Abstract
Development of a live pediatric vaccine against human respiratory syncytial virus (RSV) is complicated by the need to immunize young infants and the difficulty in balancing attenuation and immunogenicity. The ability to introduce desired mutations into infectious virus by reverse genetics provides a method for identifying and designing highly defined attenuating mutations. These can be introduced in combinations as desired to achieve gradations of attenuation. Attenuation is based on several strategies: multiple independent temperature-sensitive point mutations in the polymerase, a temperature-sensitive point mutation in a transcription signal, a set of non–temperature-sensitive mutations involving several genes, deletion of a viral RNA synthesis regulatory protein, and deletion of viral IFN α/β antagonists. The genetic stability of the live vaccine can be increased by judicious choice of mutations. The virus also can be engineered to increase the level of expression of the protective antigens. Protective antigens from antigenically distinct RSV strains can be added or swapped to increase the breadth of coverage. Alternatively, the major RSV protective antigens can be expressed from transcription units added to an attenuated parainfluenza vaccine virus, making a bivalent vaccine. This would obviate the difficulties inherent in the fragility and inefficient in vitro growth of RSV, simplifying vaccine design and use.
Keywords: paramyxoviruses, pediatric vaccines, recombinant DNA vaccines, respiratory infections, vaccine development
Viral reverse genetics involves producing infectious virus in cell culture completely from cloned cDNAs (1, 2). By introducing desired changes into the cDNA by standard recombinant DNA methods, one can engineer the virus and produce “designer” live viral vaccines. This article describes how human respiratory syncytial virus (RSV) presents a particularly difficult challenge to vaccine development, and how reverse genetics can be applied to this problem.
RSV is the most important cause of serious acute respiratory tract illness in infants and children worldwide (3, 4). Nearly everyone worldwide has been infected at least once by the age of 2 years, with illness ranging from a cold to bronchiolitis or pneumonia. This represents a tremendous disease burden; in the United States alone, 75,000 to 125,000 infants are hospitalized yearly for RSV disease (5). A safe and immunogenic RSV vaccine would have a tremendous beneficial impact on human health worldwide but is not yet available.
RSV MOLECULAR BIOLOGY
RSV is an enveloped, RNA-containing, cytoplasmic virus that is classified in the Paramyxovirus family of the nonsegmented negative-strand viruses (Mononegavirus order) (3). Its genome (Figure 1) is a single negative-sense strand of RNA of 15.2 kb that is tightly coated by the viral nucleocapsid N protein. The genome contains 10 genes that are transcribed sequentially from the 3′ end by a virally encoded polymerase to yield 10 separate mRNAs. The production of each mRNA is guided by short transcription signals (called gene-start and gene-end signals) that flank each gene. RNA replication involves the production of a complete positive-sense copy called the antigenome that serves, in turn, as the template for progeny genomes.
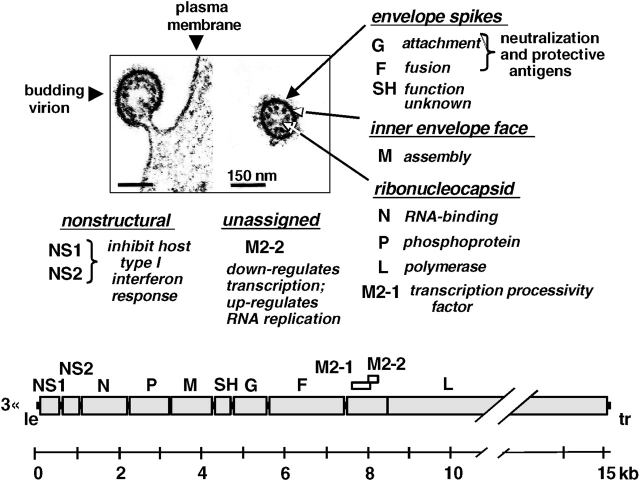
Figure 1.
Respiratory syncytial virus (RSV) genome and gene products. The electron micrographs (50) show an RSV virion budding from the plasma membrane of an infected cell (left) and a free virion (right), with the viral proteins indicated. The viral genome is diagramed at the bottom. Each large shaded rectangle indicates a gene encoding a separate mRNA. The two overlapping open reading frames (ORF) of the M2 mRNA are indicated. “Le” and “tr” refer to leader and trailer, respectively, which are small extragenic regions at each end of the genome. The leader region contains the genome promoter. The amino acid lengths of the RSV proteins are: NS1, 139; NS2, 124; N, 391; P, 241; M, 256; SH, 64; G, 298; F, 574; M2-1, 194; M2-2, 90; L, 2,165.
The viral polymerase consists of the polymerase L protein, phosphoprotein P, and transcription elongation factor M2-1 protein (3, 6). The mRNA that encodes the M2-1 protein has a second open reading frame that encodes a small protein, M2-2, which helps regulate RNA synthesis. The viral envelope is derived from the plasma membrane of the host cell during budding and contains three virally encoded transmembrane surface proteins: the attachment G protein, the fusion F protein, and the small hydrophobic SH protein of unknown function. There is an internal matrix M protein involved in viral assembly and two nonstructural proteins, NS1 and NS2, which have been shown to antagonize the host cell IFN α/β system (7, 8). The G and F proteins are the neutralization and major protective antigens.
An RSV vaccine likely would be administered at the same time as live intranasal vaccines that are presently being developed against several related paramyxoviruses that are also important agents of pediatric respiratory tract disease, namely, human parainfluenza virus (HPIV) serotypes 1, 2, and 3 (HPIV1, 2, and 3) and human metapneumovirus (9, 10). These are being developed following some of the same general principles described here.
RSV VACCINES
The peak of the most serious RSV disease is at 2–6 months of age, and so vaccination should initiate within the first few weeks of life. A major obstacle to developing a pediatric RSV vaccine is that immune responses in the young infant are reduced compared to older children and adults. One factor is immunologic immaturity, an effect that is evident through the first year of life and especially the first 6 months (11). A second factor is the immunosuppressive effect of maternally derived serum antibodies, with the greatest impact in the first few months of life (12). In addition, evaluation of pediatric vaccines is multistep for safety reasons, and involves progression from adults to seropositive children to seronegative children to RSV-naive infants of 1 to 2 months of age, approximating the target population (13, 14).
