Abstract
Sphingomyelin plays a very important role both in cell membrane formation that may well have an impact on the development of diseases like atherosclerosis and diabetes. However, the molecular mechanism that governs intracellular and plasma membrane SM levels is largely unknown. Recently, two isoforms of sphingomyelin synthase (SMS1 and SMS2), the last enzyme for SM de novo synthesis, have been cloned. We have hypothesized that SMS1 and SMS2 are the two most likely candidates responsible for the SM levels in the cells and on the plasma membrane. To test this hypothesis, cultured cells were treated with tricyclodecan-9-yl-xanthogenate (D609), an inhibitor of SMS, or with SMS1 and SMS2 siRNAs. Cells were then pulsed with [14C]-L-serine (a precursor of all sphingolipids). SMS activity and [14C]-SM in the cells were monitored. We found that SMS activity was significantly decreased in cells after D609 or SMS siRNA treatment, compared with controls. SMS inhibition by D609 or SMS siRNAs significantly decreased intracellular [14C]-SM levels. We measured cellular lipid levels, including SM, ceramide, phosphatidylcholine, and diacylglycerol and found that SMS1 and SMS2 siRNA treatment caused a significant decrease of SM levels (20% and 11%, respectively), compared to control siRNA treatment; SMS1 but not SMS2 siRNA treatment caused a significant increase of ceramide levels (10%). There was a decreasing tendency for diacylglycerol levels after both SMS1 and SMS2 siRNA treatment, however, it was not statistical significant. As shown by lipid rafts isolation and lipid determination, SMS1 and SMS2 siRNA treatment led to a decrease of SM content in detergent-resistant lipid rafts on the cell membrane. Furthermore, SMS1 and SMS2 siRNA-treated cells had a stronger resistance than did control siRNA-treated cells to lysenin (a protein that causes cell lysis due to its affinity for plasma membrane SM). These results indicate that both SMS1 and SMS2 contribute to SM de novo synthesis and control SM levels in the cells and on the cell membrane including plasma membrane, implying an important relationship between SMS activity and cell functions.
INTRODUCTION
Significant evidence has been presented to prove the existence of lipid rafts in membranes enriched with sphingolipids and cholesterol in the liquid-ordered phase (1, 2). Sphingomyelin (SM) is a major component of sphingolipids. However, little is known about the organization of SM in biological membranes. Raft domains have recently drawn extensive attention, for they may play an important role as a platform for signal transduction and protein sorting in these membranes (3, 4). Therefore, understanding the molecular mechanisms by which these domains are formed, maintained, and disintegrated has become one of the central issues in membrane biophysics and cell biology today (5,6).
Sphingomyelin synthase (SMS) is the last enzyme involved in the SM biosynthesis that transfers the phosphorylcholine moiety from phosphatidylcholine (PC) onto the primary hydroxyl of ceramide producing SM and diacylglycerol (DAG) (7). Evidence from the literature supports the belief that SM can be synthesized at more than one subcellular site. Many studies indicate that SMS is mainly located in the cis-, medial-Golgi (7–10), and plasma membrane (11–13). In addition, SMS activity has been found in chromatin, and chromatin-associated SMS is known to modify SM content (14–16). Despite the biological importance of SMS, understanding of the molecular mechanisms of its regulation and its relationship with plasma and cellular SM levels is limited by the fact that no successful purification of this protein has been achieved, and only recently the gene(s) encoding for this activity have been cloned (17,18). There are two isoforms of mammalian SMS genes, SMS1 and SMS2. The former is located on cis-, medial-Golgi, while the latter is on plasma membrane (17,19). A recent report revealed that downregulation of SMS1 results in SM-cholesterol deficiency in lipid rafts and attenuate apoptosis induced by alkyl-lysophospholipid (20), indicating a linkage between SMS1 activity and a biological function.
In this study we utilized two approaches, pharmacological and siRNA inhibition, to investigate the relationship between SMS and SM metabolism. We found that SMS inhibition by D609 (21), or by SMS1 and SMS2 siRNAs, significantly decreased intracellular, lipid rafts, and plasma membrane SM levels. This suggests that both SMS1 and SMS2 are key enzymes that control SM levels within the cells and on the membrane.
MATERIALS AND METHODS
Reagents
Potassium tricyclodecan-9-yl-xanthogenate (D609) was obtained from Calbiochem and dissolved in DMEM medium (pH 6.9). Bovine brain L-α-phosphatidylcholine (PC), NBD-C6-ceramide, and lysenin were purchased from Sigma. [14C]-L-serine was from Amersham. WST-1 cell proliferation reagent was from Roche. Polyclonal antibodies for Lyn and CD71 were from Santa Cruz Biotechnology. 16:0, 18:0, 20:0, 24:0, 24:1 Ceramides, 17:0 Sphingomyelin, and 14:0 phosphatidylcholine were from Avanti. Labeled 18:0 and 24:0 Ceramides were synthesized internally at Eli Lilly and Company. 15:0 1,3-Dipentadecanoin was from Sigma.
Cell Culture
Huh7 cells (a gift from Dr. Yi Luo, Pharmacia), human embryonic kidney (HEK) 293 cells, and Chinese hamster ovary (CHO) cells were cultured in complete medium (DMEM medium supplemented with 10% fetal bovine serum (FBS), 2 mM -glutamine, and 100 U/ml penicillin and streptomycin). HepG2 (ATCC) were cultured in complete medium (MEM medium supplemented with 10% FBS, 2 mM -glutamine, and 100 U/ml penicillin and streptomycin).
D609 treatment and SM analysis
Two doses of D609 (300 μM and 600 μM) were added to the cell culture medium, together with 0.2 mM oleic acid and 0.2 μci/ml of [14C]-L-serine. After 24 h of incubation, the cells were harvested and the medium collected. Lipids were extracted in chloroform: methanol (2:1 v/v), dried under N2 gas, and then separated by thin layer chromatography (TLC) in chloroform/methanol/20% ammonium hydroxide (14:6:1 v/v). Intracellular [14C]-SM levels were scanned with a Phosphorimager and the intensity of each spot was measured by Image–Pro Plus version 4.5 software (Media Cybernetics Inc.).
siRNA treatment and SM analysis
Two 21-mer siRNAs (Qiagen) were used to target each gene of SMS, (SMS1 or SMS2). The two target sequences for the SMS1 siRNA were: 5′-ACCTGTTGCACCGATATTCAA-3′ and 5′-TTGACTTAACCTATTGAGTTA-3′, and the two target sequences for SMS2 siRNA were: 5′-ACCGTCATGATCACAGTTGTA-3′, and 5′-ACCGTCATGATCACAGTTGTA-3′. The siRNAs were diluted in Opti-MEM (Invitrogen) medium and transfected into cells grown to 70–90% confluence using Lipofectamine 2000 reagent (Invitrogen). For each transfection, 50 nM concentrations of siRNA targeting either SMS1 or SMS2 were used, and where both genes were simultaneously targeted a total of 100 nM of siRNA (50 nM each) was cotransfected into a single well. After transfection, cells were incubated at 37°C and 5% CO2 in DMEM medium supplemented with 10% FBS and 1% glutamine in the absence of antibiotics. Cells were harvested after 24 h of siRNA transfection for mRNA quantitation and after 48 h for SMS activity assay. To quantify intracellular [14C]-SM levels, 0.2 mM oleic acid and 0.2 μci/ml of [14C]-L-serine were added to the cell culture medium after 24 h of siRNA transfection. After another 24 h of incubation, the cells were harvested, and [14C]-SM quantified as described above.
