Abstract
Malignant gliomas are the most common primary brain tumors. Despite efforts to find effective treatments, these tumors remain incurable. The failure of malignant gliomas to respond to conventional cancer therapies may reflect the unique biology of these tumors, underscoring the need for new approaches in their investigation. Recently, progress has been made in characterization of the molecular pathogenesis of glioblastoma using a developmental neurobiological perspective, by exploring the role of signaling pathways that control the differentiation of neural stem cells along the glial lineage. The transcription factor STAT3, which has an established function in neural stem cell and astrocyte development, has been found to play dual tumor suppressive and oncogenic roles in glial malignancy depending on the mutational profile of the tumor. These findings establish a novel developmental paradigm in the study of glioblastoma pathogenesis and provide the rationale for patient-tailored therapy in the treatment of this devastating disease.
INTRODUCTION
Malignant gliomas are the most prevalent primary tumors in the nervous system. Incidence rates place these tumors, including glioblastoma, which is the most common and aggressive malignant glioma, at much lower rates than solid tumors in other organ systems. However, their very low survival rates make these tumors one of the leading causes of cancer-related deaths in the young and middle-aged populations [1-4].
Most cases of malignant glioma have an insidious onset, with a clinical presentation that can be generalized—consisting of headaches, seizures, syncope, papilledema, nausea and vomiting, cognitive dysfunction—or focal with motor weakness, sensory loss, or speech impairment [5]. Imaging and pathological studies indicate that these tumors arise in the cerebral hemispheres, typically at the cortical/subcortical interface [4]. Often, malignant glioma tumors appear to be spreading through the white matter, occasionally to the opposite hemisphere through the corpus callosum, giving rise to the appearance of a butterfly tumor [4]. Histologically, these tumors have the characteristic features of malignancy, including increased mitoses and necrosis [4]. Malignant glioma tumor cells are extremely infiltrative, often migrating along the basement membrane of blood vessels or along myelinated white matter. However, tumor cells rarely metastasize outside the central nervous system [4, 6].
Malignant gliomas and glioblastoma in particular are incurable. For decades, the mainstay of therapy has been a combination of surgery, radiotherapy, and chemotherapy, the latter gaining more utility in the last decade with the introduction of more effective agents, such as temozolomide [7, 8]. However, despite these intense efforts, there has been negligible progress in the median survival of patients diagnosed with glioblastoma [9]. Clearly, there is an urgent need for more treatments. In this regard, a widely held view is that effective treatments can be developed once the genetic and molecular mechanisms underlying these tumors are elucidated.
Characterization of genetic alterations in malignant glioma has revealed that, just as in other tumors, abnormal activation of oncogenic proteins and inhibition of tumor suppressors are common in glial transformation [4, 10-12]. Oncogenic and tumor suppressive proteins often constitute components of the core cell cycle machinery or components of growth factor-regulated signal transduction pathways [13]. Among these alterations, inactivation of the tumor suppressor PTEN and expression of the oncogenic truncated epidermal growth factor receptor EGFRvIII play prominent roles in glioblastoma pathogenesis [4, 14-17]. Many more genetic alterations have also been described and reviewed [10-12, 18].
A recent approach to unravel the molecular basis of glioblastoma tumors consists of determining the role of deregulation of glial developmental signaling pathways in the pathogenesis of these tumors. The majority of glioblastoma tumors may arise from astrocytes or neural stem cells [12, 19-22]. During normal brain development, neural stem cells represent the precursors of all cellular elements of the brain including astrocytes. Progress has been made over the past decade in elucidation of the mechanisms that control neural stem cell maintenance as well as their differentiation into astrocytes [23-25]. These studies have revealed that the transcription factor STAT3 plays a central role in neural stem cell and astrocyte development [26-28]. Using this developmental view of STAT3 in gliogenesis as a starting point in the investigation of brain tumors, recent studies have uncovered that STAT3 plays distinct and opposing tumor suppressive and oncogenic roles in glioblastoma tumor pathogenesis depending on the genetic background of the tumor [29]. Because STAT3 was previously thought to promote tumor formation outside the brain [30-34], these studies have shifted our view of STAT3 functions in malignancy, and in the process, have suggested a new developmental paradigm in the study of glioblastoma pathogenesis.
In this review article, we will focus on the role of STAT3 in glioblastoma pathogenesis, preceded by a brief overview of STAT3 signaling and STAT3’s role as an oncogenic protein in tumors outside the brain. A better understanding of the role of STAT3 in brain tumors may provide a foundation for patient-tailored approaches to their treatment in the future.
STAT3 SIGNALING
STAT3 is a member of the STAT (Signal Transducers and Activators of Transcription) family of transcription factors [35]. The STATs relay information from the plasma membrane to the nucleus upon activation of several families of cytokine and growth factor receptors. The STAT proteins were first discovered as mediators of interferon signaling, but were quickly shown to couple signals from a diverse set of cytokines and growth factors to the transcriptional machinery [35, 36]. Activation of ciliary neurotrophic factor (CNTF) family cytokine receptors in neural cells triggers the activation of JAKs, a family of cytokine receptor-associated tyrosine kinases, which in turn phosphorylate the cytoplasmic tail of the receptor [36-39]. Upon phoshorylation of the cytokine receptor, STAT3 and STAT1 are recruited to phosphotyrosine motifs on the receptor via the Src homology 2 domain (SH2) of the STAT protein [35, 36, 39-41]. Recruitment of STAT3 and STAT1 to the activated cytokine receptor in turn leads to the phosphorylation of the STAT protein on specific tyrosine residues, Tyr 705 in STAT3 and Tyr 701 in STAT1, by JAK tyrosine kinase activity [42, 43]. STAT3 and STAT1 can also become directly phosphorylated by the intrinsic tyrosine kinase activity of growth factor receptors, such as the epidermal growth factor receptor (EGFR) [35, 44]. Phosphorylated STATs dimerize through reciprocal phosphotyrosine-SH2 interactions, then translocate to the nucleus and bind to promoters of cytokine- or growth factor-responsive genes [45-50].
Among the STAT proteins, STAT3 has received the most scrutiny because of its pleiotropic functions in diverse biological settings [51, 52]. Within the nervous system, STAT3 signaling plays an instructive role in astrocyte differentiation. Activation of the cytokine receptors LIFRβ and gp130 promotes the differentiation of cortical neural progenitors along the astrocytic lineage, while restricting neuronal differentiation [26, 27]. Phosphorylation of STAT3-recruiting Tyr residues within LIFRβ and gp130 are required for astrocyte differentiation of neural progenitor cells [26]. In addition, inhibition of STAT3 by several different means inhibits astrocyte differentiation [23, 26, 27, 53, 54]. Genetic ablation of the LIFRβ gene results in a reduction in the population of astrocytes in the mouse brain [55]. Other factors such as bone morphogenetic protein 4 (BMP-4) can also drive astrocytic differentiation at high cell densities by activating STAT3 signaling [56]. In recent studies, STAT3 has also been implicated in the self-renewal capacity of neural stem cells [28]. These findings suggest that STAT3 is a versatile transcription factor that has multiple functions in the development of neural stem cells and astrocytes in the mammalian brain.