Research to develop a pediatric RSV vaccine presently is focused on live attenuated strains for intranasal administration. Live intranasal vaccines induce local mucosal as well as systemic immunity. The intranasal route partially escapes the immunosuppressive effects of maternal serum antibodies (15). Clinical experience with influenza virus vaccines indicates that primary immunization with a live intranasal vaccine is more immunogenic and more broadly protective than with an inactivated vaccine (16). Finally, primary immunization of infants or experimental animals with nonreplicating killed-virus or purified-protein RSV vaccines can prime for immune-mediated enhanced disease upon subsequent exposure to RSV (3). This unexpected phenomenon was first observed in clinical trials of a formalin-inactivated RSV vaccine in the 1960's (3). Vaccine-enhanced RSV disease seems to be peculiar to the use of killed-virus or purified-protein vaccines in primary immunization. It appears to be due to a combination of factors, including the induction of poorly neutralizing antibodies due to denatured antigen, as well as induction of an imbalanced cell-mediated response due to the inactivated nature of the vaccine. Enhanced disease is not associated with infection by natural exposure or with attenuated RSV vaccine candidates, representing an important advantage of a live pediatric RSV vaccine strategy.
A number of live-attenuated RSV vaccine candidates were developed over several decades by conventional methods (13, 17, 18). The most promising set of candidates was developed by serial passage at increasingly suboptimal temperature (cold passage [cp]) followed by chemical mutagenesis and screening to identify temperature-sensitive (ts) mutants. Preclinical and clinical analysis of a cpRSV derivative and a number of further cptsRSV derivatives showed that they were attenuated but retained an unacceptable level of residual virulence (13, 18). The most extensively characterized candidate, called cpts248/404, caused nasal congestion when evaluated in 1- to 2-month-old infants (13). However, this study provided encouraging evidence that the vaccine virus replicated and was immunogenic in this young age group and induced protection against a second dose of vaccine virus administered 2 months later.
The experience over several decades of evaluating RSV vaccine candidates in seronegative chimpanzees (the animal model that most closely resembles humans with regard to RSV replication and disease) and humans indicates that attenuation of RSV correlates with a reduction in the level of virus replication, monitored by measuring virus titers in nasal secretions. These studies also showed that a moderate degree of replication is necessary for a live RSV vaccine to be satisfactorily immunogenic. Thus, a vaccine must achieve a balance between attenuation and immunogenicity, and this window appears to be narrow.
REVERSE GENETICS
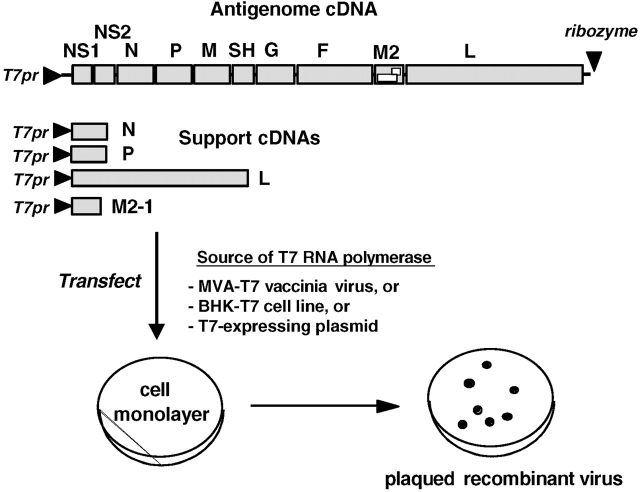
The laborious hit-or-miss conventional methods for developing attenuated strains have now been superseded by reverse genetics. The recovery of RSV from cDNA (Figure 2) requires coexpression in cultured cells of five viral components from transfected plasmids, namely (1) a complete copy of the viral RNA genome and (2) the N, P, M2-1, and L proteins (1). These are the constituents of the nucleocapsid and polymerase complex, which is the minimum unit of infectivity for the mononegaviruses. These cDNAs are engineered to be expressed by bacteriophage T7 RNA polymerase. For the recovery of viruses that will be evaluated in humans, the polymerase is supplied by a cotransfected eukaryotic expression plasmid, and recovery is performed in Vero cells that are approved for human product use. The expressed viral components assemble and launch a productive infection. This yields infectious recombinant virus that, thereafter, is indistinguishable from biologically derived virus except for whatever mutations might have been intentionally introduced. The applications of reverse genetics in designing an RSV vaccine are outlined in Table 1.
Figure 2.
Recovery of infectious recombinant RSV entirely from cloned cDNAs. The five cDNAs are indicated by shaded rectangles and encode a positive-sense copy of the viral genome (antigenome) and mRNAs for the N, P, L, and M2-1 proteins of the nucleocapsid/polymerase complex. Expression of the cDNAs is under the control of a promoter for bacteriophage T7 RNA polymerase (T7pr). The plasmid backbones that bear the cDNAs are not shown. The indicated ribozyme encoded at the downstream end of the antigenome executes self-cleavage to produce a correct 3′ end. The T7 RNA polymerase can be provided intracellularly using one of the three indicated strategies.
TABLE 1.
Applications of reverse genetics to developing a respiratory syncytial virus vaccine
| 1. Develop a panel of defined attenuating mutations | |
| a. Identify mutations in existing attenuated strains | |
| b. Develop new mutations empirically | |
| 2. Assemble desired combinations of mutations | |
| 3. Increase genetic stability | |
| a. Gene deletions | |
| b. Increase the number of attenuating mutations | |
| c. Choose “attenuating” codons that cannot readily revert | |
| 4. Increase the level of expression of the G and F protective antigens | |
| a. Promoter-proximal G and F genes | |
| b. The M2-2 deletion upregulates gene expression | |
| 5. Increased breadth of coverage against heterologous RSV strains with added or swapped genes | |
| 6. Use an attenuated HPIV vaccine virus to vector the protective antigens of RSV |
Definition of abbreviations: HPIV = human parainfluenza virus; RSV = respiratory syncytial virus.