SMS1 and SMS2 mRNA measurements
Total RNA was isolated from cells with TriZol reagent (Invitrogen). SMS1 and SMS2 mRNA levels were measured by real-time polymerase chain reaction (PCR) on the ABI Prism 7000T Sequence Detection system (Applied Biosystems). For probes and primers, the Taqman® Gene Expression Assays were used (Applied Biosystems). The assay ID for SMS1 gene is Hs00300865_s1 and the assay ID for SMS2 gene is Hs00380453_m1. As internal control, 18S rRNA primers and probes were used (Sigma-Genosys). The forward and reverse primer sequences were: 5′-AGTCCCTGCCCTTTGTACACA-3′ and 5′-GATCCGAGGGCCTCACTAAAC-3′ respectively, and the probe sequence was 5′-CGCCCGTCGCTACTACCGATTGGT-3′. To compare the relative concentration of SMS1 and SMS2, we first determined PCR amplification efficiency of three pairs of primers (SMS1, SMS2, and 18S) and found that all three have comparable efficiency. We then obtained Ct (Ct represents the PCR cycle at which an increase in reporter fluorescence above a baseline signal can first be detected) and calculated Delta Ct for both SMS1 and SMS2 in each cell line (Delta Ct =18S Ct – SMS1 Ct or 18S Ct – SMS2 Ct). An increase in the Delta Ct value represents a decrease in mRNA expression.
Sphingomyelin synthase activity assay
Cells were homogenized in a buffer containing 50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 5% sucrose, and protease inhibitors. The homogenate was centrifuged at 5000 rpm for 10 min and the supernatant was used for SMS activity assay. The reaction system contained 50 mM Tris-HCl (pH 7.4), 25 mM KCl, C6-NBD-ceramide (0.1 μg/μl), and phosphotidylcholine (0.01 μg/μl). The mixture was incubated at 37°C for 2 h. Lipids were extracted in chloroform: methanol (2:1), dried under N2 gas, and separated by thin layer chromatography (TLC) using Chloroform:MeOH:NH4OH (14:6:1).
Lipid analysis
Ceramide Analysis
Ceramides comprised of a D-erythro-sphingosine backbone and a fatty acid (16:0, 18:0, 20:0, 24:0, 24:1) amide were determined by a 2D LC-ESI MS/MS method. Lipid extracts from cells were injected onto a normal-phase column where the polar lipids were retained. The ceramide fraction is trapped on a reversed-phase column. Ceramides are eluted, separated, and detected by using a triple quadrupole mass spectrometer equipped with positive ion electrospray ionization (ESI) and selected reaction monitoring. The method has a lower limit for quantification of 10 fmol of ceramide injected. Samples for analysis were spiked with 250ng each of 3 stable isotope- labeled 16:0, 18:0 and 24:0 ceramides prior to extraction. After a 5-fold dilution, 20μL of the sample solution was analyzed by 2D-LC/MS/MS and ceramide levels were quantified by the analyte to internal standard ratios and calibration curves obtained by serial dilution of a mixture of ceramide standards.
PC - SM Analysis
Phosphatidylcholine (PC) and sphingomyelin (SM) levels were measured via a flow injection ESI-MS/MS method, adapted from the method of Schmitz and co-workers (22), suitable for rapid monitoring of PC and SM present at μmol/L – mmol/L levels in tissue or cell extracts. Protonated molecular ions of PC/SM species are selected by precursor ion scans of m/z 184, the fragment ion containing the charged PC lipid head-group. The ion intensities across the flow injection profile are summed together and after isotope correction the quantities of each PC/SM species are then calculated relative to PC and SM internal standards. Samples were spiked with 25nmol 14:0, 14:0 PC and 12.5nmol 17:0 SM internal standards prior to extraction. A 200μL aliquot of sample extract was reconstituted in 1.00mL of 75% methanol/25% chloroform (v:v), 10mM ammonium acetate and 10μL of the sample was analyzed in duplicate. Average recovery of 21:0, 21:0 PC spiked into cell was 109.3 ± 11.5%.
Lysenin treatment and cell mortality measurement
After 48 h of siRNA transfection, cells were washed twice in PBS and incubated with 200 ng/ml lysenin for 2 h. Cell viability was measured using the WST-1 cell proliferation reagent according to manufacturer’s instructions (Roche).
Lipid raft isolation and SM determination
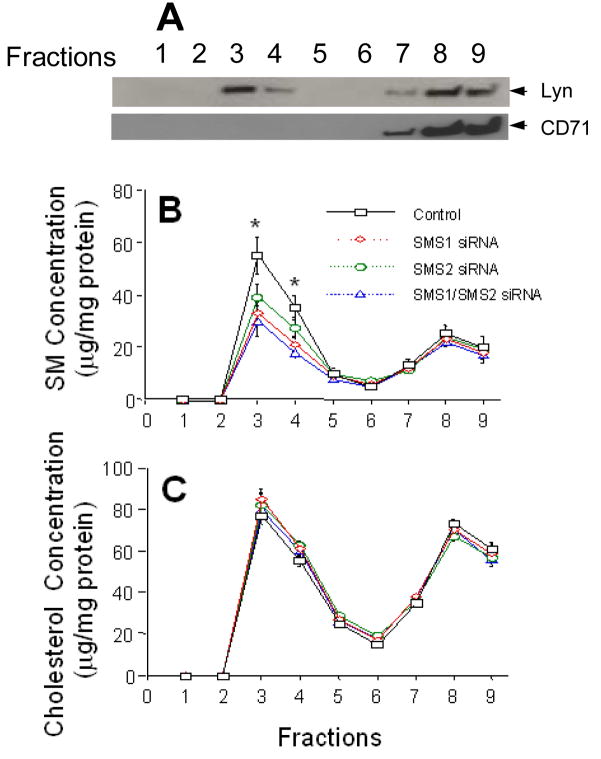
Detergent insoluble (lipid rafts) and soluble regions were isolated from HEK 293 cells according to a published approach (23). Briefly, about 1 × 107 of HEK 293 cells were lysed in 1.5 ml of hypotonic buffer, and broken by being passed through a 25-gauge needle. Nuclei were removed by centrifugation. Postnuclear supernatants were treated with 1% Triton X-100 for 20 min on ice, loaded on sucrose gradients and then centrifuged at 35,000 rpm in a Beckman SW41 Ti rotor for 18 h at 4°C. Fractions (1–9) were collected from the top of the gradient (1 ml for fraction 1 and 1.5 ml for subsequent fractions). It is known that Lyn, a tyrosine kinase, is expressed constitutively in lipid rafts region (23,24), while CD71 is expressed in non-rafts region (24). Each fraction (100 μg protein) was used for Western blot for Lyn and CD71. Lipids from each fraction were extracted as previously reported (25). SM and cholesterol levels in the extracts were determined by enzymatic assays (26).
