SUMMARY
Integral membrane proteins remain a significant challenge to structural studies by solution NMR spectroscopy. This is due not only to spectral complexity but also because the effects of slow molecular reorientation are exacerbated by the need to solublize the protein in aqueous detergent micelles. These assemblies can be quite large and require deuteration for use of the TROSY effect. In principle, another approach is to employ reverse micelle encapsulation to solublize the protein in a low viscosity solvent where the rapid tumbling of the resulting particle allows use of standard triple resonance methods. The preparation of such samples of membrane proteins is difficult. Using a 54 kDa construct of the homotetrameric potassium channel KcsA we demonstrate a strategy that employs a hybrid surfactant to transfer the protein to the reverse micelle system.
INTRODUCTION
Membrane proteins comprise 20–25% of fully sequenced genomes and fulfill a wide range of central functions in the cell. Membrane proteins also play important roles in tumor development, the immune response, and drug tolerance. They are common drug targets and are of significant biomedical interest. The last decade has seen an explosion in the number of high resolution structural models of proteins determined by crystallography and nuclear magnetic resonance (NMR). Yet as the number of structures deposited in the Protein Data Bank (PDB) (Berman et al., 2003) passes 50,000, only a few hundred of those are of integral membrane proteins. Clearly, multiple strategies and approaches to the study of membrane protein structure by NMR would be advantageous.
Even setting aside the general difficulty in the expression and purification of integral membrane proteins, the detailed structural characterization of these systems by NMR spectroscopy is fraught with peril. As a result, only a handful of truly integral membrane protein structures have been comprehensively characterized using solution NMR methods (e.g. Baker et al., 2007; Chill et al., 2006; Fernandez et al., 2004; Hiller et al., 2008; Oxenoid and Chou, 2005; Park et al., 2003; Zhou et al., 2008). A common strategy is to solubilize recombinantly expressed protein in detergent micelles. These assemblies are quite large, even for relatively small proteins, often reaching 80 kDa and beyond. To undertake multidimensional triple resonance NMR spectroscopy of such large and therefore slowly tumbling systems, the TROSY (Pervushin et al., 1997) effect in combination with perdeuteration must usually be employed. In addition to the overall loss of potential structural information normally derived from 1H interactions, there is a potentially debilitating barrier presented, the need to back-exchange amide sites with hydrogen following expression in 100% D2O. This is because integral membrane proteins often cannot be unfolded and refolded efficiently - if at all - which is perhaps the only way of fully back exchanging hydrogen for deuterium at amide sites. Without the amide hydrogen, implementation of current triple resonance NMR strategies is greatly compromised.
Over the past decade there has been significant progress in the preparation of soluble protein molecules encapsulated within the protective core of a reverse micelle dissolved in a low viscosity solvent such as liquid ethane (Peterson et al., 2005a; Peterson et al., 2005b; Wand et al., 1998). The low viscosity solvent allows for sufficiently fast tumbling of the reverse micelle particle to significantly improve the NMR relaxation properties of the protein (Peterson et al., 2005a; Wand et al., 1998). Solubilization of an integral membrane protein in a reverse micelle system could also provide this advantage while also avoiding the problem of back exchange of amide hydrogens because deuteration will not be required. In addition, the TROSY effect will not be needed (but could be still used) and the full power of triple resonance spectroscopy of the type suitable for small, rapidly tumbling proteins will be applicable. We envisage the reverse micelle particle containing an integral membrane protein to be quite different than that of an encapsulated soluble protein. The distinct advantage of the reverse micelle system, in contrast to simple dissolution in organic solvents like chloroform as has often been used for studies of peptides and small proteins, is that the system provides both hydrophobic (alkane solvent, surfactant tail; tightly bound lipid) and polar (surfactant head groups, water) molecules to support the structure of the protein. Here we show that employing a special class of surfactants allows the transfer of the KcsA potassium channel to a reverse micelle system with apparent functional integrity and high performance solution NMR behavior. Refinement of the solution conditions was objective and did not require prior knowledge of the structure of the protein and thereby suggests that the approach may be general.