IDENTIFYING MUTATIONS THAT ATTENUATE RSV
The first need is to develop a panel of mutations that attenuate RSV. These would then be used to systematically engineer vaccine candidates. A necessary requirement of such mutations is that they should not reduce virus yield in vitro, because RSV does not replicate to high titer and a reduction in replication fitness in vitro would interfere with vaccine manufacture. For example, the biologically derived cpts viruses described above could be grown efficiently in vitro at the reduced, permissive temperature of 32°C, whereas they were highly attenuated at the higher human body temperature of 37°C.
The existing biologically derived vaccine candidates mentioned above proved to be a rich source of attenuating mutations. Complete consensus genome sequences were determined for cpRSV and six cptsRSV derivatives. Mutations involved in attenuation were identified by insertion into recombinant virus and evaluation in experimental animals. CpRSV contained five amino acid substitutions in three proteins (Val-267-Ile in N, Glu-218-Ala and Thr-523-Ile in F, and Cys-319-Tyr and His-1690-Tyr in L) that, in combination, are responsible for its attenuation phenotype (19). Analysis of the other viruses identified six point mutations, each of which is an independent ts and attenuation mutation (20–22). Five of these mutations involved amino acid substitutions in the L protein (mutation 248 involved the substitution Gln-831-Leu, mutation 530 involved Phe-521-Leu, mutation 955 involved Asn-43-Ile, mutation 1009 involved Met-1169-Val, and mutation 1030 involved Tyr-1321-Asn). The sixth mutation involved a nucleotide substitution in the gene-start transcription signal of the M2 gene.
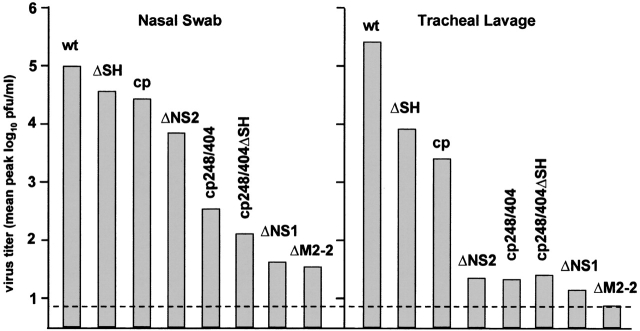
Additional attenuating mutations have been identified empirically. One method was to identify and delete nonessential viral accessory genes (23–28). Although deletion of many of the viral genes would be expected to be lethal, it was possible to delete the coding sequences for 5 of the 11 RSV proteins individually or in various combinations and nonetheless recover infectious virus; the five dispensable proteins are: NS1, NS2, SH, G, and M2-2. Each of these gene-deletion viruses replicated in cell culture to a titer comparable to that of wild-type RSV, but each was attenuated in experimental animals. In particular, evaluation of the single-gene deletion viruses in seronegative chimpanzees showed that they exhibited a range of attenuation (Figure 3). The ΔSH and ΔNS2 viruses were less attenuated than the cpts248/404 benchmark virus (representing a vaccine candidate that needs a small amount of additional attenuation) and would be suitable to combine with other attenuating mutations. The ΔNS1 and ΔM2-2 viruses were somewhat more attenuated than cpts248/404 and are suitably attenuated for clinical evaluation without further modification. Each of the viruses was highly immunogenic in chimpanzees and induced a high level of protection against challenge with wild-type RSV. The remaining single-gene-deletion virus, ΔG, was highly attenuated in mice (23), but was not evaluated in chimpanzees—it will not be developed as a vaccine candidate because G is a major protective antigen and should be present in an RSV vaccine.
Figure 3.
Mean peak virus titers present in nasopharyngeal swabs (left panel) and tracheal lavages (right panel) of chimpanzees that had been infected simultaneously by the intranasal and intratracheal routes with the indicated recombinant wild-type or attenuated RSVs. The cp248/404 virus is a recombinant version of the biologically derived cpts248/404 virus that retained mild residual virulence in RSV-naive infants and serves as a benchmark. The dotted line indicates the limit of detectability.
A number of other strategies are available to attenuate RSV; these will be mentioned only briefly, as they are not part of immediate plans to develop RSV vaccine candidates. One strategy is to delete only part of a protein, as exemplified with the M2-1 protein. M2-1 appears to be an essential protein, and deletion of the complete gene has not been possible. However, it was possible to recover virus in which up to 67 amino acids were deleted from the C-terminus of M2-1 (29). These viruses were attenuated in rodents.
Another strategy is to target a specific protein and replace charged amino acids with a noncharged amino acid, such as alanine. This is a common recombinant strategy to develop mutant proteins. This was done with the L protein, and a number of such mutations were successfully recovered in virus (30). Some of these were attenuated, thus adding to panel of point mutations available to attenuate RSV.
Another strategy for attenuation involves using host range-restriction elements in bovine RSV (BRSV). BRSV is the bovine counterpart of RSV and shares considerable antigenic and sequence relatedness (24). However, BRSV is highly attenuated in primates, reflecting a natural host range restriction. Using reverse genetics, the G and F genes of BRSV were replaced by their counterparts of human RSV, thereby combining the attenuated backbone of BRSV with the antigenic determinants of human RSV (24). This initial construct retained too much of the host range restriction of BRSV, and did not replicate sufficiently well in chimpanzees to be immunogenic. In future work, additional BRSV genes in this construct will be replaced with their human RSV counterparts in an effort to increase the replication and immunogenicity in primates. A similar strategy has been successful for HPIV3, which has a bovine counterpart, bovine parainfluenza virus (BPIV) type 3 that is attenuated in primates. Combining the HPIV3 antigenic determinants with backbones containing one or more BPIV3 genes has generated several attenuated derivatives suitable for clinical evaluation (31, 32).
