Abstract
This study provides the first spectroscopic characterization of the membrane-bound oxygen-tolerant [NiFe] hydrogenase (MBH) from Ralstonia eutropha H16 in its natural environment, the cytoplasmic membrane. The H2-converting MBH is composed of a large subunit, harboring the [NiFe] active site, and a small subunit, capable in coordinating one [3Fe4S] and two [4Fe4S] clusters. The hydrogenase dimer is electronically connected to a membrane-integral cytochrome b. EPR and Fourier transform infrared spectroscopy revealed a strong similarity of the MBH active site with known [NiFe] centers from strictly anaerobic hydrogenases. Most redox states characteristic for anaerobic [NiFe] hydrogenases were identified except for one remarkable difference. The formation of the oxygen-inhibited Niu-A state was never observed. Furthermore, EPR data showed the presence of an additional paramagnetic center at high redox potential (+290 mV), which couples magnetically to the [3Fe4S] center and indicates a structural and/or redox modification at or near the proximal [4Fe4S] cluster. Additionally, significant differences regarding the magnetic coupling between the Nia-C state and [4Fe4S] clusters were observed in the reduced form of the MBH. The spectroscopic properties are discussed with regard to the unusual oxygen tolerance of this hydrogenase and in comparison with those of the solubilized, dimeric form of the MBH.
Hydrogenases are metalloenzymes that catalyze the reversible cleavage of H2 into protons and electrons and play a pivotal role in the energy metabolism of many microorganisms (1). They are grouped into three phylogenetically distinct classes as follows: the di-iron [FeFe], nickel-iron [NiFe], and iron-sulfur cluster-free [Fe] hydrogenases (2–6). The basic module of [NiFe] hydrogenases consists of two subunits, a large subunit that contains the [NiFe] active site and a small subunit that accommodates one to three electron-transferring iron-sulfur clusters (2, 7, 8). The active site nickel is coordinated to the protein via the thiol groups of four invariant cysteine residues, two of which serve as bridging ligands to the iron. The active site iron carries three additional diatomic ligands, two cyanides (CN−) and one carbon monoxide (CO) (9, 10).
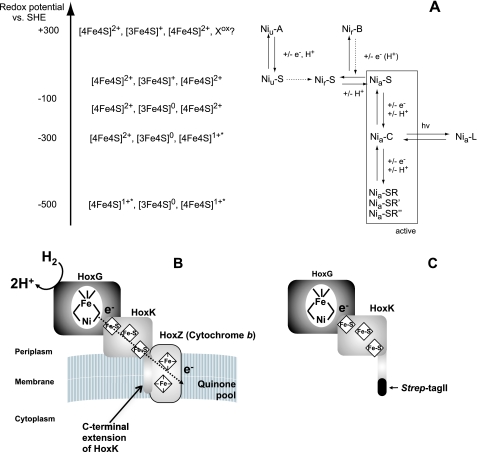
In addition to H2, the active sites of the vast majority of [FeFe] and [NiFe] hydrogenases react with dioxygen. In case of the [FeFe] hydrogenases, this usually leads to an irreversible destruction of the active site (11, 12). Most [NiFe] hydrogenases, however, are reversibly inactivated by molecular oxygen (13, 14). Under electron-rich conditions in the presence of O2 a mono-oxo species, most probably a hydroxide, is formed in the bridging position between nickel and iron (15–17). On the basis of EPR spectroscopy, this paramagnetic “ready inactive” state has been designated as Nir-B. Incubation of the enzyme with O2 under electron-poor conditions results in the so-called “unready inactive” Niu-A state. It is anticipated that a di-oxo species (e.g. hydroperoxide) binds in the bridging position (16, 17). However, the nature of this ligand is still a matter of debate (18). The Niu-A and Nir-B states differ significantly in their reactivation kinetics. H2-mediated reductive activation of the Niu-A state is a long term process and requires hours until the oxygen species is completely removed from the active site. In contrast, only seconds of incubation with H2 are required to convert the Nir-B state into the catalytically active, EPR-detectable Nia-C state in which a hydride occupies the bridging position between nickel and iron (19, 20). An overview of the different redox states of the [NiFe] active site is shown in Fig. 1A (18, 21).
FIGURE 1.