RESULTS
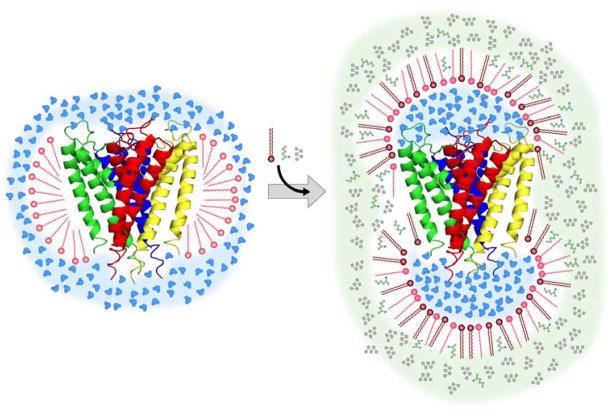
The key barrier to implementation of this approach is simply the question of how to encapsulate an integral membrane protein into the protective assembly of a reverse micelle for its subsequent dissolution into alkane solvent. Our initial foray into using reverse micelle surfactants to solubilize integral membrane proteins was guided by Montal and coworkers (Darszon et al., 1979). Essentially, protein embedded in a natural phospholipid membrane was injected at very low μM concentrations into heavy organic solutions of reverse micelle surfactant (Ramakrishnan et al., 1983). Low angle x-ray scattering studies of such complexes indicated a dumbbell like structure which is schematically illustrated in Figure 1 (Binks et al., 1989). We have termed this the “shower cap” model. We followed up on the direct injection approach with the aim of adapting it to light (low viscosity) fluids at NMR concentrations (mM). It proved not to be viable. The principle difficulty is that at high protein concentrations the amount of either natural lipids or aqueous detergents present effectively ruins the reverse micelle surfactant phase diagram, leading to aggregation. Though 15N-HSQC spectra consistent with structured protein were obtained with preparations at very low concentrations, we were unable to reach the concentrations necessary for high quality triple resonance spectroscopy. Stripping the protein of carrier lipid or aqueous detergent resulted in unfolded protein that could not be refolded in the context of the reverse micelle surfactant mixture (unpublished results). Flynn has taken this approach for a peptide oligomer where refolding is apparently not an issue (Van Horn et al., 2008). It would seem however that this strategy is not generally applicable to integral membrane proteins of significant size and topological complexity since they do not remain folded under such conditions and refolding in non-native environments can be problematic.
Figure 1.
Schematic illustration of the structure of a reverse micelle surfactant assembly hosting an integral membrane protein in a low viscosity alkane solvent. A ribbon representation of the KcsAΔC35 crystal structure (Doyle et al., 1998) is shown. Drawn with PyMol (DeLano, 2002). On the left is the aqueous detergent micelle solubilizing the integral membrane protein. A ‘hybrid’ detergent can also serve as a reverse micelle surfactant. This duality is ideally controlled by the presence of a co-surfactant. In the case of CTAB, hexanol serves to convert it to a reverse micelle surfactant. Via surfactant exchange, the integral membrane protein can be carried from the membrane to the reverse micelle surfactant system without loss of structure as shown on the right. Addition of hexanol (green) does not degrade the viscosity of the solvent (pentane in gray) indicating that it is fully incorporated into the surfactant assembly.
The key insight was to realize that the necessity of traditional aqueous detergents could be avoided by employing a special kind of surfactant: one that can act as an aqueous detergent and as a reverse micelle surfactant. Examples include lauryldimethylamine oxide (LDAO), dodecyltrimethyl ammonium bromide (DTAB), and cetyltrimethylammonium bromide (CTAB). This duality sometimes requires the addition of a co-surfactant. For example, CTAB requires the addition of hexanol as a co-surfactant to generate stable reverse micelle assemblies in alkane solvents. In some cases accessory surfactants such as double-chained dihexadecyldimethylammonium bromide (DHAB) or sodium bis(2-ethylhexyl)sulfosuccinate (AOT) may be employed. We have developed this approach with the use of the truncated KcsA potassium channel, with the basic idea illustrated in Figure 1. In this scheme the membrane protein is first solubilized in an aqueous detergent micelle. When placed into an organic solvent the detergent micelle is ‘flipped’ into a reverse micelle around the souble-domains of the protein, while its hydrophobic tails continue to protect the protein’s transmembrane domain. Here we use the large homotetrameric KcsA potassium channel to develop this approach.
The potassium channel KcsA has served as a model system for many aspects of channel biology and biophysics. A 68 kDa homotetramer from the soil bacteria Streptomyces Lividans (Schrempf et al., 1995), KcsA contains the potassium channel signature selectivity filter sequence of TVGYG (Heginbotham et al., 1992) and is a prokaryotic ancestor to Shaker and other eukaryotic channels (Derst and Karschin, 1998). The three main segments of KcsA, an outer helix, a pore region, and an inner helix supporting the selectivity filter, are homologous to regions S5, H5, and S6 in voltage gated potassium channels such as KvAP and Shaker. Like many other bacterial potassium channels, KcsA is structurally very similar to eukaryotic channels (MacKinnon et al., 1998; Ruta et al., 2003), and has been proven a robust experimental system for the study of channels from these organisms (LeMasurier et al., 2001). The 125 amino acid construct utilized in the initial crystallographic work lacks the soluble C-terminal domain and forms a stable homotetramer of 54 kDa (Doyle et al., 1998; LeMasurier et al., 2001; Walian and Jap, 2003). This protein (KcsAΔC35 ) is used here and spans residues 1 through 125. KcsA has also served as a model system for both aqueous solution NMR using detergent micelles (Baker et al., 2007; Chill et al., 2006; Takeuchi et al., 2007) and solid-state NMR using bilayers (Schneider et al., 2008).