The F protein of RSV and, indeed, of all paramyxoviruses is synthesized as a precursor that must be cleaved by a cellular protease in order to be active (3). The protease specificity and the efficiency of cleavage are determined by the sequence at the cleavage site (in the case of RSV, two cleavage sites are involved [33]), representing a determinant of virulence. Engineering one or both of these sites into forms that are less-easily cleaved would be expected to attenuate the virus, although this strategy remains to be investigated.
Mononegaviruses also can be attenuated by altering the order of the viral genes in the genome (34). Gene transcription by this type of virus has a polar gradient, such that genes that are proximal to the 3′ promoter are expressed more efficiently than downstream genes. Because the natural gradient of gene expression likely is optimal for the virus, rearrangement of the genes might yield suboptimal ratios of expression and attenuate the virus. When tested with vesicular stomatitis virus (a relative of rabies virus), this indeed proved to be the case, and attenuated derivatives were successfully produced (34). This method could be used for RSV provided that it does not reduce the yield of virus in cell culture.
The RSV G protein is produced as the full-length membrane-anchored form, as well as a truncated secreted form. The secreted form may induce oversensitization of T helper subset 2 lymphocytes, a response implicated in immune-mediated pathology (35). The G protein also contains a CX3C motif that appears to mimic the activity of the chemokine fractalkine (36). This mimicry has the potential to alter leukocyte trafficking, although exactly how this affects RSV biology remains to be defined. These are examples of RSV structural features that can be deleted from infectious virus by reverse genetics (23, 37) and might make vaccines that are more attenuated and/or less reactogenic. However, we first need a clear understanding of the roles and impact of these features on RSV immunobiology and pathogenesis.
THE FIRST RECOMBINANT VACCINE CANDIDATES
Once a panel of attenuating mutations has been identified, they can be inserted in desired combinations into the viral genome to obtain attenuated derivatives. The first recombinant vaccine candidate was made by combining the mutations found in the biologically derived cpts248/404 virus—namely, the cp, 248, and 404 mutations—with deletion of the SH gene. The resulting virus, rcp248/404ΔSH, seemed to be marginally more attenuated than its cpts248/404 parent when evaluated in seronegative chimpanzees (Figure 3) (note that the italicized “cp” and “ts” in the names of the biologically derived mutants refer to phenotypes, whereas the names of the recombinant viruses indicate mutations but not phenotypes and are not italicized). However, when rcp248/404ΔSH was evaluated in adults, seropositive children, and seronegative children, it was not distinguishable from its cpts248/404 parent (14). Thus, whereas the ΔSH mutation was attenuating on its own in mice and chimpanzees, it did not make a significant contribution to increasing the level of attenuation in humans in the context of the cp, 248, and 404 mutations. This illustrates that the level of attenuation of a given combination of mutations is not necessarily the exact sum of the individual mutations, and must be determined empirically.
The rcp248/404ΔSH virus was further modified by the insertion of the 1030 point mutation into the L gene, yielding the derivative rcp248/404/1030ΔSH (Figure 4) (14). This virus thus had five attenuating genetic elements: cp, 248, 404, ΔSH, and 1030. This virus was more ts than its predecessor. It also proved to be more attenuated when evaluated in seronegative children, and was deemed sufficiently safe to be evaluated in 1- to 2-month-old infants (14). In this population, the vaccine virus was infectious and well tolerated. A significant serum antibody response was detected in 57% of infants aged 6–24 months but in only 19% of those aged 1–2 months, reflecting the reduced immune response characteristic of very young infants as noted above. Nonetheless, replication of a second dose of vaccine administered within 2 months was highly restricted (14). This indicated that protective immunity had been induced even if the specific correlates of immunity were not identified.
Figure 4.
Current live recombinant RSV vaccine candidates based on attenuated RSV (A–D) or on the attenuated bovine/human parainfluenza virus (B/HPIV) type 3 virus expressing RSV F from an inserted gene (drawn immediately over its site of insertion). RSV genes and ORFs are represented by lightly shaded rectangles, BPIV3 genes are represented by open rectangles, and HPIV3 genes are represented by darkly shaded rectangles. Gene deletions are demarcated with dotted vertical lines. The locations of attenuating cp, 248, 404, and 1030 mutations are indicated.
However, there was some evidence of genetic instability. The rcp248/404/1030ΔSH virus is highly ts and does not form plaques above 34–35°C, compared with wild-type RSV that readily forms plaques at 40°C. Evaluation of 141 nasal wash viral specimens from vaccines identified 45 (32%) that exhibited a 1–3°C upward shift in maximum plaquing temperature (14). Sequence analysis of nine of these direct clinical specimens showed that six had sustained reversion of a single attenuating mutation, either 248 or 1030 (14). The “shifted” viruses retained at least four of the five attenuating genetic elements and remained highly attenuated. Consistent with this, the shifted viruses were not associated with illness or increased virus shedding in vaccines (14). However, it would be desirable to obtain a greater degree of genetic stability.
INCREASED GENETIC/PHENOTYPIC STABILITY
One strategy for designing a vaccine virus with increased genetic/phenotypic stability would be to include a highly attenuating gene deletion, such as ΔNS1, ΔNS2, or ΔM2-2. A gene deletion should be highly refractory to reversion, and the virtual lack of RNA recombination for mononegaviruses is further assurance against regaining a deleted gene (38).
A second expedient would be to put as many attenuating mutations into a vaccine candidate as possible, consistent with retaining a satisfactory level of immunogenicity. Adding more attenuating mutations to a vaccine candidate does not always increase its level of attenuation. This was exemplified by the addition of the ΔSH mutation to the cp248/404 background, as described above, in which the attenuating effect of ΔSH was not additive to that of the cp248/404 backbone. However, the presence of the ΔSH mutation would essentially preclude reversion to wild-type virus, and its attenuating affect should become unmasked if one or more of the other attenuating mutations were lost.