Statistical analysis
Each experiment was conducted at least five times. Data are typically expressed as mean ± S.D. Data between two groups were analyzed by Student’s t test, and among multiple groups by ANOVA followed by the Student-Newman-Keuls (SNK) test. A p value of less than 0.05 was considered significant.
RESULTS
The effect of D609 on SMS activity and de novo SM synthesis
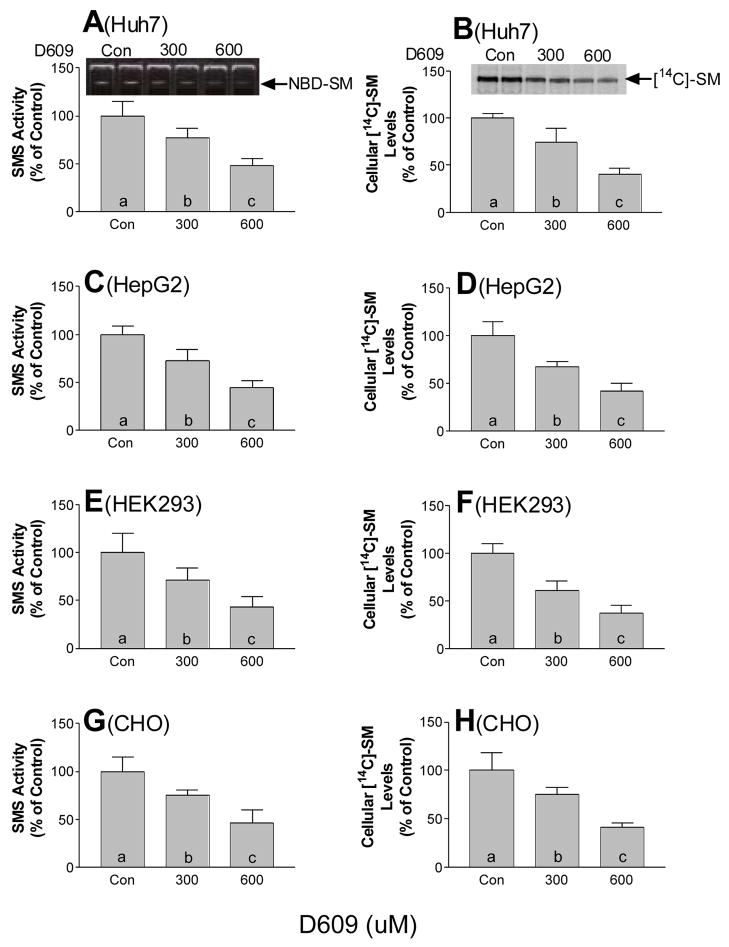
D609 is an inhibitor of SMS activity (21). To investigate the relationship between SMS activity and SM levels, Huh7 cells, a human hepatoma cell line, were treated with D609 and [14C]-serine (a precursor for all sphingolipids) was added to the medium. After 1 day of incubation, cells were collected, and lipids were extracted. Intracellular [14C]-SM was analyzed by TLC. We found that D609 treatment significantly decreased cellular SMS activity in a dose-dependent fashion, compared with the control (23% and 50%, p < 0.05 and p < 0.01, respectively) (Fig. 1A). This inhibition significantly diminished the intracellular (29% and 61%) [14C]-SM levels, compared with the control (p < 0.01, respectively) (Fig. 1B). To determine whether these observations also apply to other cell lines, we treated HepG2 cells, another human hepatoma cell line, with the D609. It also caused a significant and dose-dependent decrease in SMS activity (22% and 62%, p < 0.05 and p < 0.01, respectively) (Fig. 1C), intracellular SM levels (39% and 67%, p < 0.01, respectively) (Fig. 1D), compared with the control. We also chose two non-liver cell lines, HEK 293 and CHO, and treated them with D609, finding the same basic phenomena as in the Huh7 and HepG2 cells. In both cases, D609 treatment caused a significant and dose-dependent reduction of SMS activity (p < 0.01, respectively) (Fig. 1E and 1G), intracellular [14C]-SM levels (p < 0.01, respectively) (Figs. 1F and 1H), compared with controls. These results suggest that in all four tested cells SMS activity plays a role in newly synthesized SM pool.
FIG. 1. D609 treatment caused decrease of SMS activity and decrease of intracellular and secreted SM levels in cells.
Two doses of D609 (300 μM and 600 μM) were added to Huh7 cell (A, B), HepG2 cell (C, D), HEK 293 cell (E, F) and CHO cell (G, H) culture medium, together with 0.2 mM oleic acid and 0.2 μci/ml of [14C]-L-serine. After 24 h of incubation, cells were harvested. Lipids were extracted and intracellular [14C]-SM levels were quantitated as described in “Experimental procedure.” A, C, E, and G, Quantitative display of SMS activity. The reaction system contained 50 mM Tris-HCl (pH 7.4), 25 mM KCl, C6-NBD-ceramide (0.1 μg/μl), and phosphotidylcholine (0.01 μg/μl). The mixture was incubated at 37°C for 2 hours. Lipids were extracted in chloroform: methanol (2:1), dried under N2 gas, and separated by thin layer chromatography (TLC) using Chloroform:MeOH:NH4OH (14:6:1). B, D, F, and H, quantitative displays of intracellular [14C]-SM levels. Values are mean ± S.D., n = 5, p < 0.001 by ANOVA. Columns labeled with different lower-case letters (a-c) are statistically different by SNK test (p < 0.05).
We next sought to measure the expression levels of SMS1 and SMS2 in these cells and found that SMS1 and SMS2 mRNA levels are almost in 1:1 ratio in HEK 293 and HepG2 cells, while in Huh7 cells, it is about 5:1 (Table 1). We then utilized HEK 293 and Huh7 cells to further evaluate SMS1 and SMS2 functions on intracellular and membrane SM levels.
Table 1.
Real-time PCT analysis of cell SMS1 and SMS2 expression
| Cell | SMS1 | SMS2 |
|---|---|---|
| Mean Delta Ct | ||
| Huh7 | 17.35±0.17 | 21.12±0.08* |
| HEK 293 | 16.51±0.11 | 17.21±0.12 |
| HepG2 | 18.72±0.13 | 17.19±0.09 |
p<0.001. Value, mean±SD. n=3. An increase in the Delta Ct value represents a decrease in mRNA expression.