STAT3 AS AN ONCOGENIC PROTEIN
Numerous lines of evidence have suggested that STAT3 functions in an oncogenic capacity in tumorigenesis. STAT3 controls major cellular responses, including cell proliferation and survival, which are deregulated in malignancy [57, 58]. Expression of activated STAT3 in fibroblasts triggers malignant cell transformation [59]. A phosphorylated and constitutively active STAT3 has been found in many epithelial tumors, including head and neck, lung, prostate, and breast cancer, as well as hematopoietic malignancies such as lymphoma [30, 60]. Animal models of tumorigenesis have been instrumental in exploring the role of STAT3 in these tumors. For example, Chan and colleagues demonstrated that STAT3 is required for de novo skin tumorigenesis as assessed in a two-stage chemical carcinogenesis model [31]. Mice were treated with the tumor initiator 7,12-dimethylbenz[a]anthracene (DMBA) and the tumor promoter 12-O-tetradecanoylphorbol-13-acetate (TPA) to induce epithelial carcinogenesis. Although these agents induce skin tumors in wild-type mice, STAT3-null mice fail to develop skin tumors. In addition, injection of skin tumor-forming cells together with a STAT3 decoy into wild-type mice inhibits the growth of skin tumors [31]. Other models of tumorigenesis also emphasize the importance of STAT3 in oncogenesis. In a nucleophosmin-anaplastic lymphoma kinase (NPM-ALK) transgenic mouse model of lymphomagenesis, disruption of the STAT3 gene suggests that STAT3 is required for the maintenance of neoplastic T cells [32]. Still other studies of breast cancer describe inhibition of tumor formation in immunocompetent mice when STAT3 is inhibited by RNA interference (RNAi) [33].
Identification of an oncogenic function for STAT3 in distinct settings has fueled attempts to develop small molecule inhibitors of STAT3 for the treatment of a diverse set of tumors [61-63]. These inhibitors may have therapeutic potential in the management of tumors with constitutive STAT3 activation. For example, a STAT3 decoy oligonucleotide, that binds activated STAT3 and thus blocks DNA binding, inhibits the proliferation of head and neck cancer cells [62]. Similarly, phosphotyrosyl peptides that mimic and compete with Y705-phosphorylated STAT3 during dimerization appear to suppress Src-induced transformation [61]. More recently, large-scale chemical screens have identified compounds that inhibit the survival of STAT3-dependent breast cancer cells [63].
EARLY CLUES FOR A STAT3 TUMOR SUPPRESSIVE FUNCTION
Although most studies have highlighted the oncogenic function of STAT3 in tumors outside the brain, a number of observations have raised the possibility that STAT3 may have additional distinct roles in tumor pathogenesis. In particular, STAT3 has been shown to inhibit cell proliferation and promote differentiation in several cellular contexts [64-68]. Subversion of STAT3’s pro-differentiation and anti-proliferation effects in these cells might promote cell transformation. On the other hand, it is possible that such cytostatic functions of STAT3 might be employed as a compensatory, adaptive response by cells undergoing transformation in order to slow the neoplastic process. Accordingly, where STAT3 has been shown to promote cell differentiation or inhibit proliferation, STAT3 could have tumor suppressive functions during malignant transformation.
STAT3 appears to inhibit proliferation and promote differentiation of several distinct tumor cell types. In M1 acute myeloid leukemia cells, the pleiotropic cytokines IL-6 and LIF, which activate STAT3, promote growth arrest and terminal differentiation of leukemic cells [65]. Expression of dominant negative forms of STAT3 in these cells inhibits IL6- and LIF-induced growth arrest at G1/G0 phase and macrophage differentiation, suggesting that the effect of IL6 is mediated by STAT3 [69, 70]. Blocking STAT3 activity in prostate cells also inhibits IL6-dependent growth arrest and differentiation [66, 71], and studies in melanoma cells suggest that STAT3 mediates growth arrest in this system as well [64].
During hematopoiesis, STAT3 is instrumental in the differentiation of myeloid lineages [65, 72]. Granulocyte colony-stimulatory factor (G-CSF), an inducer of granulocyte differentiation from bone marrow myeloid progenitor cells, potently activates STAT3 signaling [73, 74]. In the murine myeloblast 32D cell line, G-CSF induces myeloid differentiation through STAT3-mediated expression of the cell cycle inhibitor p27Kip1 [75]. Homozygous transgenic mice expressing a mutant receptor of G-CSF that is unable to activate STAT3 signaling are severely neutropenic, with a dramatic accumulation of immature myeloid precursors in the bone marrow. Expression of constitutively active STAT3 in myeloid progenitors from mutant mice rescues this defect in differentiation. Conversely, dominant negative STAT3 inhibits myeloid differentiation in wild type progenitors [67]. Surprisingly, selective ablation of STAT3 in bone marrow hematopoietic progenitors does not induce neutropenia or poor granulopoiesis, suggesting that STAT3 is not necessary for myeloid differentiation in these mice [76]. A compensatory mechanism by other proteins such as STAT1 might explain the relatively impenetrant phenotype in these STAT3 ablated progenitors.
The role of STAT3 in differentiation has also been characterized in keratinocytes. STAT3 activation tightly correlates with growth arrest and differentiation in primary keratinocytes. In contrast, immortalized keratinocyte MK cells, which fail to undergo differentiation, do not contain activated STAT3 under conditions that normally favor differentiation [77]. Furthermore, expression of constitutively active STAT3 in normal laryngeal epithelium enhances the expression of keratin 13, a keratinocyte marker, while a dominant negative mutant of STAT3 (Y705F) inhibits keratin 13 expression. Taken together, these observations suggest that STAT3 is required for keratinocyte differentiation [68].
STAT3 is also necessary for the hepatocyte growth factor (HGF)-dependent differentiation of epithelial cells into tubular structures [78]. Epithelial differentiation is driven by HGF in three well differentiated phases: scattering, growth and tubulogenesis. The last step is STAT3-dependent and occurs when the cell receives signals to stop growing and start differentiating. HGF results in the phosphorylation and translocation of STAT3 to the nucleus, where it stimulates transcription of the cell cycle inhibitor waf-1. Inhibition of STAT3 in HGF-treated cells with a phosphorylated peptide that mimics the STAT3 phosphorylation site or with a STAT3-binding decoy oligonucleotide specifically abrogates tubulogenesis [78].
STAT3 may also contribute to cell death in certain contexts, thus providing another basis for its potential tumor-suppressive function. In particular, STAT3 activation precedes extensive epithelial apoptosis required for mammary gland involution [79, 80]. Conditional deletion of STAT3 in the mammary epithelia results in a strong inhibition of apoptosis and a consequent delay of mammary gland involution [81].
TUMOR SUPPRESSIVE STAT3 ROLE IN PTEN-DEFICIENT BRAIN TUMORS
Although the above studies have raised the possibility of a tumor suppressive STAT3 function, such a role in cell transformation has only been established in recent investigations of glioblastoma pathogenesis. The hypothesis that STAT3 might operate as a tumor suppressor in the brain has been based on the observation that STAT3 promotes the differentiation of neural stem cells into astrocytes [26, 27]. Glioblastoma tumors display heterogeneity in the expression of markers of different cell lineages, but the preponderance of evidence suggests that the majority of glioblastoma tumors express markers along the glial lineages, including astrocytic and oligodendrocytic lineages [4]. Therefore, these tumors might arise from a normal neural stem cell or a differentiated cell, such as an astrocyte, that acquires properties of a neural stem cell in the process of malignant transformation, a view that is supported in several studies of glioblastoma [18-21]. These observations have led to the hypothesis that gliomagenesis might be promoted by deregulation of signaling molecules such as STAT3 that promote astrocyte differentiation during normal brain development. Since STAT3 promotes cell differentiation along the astrocytic lineage, it seemed plausible that as a differentiation factor STAT3 might suppress glial malignancy.