The particular combination of surfactants that can successfully solubilize the KcsAΔC35 protein was determined empirically. The approach effectively rests on the thermodynamic hypothesis for protein folding and stability, that the lowest free energy state of the protein is the native folded structure. It is well established that the effect of denaturants and other structural perturbations, such as hydrostatic pressure and temperature, is to compress the free energy gap between the native structure and (partially) unfolded states. The idea is simply that “native like” spectra are comprised of peak counts consistent with the amino acid sequence and possess sharp resonance lines without a significant population of minor conformers. Non-native states will be characterized by line broadening, multiple conformations, low peak counts, and other spectral features consistent with the presence of interconversion between low lying states. Although not phrased this way, this is not unlike Girvin’s (Krueger-Koplin et al., 2004) and Sander’s (Sanders and Oxenoid, 2000) approach to optimization of aqueous detergent solubilization conditions of membrane proteins.
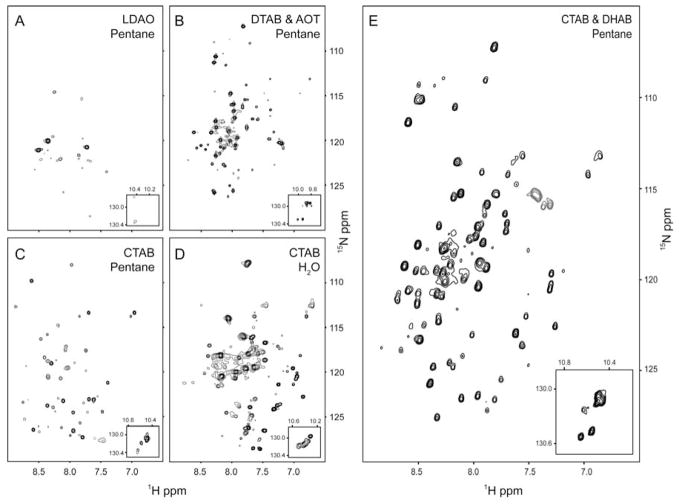
Using this guiding principle, a two-part screening procedure was utilized to determine which water-soluble surfactants (LDAO, CTAB, DTAB) gave reasonably dispersed (for a helical protein) 15N-HSQC spectra in water with an appropriate number of amide crosspeaks. We dub these ‘hybrid’ surfactants or carrier detergents. Detergent-protein samples showing the best water-micelle spectra were then dehydrated and prepared for conversion to reverse micelles. A successive screening procedure took place in which the membrane protein reverse micelle preparations were studied for crosspeak number, dispersion, and signal-to-noise. In the case of KcsAΔC35, the most-promising surfactant proved to be CTAB. Figure 2 displays a sequence of different reverse micelle surfactants that were initially attempted. Subsequent refinement of solubilization conditions included the addition of the accessory surfactant DHAB. The 15N-HSQC spectrum of KcsAΔC35 solubilized in the CTAB:DHAB:hexanol reverse micelle surfactant system is well-dispersed, though it retains the crowd of peaks about its center typical of helical membrane proteins. The spectrum is comparable in quality and dispersion to aqueous detergent preparations of KcsA (Baker et al., 2007; Chill et al., 2006; Takeuchi et al., 2007). Other attempts at encapsulation of KcsAΔC35 included the use of CTAB as a hybrid surfactant combined with the anionic double-chained surfactant AOT, as well as the zwitterionic deteregent LDAO which, despite its success forming crystals of KcsAΔC35 (Doyle et al., 1998), failed to result in spectra of sufficient quality.
Figure 2.
Surfactant optimization. 15N-HSQC spectra of KcsAΔC35 solubilized in four different reverse micelle surfactant systems. Detergents were tested as aqueous samples first before preparation as reverse micelles. All reverse micelle samples were ~0.15 mM in KcsAΔC35, optimized water loadings and solubilized in pentane. Spectra were obtained at 25 °C. Panel A: 200 mM LDAO reverse micelles in pentane with 320 mM hexanol. B: 195 mM AOT and 195 mM DTAB. C: 200 mM CTAB reverse micelles in pentane with 800 mM hexanol. E: 125 mM CTAB and 125 mM DHAB with 390 mM hexanol. Insets show the spectral region containing the Trp indole N-H correlations. Panel D shows the 15N-HSQC spectrum of KcsAΔC35 solubilized in 100 mM CTAB aqueous buffer.