A, different redox states of the [NiFe] active site and the [FeS] clusters in oxygen-sensitive standard [NiFe] hydrogenases (adapted from Refs. 18, 21). See text for further details. No Niu-A is observed for the R. eutropha MBH. B, schematic of MBH of R. eutropha attached to the membrane via the cytochrome b. C, schematic of solubilized dimeric R. eutropha MBH harboring a Strep affinity tag at the C-terminal end of the small subunit.
Formation of the Niu-A state prevents most of the [NiFe] hydrogenases, predominantly those from anaerobic microorganisms, from being catalytically active even in the presence of traces of O2. However, the so-called Knallgasbacteria contain [NiFe] hydrogenases that enable these microorganisms to gain energy from H2 oxidation even in the presence of atmospheric oxygen concentrations (22–24). Prominent examples are β-proteobacteria of the genus Ralstonia, including the well studied chemolithoautotrophic model organism Ralstonia eutropha H16. R. eutropha harbors three distinct [NiFe] hydrogenases that catalyze H2 oxidation in the presence of ambient oxygen concentrations (11, 25–27). Their O2 tolerance is based on at least two molecular mechanisms. For the cytoplasmic, hexameric NAD+-reducing hydrogenase, a modified composition of the active site nickel has been described. It is proposed that an extra CN− ligand bound to the nickel protects the enzyme from O2 inactivation (25, 28, 29). O2 resistance of the H2-sensing regulatory [NiFe] hydrogenases from R. eutropha (27) and Rhodobacter capsulatus (30) is likely based on a narrow gas channel that prevents access of O2 to the active site.
The third H2-converting catalyst in R. eutropha is a membrane-bound [NiFe] hydrogenase (MBH)3 composed of the large subunit HoxG (67.1 kDa) and the small subunit HoxK (34.6 kDa) (31), which are anchored to the membrane-integral b-type cytochrome HoxZ via the hydrophobic C terminus of HoxK (Fig. 1B) (31–33). Electrochemical experiments revealed that the isolated MBH dimer (Fig. 1C), attached to a graphite electrode, retains more than 20% of its H2-oxidizing activity in the presence of 0.21 bar O2, compared with the activity under completely anaerobic conditions (26, 34). This result impressively demonstrates that O2 reacts with the MBH active site in a way that still allows the catalytic conversion of H2. FTIR and EPR spectroscopy are valuable techniques to get insights into the composition and redox state of cofactors as well as on structure-function relationships even if a crystal structure of the enzyme is not yet available (for recent reviews see Refs. 18, 35). Earlier data obtained by Fourier transform infrared (FTIR) spectroscopy indicate that the active site iron exhibits the standard set of diatomic ligands, two CN− and one CO (26).
EPR allows the detection of redox-dependent changes of the electronic structure of both the [NiFe] active site in the large subunit and the Fe-S clusters in the small subunit. However, only paramagnetic metal species can be traced by EPR, whereas divalent nickel species that are constituents of the hydrogenase catalytic cycle are invisible by this technique.
The dimeric form of the MBH has been used successfully as a constituent in enzymatic fuel cells and for light-driven H2 production (36, 37). These applications crucially depend on oxygen-tolerant hydrogen catalysts. A very recent biochemical and electrochemical study revealed that O2 tolerance of the R. eutropha MBH is ultimately linked to a high affinity for hydrogen with a concomitant extremely low affinity for oxygen (34). To get insights into the electronic structure of the active site and the iron-sulfur clusters, which underlies the oxygen tolerance, we have re-investigated the spectroscopic properties of the MBH. In addition to earlier studies (38, 39), which are discussed below and which employed solely EPR spectroscopy, we have also used FTIR spectroscopy that probes the intra-ligand stretching modes of the diatomic active site ligands CO and CN−. The frequencies of these modes are known to be sensitive to the electron density distribution in the catalytic center and thus can be used to monitor redox transitions (35, 40). The CO stretching frequency has been shown to respond also to changes in the amino acid composition of the protein backbone in the vicinity of the active site (21, 41, 42). Importantly, FTIR spectroscopy does not only rely on paramagnetic states of metal centers and, hence, is a useful tool to investigate all O2-mediated noncatalytic states as well as all redox states involved in H2 conversion. In this study we show that the MBH was never found in the oxygen-inactivated Ni-A state, which is in perfect agreement with recent electrochemical studies that revealed an extremely fast reactivation of the MBH dimer after treatment with dioxygen (34). However, our studies also present evidence that a significant portion of the MBH dimer that has been removed from the cytoplasmic membrane resided in a catalytically inactive form. Therefore, we investigated for the first time the spectroscopic properties of the trimeric form of the MBH, including HoxZ, tightly bound to the cytoplasmic membrane (Fig. 1B). The trimeric MBH turned out to form only the Nir-B state and a small amount of ready or unready Ni-S states upon oxidation, which both reacted completely and reversibly with H2. The spectroscopic properties of the MBH are discussed with regard to the O2 tolerance of this unusual catalyst.