To directly test the hypothesis that STAT3 might suppress glioblastoma tumors, the effect of STAT3 gene disruption has been studied in astrocytes [29]. Immortalized Stat3-/- astrocytes display increased proliferation and invasiveness as compared to Stat3loxP/loxP astrocytes, indicating that STAT3 inhibits astrocyte proliferation and invasiveness. However, disruption of the STAT3 gene alone is not sufficient to induce transformation of these cells. Combining STAT3 loss with other oncogenic backgrounds has revealed that STAT3 loss enhances the ability of PTEN knockdown astrocytes to undergo malignant transformation in an orthotopic transplantation model of tumor formation in immunocompromised SCID mice. These results demonstrate that STAT3 suppresses the malignant transformation of PTEN-deficient astrocytes in vivo [29].
The identification of STAT3’s tumor suppressive function in astrocytes and its intimate functional relationship with the major tumor suppressor PTEN raises the important question of how STAT3 might be regulated in the PTEN pathway. Investigation of this question indicates that PTEN controls a signaling cascade that robustly influences STAT3 in astrocytes [29]. Knockout of the PTEN gene in astrocytes triggers the downregulation of cytokine receptor LIFRβ. Since LIFRβ activates the JAK-STAT signaling pathway leading to the phosphorylation and activation of STAT3, the downregulation of LIFRβ gene expression provides the basis for the inhibition of STAT3 in PTEN-deficient cells. How does PTEN loss inhibit LIFRβ transcription in astrocytes? PTEN loss leads to activation of Akt and hence the inhibition of the transcription factor FOXO3 by cytoplasmic sequestration. Importantly, transcription of the LIFRβ gene has been found to be directly activated by FOXO3. Consequently, PTEN knockout leads to the downregulation of LIFRβ expression via Akt-inhibition of FOXO3 [29].
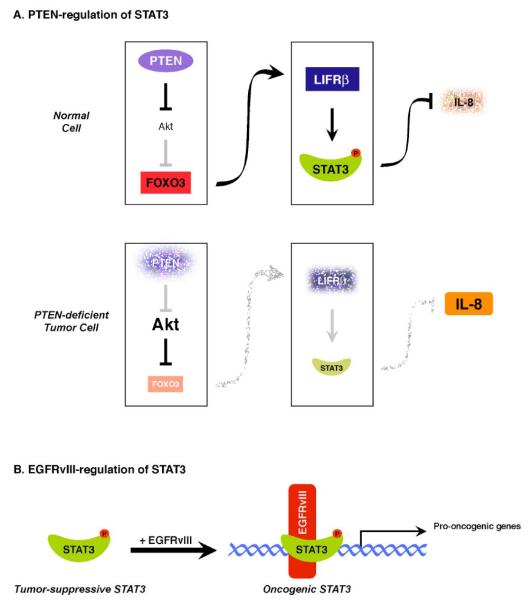
The elucidation of a link between the PTEN-Akt-FOXO3 axis and the LIFRβ-STAT3 signaling pathway together with results of STAT3-suppression of transformation of PTEN knockdown astrocytes suggest an intriguing to-and-fro model of PTEN and STAT3 in glial malignancy. According to this model, as PTEN becomes deficient in astrocytes and pushes these cells toward malignancy, STAT3 suppresses the process of cell transformation. However, as PTEN becomes more disrupted, the ensuing biochemical cascade relieves the STAT3 brake and drives malignant glial transformation (Fig. 1A).
Fig. (1). Dual role of STAT3 in glial tumorigenesis.
A. STAT3 as a PTEN-regulated tumor suppressor: In a normal cell with intact PTEN function, the protein kinase Akt is inhibited, allowing FOXO3 to activate transcription of the LIFRβ gene. In this scenario, STAT3 is activated by phosphorylation—downstream of LIFRβ—and represses transcription of the IL8 gene, thereby inhibiting glioma cell proliferation and invasiveness. Conversely, in a PTEN-deficient tumor cell, Akt is constitutively active, thus inhibiting FOXO3 function and leading to LIFRβ downregulation. STAT3 is no longer active and its repression of the IL8 gene is relieved, leading to upregulation of IL8, which drives glioma cell proliferation and invasiveness.
B. EGFRvIII induces an oncogenic switch in STAT3 function: STAT3 behaves as a tumor suppressor. However, in the presence of EGFRvIII, STAT3 and EGFRvIII form a physical complex in the nucleus that stimulates the transcription of oncogenic gene targets.
(Modified with permission from de la Iglesia et al. (2008). Genes & development 22, 449-462).
The relevance of STAT3’s tumor suppressive role has been confirmed in human PTEN-deficient tumors. In a panel of human brain tumors, PTEN loss correlates tightly with downregulation of LIFRβ and low levels of Tyr705 phosphorylation of STAT3. These findings provide correlative evidence that PTEN loss and inhibition of LIFRβ-STAT3 signaling are linked in human tumors [29]. In cultured human glioblastoma cells, this link is further corroborated by the finding of differential activation of STAT3 in PTEN-positive and PTEN-deficient glioblastoma cells [82]. The cytokine LIF stimulates the phosphorylation of STAT3 in wild-type PTEN-expressing glioblastoma cells but fails to induce STAT3 phosphorylation in PTEN-deficient human glioblastoma cells. Interestingly, in some of the PTEN-deficient human tumor cells LIFRβ levels are very low, and these are restored upon inhibition of the PI3K-Akt signaling pathway. These results suggest a consistent underlying mechanism of PTEN-regulation of the LIFRβ-STAT3 pathway as established in genetic studies of mouse astrocytes [82]. However, in other human glioblastoma cells, LIFRβ levels appear high despite the inability of LIF to induce STAT3 tyrosine phosphorylation. This observation suggests that there might be additional abnormalities in PTEN-deficient glioblastoma cells, either within the receptor or downstream kinases that couple LIFRβ phosphorylation to STAT3 activation [82]. It will be interesting to investigate these questions in future studies both in human glioblastoma cells as well as mouse astrocytes.
The inhibition of STAT3 phosphorylation specifically in human PTEN-deficient but not PTEN-positive glioblastoma cells suggests that, as in mouse astrocytes, STAT3 might play a tumor suppressive role in human glioblastoma cells. Consistent with this conclusion, expression of a constitutively active form of STAT3—that dimerizes independently of tyrosine phosphorylation (S3C)—suppresses the proliferation of PTEN-deficient glioblastoma cells [82]. By contrast, expression of S3C in other cells, such as fibroblasts, has been shown to promote their transformation [59]. These observations, along with immunoblotting analyses of human glioblastoma tumors, represent an important step forward in validating the tumor-suppressive role of STAT3 in PTEN-deficient human tumors.
How does STAT3 mediate a tumor-suppressive effect in glial cells? Gene-profiling analyses of glioblastoma cells expressing S3C have identified the chemokine IL8 as a direct target of STAT3 that is directly repressed by STAT3 in glioblastoma cells. STAT3 occupies the IL8 promoter and directly represses IL8 transcription in glial cells. IL8 knockdown in glioblastoma cells inhibits their proliferation and invasiveness, phenocopying the effect of activated STAT3 expression in these cells [82]. Importantly, IL8 is not expressed in normal brain but is expressed in human glioblastoma tumors [83], and in particular, in tumors with PTEN loss [82]. These findings suggest that IL8 derepression represents an important and functionally-relevant consequence of deregulation of the PTEN-STAT3 tumor suppressive pathway in glial cells (Fig. 1A).