The effect SMS1 and SMS2 siRNAs on SMS activity and de novo SM synthesis
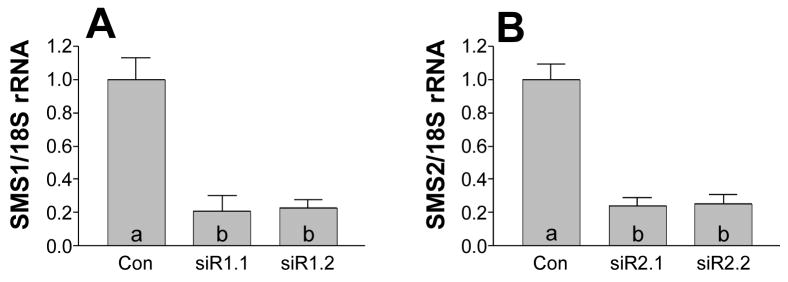
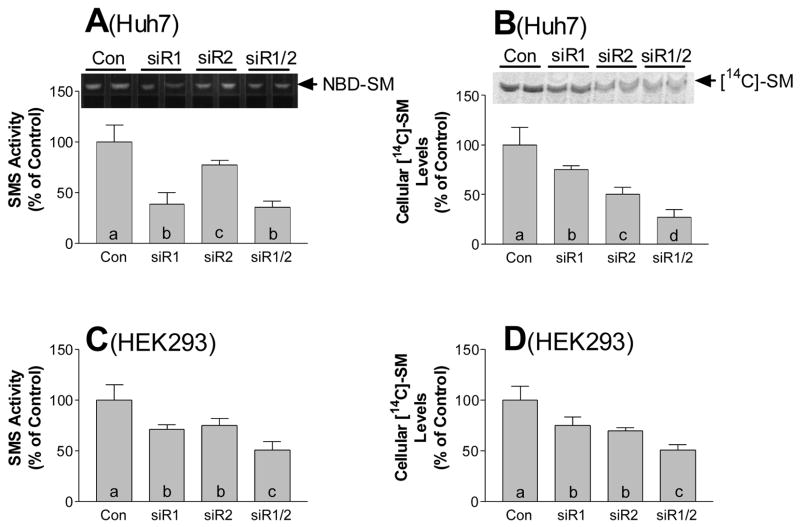
For further investigation of the relationship between SMS inhibition and SM levels, and dissecting potential differences between SMS1 and SMS2 genes, we utilized the siRNA approach. Six SMS1- and SMS2-specific siRNAs were designed and synthesized. Real-time PCR analysis demonstrated that two specific siRNAs for SMS1 (siR1.1 and siR1.2) caused an approximately 70% reduction of SMS1 mRNA levels, and two specific siRNAs for SMS2 (siR2.1 and siR2.2) also caused approximately the same 70% reduction of SMS2 mRNA levels in Huh7 cells, in comparison with control siRNA treatments (p < 0.0001) (Figs. 2A and 2B). We chose siR1.1 and siR2.1 for further study, finding that siR1.1 diminished Huh7 cellular SMS activity by about 70%, while siR2.1 diminished it only 20%, and the combination of both siRNAs diminished it about 75%, in comparison with control siRNA treatments (p < 0.001, p < 0.05, p < 0.001, respectively) (Fig. 3A). To investigate the consequence of SMS inhibition, we incubated the siRNA-transfected Huh7 cells with [14C]-L-serine for 24 h. We found that although both siR1.1 and siR2.1 significantly decreased intracellular [14C]-SM levels, compared with control (p < 0.02 and p < 0.001, respectively), the inhibition of SMS1 had less influence on intracellular SM levels than that of SMS2 (26% vs. 50%, p < 0.01) (Fig. 3B), and the combination of both siRNAs had an additive effect (70%) (Fig. 3B). These results indicated that, although SMS2 makes less contribution to total SMS activity than SMS1 (Fig. 3A) (Table 1) in Huh7 cells, it makes at least equal contribution to de novo SM biosynthesis. Following this, we treated HEK 293 cells with both siRNAs. This also caused a significant decrease in SMS activity (23% and 19%, p < 0.05, respectively) (Fig. 3C) and intracellular SM levels (19% and 26%, p < 0.05, respectively) (Fig. 3D), compared with controls. The combination of both siRNAs had an additive effect on SMS activity and intracellular [14C]-SM levels (Fig. 3C and 3D). These results revealed that both SMS1 and SMS2 also make contribution to the de novo SM synthesis in HEK 293 cells.
FIG. 2. siRNA treatment decreased SMS1 and SMS2 mRNA levels in Huh7 cells.
SMS1 and SMS2 siRNAs were utilized to transfect Huh7 cells. After 24 h of transfection, total RNA was extracted from the cells. A, SMS1 mRNA in Huh7 cells was measured by quantitative real-time PCR. B, SMS2 mRNA in Huh7 cells was measured by quantitative real-time PCR. Expression was described as the ratio of SMS1 or SMS2 mRNA to 18S rRNA. Values are mean ± S.D., n = 3, p < 0.001 by ANOVA. Columns labeled with different lower-case letters (a-c) are statistically different by SNK test (p < 0.0001). siR1.1, SMS1 siRNA1; siR1.2, SMS1 siRNA2; siR2.1, SMS2 siRNA1; siR2.2, SMS2 siRNA2.
FIG. 3. . The effect of SMS1 and SMS2 siRNAs on SMS activity and intracellular [14C]-SM levels.
SMS1 and SMS2, or SMS1 plus SMS2 siRNAs, were utilized to transfect Huh7 (A, B) and HEK 293 (C, D) cells. After 24 h of transfection, 0.2 mM oleic acid and 0.2 μci/ml of [14C]-L-serine were added to the cell culture medium. Intracellular [14C]-SM levels were quantitated as described in “Experimental procedure.” A and C, quantitative display of SMS activity. B and D, quantitative displays of intracellular [14C]-SM levels. Values are mean ± S.D., n=5, p < 0.01 by ANOVA. Columns labeled with different lower-case letters are statistically different by SNK test (p < 0.05).