MATERIALS AND METHODS
Preparation of the Cytoplasmic Membrane
The R. eutropha MBH was overexpressed in a nontagged version using plasmid pLO6, harboring the complete MBH operon, in R. eutropha strain HF631, a megaplasmid-free derivative of R. eutropha (43). The cytoplasmic membrane was separated by osmotic shock to remove the outer membrane according to a modified protocol of Witholt et al. (44).
Cells were cultivated lithoautotrophically in mineral salts medium (45) under an atmosphere of 75% H2, 15% O2, and 10% CO2 at 30 °C and continuous shaking at 120 rpm. Cells from 0.5 liter of culture volume were harvested at an A436 of 18–20 (centrifugation at 5000 × g at 4 °C for 20 min) and resuspended in 1 liter of 50 mm Tris/HCl buffer at pH 7.8. The cells were collected by centrifugation (5000 × g, 20 min, 4 °C) and resuspended in 400 ml of sucrose buffer (192 g of sucrose, 50 mm Tris/HCl buffer at pH 7.8 in 500 ml, 1 mm EDTA) and incubated at room temperature for 5 min. Subsequently, the suspension was again centrifuged (32,000 × g, 20 min, 4 °C), and the cells were exposed to osmotic shock by resuspending the pellet in 200 ml of deionized H2O. The resulting spheroplasts were collected by centrifugation (4000 × g, 20 min, 4 °C), resuspended in 100 ml of 50 mm K-PO4 buffer (pH 7.0), and subsequently disrupted using a French pressure cell (Constant Cell Disruption Systems) and ultrasonication (2 min, level 2.5, 75%) (Branson Sonifier). The cell extract was subjected to ultracentrifugation (100,000 × g, 60 min 4 °C), and the membrane fraction was isolated by removing the upper brownish part of the pellet with a spatula. The cytoplasmic membrane was homogenized in an appropriate volume of 50 mm K-PO4 buffer (pH 7.0), ultracentrifuged (100,000 × g, 30 min, 4 °C), and finally homogenized in about 1 ml of 50 mm K-PO4 buffer (pH 7.0).
Enzyme Purification
For purification of the MBH, a derivative carrying a Strep Tag II at the C terminus of the small subunit (HoxK) encoded on plasmid pGE636 in R. eutropha strain HF631 was used (46). The cells were grown lithoautotrophically in a fermentor (type NLF 22, Bioengineering, Wald, Switzerland) in mineral salts medium (45) under an atmosphere of 75% H2, 15% O2, and 10% CO2 at 30 °C. The cells were harvested by centrifugation (5000 × g, 20 min 4 °C) at an A436 of about 20. The cell pellet was washed with an appropriate amount of phosphate buffer (90 g liter−1 Na2HPO4 × 12 H2O, 15 g liter−1 KH2PO4), frozen in liquid nitrogen, and stored at −80 °C. The MBH dimer was purified as described previously (34). The protein was concentrated using centrifugal filter devices (Amicon Ultra15 (PL-30), Amicon Microcon (YM-30), Millipore) to volumes with protein concentrations (200–500 μm) appropriate for IR and EPR spectroscopy. The protein concentration was determined by the Bradford method (47) using bovine serum albumin as standard. The purity of the samples was examined by SDS-PAGE (48).
Enzyme Assay
Hydrogenase assays were performed as described previously (32) in 50 mm K-PO4 buffer at pH 7.0 for membrane-attached MBH and at pH 5.5 for solubilized MBH. Methylene blue was used as artificial electron acceptor, and its absorption was measured photometrically at 570 nm.