In other studies, IL8 has been implicated in angiogenesis in glioblastoma tumors through a mechanism in which the tumor suppressor ING4 also represses IL8 transcription [84]. In this case, ING4 inhibits the IL8 promoter via the interaction of ING4 with NFkB, a transcriptional activator that stimulates IL8 transcription [84]. Intriguingly, STAT3 has also been found to repress NFkB-induced transcription [85, 86]. These results suggest that STAT3 might collaborate with ING4 to inhibit IL8 expression in glial cells. It will be interesting to determine if STAT3 forms a physical complex with ING4 and thereby regulates NFkB-dependent IL8 transcription in glial cells.
ONCOGENIC STAT3 ROLE IN EGFRVIII-POSITIVE BRAIN TUMORS
The finding that STAT3 operates in a tumor suppressive capacity in glial cells under normal circumstances and in the context of PTEN deficiency raises the important question of why STAT3 can behave in an oncogenic manner in tumors outside the brain. This paradox suggests two models. 1) STAT3 behaves in a tumor suppressive or oncogenic capacity depending on the cell type—tumor suppressive in glial cells and oncogenic in cells outside the nervous system. 2) STAT3 behaves in distinct manners in tumorigenesis depending on the mutational background of the tumor.
STAT3 reportedly promotes the survival of some glioblastoma cells in vitro [87, 88], and STAT3 activation as reflected by Tyr705 phosphorylation has been described in human gliomas [89]. Although other studies have provided evidence of the absence of STAT3 activity in a large percentage of these tumors, including glioblastoma [90, 91], this set of studies has suggested that STAT3 might behave in an oncogenic manner under certain circumstances in glial cells.
Genetic investigation of how STAT3 functions in tumors that are not deficient in PTEN but have other genetic alterations have led to the finding that STAT3 plays an essential oncogenic role in malignant astrocyte transformation in response to expression of the oncogenic protein EGFRvIII [29]. Expression of the oncogenic variant III of the epidermal growth factor receptor (EGFRvIII) in Stat3loxP/loxP astrocytes results in their malignant transformation in SCID mice. However, Stat3-/-;EGFRvIII astrocytes do not generate tumors when injected subcutaneously in immunocompromised mice. These findings suggest that the genetic make-up of the tumor specifies how STAT3 behaves in glial transformation [29].
Investigation of the molecular basis of STAT3’s oncogenic function in EGFRvIII-expressing astrocytes suggests that EGFRvIII forms a complex with STAT3 in the nucleus and thereby converts STAT3 from a tumor-suppressive protein into an oncogenic protein (Fig. 1B). How EGFRvIII, a transmembrane protein, finds its way to the nucleus, is unknown. Interestingly, ligated EGFR in breast cancer cells also localizes in the nucleus and associates with STAT3, thereby stimulating the malignant potential of these tumor cells [92, 93].
Collectively, studies of STAT3 in glial transformation suggest that whereas STAT3 is tumor suppressive in PTEN-deficient glioblastoma tumors, STAT3 behaves in an oncogenic manner in EGFRvIII-expressing tumors. The dichotomous role of STAT3 function in glial transformation is buttressed by analyses of PTEN loss and EGFRvIII in human glioblastoma, which suggest, remarkably, that these two major glioblastoma-associated genetic alterations stratify into distinct subsets of tumors [29, 94, 95]. Moreover, whereas LIFRβ-STAT3 signaling is inhibited in PTEN-deficient glioblastoma tumors, EGFRvIII expressing tumors display evidence of activated STAT3 [29]. Interestingly, a small subset of human tumors harbor both PTEN loss and EGFRvIII expression [94, 95]. In these tumors, STAT3 is anticipated to play an oncogenic role based on genetic evidence in astrocyte transformation in which both mutations are introduced [29]. These results further support the conclusion that EGFRvIII converts STAT3 from a tumor suppressive protein to an oncogenic protein.
POTENTIAL FOR PERSONALIZED THERAPY OF BRAIN TUMORS BASED ON STAT3 FUNCTION
The dual role of STAT3 as a tumor suppressive and oncogenic protein depending on the genetic background of a tumor raises exciting possibilities for patient-tailored treatment of brain tumors. A number of studies have focused on STAT3 inhibitors because of their potential in treating a wide variety of tumors [96-99]. Identification of STAT3 small molecule inhibitors has been a major goal of these efforts, with several classes of inhibitors emerging from attempts at drug development [100, 101]. In addition, oligonucleotide decoys and RNA interference have also been suggested as potential biological tools for inhibiting STAT3 in human cancer models [102]. In brain tumors expressing the EGFRvIII mutation, STAT3 is a promising potential target for these therapies. STAT3 inhibition may also sensitize these tumors to other chemotherapeutic agents [103].
Although STAT3 inhibitors may turn out to be useful in the treatment of EGFRvIII-expressing glioblastoma tumors, STAT3 inhibitors may not be efficacious and may be even potentially harmful in the treatment of PTEN-deficient tumors. Because STAT3 acts as a PTEN-regulated tumor suppressor, STAT3 inhibitors may presumably accelerate tumor growth by further derepressing IL-8 expression in these tumors. For PTEN-deficient glioblastoma tumors, agents should be developed that target IL-8 or the IL-8 receptor to limit tumor growth and invasion. Alternatively, activation of STAT3 itself may be potentially useful for these tumors. This approach may be inherently limited by the endogenous suppression of STAT3 activation via PTEN loss and LIFRβ downregulation, and no small molecule activators of STAT3 have been identified to date. However, small molecule inhibitors of the negative regulators of LIFRβ-STAT3 signaling, including the phosphatase Shp2 [104, 105] or the protein SOCS3 [106, 107], could be potentially useful in these cases.
Collectively, these studies offer the possibility of a patient-tailored approach in treatment that would depend on the genetic background of the tumor. How would such patient-tailored approaches be practically employed in the clinical setting? A plausible strategy would be to genotype and analyze surgically-excised tumors. In the future, this approach may facilitate the use of STAT3 or IL-8 inhibitors to complement standard treatment of surgery, radiation, and conventional chemotherapy.
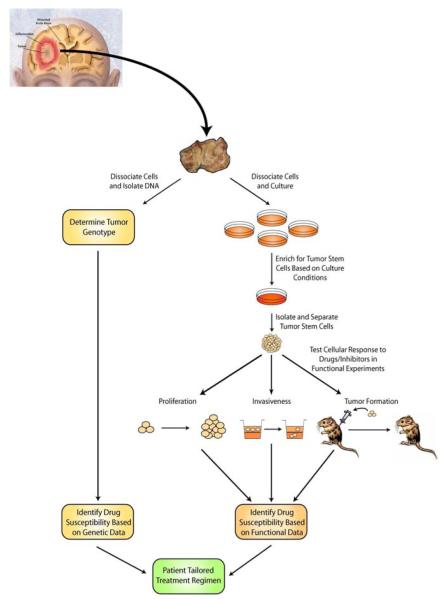
Although simple genetic analysis of primary tumor samples followed by mutation-based treatment may be effective, this management model may be complicated by the genetic diversity of the tumor. Although PTEN loss and STAT3’s function as a tumor suppressor appear to be tightly correlated [29], many exceptions to this correlation are likely to arise, depending on the presence of additional mutations in other pathways that may ultimately converge on STAT3 and its targets. To overcome this problem, the future management of brain tumors may also entail culturing glioma stem cells, followed by direct analyses of STAT3’s function in these cells. Initially, simple STAT3 RNA interference may help elucidate the role of STAT3 as a tumor suppressor or oncogene in the patient-specific tumor cells, and thereby suggest a specific management course. However, with technological advances in large scale screens, glioma stem cells could also be assayed for their responsiveness to chemical or biological agents. Once an appropriate combination of inhibitors is identified via high throughput screens for proliferation, invasiveness, and validated in orthotopic tumor formation experiments, a patient could be given the appropriate drug regimen (Fig. 2). Because recurrence in malignant glioma is the most persistent challenge in curing the disease, this approach could offer a new and potentially dramatic method of targeting tumor stem cells that remain in the parenchyma following surgery and substantially improve patient morbidity and mortality.