Additional optimization of the CTAB:DHAB:hexanol system included a survey of water-loading (i.e. molar ratio of water to surfactants), overall surfactant levels for a balance of T2 times and signal-to-noise, variation of co-surfactant (hexanol) concentration, co-solvents (such as diethyl ether), and refinement of preparation procedures to yield consistent, reproducible 15N-HSQC spectra. This optimization process revealed that the double-tailed surfactant DHAB, mixed in a 1:1 ratio to CTAB, heightened the quality of KcsA spectra, perhaps stabilizing the homotetrameric structure’s transmembrane domains by contributing to the lateral pressure with its cylindrical structure (van den Brink-van der Laan et al., 2004). We conclude that this is an optimal sample preparation.
We interpret the sparse nature of the CTAB:AOT and LDAO preparations of KcsA as being due to an inhomogeneous population of (partially) denatured proteins that is not interconverting at a rate greater than the chemical shift time scale. The nature of the energy landscape of proteins is such that perturbations away from the native structure necessarily compress the free energy gap between partially unfolded forms. This has been observed many times in the study of protein stability using the so-called native state hydrogen exchange method (Bai et al., 1995). This leads to broadening effects in the NMR spectrum due to interconversion between states. In contrast, the 15N-HSQC spectrum of KcsAΔC35 in CTAB:DHAB is characteristic of a folded, homogenous, helical protein. As surveyed by a high resolution HNCO spectrum, all but a few residues are represented, with several more showing minor conformers in slow exchange. The spectra presented here were obtained in liquid pentane and their relatively high quality suggests that the protein has an effective molecular reorientation time (τm) of 10–15 ns. Measurement of the backbone 15N-relaxation confirms this. While the average T2 of 15N amide sites in KcsA solubilized in aqueous detergent micelles is on the order of 20 ms, those in the reverse micelle system are considerably longer with amide NH identified with residues in core regions of helical secondary structure averaging 80 ms. This is a critically important result as it suggests that the standard triple resonance experiments can be applied without the limitations imposed by extensive deuteration even in liquid pentane. Use of even lower viscosity solvents, such as propane or ethane, should significantly improve the hydrodynamic behavior of the reverse micelle assembly.
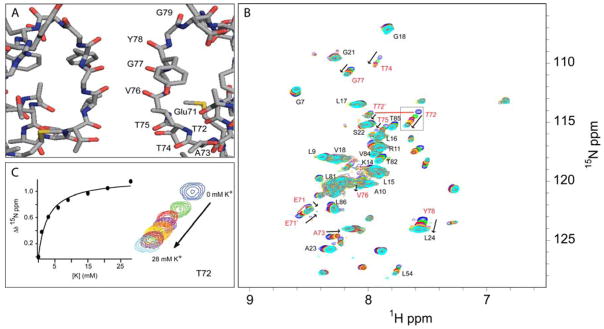
The backbone resonance assignments of the selectivity filter of KcsAΔC35 solubilized in CTAB:DHAB:hexanol in pentane have been largely completed. The highly conserved potassium channel TVGYG selectivity filter coordinates passage of potassium ions through the channel at near diffusion rates by the backbone carbonyls of 75TVGY and the sidechain hydroxyl of threonine 75 (Zhou et al., 2001). To test for both the proper tetrameric structure of KcsA and its competence in binding potassium in the reverse micelle surfactant system, we titrated potassium ions into a KcsAΔC35 sample prepared without potassium in a background of 50 mM sodium ion (Figure 3). Selective chemical shift perturbations localized to resonances associated with the selectivity filter are observed such as residues E71, T72, A73, T74, T75, V76, and G77. The chemical shift perturbations show a smooth saturation consistent with specific binding and a corresponding KD of 2.9 ± 0.1 mM for T72, assuming that all binding sites in the selectivity filter are equal. Calculations of bindings performed for T74 and G77, selectivity filter residues without appreciable overlap in HSQC spectra, have similar dissociation constants. These results match previous KcsA potassium binding studies by NMR (Chill et al., 2006) and strongly indicate that the physiologically relevant tetrameric pore structure is retained in the reverse micelle surfactant system.
Figure 3.
Functional competence. KcsAΔC35 solubilized in 210 mM 1:1:0.1 CTAB:DHAB:PG with 390 mM hexanol and a water loading of 7 in pentane at 25 °C. Panel A: Structure of KcsA’s selectivity filter with select residues labeled at their carbonyl oxygens (Doyle et al., 1998). Panel B: Overlay of eight potassium titration 15N-HSQC spectra, from 0 to 28 mM K+. Selectivity filter-associated residues are labeled in red, with select other residues assignments labeled black. The presence of multiple conformers in residues T72 and E71 are shown by red lines connecting the two sets of peaks. Panel C: Thr-72 crosspeak overlays from 0 to 28 mM potassium. The corresponding binding curve was fitted to a simple binding isotherm. Similar binding curves are seen for other selectivity filter residues.