EPR Spectroscopy
9.5 GHz X-band EPR spectroscopy has been carried out using a Bruker ESP300E spectrometer equipped with a rectangular microwave cavity in the TE102 mode. For low temperature measurements the sample was kept in an Oxford ESR 900 helium flow cryostat that allows for temperature control between 6 and 100 K (Oxford ITC4). The microwave frequency was detected with an EIP frequency counter (Microwave Inc.). For determination of g values, the magnetic field was calibrated with an external Li/LiF standard with a known g value of 2.002293 (49). Spin quantifications have been performed by comparing the double integrated signal with the signal of a CuSO4 standard of known concentration. Base-line corrections, if required, were performed by subtracting a background spectrum, obtained under the same experimental conditions from a sample containing only a buffer solution. Simulations of the EPR spectra have been performed with the program EasySpin (50) that diagonalizes the Spin-Hamiltonian and calculates transition probabilities.
Fourier Transform Infrared Spectroscopy
FTIR spectra were recorded on a Bruker Tensor 27 spectrometer equipped with a liquid nitrogen-cooled MCT detector with a spectral resolution of 2 cm−1. The sample compartment was purged with dried air, and the sample (0.2–0.5 mm isolated protein, ∼0.05 mm protein attached to the cytoplasmic membrane) was held in a temperature-controlled (10 °C) gas-tight IR-cell for liquid samples (volume ∼7 μl, path length = 50 μm) with CaF2 windows. Spectra were base-line corrected by using a spline function implemented within OPUS 4.2 software of the spectrometer. Reduced protein samples were prepared through incubation under 1 bar H2 or under 0.05 bar H2 in 1 bar gas mixture (95% nitrogen, 5% hydrogen) atmosphere for 20–30 min at room temperature.
Metal and Cyanide Determination
The iron and nickel contents of solubilized dimeric R. eutropha MBH preparations were quantified by ICP-OES analysis with an Optima 2100 DV from PerkinElmer Life Sciences. The multiple element standard solution XVI (Merck) was used as reference. The cyanide content was determined chemically by distillation and subsequent UV/visible spectroscopic quantification of the formed polymethine according to Ref. 10.
Potential Measurements
The redox potentials of bulk protein samples in the “as-isolated” state or after different chemical treatments were measured in a homemade, low volume cell by means of a miniaturized Pt/Ag/AgCl “single-rod” redox electrode (Pt5900, Schott) at room temperature. All potentials cited in the text refer to the standard hydrogen electrode.
RESULTS
Determination of the Nickel, Iron, and Cyanide Content
The MBH dimer was solubilized from the membrane and purified by affinity chromatography as described previously (34). According to SDS-PAGE and subsequent protein staining, the purity of the MBH subunits was estimated to be ∼99%. Metal analysis using ICP-OES revealed a nickel content of 0.7 nickel per protein molecule and an iron content of about 10 ± 2 iron. This is in good agreement with the results of an earlier study (38) and consistent with the composition of one [NiFe], one [3Fe4S], and two [4Fe4S] as found in oxygen-sensitive standard [NiFe] hydrogenases, e.g. that from Desulfovibrio vulgaris (18, 41). The metal content reflects an occupancy of 70% for all metal cofactors. Nevertheless, the ratio of iron and nickel meets the expectation. Determination of the cyanide content in the purified MBH revealed 1.8 CN− per protein molecule indicating a standard-like coordination of the iron in the active site, as observed by FTIR spectroscopy, i.e. two cyanides and one CO.