Fig. (2). Future potential patient-tailored therapies for clinical management of malignant gliomas.
Following surgical excision of the tumor, samples can be characterized genetically or assayed in functional experiments. Samples can be examined for known mutations associated with glioma, such as PTEN loss or the EGFRvIII truncation, and appropriate therapy can be given based on empiric data. Alternatively, samples can be cultured and enriched for the glioma stem cell population, which can then be analyzed functionally in the presence of an array of drugs and inhibitors to identify the unique drug susceptibilities of a given tumor (modified from Mayo Foundation for Medical Education and Research).
PERSPECTIVES
In recent years, the problem of malignant gliomas has attracted the attention of developmental neurobiologists. Characterization of the molecular mechanisms underlying glial cell fate specification in the developing mammalian brain has the raised the hope that investigation of these fundamental developmental pathways may lead to novel insights into glioblastoma tumor pathogenesis. As summarized in this review article, this approach has led to unexpected findings on the crucial role of the transcription factor STAT3 in the biology of glioblastoma tumors. These studies raise several key questions for future research on STAT3-regulation of glioblastoma pathogenesis, as well as provide a template for investigations of additional signaling molecules at the interface of developmental and cancer biology of the brain.
The finding that STAT3 exerts distinct tumor suppressive and oncogenic functions in glial malignancy depending on PTEN and EGFRvIII status of the tumor raises the fundamental question of the mechanism that underlies STAT3’s distinct function in these two different contexts. Although repression of the chemokine IL8 contributes to STAT3-suppression of PTEN-deficient glioblastoma cell proliferation and invasiveness [82], it will be important to identify additional targets of STAT3 that suppress the transformation of PTEN-deficient glial cells. In addition, the target genes of STAT3 that mediate EGFRvIII-induced astrocyte transformation remain to be identified.
Another question that requires attention is the role of STAT3 in the transformation of neural stem cells, which besides astrocytes represent the other presumed cells-of-origin of glioblastoma. Does STAT3 have tumor suppressive and oncogenic effects in PTEN-deficient and EGFRvIII-expressing neural stem cells respectively, or does STAT3 regulate the transformation of neural stem cells independently of PTEN and EGFRvIII status? Interestingly, recent studies suggest that distinct subpopulations of neural stem cells in the subventricular zone have unique capabilities in differentiation [108]. This raises the possibility that specific subsets of these stem cells might generate tumors in response to activation of specific signaling pathways. Thus, the cell intrinsic programs that govern heterogeneous stem cell behavior could also account for the dichotomous role of deregulated STAT3 in tumorigensis.
Beyond STAT3’s role in transformation of mouse astrocytes and neural stem cells, it will be also important to fully elucidate STAT3’s function in the pathogenesis of human glioblastoma tumors. The characterization of STAT3 activity in EGFRvIII-positive and PTEN-deficient human tumors as well as demonstration of STAT3’s suppression of the malignant phenotype of PTEN-deficient human glioblastoma cells are positive steps in this direction. However, STAT3’s function in the malignant behavior of EGFRvIII-expressing human glioblastoma cells and tumors requires further study.
Growing recent evidence supports the concept that malignant gliomas contain a population of cells that have stem cell-like properties and represent a small fraction of the total bulk of the tumor [109-111]. These cells, termed glioma stem cells or, more accurately, brain tumor initiating cells (BTICs), reportedly express the cell surface marker CD133 and can form neurospheres in vitro [109, 112]. BTICs are thought to have the potential to repopulate the tumor. A small number of BTICs, but not CD133 negative cells, when injected intracranially in immunocompromised mice, can form tumors that have pathological features of glioblastoma [109]. Although the cellular and biological properties of BTICs have been the subject of intense scrutiny, the critical molecular players that control the biological responses of these cells remain largely unknown. The discovery of STAT3’s role in glial transformation in mouse genetic studies provides the exciting opportunity to assess STAT3 function in BTICs, including from EGFRvIII-expressing or PTEN-deficient tumors.
Although studies of STAT3’s dual roles in tumor pathogenesis have focused on glial malignancy, whether the dichotomy in STAT3 function might also apply to tumors outside the nervous system should be explored. EGFRvIII expression and PTEN loss, in particular, are featured in other malignancies including prostate and breast cancer [14, 15, 113]. Preliminary studies suggest that expression of constitutively active STAT3 in PTEN-deficient prostate cancer cells inhibits their proliferation [114]. These experiments are consistent with the possibility that STAT3’s dual role in tumor pathogenesis may be extended to more common forms of cancer. Such a scenario would significantly broaden the implications of potential STAT3-based patient-tailored therapy of cancer.
Although the LIFRβ-STAT3 pathway is one of the first signaling mechanisms implicated in the development of astrocytes and neural stem cells, several other signaling molecules have been demonstrated to play key roles in cell fate specification in the developing brain. Therefore, studies of STAT3 signaling in glial malignant transformation can be used as a model in investigations of other developmental signals in brain tumors. Studies of the transcription factor Olig2, which plays a critical role in oligodendrocyte and neural stem cell development, as well as the bone morphogenetic (BMP)-Smad signaling pathway, which promotes an astrocytic fate, also support this approach. These investigations have revealed that Olig2 contributes to EGFRvIII-dependent neural stem cell transformation [115]. In addition, activation of BMP-Smad signaling inhibits the tumorigenic potential of human glioblastoma BTICs [111, 116]. It will be interesting to determine the role of other mechanisms regulating gliogenesis in neural stem cell or astrocyte transformation.
In the years ahead, investigations of developmental signaling mechanisms including the STAT3 pathway will continue to advance our understanding of glioblastoma pathogenesis. In addition, these efforts are likely to provide important clues for the potential development of novel treatments. As studies of STAT3 suggest, these treatments may turn out to be more effective if targeted in a personalized fashion. Although malignant gliomas remain deadly, a deep understanding of the basic biology of these tumors provides a ray of hope for the future management of this devastating disease.
ACKNOWLEDGEMENT
This work was supported by NIH grants to A.B. (NS041021, NS047188, and NS051255).
REFERENCES
- [1].Central Brain Tumor Registry of the United States (CBTRUS) report (2002-2003) http://www.cbtrus.org.