The location of chemical shift perturbations are consistent with KcsA’s physiological role to conduct potassium and agree with crystallographic studies showing different conformations of the KcsA selectivity filter based on potassium levels (Zhou and MacKinnon, 2004). Thus it appears that the tetrameric structure, essential to the formation of the ion channel, is both preserved and functional in the reverse micelle solubilized protein. Additionally, two residues closely associated with the selectivity filter, E71 and T72, have multiple conformers in slow exchange on the NMR chemical shift time scale. It is possible that the distinct conformers of these residues reflect the fluctuation, on a timescale of less than 100 s−1, of the channel as it undergoes the transition from low- to high-potassium occupancy states (Zhou and MacKinnon, 2004). Alternatively, the presence of these conformers may represent a slow exchange between the 1,3 and 2,4 occupancy states of potassium ions in the selectivity filter’s four ion-binding sites (Zhou and MacKinnon, 2004), visible here because of the lack of a net flux through the channel in the artificial conditions of presumably identical ionic solutions on both sides of the channel. The existence of such conformers in channel-associated residues is thus expected. Other residues having slight chemical shift changes upon titration with potassium include L54 and A23 that are in regions that flank the selectivity filter. A more comprehensive analysis awaits the completion of the resonance assignments and determination of the structure of the protein.
DISCUSSION
The reverse micelle solubilization strategy outlined above appears to have been successful for the truncated form of the KcsA potassium channel used here. Importantly, the search used for optimal solubilization conditions was objective and did not require prior knowledge of the structure. The strategy simply relies on a fundamental property of proteins that arises from the thermodynamic hypothesis for protein folding and stability. This view comes from the response of the protein energy landscape to perturbations from chemical denaturants (Fuentes and Wand, 1998a) and very high hydrostatic pressure (Fuentes and Wand, 1998b) as assayed by the “native state” hydrogen exchange method (Bai et al., 1995) and low temperature unfolding studies (Babu et al., 2004). The basic approach is therefore to begin with those surfactants that are capable of the water-alkane micelle-reverse micelle switch (e.g. LDAO, CTAB) and refine by adjustment of co-surfactant-surfactant combinations with the criteria of more “native-like” 15N-HSQC spectra as the guide. Native-like spectra are characterized by an appropriate number of correlations (cross peaks), uniformity of line shape and intensity, appropriate chemical shift dispersion and so on. The ability to employ objective criteria in the refinement of solubilization conditions would begin to suggest that it could be generally applicable to integral membrane proteins. However, only empirical experience will confirm this optimistic view.
The backbone 15N relaxation at rigid sites of solubilized KcsAΔC35 dissolved in pentane are significantly longer due to faster tumbling times than those of the aqueous detergent-solubilized protein. It is anticipated that even further improvement will be obtained for preparations of the protein in liquid ethane. The macromolecular tumbling of the revere micelle particle in liquid pentane is sufficiently rapid to allow for triple resonance spectroscopy without the need for extensive deuteration. Avoidance of extensive deuteration by recombinant expression preserves access to the rich structural information contained in the 1H-1H nuclear Overhauser effect and eliminates the need to exchange amide deuterons with hydrogen. Finally, it should be noted that reverse micelle preparations of proteins are perhaps ideal NMR samples for cold probe technology in the sense that they are non-conductive and do not result in the degradation of cryogenic probe sensitivity normally associated with salty aqueous protein samples (Flynn et al., 2000).
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Full length 13C,15N-KcsA was grown using a pQE-60 plasmid vector in M15 cells in M9 minimal media. Cell cultures were grown to an OD600 of 0.8 at 37 °C, allowed to cool to room temperature, and induced with IPTG to a final concentration of 1 mM overnight at 27 °C (Chill et al., 2006). Cells pellets were resuspended in 150 mM KCl, 50 mM Tris, pH 7.5 buffer, sonicated, and then solublized with n-decyl-β-D-maltopyranoside (DM) at a concentration of 40 mM and incubated for 3 hours. Following centrifugation at 35,000 g for 45 min, the supernatant containing the His-tagged full length KcsA was bound to TALON resin (Clontech) and washed first with 500 mM KCl, 50 mM Tris, pH 7.5, 10 mM DM high salt buffer and then with 150 mM KCl, 50 mM Tris, 50 mM imidazole, pH 7.5, 10 mM DM buffer. Transfer from the cell extraction detergent DM to a different detergent was accomplished by exchanging the detergent in place on the TALON resin with the protein still bound. Initial extraction with DM and then exchange to another detergent gave better overall yields than extracting directly with another detergent. For KcsA, after the aforementioned high salt and low imidazole washes, a solution of 10 mM CTAB, 150 mM KCl, 50 mM Tris, pH 7.5 was run over the column with a 10-fold greater volume than the resin. The resin was then placed with a 10-fold excess of fresh CTAB buffer and allowed to mix overnight on a rotisserie at room temperature. The resin was repacked and washed with an additional 20-fold volume of 10 mM CTAB buffer, then with 30-fold volume of 2.5 mM CTAB buffer. The switch from a higher concentration of CTAB to a lower concentration helps avoid accumulation of excessive amounts of CTAB detergent. In the case of using a different detergent such as LDAO, it would simply be substituted for CTAB. When evaluating different detergents in the case of screening for optimal conditions, the TALON resin can be split into equal portions after protein loading and high salt and low imidazole wash steps, and rinsed and eluted with different detergent solutions in parallel.