The attenuation phenotype associated with amino acid point mutations also can be “stabilized” by judicious choice of the codon that is engineered into the vaccine candidate. Each of the amino acid point mutations identified in the biologically derived vaccine candidates described above involved a single nucleotide substitution. Because the mutation rate of RSV and other mononegaviruses is approximately 10−4 (39), reversion can readily occur. However, a strategy has been developed to optimize the genetic stability of an amino acid point mutation, exemplified with a ts attenuating point mutation in the L protein of HPIV1 (40). The first step is to construct recombinant viruses representing all 20 possible amino acid assignments at the locus of interest. Among the assignments that yield viable virus, the next step is to characterize their attenuation phenotype. Next, all of the possible codons encoding attenuating versus nonattenuating assignments are examined on paper, taking into account the degeneracy of the genetic code. The aim is to identify an “attenuating” codon that differs by two or preferably three nucleotides from any possible “nonattenuating” codon. This provides for stabilization of the phenotype, as the estimated frequency with which an “attenuating” codon could change to a “nonattenuating” codon decreases dramatically with the number of nucleotides necessary (the estimated rate of single, double, or triple nucleotide substitution is 10−4, 10−8, or 10−12, respectively).
Increased phenotypic and genetic stability was confirmed for the HPIV1 mutation by monitoring the stability of the ts phenotype and the sequence of the point mutation during passage in vitro at increasing temperature: reversion was readily observed when only a single change was needed, but was not observed when three were needed. This strategy also offers the possibility of optimization of the attenuation phenotype, because alternative amino acid substitutions sometimes offer increased (or decreased) attenuation phenotypes, which might provide for fine-tuning. However, in some situations, as exemplified by a second mutation in HPIV1 L (40), there were so many “nonattenuating” codon possibilities that stabilization was limited. Also, stabilization of a mutant locus does not preclude second-site mutations that might alter phenotype. Given the high mutation rate and genetic variability of RNA viruses, it is unrealistic to expect that absolute genetic stability can be achieved for any vaccine virus.
IMPROVED VACCINE IMMUNOGENICITY?
Attenuated vaccine viruses typically are less immunogenic than the wild-type parent, which likely is due, in large part, to reduced replication and antigen production in vivo.
One strategy to increase the level of expression of G and F protective antigens is to move their genes so as to be immediately downstream of the viral promoter. This was done for F and G individually and in combination, and resulted in two- to threefold increase in their synthesis without changing the virulence of the virus (41). Although this is a modest effect, it would be an important improvement if it were reflected in a comparable increase in immunogenicity in young infants. A permutation of this strategy is to insert additional promoter-proximal copies of the G and F genes while leaving the original copies in their original locations. When this was done for recombinant human metapneumovirus, there was an increase of 6- to 14-fold in the expression of mRNA, although the effect on protein expression remains to be investigated (42). It might also be possible to achieve higher levels of expression by reconstructing the F and G genes with codon choices optimized for efficient translation.
Another way to increase antigen expression was identified serendipitously with the ΔM2-2 deletion (26). In cell culture, this virus exhibits a substantial delay and decrease in RNA replication—which likely is the basis for its attenuation in vivo—and a substantial increase in gene transcription. Thus, M2-2 appears to be involved in the balance between transcription and RNA replication. The increase in transcription was associated with a concomitant increase in the expression of viral antigen, including the G and F proteins. The phenotype of attenuation coupled with increased gene expression is highly desirable for a vaccine virus.
Deletion of the NS1 and/or NS2 genes offers another way to increase immunogenicity. As noted above, these proteins inhibit the host IFN α/β response, which otherwise stimulates the innate and adaptive immune responses. Augmented IFN production associated with the ΔNS1 and ΔNS2 mutations might provide for increased immunogenicity. This would be highly desirable, and, indeed, recombinant BRSV in which both the NS1 and NS2 genes were deleted exhibited increased immunogenicity in calves (43).
BROADENING VACCINE COVERAGE BY ADDING AND SWAPPING GENES
RSV has a single serotype with two antigenic subgroups (RSV-A and -B). These have extensive sequence and antigenic heterogeneity in the G glycoprotein, whereas the F glycoprotein exhibits a substantially greater level of sequence and antigenic conservation (3). Cross-protection between the HRSV antigenic subgroups is incomplete, and, ideally, both subgroups should be represented in a vaccine, at least for the G protein. One strategy to achieve this was to modify the present RSV-A recombinant virus to express the RSV-B G protein from an added gene, such that G of both subgroups is represented in a single virus (44). Another strategy was to replace the G and F protective antigens of the present recombinant RSV-A virus with their counterparts from RSV-B (45). This virus replicated with wild-type-like efficiency in chimpanzees, and could be attenuated with the panel of mutations developed for the RSV-A background (45). Thus, the resulting RSV vaccine would have two components: an attenuated RSV-A virus and an attenuated RSV-A/RSV-B chimera. A third strategy would be to develop a complete recombinant RSV-B virus and develop a panel of attenuating mutations based on those identified for RSV-A.
NEW RECOMBINANT VACCINE CANDIDATES
The ability to produce vaccine candidates by reverse genetics far outstrips the capacity to evaluate them clinically. Of necessity, the process involves evaluating a few carefully chosen candidates at a time. The five current candidates described below represent a variety of strategies: point mutations in RNA synthetic machinery, ts and non-ts mutations, deletion of IFN antagonists, alteration of the viral RNA synthetic program, and the use of vectored antigens.
One candidate (Figure 4A) is a version of the above-mentioned rcp248/404/1030ΔSH virus, in which the 248 and 1030 amino acid point mutations will be “stabilized” by the strategy described above, a modification that is presently in progress. In a respiratory virus, the ts phenotype is thought to preferentially restrict replication in the lower, warmer respiratory tract, thereby conferring increased safety.
A second candidate (Figure 4B) is virus in which the NS1 gene has been deleted; namely, rΔNS1. As noted above, the rΔNS1 virus might be satisfactorily attenuated without further modification—this can only be determined in clinical studies. Deletion of this IFN antagonist might provide for increased immunogenicity, as already noted. Conversely, this attenuation strategy would be dependent on the host IFN system, raising issues concerning the IFN competency of young infants as well as natural diversity in the population.
A third vaccine candidate (Figure 4C) is the rΔM2-2 virus, lacking the M2-2 open reading frame of the M2 mRNA. This virus was similar to rΔNS1 in its degree of attenuation in chimpanzees, and thus also might be satisfactorily attenuated without further modification. As noted above, the increased expression of viral protein associated with the ΔM2-2 mutation also might provide for increased immunogenicity.