- [2].Ries LAG,EM, Kosary CL, Hankey BF, Miller BA, Clegg L, Mariotto A, Fay MP, Feuer EJ, Edwards BK, editors. SEER Cancer Statistics Rev., 1975-2001. 2004 [Google Scholar]
- [3].Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. CA Cancer J. Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- [4].Louis DN, Ohgaki H, Wiestler OD, Cavenee WKE. World Health Organization Histological Classification of Tumours of the Central Nervous System. IARC Press. International Agency for Research on Cancer (IARC); 2007. [Google Scholar]
- [5].Jaeckle KA, Cohen ME, Duffner PK. Clinical Presentation and Therapy of Nervous System Tumors. Butterworth-Heinemann; 1996. pp. 1131–49. [Google Scholar]
- [6].Bernstein JJ, Woodard CA. Neurosurgery. 1995;36:124–32. doi: 10.1227/00006123-199501000-00016. discussion 32. [DOI] [PubMed] [Google Scholar]
- [7].Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO. N. Engl. J. Med. 2005;352:987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- [8].Athanassiou H, Synodinou M, Maragoudakis E, Paraskevaidis M, Verigos C, Misailidou D, Antonadou D, Saris G, Beroukas K, Karageorgis P. J. Clin. Oncol. 2005;23:2372–7. doi: 10.1200/JCO.2005.00.331. [DOI] [PubMed] [Google Scholar]
- [9].Lamborn KR, Chang SM, Prados MD. Neuro. Oncol. 2004;6:227–35. doi: 10.1215/S1152851703000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK. Genes Dev. 2007;21:2683–710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- [11].Konopka G, Bonni A. Curr. Mol. Med. 2003;3:73–84. doi: 10.2174/1566524033361609. [DOI] [PubMed] [Google Scholar]
- [12].Holland EC. Nat. Rev. Genet. 2001;2:120–9. doi: 10.1038/35052535. [DOI] [PubMed] [Google Scholar]
- [13].Hanahan D, Weinberg RA. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- [14].Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, Frye C, Hu R, Swedlund B, Teng DH, Tavtigian SV. Nat. Genet. 1997;15:356–62. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- [15].Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, Ittmann M, Tycko B, Hibshoosh H, Wigler MH, Parsons R. Science. 1997;275:1943–7. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- [16].Nagane M, Lin H, Cavenee WK, Huang HJ. Cancer Lett. 2001;162(Suppl):S17–S21. doi: 10.1016/s0304-3835(00)00648-0. [DOI] [PubMed] [Google Scholar]
- [17].Friedman HS, Bigner DD. N. Engl. J. Med. 2005;353:1997–9. doi: 10.1056/NEJMp058186. [DOI] [PubMed] [Google Scholar]
- [18].Louis DN. Annu. Rev. Pathol. 2006;1:97–117. doi: 10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- [19].Bachoo RM, Maher EA, Ligon KL, Sharpless NE, Chan SS, You MJ, Tang Y, DeFrances J, Stover E, Weissleder R, Rowitch DH, Louis DN, DePinho RA. Cancer Cell. 2002;1:269–77. doi: 10.1016/s1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- [20].Uhrbom L, Dai C, Celestino JC, Rosenblum MK, Fuller GN, Holland EC. Cancer Res. 2002;62:5551–8. [PubMed] [Google Scholar]
- [21].Bajenaru ML, Hernandez MR, Perry A, Zhu Y, Parada LF, Garbow JR, Gutmann DH. Cancer Res. 2003;63:8573–7. [PubMed] [Google Scholar]
- [22].Kwon CH, Zhao D, Chen J, Alcantara S, Li Y, Burns DK, Mason RP, Lee EY, Wu H, Parada LF. Cancer Res. 2008;68:3286–94. doi: 10.1158/0008-5472.CAN-07-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bonni A. STATs in the central nervous system. Vol. 42. Kluwer Academic Publishers; 2003. pp. 663–85. [Google Scholar]
- [24].Guillemot F. Prog. Neurobiol. 2007;83:37–52. doi: 10.1016/j.pneurobio.2007.02.009. [DOI] [PubMed] [Google Scholar]
- [25].Shi Y, Sun G, Zhao C, Stewart R. Crit. Rev. Oncol. Hematol. 2008;65:43–53. doi: 10.1016/j.critrevonc.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bonni A, Sun Y, Nadal-Vicens M, Bhatt A, Frank DA, Rozovsky I, Stahl N, Yancopoulos GD, Greenberg ME. Science. 1997;278:477–83. doi: 10.1126/science.278.5337.477. [DOI] [PubMed] [Google Scholar]
- [27].Rajan P, McKay RD. J. Neurosci. 1998;18:3620–9. doi: 10.1523/JNEUROSCI.18-10-03620.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Yoshimatsu T, Kawaguchi D, Oishi K, Takeda K, Akira S, Masuyama N, Gotoh Y. Development. 2006;133:2553–63. doi: 10.1242/dev.02419. [DOI] [PubMed] [Google Scholar]
- [29].de la Iglesia N, Konopka G, Puram SV, Chan JA, Bachoo RM, You MJ, Levy DE, Depinho RA, Bonni A. Genes Dev. 2008;22:449–62. doi: 10.1101/gad.1606508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bromberg J. J. Clin. Invest. 2002;109:1139–42. doi: 10.1172/JCI15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chan KS, Sano S, Kiguchi K, Anders J, Komazawa N, Takeda J, DiGiovanni J. J. Clin. Invest. 2004;114:720–8. doi: 10.1172/JCI21032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chiarle R, Simmons WJ, Cai H, Dhall G, Zamo A, Raz R, Karras JG, Levy DE, Inghirami G. Nat. Med. 2005;11:623–9. doi: 10.1038/nm1249. [DOI] [PubMed] [Google Scholar]
- [33].Ling X, Arlinghaus RB. Cancer Res. 2005;65:2532–6. doi: 10.1158/0008-5472.CAN-04-2425. [DOI] [PubMed] [Google Scholar]
- [34].Schlessinger K, Levy DE. Cancer Res. 2005;65:5828–34. doi: 10.1158/0008-5472.CAN-05-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Darnell JE., Jr. Science. 1997;277:1630–5. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- [36].Darnell JE, Jr., Kerr IM, Stark GR. Science. 1994;264:1415–21. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- [37].Frank DA, Greenberg ME. Perspect. Dev. Neurobiol. 1996;4:3–18. [PubMed] [Google Scholar]
- [38].Horvath CM, Darnell JE. Curr. Opin. Cell Biol. 1997;9:233–9. doi: 10.1016/s0955-0674(97)80067-1. [DOI] [PubMed] [Google Scholar]
- [39].Saharinen P, Silvennoinen O. JAK Kinases. Kluwer Academic Publishers; 2003. pp. 27–39. [Google Scholar]
- [40].Wegenka UM, Buschmann J, Lutticken C, Heinrich PC, Horn F. Mol. Cell Biol. 1993;13:276–88. doi: 10.1128/mcb.13.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wegenka UM, Lutticken C, Buschmann J, Yuan J, Lottspeich F, Muller-Esterl W, Schindler C, Roeb E, Heinrich PC, Horn F. Mol. Cell Biol. 1994;14:3186–96. doi: 10.1128/mcb.14.5.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Shuai K, Stark GR, Kerr IM, Darnell JE., Jr. Science. 1993;261:1744–6. doi: 10.1126/science.7690989. [DOI] [PubMed] [Google Scholar]