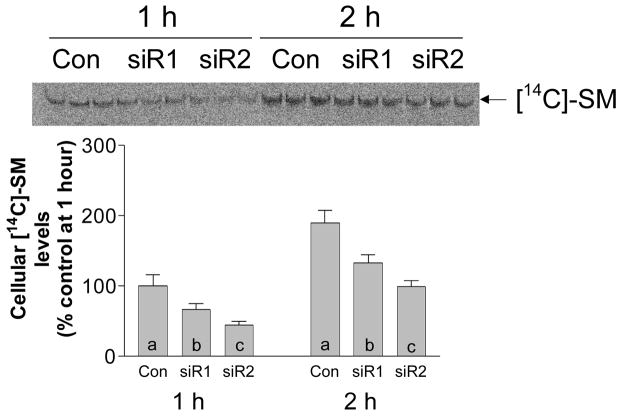
It has been reported that SMS1 is involved in SM biosynthesis while SMS2 is involved in remodeling (19). It would therefore be interesting to see an early time-course of the synthesis of radiolabeled SM for the SMS2 siRNA treated cells as compared to SMS1 siRNA treated ones. We found that, within 1–2 h, both SMS1 and SMS2 knockdown cells have significantly less newly synthesized SM pool than that of controls (Fig. 4). Moreover, SMS2 deficiency have a stronger effect than SMS1 deficiency (Fig. 4), indicating that, at least in Huh7 cells, SMS2 is as important as SMS1 in SM de novo synthesis.
FIG. 4. A time course of SMS1 and SMS2 siRNAs on intracellular [14C]-SM levels.
SMS1 and SMS2 siRNAs were utilized to transfect Huh7 cells. After 24 h of transfection, 0.2 mM oleic acid and 0.2 μci/ml of [14C]-L-serine were added to the cell culture medium. Intracellular [14C]-SM levels were quantitated as described in “Experimental procedure” after 1 and 2 hour incubation. Values are mean ± S.D., n=3, p < 0.01 by ANOVA. Columns labeled with different lower-case letters are statistically different by SNK test (p < 0.05).
SMS gene knockdown influences cellular SM and ceramide levels
To investigate whether a reduction of SMS1 and SMS2 mRNA by siRNA had any impact on cellular sphingolipid levels, including SM, PC, Ceramide, and diacylglycerol (DAG), the mass spectrometer (MS) was utilized. As indicated in Table 2, cells transfected with SMS1, SMS2, and combined siRNAs contained significantly less total SM than control siRNA-transfected cells (19.2%, 11.5%, and 19.2%, p<0.01, p<0.05, and p<0.01, respectively). SMS1 and SMS1/SMS2 siRNA treatment significantly increased cellular ceramide contents (9.6% and 7.8%, p<0.05, respectively), while SMS2 siRNA did not cause same effect. Although, there was a decreasing tendency, the changes of cellular DAG contents did not reach statistical significant (data not shown). There was no significant difference of cellular PC levels among the different group of cells (data not shown).
Table 2.
Lipid measurement in SMS1 and SMS2 gene knockdown Huh7 cells
| SM | PC | Cer | DAG | |
|---|---|---|---|---|
| (nmol/mg protein) | ||||
| Control | 40±3a | 334±25 | 0.83±0.03a | 3.45±1.5 |
| SMS1 siRNA | 32±5b | 330±27 | 0.92±0.04b | 2.11±0.6 |
| SMS2 siRNA | 35±2b | 332±16 | 0.82±0.04a | 2.14±0.5 |
| SMS1/2 siRNA | 32±4b | 325±31 | 0.94±0.03b | 2.09±0.5 |
In sphingomyelin and Ceramide columns, P<0.01 by ANOVA. In phosphatidylcholine and diacylglycerol columns, P>0.05 by ANOVA. Within Columns labeled with different lower-case letters (a and b) are statistically different by the SNK test (P<0.05). Value, mean±SD. n=4. SMS, sphingomyelin synthase. SM, Sphingomyelin; PC, Phosphatidylcholine; Cer, Ceramide; DAG, Diacylglycerol.
SMS gene knockdown influences SM levels in isolated membrane lipid rafts
In order to study the impact of SMS knockdown on lipid rafts, we isolated detergent insoluble (lipid rafts) and soluble regions from HEK 293 cells according to a published approach (23). It is known that Lyn, a tyrosine kinase, is expressed constitutively in lipid rafts region (23,24), while CD71 is expressed in non-rafts region (24). We utilized Lyn and CD71 as raft and non-raft markers, respectively, to perform Western blot in each fraction. As shown in Figure 5A, fraction 3 and 4 were isolated rafts, since they contained high levels of Lyn, and fraction 7 to 9 were non-rafts, since they contained high levels of CD71. We next sought to determine SM and cholesterol levels in isolated rafts and non-rafts. We found that 1) lipid raft fractions contain 2.5-fold higher SM than non-raft fractions (Fig. 5B); 2) siR1.1, siR2.1, and combined treatment significantly decrease SM levels in lipid raft fractions (29%, 17%, 37%, p < 0.01, respectively) but not in non-lipid raft fractions (Fig. 5B); and 3) the siRNA treatment have no influence on both raft and non-raft cholesterol levels (Fig. 5C). These results suggest that both SMS gene knockdown significantly and specifically decrease SM levels in lipid rafts on the cell membrane.
FIG. 5. Isolation of lipid rafts and non-rafts region from HEK 293 cells.
SMS1 and SMS2, or combined siRNAs were utilized to transfect HEK 293 cells. After 48 h of transfection, detergent insoluble and soluble membrane domains were separated by sucrose gradients. Fractions (1–9) were collected from the top of the gradient. Each fraction (100 μg protein) was used for Western blot for Lyn and CD71. SM and cholesterol in each fraction were determined by enzymatic assays. A, Western blot for Lyn and CD71 on lipid raft and non-raft regions. B, SM measurement in fractions. C, cholesterol measurement in fractions. Values are mean ± S.D., n = 4, * p < 0.001 by ANOVA. For SM measurement: control vs. siR1, p < 0.01, control vs. siR2, p < 0.05, and control vs. siR1/siR2, p < 0.01 in fractions 3 and 4, respectively. siR1 and siR2, siRNAs for SMS1 and SMS2, respectively; siR1/R2, siRNAs for SMS1 plus SMS2.