EPR Spectroscopic Analysis of the Solubilized MBH Dimer
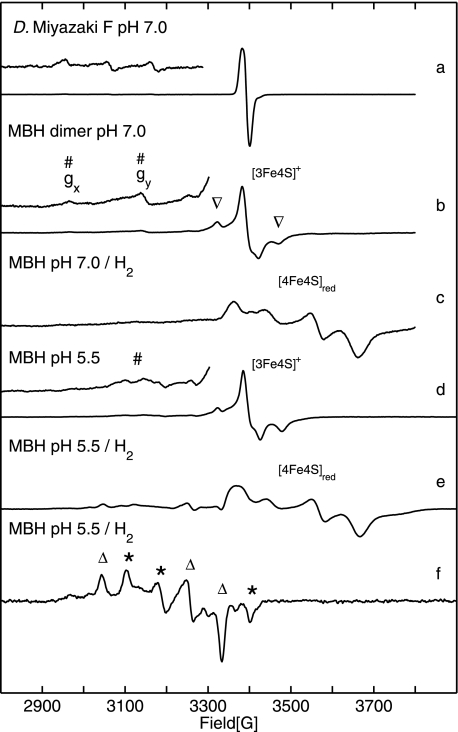
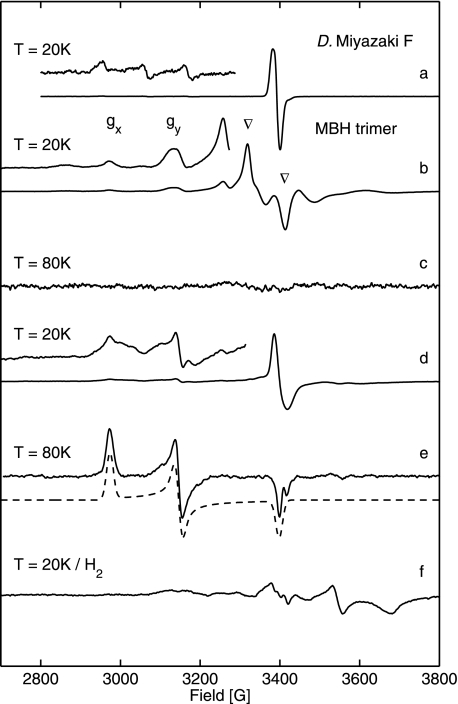
Fig. 2 shows 9.5 GHz EPR spectra obtained for the solubilized, dimeric MBH in its oxidized (as isolated) and reduced forms. The spectra taken at 20 K from samples buffered at pH 5.5 and pH 7.0 (Fig. 2, traces b and d) exhibit a complex line shape. The prominent signal at g = 2.0 results from an oxidized [3Fe-4S]+ cluster with an S = 1/2 ground state. The signal consists of a narrow component, similar to that observed for standard [NiFe] hydrogenases (see Fig. 2, trace a), and a superimposed complex broad “split” component (labeled by ▿). The intensity of this broad signal varied between different enzyme preparations, and in some samples, it even exceeded that of the narrow signal. It is interesting to note that the broad signal component was never observed in the spectra of most well studied periplasmic standard hydrogenases such as that from D. vulgaris Miyazaki F (Fig. 2, trace a).
FIGURE 2.
EPR spectra of solubilized MBH dimer showing signals from the [NiFe] and [FeS] centers. The enhanced traces of the spectra show the range of nickel signals with five times increased amplification. Definition of symbols is as follows: #, Nir-B; Δ, Nia-L; *, Nia-C; ▿, split signal from [3Fe4S]+. Trace a, oxidized (as isolated) D. vulgaris Miyazaki F hydrogenase. The enhanced trace shows signals from superimposed g x and gy components of Niu-A and Nir-B. Trace b, oxidized (as isolated) R. eutropha MBH protein solution at pH 7.0 saturated with air, showing signals of the [3Fe4S]+ cluster and Nir-B state. Trace c, H2-reduced (1 bar H2) R. eutropha MBH sample (pH 7.0) displaying signals from reduced [4Fe4S] centers. Trace d, oxidized (as isolated), and trace e, H2-reduced (1 bar H2) R. eutropha MBH samples at pH 5.5. f, H2-reduced (1 bar H2) R. eutropha MBH at T = 80 K, showing a superposition of signals from the Nia-C and Nia-L states. Experimental conditions for spectra (traces a–e) T = 20 K, (f) T = 80K; 1 milliwatt microwave power; microwave frequency 9.56 GHz; 1 mT modulation amplitude, 12.5 kHz modulation frequency.
Upon mild reduction of the as-isolated MBH dimer sample with 5 mm β-mercaptoethanol, the redox potential decreased to +40 mV resulting in a transformation of the complex broad and split EPR signal into a narrow signal, as it is usually observed for an uncoupled [3Fe4S]+ cluster (see supplemental Fig. S1). This narrow signal at g = 2.0 is very similar to that found for the [3Fe4S]+ center in the D. vulgaris Miyazaki F standard [NiFe] hydrogenase (Fig. 2, trace a).