- [43].Kaptein A, Paillard V, Saunders M. J. Biol. Chem. 1996;271:5961–4. doi: 10.1074/jbc.271.11.5961. [DOI] [PubMed] [Google Scholar]
- [44].Shao H, Cheng HY, Cook RG, Tweardy DJ. Cancer research. 2003;63:3923–30. [PubMed] [Google Scholar]
- [45].Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D’Andrea A, Livingston DM. Nature. 1996;383:344–7. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- [46].Zhang JJ, Vinkemeier U, Gu W, Chakravarti D, Horvath CM, Darnell JE., Jr. Proc. Natl. Acad. Sci. USA. 1996;93:15092–6. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Gingras S, Simard J, Groner B, Pfitzner E. Nucleic Acids Res. 1999;27:2722–9. doi: 10.1093/nar/27.13.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].McDonald C, Reich NC. J. Interferon Cytokine Res. 1999;19:711–22. doi: 10.1089/107999099313550. [DOI] [PubMed] [Google Scholar]
- [49].Paulson M, Press C, Smith E, Tanese N, Levy DE. Nat. Cell Biol. 2002;4:140–7. doi: 10.1038/ncb747. [DOI] [PubMed] [Google Scholar]
- [50].Lau JF, Nusinzon I, Burakov D, Freedman LP, Horvath CM. Mol. Cell Biol. 2003;23:620–8. doi: 10.1128/MCB.23.2.620-628.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Poli V, Alonzi T. STAT3 functions in vivo. Kluwer Academic Publishers; 2003. pp. 493–512. [Google Scholar]
- [52].Takeda K, Akira S. Tissue-specific function of STAT3. Kluwer Academic Publishers; 2003. pp. 513–23. [Google Scholar]
- [53].Aberg MA, Ryttsen F, Hellgren G, Lindell K, Rosengren LE, MacLennan AJ, Carlsson B, Orwar O, Eriksson PS. Mol. Cell Neurosci. 2001;17:426–43. doi: 10.1006/mcne.2000.0947. [DOI] [PubMed] [Google Scholar]
- [54].Ge W, Martinowich K, Wu X, He F, Miyamoto A, Fan G, Weinmaster G, Sun YE. J. Neurosci. Res. 2002;69:848–60. doi: 10.1002/jnr.10364. [DOI] [PubMed] [Google Scholar]
- [55].Ware CB, Horowitz MC, Renshaw BR, Hunt JS, Liggitt D, Koblar SA, Gliniak BC, McKenna HJ, Papayannopoulou T, Thoma B, et al. Development. 1995;121:1283–99. doi: 10.1242/dev.121.5.1283. [DOI] [PubMed] [Google Scholar]
- [56].Rajan P, Panchision DM, Newell LF, McKay RD. J. Cell Biol. 2003;161:911–21. doi: 10.1083/jcb.200211021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Bromberg J, Chen X. Methods Enzymol. 2001;333:138–51. doi: 10.1016/s0076-6879(01)33052-5. [DOI] [PubMed] [Google Scholar]
- [58].Levy DE, Lee CK. J. Clin. Invest. 2002;109:1143–8. doi: 10.1172/JCI15650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- [60].Turkson J, Jove R. Oncogene. 2000;19:6613–26. doi: 10.1038/sj.onc.1204086. [DOI] [PubMed] [Google Scholar]
- [61].Turkson J, Ryan D, Kim JS, Zhang Y, Chen Z, Haura E, Laudano A, Sebti S, Hamilton AD, Jove R. J. Biol. Chem. 2001;276:45443–55. doi: 10.1074/jbc.M107527200. [DOI] [PubMed] [Google Scholar]
- [62].Leong PL, Andrews GA, Johnson DE, Dyer KF, Xi S, Mai JC, Robbins PD, Gadiparthi S, Burke NA, Watkins SF, Grandis JR. Proc. Natl. Acad. Sci. USA. 2003;100:4138–43. doi: 10.1073/pnas.0534764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Song H, Wang R, Wang S, Lin J. Proc. Natl. Acad. Sci. USA. 2005;102:4700–5. doi: 10.1073/pnas.0409894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Kortylewski M, Heinrich PC, Mackiewicz A, Schniertshauer U, Klingmuller U, Nakajima K, Hirano T, Horn F, Behrmann I. Oncogene. 1999;18:3742–53. doi: 10.1038/sj.onc.1202708. [DOI] [PubMed] [Google Scholar]
- [65].Smithgall TE, Briggs SD, Schreiner S, Lerner EC, Cheng H, Wilson MB. Oncogene. 2000;19:2612–8. doi: 10.1038/sj.onc.1203477. [DOI] [PubMed] [Google Scholar]
- [66].Spiotto MT, Chung TD. Prostate. 2000;42:186–95. doi: 10.1002/(sici)1097-0045(20000215)42:3<186::aid-pros4>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- [67].McLemore ML, Grewal S, Liu F, Archambault A, Poursine-Laurent J, Haug J, Link DC. Immunity. 2001;14:193–204. doi: 10.1016/s1074-7613(01)00101-7. [DOI] [PubMed] [Google Scholar]
- [68].Wu R, Sun S, Steinberg BM. Mol. Med. 2003;9:77–84. doi: 10.2119/2003-00001.wu. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Nakajima K, Yamanaka Y, Nakae K, Kojima H, Ichiba M, Kiuchi N, Kitaoka T, Fukada T, Hibi M, Hirano T. EMBO J. 1996;15:3651–8. [PMC free article] [PubMed] [Google Scholar]
- [70].Minami M, Inoue M, Wei S, Takeda K, Matsumoto M, Kishimoto T, Akira S. Proc. Natl. Acad. Sci. USA. 1996;93:3963–6. doi: 10.1073/pnas.93.9.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Spiotto MT, Chung TD. Prostate. 2000;42:88–98. doi: 10.1002/(sici)1097-0045(20000201)42:2<88::aid-pros2>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- [72].Ward AC, Touw I, Yoshimura A. Blood. 2000;95:19–29. [PubMed] [Google Scholar]
- [73].Tian SS, Lamb P, Seidel HM, Stein RB, Rosen J. Blood. 1994;84:1760–4. [PubMed] [Google Scholar]
- [74].Tian SS, Tapley P, Sincich C, Stein RB, Rosen J, Lamb P. Blood. 1996;88:4435–44. [PubMed] [Google Scholar]
- [75].de Koning JP, Soede-Bobok AA, Ward AC, Schelen AM, Antonissen C, van Leeuwen D, Lowenberg B, Touw IP. Oncogene. 2000;19:3290–8. doi: 10.1038/sj.onc.1203627. [DOI] [PubMed] [Google Scholar]
- [76].Lee CK, Raz R, Gimeno R, Gertner R, Wistinghausen B, Takeshita K, DePinho RA, Levy DE. Immunity. 2002;17:63–72. doi: 10.1016/s1074-7613(02)00336-9. [DOI] [PubMed] [Google Scholar]
- [77].Hauser PJ, Agrawal D, Hackney J, Pledger WJ. Cell Growth Differ. 1998;9:847–55. [PubMed] [Google Scholar]
- [78].Boccaccio C, Ando M, Tamagnone L, Bardelli A, Michieli P, Battistini C, Comoglio PM. Nature. 1998;391:285–8. doi: 10.1038/34657. [DOI] [PubMed] [Google Scholar]
- [79].Philp JC, Burdon TG, Watson CJ. FEBS Lett. 1996;396:77–80. doi: 10.1016/0014-5793(96)01069-1. [DOI] [PubMed] [Google Scholar]
- [80].Liu XW, Robinson GW, Hennighausen L. Mol. Endocrinol. 1996:1496–506. doi: 10.1210/mend.10.12.8961260. [DOI] [PubMed] [Google Scholar]