SMS gene knockdown influences plasma membrane SM organization
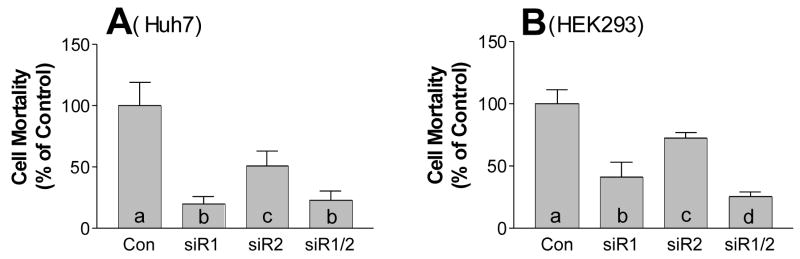
Since isolated membrane lipid rafts is a mixture of such microdomains in all the membranes, including plasma membrane, ER membrane, Golgi complex membrane, and so on. We still do not know whether SMS gene knockdown have an impact on SM levels on plasma membrane, where all the signal transduction is initiated. Lysenin is a recently discovered SM-specific cytotoxin (27). Lysenin recognizes SM only when it forms aggregates or microdomains (28). Based on our results above, we expected that SMS gene knockdown would reduce plasma membrane SM levels and influence the formation of aggregates or microdomains that are recognizable by lysenin. To investigate the effect of SMS gene knockdown on the formation of these microdomains, we tested siRNA-transfected Huh7 cells for sensitivity to lysenin-mediated cytolysis. As indicated in Figure 6A, cells transfected with SMS1, SMS2, or combined siRNAs showed significantly less sensitivity to lysenin-mediated cytolysis than control siRNA-transfected cells. Consistent with the relative contribution to the total cellular SMS activity in Huh7 cells (Fig. 3A), SMS1 knockdown provided the maximum protection from lysenin (82% survival, siR1.1 vs. control, p < 0.0001), while SMS2 knockdown provided less (43% survival, siR2.1 vs. control, p < 0.01). No additive effect was provided by the combined knockdowns of both genes (77% survival, siR1.1/siR2.1 vs. control, p < 0.0001) (Fig. 6A). We did same experiment on HEK 293 cells, although the SMS activity only decrease about 20% (Fig. 3C) in both SMS1 and SMS2 knockdown cells, the protection from lysenin-mediated cell lysis is very obvious (60% and 37%, p < 0.001, respectively), compared with controls (Fig. 6B). Moreover, there is an additive effect when combined siRNAs were used (Fig. 6B). These results suggest that the knockdown of both SMS1 and SMS2 mRNAs not only significantly decreases SM levels in the lipid rafts of the cell membrane, but also significantly alters SM-rich microdomains (probably lipid rafts) on the plasma membrane.
FIG. 6. SMS1 and SMS2 gene knockdown decreased lysenin-mediated cell mortality.
A, SMS1 and SMS2, or combined siRNAs, were utilized to transfect Huh7 cells. After 24 h of transfection, lysenin (200 ng/ml) was added to the cell culture medium and cell mortality was monitored by WST-1 Cell Proliferation Reagent (Roche). B, SMS1 and SMS2, or siRNAs were utilized to transfect HEK 293 cells. The rest was same as the Huh7 cell experiment. Values are mean ± S.D., n = 5, p < 0.001 by ANOVA. Columns labeled with different lower-case letters are statistically different by Student t test (p < 0.05). siR1 and siR2, siRNAs for SMS1 and SMS2, respectively; siR1/R2, siRNAs for SMS1 plus SMS2.
DISCUSSION
In this study, we have demonstrated that: 1) cells treated with D609 showed a significant decrease in SMS activity, and this treatment decreased SM de novo synthesis; 2) SMS1 and SMS2 siRNAs treatment significantly decreased cellular SMS activity and SM de novo synthesis; 3) both SMS1 and SMS2 gene knockdown cells had significantly lower cellular SM levels than controls; 4) both SMS1 and SMS2 deficiency significantly decreased SM levels in lipid rafts on cell membrane, and 5) both SMS1 and SMS2 siRNA-treated cells had significant stronger lysenin resistant potential than controls, indicating a decrease of SM levels on plasma membrane.
SM is a ubiquitous structural component of mammalian cell membranes and lipoproteins, and its cellular and plasma levels are regulated by both synthetic and catabolic pathways. In particular, the biochemical synthesis of SM occurs through the action of a serine palmitoyl-CoA transferase (SPT, the first enzyme of SM biosynthesis), 3-ketosphinganine reductase, ceramide synthase, dihydroceramide desaturase, and sphingomyelin synthase (SMS, the last enzyme of SM biosynthesis) (7). Many reports indicate that SPT is the key enzyme for all sphingolipid biosynthesis (7).
There is, however, some evidence that SMS is the key enzyme for SM biosynthesis. Cells treated with D609 had significantly decreased SMS activity, which in turn significantly decreased intracellular levels of SM (29). In this study we found that, in a variety of cell lines, D609 treatment caused a significant inhibition of SMS activity, leading to a significant decrease of SM levels within the cells (Fig. 1, Table 2). Moreover, SMS activity can be regulated. It has been shown that 25-hydroxycholesterol stimulates SM synthesis in CHO cells (30,31). It has also been demonstrated that the activity of SMS is enhanced under conditions of increased proliferation, such as regenerating rat liver (32), SV-40 transformation of human fibroblasts (20), highly malignant hepatoma (33), and the treatment of astrocytes with bFGF (34). Additionally, it has been reported that SM synthase activity is inhibited by TNFα in Kym-1 rhabdomyosarcoma cells before the onset of TNFα-induced apoptosis, and that this inhibition is caspase-dependent (35).
It has been reported that SMS1 is involved in SM biosynthesis while SMS2 is involved in remodeling (19). The finding that SMS2 gene knockdown results in a significant reduction in newly synthesized SM pool is unexpected. A time course of SM synthesis on a scale of the initial 1–2 hours provided a direct evidence that SMS2 is involved in SM de novo, and its role could be as important as SMS1, although SMS2 mRNA levels is only about 20% of that of SMS1 (Table 1) and SMS2 make minor contribution to the total SMS activity in Huh7 cells (Fig. 3A). We still do not completely understand the bases for the discrepancy between the results obtained after metabolic labeling (Fig. 3B), showing a robust decrease in SM, and the data on SMS enzyme activity (Fig. 3A) or mass measurements (Table 2), showing a modest decrease, however, we believe that both SMS1 and SMS2 utilize different cellular compartment for SM de novo biosynthesis. This observation deserves further investigation, since a method specific for SMS2 activity measurement seems to be available (36).
SMS activity may make an important contribution to the cell membrane structure. The interaction of SM and cholesterol drives the formation of plasma membrane rafts (1). As much as 70% of all cellular SM are found in such rafts (37). Our result indicated that about 65% of cell membrane SM is located in lipid rafts (Fig. 5). A general consensus has developed over the last few years that plasma membrane rafts represent signaling microdomains. Indeed, Van der Luit et al. reported that downregulation of SMS1 decrease SM in lipid rafts and diminish cell apoptosis induced by alkyl-lysophospholipid (20). Luberto et al. reported that D609 (a SMS inhibitor) treatment inhibits TNFα-(38,39) or phorbol ester-mediated (39) NF-κB activation. The question remaining to be answered is: are both SMS1 and SMS2 responsible for plasma membrane SM? We found, at least in Huh7 and HEK293 cells, that both SMS1 and SMS2 are responsible for plasma membrane SM levels. We have the following evidence to support this contention: 1) SMS1 and SMS2 siRNAs significantly decrease intracellular SM levels (Table 2); 2) SMS1 and SMS2 siRNA treatment led to a decrease of SM levels in lipid rafts on cell membrane (Fig. 5); and 3) both siRNA-treated cells had a stronger lysenin resistant potential than that of controls (Fig. 6). Since lysenin recognizes SM only when it forms aggregates or domains (27,28), our data suggest that both SMS1 and SMS2 activities are responsible for the level of plasma membrane SM, as well as the formation or maintenance of sub domains on the membrane.