Protein was eluted from the column with a solution of 2 mM CTAB, 150 mM KCl, 50 mM Tris, 500 mM imidazole, pH 7.5. Imidazole was removed by repetitive ultrafiltration or by overnight dialysis. The C-terminal cytoplasmic domain and His-tag were cleaved with α–chymotrypsin and purified by gel exclusion chromatography (Superdex-200) in 3 mM CTAB, 150 mM KCl and 50 mM Tris, pH 7.0, concentrated, and subsequently dialyzed against water containing 5 mM CTAB. Samples were frozen and lyophilized overnight in a small glass tube to reduce the water content to that required for optimal encapsulation.
Reverse Micelle Sample Preparation
For KcsAΔC35, typical optimized reverse micelle conditions utilized a mixture of 1:1 CTAB:DHAB with a total surfactant concentration of 200–250 mM, and a molar ratio of water to surfactant (“water loading” or W0) of 6 to 8. The aqueous buffer of a typical sample consisted of 50 mM KCl, 50 mM Bis-Tris, pH 7.0, and 50 mM Sodium 3-(trimethylsilyl)-1-propanesulfonate (DSS) for chemical shift referencing. The bulk solvent consisted of 94% volume d12-pentane. Hexanol at a volume of 6%, or about a 5:1 molar ratio of hexanol to CTAB surfactant, was added as a co-surfactant. Typically a co-surfactant such as DHAB is added to the lyophilized protein-hybrid detergent mixture, followed by the bulk solvent (i.e. pentane), hexanol, and finally aqueous buffer.
Additionally, native lipids can be added to preparations if deemed necessary. In some KcsA reveres micelle preparations this included phosphatidylglycerol (PG) (Avanti Polar Lipids) at 5% of the total molar amount of CTAB and DHAB. PG is co-purified with KcsA and is believed to be important for proper gating of the channel (Valiyaveetil et al., 2002). Addition of PG did not appreciably change HSQC spectra of KcsA in reverse micelles.
A co-surfactant such as hexanol is required for inverted conical surfactants such as CTAB and LDAO. Cylindrical surfactants such as double-chained DHAB require less hexanol, and conical surfactants such as AOT require no hexanol. It is believed that hexanol packs into the spaces of the hydrophobic tails that are generated by the reverse curvature of reverse micelles. For inverted conical surfactants these gaps are greater and require higher levels of hexanol as a co-surfactant.
KcsA Potassium Titration Experiments
Encapsulated KcsAΔC35 was prepared as described above except using 50 mM NaCl in place of 50 mM KCl for all post-TALON resin buffers, to an initial W0 of 7.0. 15N-HSQC spectra were collected between sample titrations with 50 mM bis-TRIS, pH 7.0, with differing amounts of KCl in 0.5 μL volumes that resulted in small (0.2) increases in water loading while raising the K+ levels in increments of 1 to 7 mM. This ultimately resulted in an overall water loading increase of 2 during a full titration as measured by integration of one dimensional 1H NMR spectra, which is not enough to appreciably affect the spectra or particle tumbling time.
NMR Data Collection and Analysis
All samples were prepared in liquid pentane. NMR data was collected at 25 °C in Wilmad screw cap NMR tubes at 600 MHz (1H) with a Varian Inova NMR spectrometer equipped with an early generation triple resonance cryogenic probe (EB S/N 3600:1) or a Bruker Avance III NMR spectrometer equipped with a current generation triple resonance cryogenic probe (EB S/N 5700:1). Resonance assignments for 1H, 15N, 13Cα, and 13Cβ were obtained by standard HNCACB and CBCA(CO)NH experiments (Grzesiek and Bax, 1992; Kay et al., 1994; Muhandiram and Kay, 1994; Wittekind and Mueller, 1993). DSS in the aqueous buffer was used as an internal reference (0.0 ppm). FELIX was used to process the data and Sparky (Goddard and Kneller, 2004) was used for data analysis.