- [81].Chapman RS, Lourenco PC, Tonner E, Flint DJ, Selbert S, Takeda K, Akira S, Clarke AR, Watson CJ. Genes Dev. 1999;13:2604–16. doi: 10.1101/gad.13.19.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].de la Iglesia N, Konopka G, Lim KL, Nutt CL, Bromberg JF, Frank DA, Mischel PS, Louis DN, Bonni A. J. Neurosci. 2008;28:5870–8. doi: 10.1523/JNEUROSCI.5385-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Yamanaka R, Tanaka R, Saitoh T, Okoshi S. J. Neurooncol. 1994;21:243–7. doi: 10.1007/BF01063773. [DOI] [PubMed] [Google Scholar]
- [84].Garkavtsev I, Kozin SV, Chernova O, Xu L, Winkler F, Brown E, Barnett GH, Jain RK. Nature. 2004;428:328–32. doi: 10.1038/nature02329. [DOI] [PubMed] [Google Scholar]
- [85].Yu Z, Zhang W, Kone BC. Biochem. J. 2002;367:97–105. doi: 10.1042/BJ20020588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Yu Z, Kone BC. J. Am. Soc. Nephrol. 2004;15:585–91. doi: 10.1097/01.asn.0000114556.19556.f9. [DOI] [PubMed] [Google Scholar]
- [87].Rahaman SO, Harbor PC, Chernova O, Barnett GH, Vogelbaum MA, Haque SJ. Oncogene. 2002;21:8404–13. doi: 10.1038/sj.onc.1206047. [DOI] [PubMed] [Google Scholar]
- [88].Konnikova L, Kotecki M, Kruger MM, Cochran BH. BMC Cancer. 2003;3:23. doi: 10.1186/1471-2407-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Weissenberger J, Loeffler S, Kappeler A, Kopf M, Lukes A, Afanasieva TA, Aguzzi A, Weis J. Oncogene. 2004;23:3308–16. doi: 10.1038/sj.onc.1207455. [DOI] [PubMed] [Google Scholar]
- [90].Schaefer LK, Ren Z, Fuller GN, Schaefer TS. Oncogene. 2002;21:2058–65. doi: 10.1038/sj.onc.1205263. [DOI] [PubMed] [Google Scholar]
- [91].Wang H, Zhang W, Huang HJ, Liao WS, Fuller GN. Lab Invest. 2004 doi: 10.1038/labinvest.3700123. [DOI] [PubMed] [Google Scholar]
- [92].Lin SY, Makino K, Xia W, Matin A, Wen Y, Kwong KY, Bourguignon L, Hung MC. Nat. Cell Biol. 2001;3:802–8. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- [93].Lo HW, Hsu SC, Ali-Seyed M, Gunduz M, Xia W, Wei Y, Bartholomeusz G, Shih JY, Hung MC. Cancer Cell. 2005;7:575–89. doi: 10.1016/j.ccr.2005.05.007. [DOI] [PubMed] [Google Scholar]
- [94].Choe G, Horvath S, Cloughesy TF, Crosby K, Seligson D, Palotie A, Inge L, Smith BL, Sawyers CL, Mischel PS. Cancer Res. 2003;63:2742–6. [PubMed] [Google Scholar]
- [95].Mellinghoff IK, Wang MY, Vivanco I, Haas-Kogan DA, Zhu S, Dia EQ, Lu KV, Yoshimoto K, Huang JH, Chute DJ, Riggs BL, Horvath S, Liau LM, Cavenee WK, Rao PN, Beroukhim R, Peck TC, Lee JC, Sellers WR, Stokoe D, Prados M, Cloughesy TF, Sawyers CL, Mischel PS. N. Engl. J. Med. 2005;353:2012–24. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- [96].Weerasinghe P, Li Y, Guan Y, Zhang R, Tweardy DJ, Jing N. Prostate. 2008 doi: 10.1002/pros.20807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].van Kester MS, Out-Luiting JJ, von dem Borne PA, Willemze R, Tensen CP, Vermeer MH. J. Invest. Dermatol. 2008;128:1691–5. doi: 10.1038/sj.jid.5701246. [DOI] [PubMed] [Google Scholar]
- [98].Xiong H, Zhang ZG, Tian XQ, Sun DF, Liang QC, Zhang YJ, Lu R, Chen YX, Fang JY. Neoplasia. 2008;10:287–97. doi: 10.1593/neo.07971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Dowlati A, Kluge A, Nethery D, Halmos B, Kern JA. Anticancer Drugs. 2008;19:9–16. doi: 10.1097/CAD.0b013e3282f1a908. [DOI] [PubMed] [Google Scholar]
- [100].Deng J, Grande F, Neamati N. Curr. Cancer Drug Targets. 2007;7:91–107. doi: 10.2174/156800907780006922. [DOI] [PubMed] [Google Scholar]
- [101].Bhasin D, Cisek K, Pandharkar T, Regan N, Li C, Pandit B, Lin J, Li PK. Bioorg. Med. Chem. Lett. 2008;18:391–5. doi: 10.1016/j.bmcl.2007.10.031. [DOI] [PubMed] [Google Scholar]
- [102].Zhang X, Zhang J, Wang L, Wei H, Tian Z. BMC Cancer. 2007;7:149. doi: 10.1186/1471-2407-7-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Gariboldi MB, Ravizza R, Molteni R, Osella D, Gabano E, Monti E. Cancer Lett. 2007;258:181–8. doi: 10.1016/j.canlet.2007.08.019. [DOI] [PubMed] [Google Scholar]
- [104].Hellmuth K, Grosskopf S, Lum CT, Wurtele M, Roder N, von Kries JP, Ros ario M, Rademann J, Birchmeier W. Proc. Natl. Acad. Sci. USA. 2008;105:7275–80. doi: 10.1073/pnas.0710468105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Chen L, Sung SS, Yip ML, Lawrence HR, Ren Y, Guida WC, Sebti SM, Lawrence NJ, Wu J. Mol. Pharmacol. 2006;70:562–70. doi: 10.1124/mol.106.025536. [DOI] [PubMed] [Google Scholar]
- [106].Auernhammer CJ, Isele NB, Kopp FB, Spoettl G, Cengic N, Weber MM, Senaldi G, Engelhardt D. Endocrinology. 2003;144:1202–10. doi: 10.1210/en.2002-220933. [DOI] [PubMed] [Google Scholar]
- [107].Yadav A, Kalita A, Dhillon S, Banerjee K. J. Biol. Chem. 2005;280:31830–40. doi: 10.1074/jbc.M501316200. [DOI] [PubMed] [Google Scholar]
- [108].Merkle FT, Mirzadeh Z, Alvarez-Buylla A. Science. 2007;317:381–4. doi: 10.1126/science.1144914. [DOI] [PubMed] [Google Scholar]
- [109].Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- [110].Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Nature. 2006;444:756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- [111].Piccirillo SG, Reynolds BA, Zanetti N, Lamorte G, Binda E, Broggi G, Brem H, Olivi A, Dimeco F, Vescovi AL. Nature. 2006;444:761–5. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- [112].Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Cancer Res. 2003;63:5821–8. [PubMed] [Google Scholar]
- [113].Wikstrand CJ, Hale LP, Batra SK, Hill ML, Humphrey PA, Kurpad SN, McLendon RE, Moscatello D, Pegram CN, Reist CJ, Traweek ST, Wong AJ, Zalutsky MR, Bigner DD. Cancer Res. 1995;55:3140–8. [PubMed] [Google Scholar]
- [114].de la Iglesia N, Puram SV, Bonni A. unpublished observations.
- [115].Ligon KL, Huillard E, Mehta S, Kesari S, Liu H, Alberta JA, Bachoo RM, Kane M, Louis DN, Depinho RA, Anderson DJ, Stiles CD, Rowitch DH. Neuron. 2007;53:503–17. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Lee J, Son MJ, Woolard K, Donin NM, Li A, Cheng CH, Kotliarova S, Kotliarov Y, Walling J, Ahn S, Kim M, Totonchy M, Cusack T, Ene C, Ma H, Su Q, Zenklusen JC, Zhang W, Maric D, Fine HA. Cancer Cell. 2008;13:69–80. doi: 10.1016/j.ccr.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]