Important biological roles have been clearly established for ceramide (one substrate of SMS) in the regulation of fundamental cellular functions such as proliferation and apoptosis (40–42). It has therefore been hypothesized that the cellular role of SMS goes beyond the production of SM. In fact, SMS could represent a key mechanism in the control of the cellular levels of ceramide, and would therefore influence functions mediated by this bioactive lipid. In this study, we found that SMS1 but not SMS2 knockdown significantly increased cellular ceramide levels (Table 2), suggesting at least in Huh7 cells, SMS1 but not SMS2 activity closely related to cellular ceramide levels. This observation deserves further investigation.
In summary, SMS inhibition mediated by D609 or SMS siRNAs significantly decreases SM levels within the cells. These results suggest that SMS1 and SMS2 are responsible for intracellular SM levels, and hence may contribute to the alteration of lipid rafts on plasma membrane observed in certain disease states (43).
Footnotes
This work was supported by grants from the National Institutes of Health-USA (HL-64735 and HL-69817).
The abbreviations used are: SM, sphingomyelin; SMS, sphingomyelin synthase; siRNA, short interfering RNAs; D609, tricyclodecan-9-yl-xanthogenate.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 2.Simons K, van Meer G. Lipid sorting in epithelial cells. Biochemistry. 1988;27:6197–6202. doi: 10.1021/bi00417a001. [DOI] [PubMed] [Google Scholar]
- 3.Brown DA, Rose JK. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 4.Parton RG, Simons K. Digging into caveolae. Science. 1995;269:1398–1399. doi: 10.1126/science.7660120. [DOI] [PubMed] [Google Scholar]
- 5.Harder T, Scheiffele P, Verkade P, Simons K. J Cell Biol. 1998;141:929–942. doi: 10.1083/jcb.141.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson RG. The caveolae membrane system. Annu Rev Biochem. 1998;67:199–225. doi: 10.1146/annurev.biochem.67.1.199. [DOI] [PubMed] [Google Scholar]
- 7.Merrill AH, Jones DD. An update of the enzymology and regulation of sphingomyelin metabolism. Biochim Biophysi ACTA. 1990;1044:1–12. doi: 10.1016/0005-2760(90)90211-f. [DOI] [PubMed] [Google Scholar]
- 8.Futerman AH, Stieger B, Hubbard AL, Pagan RE. Sphingomyelin synthesis in rat liver occurs predominantly at the cis and medial cisternae of the Golgi apparatus. J Biol Chem. 1990;265:8650–8657. [PubMed] [Google Scholar]
- 9.Jeckel D, Karrenbauer A, Birk R, Schmidt RR, Wieland F. Sphingomyelin is synthesized in the cis Golgi. FEBS Lett. 1990;261:155–157. doi: 10.1016/0014-5793(90)80659-7. [DOI] [PubMed] [Google Scholar]
- 10.Schweizer A, Clausen H, van Meer G, Hauri HP. Localization of O-glycan initiation, sphingomyelin synthesis, and glucosylceramide synthesis in Vero cells with respect to the endoplasmic reticulum-Golgi intermediate compartment. J Biol Chem. 1994;269:4035–4041. [PubMed] [Google Scholar]
- 11.van Helvoort A, van’t Hof W, Ritsema T, Sandra A, van Meer G. Conversion of diacylglycerol to phosphatidylcholine on the basolateral surface of epithelial (Madin- Darby canine kidney) cells. Evidence for the reverse action of a sphingomyelin synthase. J Biol Chem. 1994;269:1763–1769. [PubMed] [Google Scholar]
- 12.Moreau P, Cassagne C. Phospholipid trafficking and membrane biogenesis. Biochim Biophys ACTA. 1994;1197:257–290. doi: 10.1016/0304-4157(94)90010-8. [DOI] [PubMed] [Google Scholar]
- 13.Obradors MJ, Sillence D, Howitt S, Allan D. The subcellular sites of sphingomyelin synthesis in BHK cells. Biochim Biophys ACTA. 1997;1359:1–12. doi: 10.1016/s0167-4889(97)00088-8. [DOI] [PubMed] [Google Scholar]
- 14.Albi E, Magni MV. Sphingomyelin synthase in rat liver nuclear membrane and chromatin. FEBS Lett. 1999;460:369–372. doi: 10.1016/s0014-5793(99)01378-2. [DOI] [PubMed] [Google Scholar]
- 15.Albi E, Peloso I, Magni MP. Nuclear membrane sphingomyelin-cholesterol changes in rat liver after hepatectomy. Biochem Biophys Res Commun. 1999;262:692–695. doi: 10.1006/bbrc.1999.1188. [DOI] [PubMed] [Google Scholar]
- 16.Albi E, Pieroni S, Magni MV, Sartori C. Chromatin sphingomyelin changes in cell proliferation and/or apoptosis induced by ciprofibrate. J Cell Physiol. 2003;196:354–361. doi: 10.1002/jcp.10314. [DOI] [PubMed] [Google Scholar]
- 17.Huitema K, van den Dikkenberg J, Brouwers JF, Holthuis JC. Identification of a family of animal sphingomyelin synthases. EMBO J. 2004;23:33–44. doi: 10.1038/sj.emboj.7600034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamaoka S, Miyaji M, Kitano T, Umehara H, Okazaki T. Expression cloning of a human cDNA restoring sphingomyelin synthesis and cell growth in sphingomyelin synthase-defective lymphoid cells. J Biol Chem. 2004;279:18688–18693. doi: 10.1074/jbc.M401205200. [DOI] [PubMed] [Google Scholar]
- 19.Tafesse FG, Ternes P, Holthuis JC. The multigenic sphingomyelin synthase family. J Biol Chem. 2006;281:29421–29425. doi: 10.1074/jbc.R600021200. [DOI] [PubMed] [Google Scholar]
- 20.van der Luit AH, Budde M, Zerp S, Caan W, Klarenbeek JB, Verheij M, van Blitterswijk WJ. Resistance to alkyl-lysophospholipid-induced apoptosis due to downregulated sphingomyelin synthase 1 expression with consequent sphingomyelin- and cholesterol-deficiency in lipid rafts. Biochem J. 2007;401:541–549. doi: 10.1042/BJ20061178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luberto C, Hannun YA. Sphingomyelin synthase, a potential regulator of intracellular levels of ceramide and diacylglycerol during SV40 transformation. Does sphingomyelin synthase account for the putative phosphatidylcholine-specific phospholipase C? J Biol Chem. 1998;273:14550–14559. doi: 10.1074/jbc.273.23.14550. [DOI] [PubMed] [Google Scholar]