At pH 7.0 signals of a Ni(III) species in the active site of the oxidized MBH dimer were detected. The gx and gy values (2.30 and 2.17) agree with those of the Nir-B state observed for standard [NiFe] hydrogenases (Fig. 2, traces a and b) (51–54). The Nir-B signals were broadened in protein samples at pH 5.5 (Fig. 2, trace d). This result indicates heterogeneity of the underlying species, hampering a reliable quantification at this low pH. Relative quantification of the integrated EPR signal intensities for a variety of samples at pH 7.0 showed that the amount of the Nir-B relative to the [3Fe4S]+ signal was about 30%. Absolute quantification of the [3Fe4S]+ signal (narrow signal component, Fig. 2, traces b and d) using a CuSO4 standard sample revealed ∼1 spin per protein indicating that the oxidized [3Fe4S]+ cluster is present in essentially 100% of the protein. Notably, in samples with a high contribution of the [3Fe4S]+ split component, spin quantification yielded up to 1.8 spins per protein for this signal (see supplemental Table S1).
EPR signals attributable to the Niu-A state were not observed in any R. eutropha MBH preparations, neither at high redox potential in as-isolated samples nor in samples reoxidized after previous treatment with H2. This is in sharp contrast to the situation in standard [NiFe] hydrogenases, where usually a mixture of Niu-A and Nir-B states is observed in the as-isolated enzyme (18, 41).
Upon incubation with 1 bar H2, the signal of the [3Fe4S]+ cluster disappeared, whereas a complex spectrum of reduced [4Fe4S]+ clusters emerged (g value range 2.0 to 1.82, see Fig. 2, trace c). Absolute quantification of the broad EPR signal, using a CuSO4 spin standard sample, revealed 0.5–0.8 spins per protein for the reduced iron-sulfur centers. This value corresponds to one reduced [4Fe4S] center in only 50–80% of the isolated MBH protein. The shape of the spectrum indicates coupling to another paramagnetic center, which could be, according to an earlier proposal (38), the reduced [3Fe4S]0 cluster with a spin S = 2.
EPR spectra of H2-reduced samples at pH 5.5 display signals at lower field that are in the range expected for Nia-C and Nia-L (Fig. 2, trace e). At higher temperature (T = 80 K), the reduced iron-sulfur centers are broadened beyond detection, and the Nia-C/Nia-L signals became narrow and well resolved (Fig. 2, trace f). The g values obtained by simulations (Table 1) are in good agreement with those of Nia-C and Nia-L from standard [NiFe] hydrogenases (51). The spectra clearly show a superposition of nearly equal amounts of Nia-C and Nia-L. It should be emphasized that this Nia-L signal is observed without any extra illumination of the samples, except normal light conditions in the laboratory. It appears to be that Nia-L is easily formed in the isolated dimeric R. eutropha MBH, which is in contrast to standard [NiFe] hydrogenases, where prolonged illumination at low temperature is required to induce the transition from Nia-C to Nia-L. Contrary to the latter, the Nia-L state of the solubilized MBH dimer was stable under normal daylight conditions and did not convert to the Nia-C state at elevated temperature in the dark (20, 55), as observed in standard [NiFe] hydrogenases. This finding indicates that the catalytic active part of the solubilized enzyme shows a functional modification compared with standard [NiFe] hydrogenases.
TABLE 1.