A fourth candidate (Figure 4D) is a mutant called r248ΔNS2. This combines the ΔNS2 mutation, which, by itself, was insufficiently attenuating in chimpanzees, with a single attenuating point mutation, 248, which will be “stabilized” as described above. This virus combines the potential of increased immunogenicity associated with deletion of an IFN antagonist and the greater restriction in the lower respiratory tract associated with a ts mutation.
The fifth candidate (Figure 4E) involves using a B/HPIV3-vectored vaccine, as described below.
VECTORING TO COMBINE PEDIATRIC VACCINE VIRUSES
One of the major challenges of preparing a live RSV vaccine is that the virus does not grow robustly in cell culture and can readily lose infectivity. This might prove to be a major obstacle to vaccine formulation, storage, and administration. To obviate these limitations, we took advantage of the substantially greater growth and stability of the HPIVs to use them as vectors to express the RSV G and F protective antigens from added transcriptional units. The expression of RSV antigens from an attenuated HPIV makes a bivalent pediatric vaccine virus.
For example, we used B/HPIV3, an attenuated HPIV3 vaccine candidate, as a vector to express RSV G and F individually or in combination. B/HPIV3 consists of the attenuated BPIV3 backbone, in which the fusion F and hemagglutinin-neuraminidase HN protective antigen genes have been replaced with their counterparts from HPIV3 (Figure 4E) (32). The magnitude of expression of the vectored RSV genes in vitro was greater than that of RSV, especially for the single-gene inserts. The resulting viruses were suitably attenuated and highly immunogenic against both viruses when evaluated in rhesus monkeys (46). The inserted genes were surprisingly stable and suitable for vaccine manufacture. This bivalent vaccine would be suitable for administration to infants aged 1 to 5 months because vaccination against both RSV and HPIV3 should begin at this time.
HPIV1 and HPIV2 also can serve as attenuated vectors for expressing RSV G and F. Wild-type HPIV1 has already been used to express the human metapneumovirus F protein (47), and attenuated versions of HPIV1 and HPIV2 are being developed (48, 49). HPIV1 and HPIV2 infect and cause disease beginning at approximately 6 months of age and, thus, these vectors would be ideal for use beginning at 5 or 6 months of age. In this capacity, HPIV1 or 2 vectors expressing RSV antigens could be used to boost earlier immunization using attenuated RSV or B/HPIV3-RSV. In summary, this strategy employs the more robust pediatric vaccine viruses (HPIV1, 2, and 3) to express the protective antigens of a more fragile one (RSV).
CONCLUSIONS
Current strategies for developing a pediatric RSV vaccine are focused on (1) live attenuated RSV strains and (2) RSV antigens vectored from a live attenuated HPIV strain. Live attenuated RSV strains have the advantage of being the closest facsimile of natural RSV infection, which is known to provide resistance against a subsequent infection without disease enhancement. Conversely, a vectored vaccine would avoid the problems of poor growth and fragility of RSV, and has other advantages, such as increased antigen expression and the potential for improved boosts. However, a vectored vaccine delivers only a subset of RSV antigens. Whether this would affect efficacy and safety remains to be determined. It also would be possible to combine the two approaches: for example, an RSV/HPIV3 vaccine that could be given beginning at 1 month of age could include both a live attenuated RSV component and a B/HPIV3-vectored RSV component.
Supported by intramural National Institutes of Health funding and, in part, by a research and development program with Wyeth Vaccines that has since been terminated.
Conflict of Interest Statement: P.L.C. is a United States Government employee and performed this work as part of his official duties. U.S. Government employees are directed to patent any inventions and to have collaborative research and development agreements (CRADAs) with commercial companies; these patent rights are transferred to the U.S. Government, and U.S. Government employees can receive royalties that are distributed through the Government as per U.S. law. P.L.C. has not received royalties based on the work described in this article during the past 3 years. The National Institutes of Heath (NIH) did have a CRADA with Wyeth that terminated in 2002, and this project presently does not have support from a commercial company; B.R.M. is a U.S. Government employee and performed this work as part of his official duties as a U.S. Government employee. U.S. Government employees are required to patent inventions and to have CRADAs with commercial companies, and royalties are distributed to NIH and inventors as per U.S. law.
References
- 1.Collins PL, Hill MG, Camargo E, Grosfeld H, Chanock RM, Murphy BR. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc Natl Acad Sci USA 1995;92:11563–11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conzelmann KK. Reverse genetics of mononegavirales. Curr Top Microbiol Immunol 2004;283:1–41. [DOI] [PubMed] [Google Scholar]
- 3.Collins PL, Chanock RM, Murphy BR. Respiratory syncytial virus. In: Knipe DM, Howley, PM, editors. Fields Virology, 4th ed. Philadelphia, PA: Lippincott, Williams and Wilkins; 2001. p. 1443–1485.