Acknowledgments
This work was supported by the Mathers Foundation and NIH grant GM 085120. JMK is the recipient of a National Science Foundation Graduate Research Fellowship (2004016452) and is an NIH pre-doctoral trainee (GM 08275). CRB is the recipient of an NIH NRSA postdoctoral fellowship (GM20806). We thank Professor Benoit Roux for the KcsA expression vector and Professor Zhe Lu, Dr. Ronald W. Peterson, Dr. Sabrina Bédard, and Professor Brian G. Lefebvre for helpful discussion. AJW declares a competing financial interest as a Member of Daedalus Innovations, LLC, a manufacturer of reverse micelle NMR apparatus.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Babu CR, Hilser VJ, Wand AJ. Direct access to the cooperative substructure of proteins and the protein ensemble via cold denaturation. Nat Struct Mol Biol. 2004;11:352–357. doi: 10.1038/nsmb739. [DOI] [PubMed] [Google Scholar]
- Bai Y, Sosnick TR, Mayne L, Englander SW. Protein folding intermediates: native-state hydrogen exchange. Science. 1995;269:192–197. doi: 10.1126/science.7618079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KA, Tzitzilonis C, Kwiatkowski W, Choe S, Riek R. Conformational dynamics of the KcsA potassium channel governs gating properties. Nat Struct Mol Biol. 2007:1089–1095. doi: 10.1038/nsmb1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat Struct Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- Binks BP, Chatenay D, Nicot C, Urbach W, Waks M. Structural parameters of the myelin transmembrane proteolipid in reverse micelles. Biophys J. 1989;55:949–955. doi: 10.1016/S0006-3495(89)82893-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chill JH, Louis JM, Miller C, Bax A. NMR study of the tetrameric KcsA potassium channel in detergent micelles. Prot Sci. 2006;15:684–698. doi: 10.1110/ps.051954706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darszon A, Strasser RJ, Montal M. Rhodopsin--phospholipid complexes in apolar environments: photochemical characterization. Biochemistry. 1979;18:5205–5213. doi: 10.1021/bi00590a027. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL molecular graphics system. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- Derst C, Karschin A. Evolutionary link between prokaryotic and eukaryotic K+ channels. J Exp Biol. 1998;201:2791–2799. [PubMed] [Google Scholar]
- Doyle DA, Cabral JM, Pfuetzner RA, Kuo AL, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- Fernandez C, Hilty C, Wider G, Gunterr P, Wuthrich K. NMR structure of the integral membrane protein OmpX. J Mol Biol. 2004;336:1211–1221. doi: 10.1016/j.jmb.2003.09.014. [DOI] [PubMed] [Google Scholar]
- Flynn PF, Mattiello DL, Hill HDW, Wand AJ. Optimal use of cryogenic probe technology in NMR studies of proteins. J Am Chem Soc. 2000;122:4823–4824. [Google Scholar]
- Fuentes EJ, Wand AJ. Local dynamics and stability of apocytochrome b562 examined by hydrogen exchange. Biochemistry. 1998a;37:3687–3698. doi: 10.1021/bi972579s. [DOI] [PubMed] [Google Scholar]
- Fuentes EJ, Wand AJ. Local stability and dynamics of apocytochrome b562 examined by the dependence of hydrogen exchange on hydrostatic pressure. Biochemistry. 1998b;37:9877–9883. doi: 10.1021/bi980894o. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG. SPARKY 3. University of California; San Fransisco: 2004. [Google Scholar]
- Grzesiek S, Bax A. Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J Am Chem Soc. 1992;114:6291–6293. [Google Scholar]
- Heginbotham L, Abramson T, MacKinnon R. A functional connection between the pores of distantly related ion channels as revealed by mutant K+ channels. Science. 1992;258:1152–1155. doi: 10.1126/science.1279807. [DOI] [PubMed] [Google Scholar]
- Hiller S, Garces RG, Malia TJ, Orekhov VY, Colombini M, Wagner G. Solution structure of the integral human membrane protein VDAC-1 in deteregent micelles. Science. 2008;321:1206–1210. doi: 10.1126/science.1161302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay LE, Xu GY, Yamazaki T. Enhanced-sensitivity triple-resonance spectroscopy with minimal H2O saturation. J Magn Reson Ser A. 1994;109:129–133. [Google Scholar]
- Krueger-Koplin RD, Sorgen PL, Krueger-Koplin ST, Rivera-Torres AO, Cahill SM, Hicks DB, Grinius L, Krulwich TA, Girvin ME. An evaluation of detergents for NMR structural studies of membrane proteins. J Biomol NMR. 2004;28:43–57. doi: 10.1023/B:JNMR.0000012875.80898.8f. [DOI] [PubMed] [Google Scholar]