- 22.Liebisch G, Lieser B, Rathenberg J, Drobnik W, Schmitz G. High-throughput quantification of phosphatidylcholine and sphingomyelin by electrospray ionization tandem mass spectrometry coupled with isotope correction algorithm. Biochim Biophys Acta. 2004;1686:108–117. doi: 10.1016/j.bbalip.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Katzman RB, Longnecker R. LMP2A does not require palmitoylation to localize to buoyant complexes or for function. J Virol. 2004;78:10878–10887. doi: 10.1128/JVI.78.20.10878-10887.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higuchi M, Izumi KM, Kieff E. Epstein-Barr virus latent-infection membrane proteins are palmitoylated and raft-associated: protein 1 binds to the cytoskeleton through TNF receptor cytoplasmic factors. Proc Natl Acad Sci USA. 2001;98:4675–4680. doi: 10.1073/pnas.081075298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang XC, Masucci-Magoulas L, Mar J, Lin M, Walsh A, Breslow JL, Tall AR. Down-regulation of mRNA for the low density lipoprotein receptor in transgenic mice containing the gene for human cholesteryl ester transfer protein. Mechanism to explain accumulation of lipoprotein B particles. J Biol Chem. 1993;268:27406–27412. [PubMed] [Google Scholar]
- 26.Hojjati MR, Jiang XC. Rapid, specific, and sensitive measurements of plasma sphingomyelin and phosphatidylcholine. J Lipid Res. 2006;47:673–676. doi: 10.1194/jlr.D500040-JLR200. [DOI] [PubMed] [Google Scholar]
- 27.Yamaji A, Sekizawa Y, Emoto K, Sakuraba H, Inoue K, Kobayashi H, Umeda M. Lysenin, a novel sphingomyelin-specific binding protein. J Biol Chem. 1998;273:5300–5306. doi: 10.1074/jbc.273.9.5300. [DOI] [PubMed] [Google Scholar]
- 28.Ishitsuka R, Yamaji-Hasegawa A, Makino A, Hirabayashi Y, Kobayashi T. A lipid-specific toxin reveals heterogeneity of sphingomyelin-containing membranes. Biophys J. 2004;86:296–307. doi: 10.1016/S0006-3495(04)74105-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng A, Luberto C, Meier P, Bai A, Yang X, Hannun YA, Zhou D. Sphingomyelin synthase as a potential target for D609-induced apoptosis in U937 human monocytic leukemia cells. Exp Cell Res. 2004;292:385–392. doi: 10.1016/j.yexcr.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 30.Ridgway ND. 25-Hydroxycholesterol stimulates sphingomyelin synthesis in Chinese hamster ovary cells. J Lipid Res. 1995;36:1345–1358. [PubMed] [Google Scholar]
- 31.Lagace TA, Byers DM, Cook HW, Ridgway ND. Chinese hamster ovary cells overexpressing the oxysterol binding protein (OSBP) display enhanced synthesis of sphingomyelin in response to 25-hydroxycholesterol. J Lipid Res. 1999;40:109–116. [PubMed] [Google Scholar]
- 32.Miro-Obradors MJ, Osada J, Aylagas H, Sanchez-Vegazo I, Palacios-Alaiz E. Microsomal sphingomyelin accumulation in thioacetamide-injured regenerating rat liver: involvement of sphingomyelin synthase activity. Carcinogenesis. 1993;14:941–946. doi: 10.1093/carcin/14.5.941. [DOI] [PubMed] [Google Scholar]
- 33.van den Hill A, van Heusden GP, Wirtz KW. The synthesis of sphingomyelin in the Morris hepatomas 7777 and 5123D is restricted to the plasma membrane. Biochim Biophys ACTA. 1985;833:354–357. doi: 10.1016/0005-2760(85)90210-3. [DOI] [PubMed] [Google Scholar]
- 34.Riboni L, Viani P, Bassi R, Giussani P, Tettamanti G. Basic fibroblast growth factor-induced proliferation of primary astrocytes. evidence for the involvement of sphingomyelin biosynthesis. J Biol Chem. 2001;276:12797–12804. doi: 10.1074/jbc.M011570200. [DOI] [PubMed] [Google Scholar]
- 35.Bourteele S, Hausser A, Doppler H, Horn-Muller J, Ropke C, Schwarzmann G, Pfizenmaier K, Muller G. Tumor necrosis factor induces ceramide oscillations and negatively controls sphingolipid synthases by caspases in apoptotic Kym-1 cells. J Biol Chem. 1998;273:31245–31251. doi: 10.1074/jbc.273.47.31245. [DOI] [PubMed] [Google Scholar]
- 36.Geta Tafesse F, Huitema K, Hermansson M, van der Poel S, van den Dikkenberg J, Uphoff A, Somerharju P, Holthuis JC. Both sphingomyelin synthase SMS1 and SMS2 are required for sphingomyelin homeostasis and growth in human HeLa cells. J Biol Chem. 2007 doi: 10.1074/jbc.M702423200. in press. [DOI] [PubMed] [Google Scholar]
- 37.Prinetti A, Chigorno V, Prioni S, Loberto N, Marano N, Tettamanti G, Sonnino S. Changes in the lipid turnover, composition, and organization, as sphingolipid-enriched membrane domains, in rat cerebellar granule cells developing in vitro. J Biol Chem. 2001;276:21136–21145. doi: 10.1074/jbc.M010666200. [DOI] [PubMed] [Google Scholar]
- 38.Schutze S, Potthoff K, Machleidt T, Berkovic D, Wiegmann K, Kronke M. TNF alpha activates NF-kappa B by phosphatidylcholine-specific phospholipase C-induced “acidic” sphingomyelin breakdown. Cell. 1992;71:765–776. doi: 10.1016/0092-8674(92)90553-o. [DOI] [PubMed] [Google Scholar]
- 39.Luberto C, Yoo DS, Suidan HS, Bartoli GM, Hannun YA. Differential effects of sphingomyelin hydrolysis and resynthesis on the activation of NF-kappa B in normal and SV40-transformed human fibroblasts. J Biol Chem. 2000;275:14760–14766. doi: 10.1074/jbc.275.19.14760. [DOI] [PubMed] [Google Scholar]
- 40.Hannun YA, Luberto C. Ceramide in the eukaryotic stress response. Trends Cell Biol. 2000;10:73–80. doi: 10.1016/s0962-8924(99)01694-3. [DOI] [PubMed] [Google Scholar]
- 41.Kolesnick R. The therapeutic potential of modulating the ceramide/sphingomyelin pathway. J Clin Invest. 2002;110:3–8. doi: 10.1172/JCI16127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaffrezou JP, Laurent G, Levade T. Ceramide in regulation of apoptosis. Implication in multitoxicant resistance. Subcell Biochem. 2002;36:269–284. [PubMed] [Google Scholar]
- 43.Ikonen E, Vainio S. Lipid microdomains and insulin resistance: is there a connection? Sci STKE. 2005;268:1–3. doi: 10.1126/stke.2682005pe3. [DOI] [PubMed] [Google Scholar]