- 4.McCarthy CA, Hall CB. Respiratory syncytial virus: concerns and control. Pediatr Rev 2003;24:301–309. [DOI] [PubMed] [Google Scholar]
- 5.Shay DK, Holman RC, Newman RD, Liu LL, Stout JW, Anderson LJ. Bronchiolitis-associated hospitalizations among US children, 1980–1996. JAMA 1999;282:1440–1446. [DOI] [PubMed] [Google Scholar]
- 6.Fearns R, Collins PL. Role of the M2-1 transcription antitermination protein of respiratory syncytial virus in sequential transcription. J Virol 1999;73:5852–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramaswamy M, Shi L, Monick MM, Hunninghake GW, Look DC. Specific inhibition of type I interferon signal transduction by respiratory syncytial virus. Am J Respir Cell Mol Biol 2004;30:893–900. [DOI] [PubMed] [Google Scholar]
- 8.Spann KM, Tran KC, Chi B, Rabin RL, Collins PL. Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages. J Virol 2004;78:4363–4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biacchesi S, Skiadopoulos MH, Yang L, Lamirande EW, Tran KC, Murphy BR, Collins PL, Buchholz UJ. Recombinant human metapneumovirus lacking the small hydrophobic SH and/or attachment G glycoprotein: deletion of G yields a promising vaccine candidate. J Virol 2004;78:12877–12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy BR, Collins PL. Live-attenuated virus vaccines for respiratory syncytial and parainfluenza viruses: applications of reverse genetics. J Clin Invest 2002;110:21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowe JE Jr. Host responses to respiratory virus infection and immunization. Curr Top Microbiol Immunol 1999;236:191–214. [DOI] [PubMed] [Google Scholar]
- 12.Crowe JE Jr, Firestone CY, Murphy BR. Passively acquired antibodies suppress humoral but not cell-mediated immunity in mice immunized with live attenuated respiratory syncytial virus vaccines. J Immunol 2001;167:3910–3918. [DOI] [PubMed] [Google Scholar]
- 13.Wright PF, Karron RA, Belshe RB, Thompson J, Crowe JE Jr, Boyce TG, Halburnt LL, Reed GW, Whitehead SS, Anderson EL, et al. Evaluation of a live, cold-passaged, temperature-sensitive, respiratory syncytial virus vaccine candidate in infancy. J Infect Dis 2000;182:1331–1342. [DOI] [PubMed] [Google Scholar]
- 14.Karron RA, Wright PF, Belshe RB, Thumar B, Casey R, Newman F, Polack FP, Randolph VB, Deatly A, Hackell J, et al. Identification of a recombinant live-attenuated respiratory syncytial virus vaccine candidate that is highly attenuated in infants. J Infect Dis 2005;191:1093–1104. [DOI] [PubMed] [Google Scholar]
- 15.Crowe JE Jr, Bui PT, Siber GR, Elkins WR, Chanock RM, Murphy BR. Cold-passaged, temperature-sensitive mutants of human respiratory syncytial virus (RSV) are highly attenuated, immunogenic, and protective in seronegative chimpanzees, even when RSV antibodies are infused shortly before immunization. Vaccine 1995;13:847–855. [DOI] [PubMed] [Google Scholar]
- 16.Johnson PR, Feldman S, Thompson JM, Mahoney JD, Wright PE. Immunity to influenza A virus infection in young children: a comparison of natural infection, live cold-adapted vaccine, and inactivated vaccine. J Infect Dis 1986;154:121–127. [DOI] [PubMed] [Google Scholar]
- 17.Crowe JE Jr, Bui PT, London WT, Davis AR, Hung PP, Chanock RM, Murphy BR. Satisfactorily attenuated and protective mutants derived from a partially attenuated cold-passaged respiratory syncytial virus mutant by introduction of additional attenuating mutations during chemical mutagenesis. Vaccine 1994;12:691–699. [DOI] [PubMed] [Google Scholar]
- 18.Karron RA, Wright PF, Crowe JE Jr, Clements ML, Thompson J, Makhene M, Casey R, Murphy BR. Evaluation of two live, cold-passaged, temperature-sensitive respiratory syncytial virus (RSV) vaccines in chimpanzees, adults, infants and children. J Infect Dis 1997;176:1428–1436. [DOI] [PubMed] [Google Scholar]
- 19.Whitehead SS, Juhasz K, Firestone CY, Collins PL, Murphy BR. Recombinant respiratory syncytial virus (RSV) bearing a set of mutations from cold-passaged RSV is attenuated in chimpanzees. J Virol 1998;72:4467–4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whitehead SS, Firestone CY, Karron RA, Crowe JE Jr, Elkins WR, Collins PL, Murphy BR. Addition of a missense mutation present in the L gene of respiratory syncytial virus (RSV) cpts530/1030 to RSV vaccine candidate cpts248/404 increases its attenuation and temperature sensitivity. J Virol 1999;73:871–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juhasz K, Whitehead SS, Boulanger CA, Firestone CY, Collins PL, Murphy BR. The two amino acid substitutions in the L protein of cpts530/1009, a live-attenuated respiratory syncytial virus candidate vaccine, are independent temperature-sensitive and attenuation mutations. Vaccine 1999;17:1416–1424. [DOI] [PubMed] [Google Scholar]
- 22.Whitehead SS, Firestone CY, Collins PL, Murphy BR. A single nucleotide substitution in the transcription start signal of the M2 gene of respiratory syncytial virus vaccine candidate cpts248/404 is the major determinant of the temperature-sensitive and attenuation phenotypes. Virology 1998;247:232–239. [DOI] [PubMed] [Google Scholar]
- 23.Teng MN, Whitehead SS, Collins PL. Contribution of the respiratory syncytial virus G glycoprotein and its secreted and membrane-bound forms to virus replication in vitro and in vivo. Virology 2001;289:283–296. [DOI] [PubMed] [Google Scholar]
- 24.Buchholz UJ, Granzow H, Schuldt K, Whitehead SS, Murphy BR, Collins PL. Chimeric bovine respiratory syncytial virus with glycoprotein gene substitutions from human respiratory syncytial virus (HRSV): effects on host range and evaluation as a live-attenuated HRSV vaccine. J Virol 2000;74:1187–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitehead SS, Bukreyev A, Teng MN, Firestone CY, St Claire M, Elkins WR, Collins PL, Murphy BR. Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J Virol 1999;73:3438–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bermingham A, Collins PL. The M2-2 protein of human respiratory syncytial virus is a regulatory factor involved in the balance between RNA replication and transcription. Proc Natl Acad Sci USA 1999;96:11259–11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin H, Zhou H, Cheng X, Tang R, Munoz M, Nguyen N. Recombinant respiratory syncytial viruses with deletions in the NS1, NS2, SH, and M2-2 genes are attenuated in vitro and in vivo. Virology 2000;273:210–218. [DOI] [PubMed] [Google Scholar]