- LeMasurier M, Heginbotham L, Miller C. KcsA: it’s a potassium channel. J Gen Physiol. 2001;118:303–314. doi: 10.1085/jgp.118.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKinnon R, Cohen SL, Kuo AL, Lee A, Chait BT. Structural conservation in prokaryotic and eukaryotic potassium channels. Science. 1998;280:106–109. doi: 10.1126/science.280.5360.106. [DOI] [PubMed] [Google Scholar]
- Muhandiram DR, Kay LE. Gradient-enhanced triple-resonance three-dimensional NMR experiments with improved sensitivity. J Magn Reson Ser B. 1994;103:203–216. [Google Scholar]
- Oxenoid K, Chou JJ. The structure of phospholamban pentamer reveals a channel-like architecture in membranes. Proc Nat Acad Sci USA. 2005;102:10870–10875. doi: 10.1073/pnas.0504920102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Mrse AA, Nevzorov AA, Meseleh MF, Oblatt-Montal M, Montal M, Opella SJ. Three-dimensional structure of the channel-forming trans-membrane domain of virus protein “u” (Vpu) from HIV-1. J Mol Biol. 2003;333:409–424. doi: 10.1016/j.jmb.2003.08.048. [DOI] [PubMed] [Google Scholar]
- Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Nat Acad Sci USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RW, Lefebvre BG, Wand AJ. High-resolution NMR studies of encapsulated proteins in liquid ethane. J Am Chem Soc. 2005a;127:10176–10177. doi: 10.1021/ja0526517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RW, Pometun MS, Shi Z, Wand AJ. Novel surfactant mixtures for NMR spectroscopy of encapsulated proteins dissolved in low-viscosity fluids. Prot Sci. 2005b;14:2919–2921. doi: 10.1110/ps.051535405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan VR, Darszon A, Montal M. A small angle x-ray scattering study of a rhodopsin-lipid complex in hexane. J Biol Chem. 1983;258:4857–4860. [PubMed] [Google Scholar]
- Ruta V, Jiang Y, Lee A, Chen J, MacKinnon R. Functional analysis of an archaebacterial voltage-dependent K+ channel. Nature. 2003;422:180–185. doi: 10.1038/nature01473. [DOI] [PubMed] [Google Scholar]
- Sanders CR, Oxenoid K. Customizing model membranes and samples for NMR spectroscopic studies of complex membrane proteins. Biochim Biophys Acta. 2000;1508:129–145. doi: 10.1016/s0005-2736(00)00308-4. [DOI] [PubMed] [Google Scholar]
- Schneider R, Ader C, Lange A, Giller K, Hornig S, Pongs O, Becker S, Baldus M. Solid-state NMR spectroscopy applied to a chimeric potassium channel in lipid bilayers. J Am Chem Soc. 2008;130:7427–7435. doi: 10.1021/ja800190c. [DOI] [PubMed] [Google Scholar]
- Schrempf H, Schmidt O, Kummerlen R, Hinnah S, Muller D, Betzler M, Steinkamp T, Wagner R. A prokaryotic potassium ion channel with two predicted transmembrane segments from Streptomyces lividans. EMBO J. 1995;14:5170–5178. doi: 10.1002/j.1460-2075.1995.tb00201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K, Takahashi H, Kawano S, Shimada I. Identification and characterization of the slowly exchanging pH-dependent conformational rearrangement in KcsA. J Biol Chem. 2007:15179–15186. doi: 10.1074/jbc.M608264200. [DOI] [PubMed] [Google Scholar]
- Valiyaveetil FI, Zhou Y, MacKinnon R. Lipids in the structure, folding, and function of the KcsA K+ channel. Biochemistry. 2002;41:10771–10777. doi: 10.1021/bi026215y. [DOI] [PubMed] [Google Scholar]
- van den Brink-van der Laan E, Chupin V, Killian JA, de Kruijff B. Stability of KcsA tetramer depends on membrane lateral pressure. Biochemistry. 2004;43:4240–4250. doi: 10.1021/bi036129d. [DOI] [PubMed] [Google Scholar]
- Van Horn WD, Ogilvie ME, Flynn PF. Use of reverse micelles in membrane protein structural biology. J Biomol NMR. 2008;40:203–211. doi: 10.1007/s10858-008-9227-5. [DOI] [PubMed] [Google Scholar]
- Walian P, Jap BK. A new era in membrane channel biology. Structure. 2003;11:1467–1468. doi: 10.1016/j.str.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Wand AJ, Ehrhardt MR, Flynn PF. High-resolution NMR of encapsulated proteins dissolved in low-viscosity fluids. Proc Nat Acad Sci USA. 1998;95:15299–15302. doi: 10.1073/pnas.95.26.15299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittekind M, Mueller L. HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the alpha- and beta-carbon resonances in proteins. J Magn Reson Ser B. 1993;101:201–205. [Google Scholar]
- Zhou Y, Cierpicki T, Flores Jimenez RH, Lukasik SM, Ellena JF, Cafiso DS, Kadokura H, Beckwith J, Bushweller JH. NMR solution structure of the integral membrane enzyme DsbB: Functional insights into DsbB-catalyzed disulfide bond formation. Mol Cell. 2008;31:896–908. doi: 10.1016/j.molcel.2008.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, MacKinnon R. Ion binding affinity in the cavity of the KcsA potassium channel. Biochemistry. 2004;43:4978–4982. doi: 10.1021/bi049876z. [DOI] [PubMed] [Google Scholar]