholar]
- Shumard RF, Callender ME. Monensin, a new biologically active compound. VI. Anticoccidial activity. Antimicrob Agents Chemother (Bethesda) 1967;7:369–377. [PubMed] [Google Scholar]
- Sinai AP, Joiner KA. The Toxoplasma gondii protein ROP2 mediates host organelle association with the parasitophorous vacuole membrane. The Journal of cell biology. 2001;154:95–108. doi: 10.1083/jcb.200101073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Kalla NR, Sanyal SN. Effect of monensin on the enzymes of oxidative stress, thiamine pyrophosphatase and DNA integrity in rat testicular cells in vitro. Exp Toxicol Pathol. 2006;58:203–208. doi: 10.1016/j.etp.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Smith CK, 2nd, Galloway RB. Influence of monensin on cation influx and glycolysis of Eimeria tenella sporozoites in vitro. J Parasitol. 1983;69:666–670. [PubMed] [Google Scholar]
- Smith CK, 2nd, Strout RG. Eimeria tenella: effect of narasin, a polyether antibiotic on the ultrastructure of intracellular sporozoites. Exp Parasitol. 1980;50:426–436. doi: 10.1016/0014-4894(80)90045-4. [DOI] [PubMed] [Google Scholar]
- Soete M, Camus D, Dubremetz JF. Experimental induction of bradyzoite-specific antigen expression and cyst formation by the RH strain of Toxoplasma gondii in vitro. Exp Parasitol. 1994;78:361–370. doi: 10.1006/expr.1994.1039. [DOI] [PubMed] [Google Scholar]
- Soldati D. The apicoplast as a potential therapeutic target in and other apicomplexan parasites. Parasitol Today. 1999;15:5–7. doi: 10.1016/s0169-4758(98)01363-5. [DOI] [PubMed] [Google Scholar]
- Soldati D, Boothroyd JC. Transient transfection and expression in the obligate intracellular parasite Toxoplasma gondii. Science. 1993;260:349–352. doi: 10.1126/science.8469986. [DOI] [PubMed] [Google Scholar]
- Souza AC, Machado FS, Celes MR, Faria G, Rocha LB, Silva JS, Rossi MA. Mitochondrial damage as an early event of monensin-induced cell injury in cultured fibroblasts L929. J Vet Med A Physiol Pathol Clin Med. 2005;52:230–237. doi: 10.1111/j.1439-0442.2005.00728.x. [DOI] [PubMed] [Google Scholar]
- Stephen B, Rommel M, Daugschies A, Haberkorn A. Studies of resistance to anticoccidials in Eimeria field isolates and pure Eimeria strains. Vet Parasitol. 1997;69:19–29. doi: 10.1016/s0304-4017(96)01096-5. [DOI] [PubMed] [Google Scholar]
- Tomavo S, Boothroyd JC. Interconnection between organellar functions, development and drug resistance in the protozoan parasite, Toxoplasma gondii. Int J Parasitol. 1995;25:1293–1299. doi: 10.1016/0020-7519(95)00066-b. [DOI] [PubMed] [Google Scholar]
- Umar A, Koi M, Risinger JI, Glaab WE, Tindall KR, Kolodner RD, Boland CR, Barrett JC, Kunkel TA. Correction of hypermutability, N-methyl-N'-nitro-N-nitrosoguanidine resistance, and defective DNA mismatch repair by introducing chromosome 2 into human tumor cells with mutations in MSH2 and MSH6. Cancer Res. 1997;57:3949–3955. [PubMed] [Google Scholar]
- Van Roey P, Meehan L, Kowalski JC, Belfort M, Derbyshire V. Catalytic domain structure and hypothesis for function of GIY-YIG intron endonuclease I-TevI. Nat Struct Biol. 2002;9:806–811. doi: 10.1038/nsb853. [DOI] [PubMed] [Google Scholar]
- Wilson RJ, Williamson DH. Extrachromosomal DNA in the Apicomplexa. Microbiol Mol Biol Rev. 1997;61:1–16. doi: 10.1128/mmbr.61.1.1-16.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Khalpey Z, Cascalho M. Cellular physiology of mismatch repair. Curr Pharm Des. 2004;10:4121–4126. doi: 10.2174/1381612043382468. [DOI] [PubMed] [Google Scholar]
- Yang G, Scherer SJ, Shell SS, Yang K, Kim M, Lipkin M, Kucherlapati R, Kolodner RD, Edelmann W. Dominant effects of an Msh6 missense mutation on DNA repair and cancer susceptibility. Cancer Cell. 2004;6:139–150. doi: 10.1016/j.ccr.2004.06.024. [DOI] [PubMed] [Google Scholar]
- Zenner L, Darcy F, Cesbron-Delauw MF, Capron A. Rat model of congenital toxoplasmosis: rate of transmission of three Toxoplasma gondii strains to fetuses and protective effect of a chronic infection. Infect Immun. 1993;61:360–363. doi: 10.1128/iai.61.1.360-363.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu WH, Loh TT. Effects of Na+/H+ antiport and intracellular pH in the regulation of HL-60 cell apoptosis. Biochimica et biophysica acta. 1995;1269:122–128. doi: 10.1016/0167-4889(95)00102-x. [DOI] [PubMed] [Google Scholar]
- Zuther E, Johnson JJ, Haselkorn R, McLeod R, Gornicki P. Growth of Toxoplasma gondii is inhibited by aryloxyphenoxypropionate herbicides targeting acetyl-CoA carboxylase. Proc Natl Acad Sci U S A. 1999;96:13387–13392. doi: 10.1073/pnas.96.23.13387. [DOI] [PMC free article] [PubMed] [Google Scholar]