- 28.Jin H, Cheng X, Zhou HZ, Li S, Seddiqui A. Respiratory syncytial virus that lacks open reading frame 2 of the M2 gene (M2-2) has altered growth characteristics and is attenuated in rodents. J Virol 2000;74:74–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang RS, Nguyen N, Cheng X, Jin H. Requirement of cysteines and length of the human respiratory syncytial virus M2-1 protein for protein function and virus viability. J Virol 2001;75:11328–11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang RS, Nguyen N, Zhou H, Jin H. Clustered charge-to-alanine mutagenesis of human respiratory syncytial virus L polymerase generates temperature-sensitive viruses. Virology 2002;302:207–216. [DOI] [PubMed] [Google Scholar]
- 31.Skiadopoulos MH, Schmidt AC, Riggs JM, Surman SR, Elkins WR, St Claire M, Collins PL, Murphy BR. Determinants of the host range restriction of replication of bovine parainfluenza virus type 3 in rhesus monkeys are polygenic. J Virol 2003;77:1141–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt AC, McAuliffe JM, Huang A, Surman SR, Bailly JE, Elkins WR, Collins PL, Murphy BR, Skiadopoulos MH. Bovine parainfluenza virus type 3 (BPIV3) fusion and hemagglutinin-neuraminidase glycoproteins make an important contribution to the restricted replication of BPIV3 in primates. J Virol 2000;74:8922–8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonzalez-Reyes L, Ruiz-Arguello MB, Garcia-Barreno B, Calder L, Lopez JA, Albar JP, Skehel JJ, Wiley DC, Melero JA. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc Natl Acad Sci USA 2001;98:9859–9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flanagan EB, Zamparo JM, Ball LA, Rodriguez LL, Wertz GW. Rearrangement of the genes of vesicular stomatitis virus eliminates clinical disease in the natural host: new strategy for vaccine development. J Virol 2001;75:6107–6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson TR, Johnson JE, Roberts SR, Wertz GW, Parker RA, Graham BS. Priming with secreted glycoprotein G of respiratory syncytial virus (RSV) augments interleukin-5 production and tissue eosinophilia after RSV challenge. J Virol 1998;72:2871–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tripp RA, Jones LP, Haynes LM, Zheng H, Murphy PM, Anderson LJ. CX3C chemokine mimicry by respiratory syncytial virus G glycoprotein. Nat Immunol 2001;2:732–738. [DOI] [PubMed] [Google Scholar]
- 37.Teng MN, Collins PL. The central conserved cystine noose of the attachment G protein of human respiratory syncytial virus is not required for efficient viral infection in vitro or in vivo. J Virol 2002;76:6164–6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spann KM, Collins PL, Teng MN. Genetic recombination during coinfection of two mutants of human respiratory syncytial virus. J Virol 2003;77:11201–11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beeler JA, van Wyke Coelingh K. Neutralization epitopes of the F glycoprotein of respiratory syncytial virus: effect of mutation upon fusion function. J Virol 1989;63:2941–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McAuliffe JM, Surman SR, Newman JT, Riggs JM, Collins PL, Murphy BR, Skiadopoulos MH. Codon substitution mutations at two positions in the L polymerase protein of human parainfluenza virus type 1 yield viruses with a spectrum of attenuation in vivo and increased phenotypic stability in vitro. J Virol 2004;78:2029–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krempl C, Murphy BR, Collins PL. Recombinant respiratory syncytial virus with the G and F genes shifted to the promoter-proximal positions. J Virol 2002;76:11931–11942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biacchesi S, Skiadopoulos MH, Tran KC, Murphy BR, Collins PL, Buchholz UJ. Recovery of human metapneumovirus from cDNA: optimization of growth in vitro and expression of additional genes. Virology 2004;321:247–259. [DOI] [PubMed] [Google Scholar]
- 43.Valarcher JF, Furze J, Wyld S, Cook R, Conzelmann KK, Taylor G. Role of alpha/beta interferons in the attenuation and immunogenicity of recombinant bovine respiratory syncytial viruses lacking NS proteins. J Virol 2003;77:8426–8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin H, Clarke D, Zhou HZ, Cheng X, Coelingh K, Bryant M, Li S. Recombinant human respiratory syncytial virus (RSV) from cDNA and construction of subgroup A and B chimeric RSV. Virology 1998;251:206–214. [DOI] [PubMed] [Google Scholar]
- 45.Whitehead SS, Hill MG, Firestone CY, St Claire M, Elkins WR, Murphy BR, Collins PL. Replacement of the F and G proteins of respiratory syncytial virus (RSV) subgroup A with those of subgroup B generates chimeric live attenuated RSV subgroup B vaccine candidates. J Virol 1999;73:9773–9780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidt AC, Wenzke DR, McAuliffe JM, St. Clair M, Elkins WR, Murphy BR, Collins PL. Mucosal immunization of rhesus monkeys against respiratory syncytial virus subgroups A and B and human parainfluenza virus type 3 using a live cDNA-derived vaccine based on a host range-attenuated bovine parainfluenza virus type 3 vector backbone. J Virol 2002;76:1089–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skiadopoulos MH, Biacchesi S, Buchholz UJ, Riggs JM, Surman SR, Amaro-Carambot E, McAuliffe JM, Elkins WR, St Claire M, Collins PL, et al. The two major human metapneumovirus genetic lineages are highly related antigenically, and the fusion (F) protein is a major contributor to this antigenic relatedness. J Virol 2004;78:6927–6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Newman JT, Riggs JM, Surman SR, McAuliffe JM, Mulaikal TA, Collins PL, Murphy BR, Skiadopoulos MH. Generation of recombinant human parainfluenza virus type 1 vaccine candidates by importation of temperature-sensitive and attenuating mutations from heterologous paramyxoviruses. J Virol 2004;78:2017–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skiadopoulos MH, Vogel L, Riggs JM, Surman SR, Collins PL, Murphy BR. The genome length of human parainfluenza virus type 2 follows the rule of six, and recombinant viruses recovered from non–polyhexameric-length antigenomic cDNAs contain a biased distribution of correcting mutations. J Virol 2003;77:270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalica AR, Wright PF, Hetrick FM, Chanock RM. Electron microscopic studies of respiratory syncytial temperature-sensitive mutants. Arch Gesamte Virusforsch 1973;41:248–258. [DOI] [PubMed] [Google Scholar]