g-tensor principal values observed for the various redox states of the [NiFe] center ofR. eutropha H16 MBH obtained by simulation compared with literature data for theD. vulgaris Miyazaki F hydrogenase
| Redox state | gx | gy | gz | Line width/mT | Refs. | |
|---|---|---|---|---|---|---|
| R. eutropha H16 dimer | Nir-B | 2.30 | 2.17 | 2.01 | a | This work and Ref. 38 |
| Nia-C | 2.20 | 2.14 | 2.01 | 2.2 | This work | |
| Nia-L | 2.25 | 2.10 | 2.05 | 1.8 | This work | |
| R. eutropha H16 trimer | Nir-B | 2.30 | 2.17 | 2.01 | 2.0 | This work |
| Nia-C | 2.20 | 2.14 | 2.01 | 1.8 | This work | |
| Nia-L1 | 2.30 | 2.11 | 2.05 | 1.5 | This work | |
| Nia-L2 | 2.27 | 2.11 | 2.05 | 1.5 | This work | |
| Nia-L3 | 2.24 | 2.11 | 2.05 | 1.4 | This work | |
| D. vulgaris Miyazaki F | Nir-B | 2.33 | 2.16 | 2.01 | 1.9 | This work and Ref. 59 |
| Nia-C | 2.198 | 2.142 | 2.012 | 52 | ||
| Nia-L1 | 2.26 | 2.11 | 2.05 | 60 | ||
| Nia-L2 | 2.298 | 2.116 | 2.045 | 52 | ||
| R. metallidurans CH34 | Nir-B | 2.30 | 2.17 | 2.01 | 39 | |
| Nia-C | 2.20 | 2.16 | 2.01 | 39 | ||
| Nia-L | 2.31 | 2.12 | 2.05 | 39 |
a Line widths of spectra at 20 K are typically broad and range from 2 to 3 mT.
The total amount of the Nia-C and Nia-L mixture was estimated to be up to 20%. We consider the occurrence of Nia-L in the solubilized MBH dimer without illumination as nonphysiological (see below). It is likely that this unusual property results from a non-native structure and/or heterogeneity of the protein in the solubilized state.
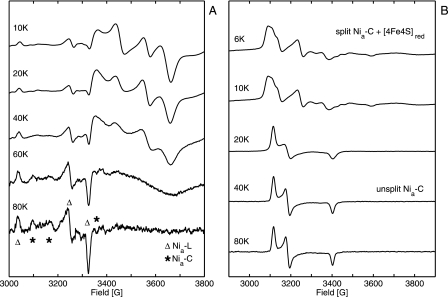
The finding that much more Nia-C/Nia-L is formed at pH 5.5 than at pH 7.0 can be interpreted in terms of the pH dependence of the redox potential in the H2-saturated protein solutions. Based on Nernst equation, redox potentials are expected to be about −320 and −410 mV at pH 5.5 and 7.0, respectively. Measurements of protein solutions at pH 7.0 using a platinum redox electrode revealed a potential at −380 mV. These findings are consistent with the results of an earlier study on the MBH from Ralstonia metallidurans CH34, which is closely related to R. eutropha H16. In that investigation, redox titration experiments have revealed a maximum amount of Nia-C at −301 mV, whereas only a small amount of Nia-C signal was observed at −420 mV (39). To investigate a possible coupling between Nia-C and the reduced [4Fe4S] cluster, the temperature dependence of the EPR spectra of H2-reduced MBH samples was studied (see Fig. 3A).
FIGURE 3.
Temperature-dependent EPR spectra of H2-reduced hydrogenase. A, signals of Nia-C, Nia-L, and reduced [4Fe4S] centers in R. eutropha MBH. B, signals of the coupled Nia-C and reduced [4Fe4S] centers in D. vulgaris Miyazaki F hydrogenase. Experimental conditions are as follows: 1 milliwatt microwave power; microwave frequency 9.56 GHz; 1 mT modulation amplitude, 12.5 kHz modulation frequency.
Between 10 and 40 K, the spectra of Nia-C and reduced [4Fe4S] remained unchanged in MBH samples. At 60 K, however, the signal of the reduced iron-sulfur cluster was significantly broadened because of spin relaxation. This behavior is an inherent property of reduced [4Fe4S] clusters. At 80 K the [4Fe4S]+ signal vanished completely, and only Nia-C/Nia-L signals were observed. This observation shows that a magnetic coupling between the Nia-C site and a reduced [4Fe4S] center is either absent or very different from that observed in standard [NiFe] hydrogenases (20, 55, 56). For comparison, Fig. 3B shows the temperature-dependent EPR spectra of the D. vulgaris Miyazaki F hydrogenase obtained after 3 h of incubation under 1 bar H2 at 37 °C. At temperatures between 25 and 80 K, the well resolved signal of Nia-C signal was visible, but below 15 K a complex spectrum was obtained that had been attributed to magnetic coupling between the Nia-C and the reduced proximal [4Fe4S] cluster (20, 55, 56).
Our results from EPR spectroscopy are generally in good agreement with previous EPR studies on Ralstonia MBH (38, 39), where a different method has been used to prepare the enzyme in its solubilized dimeric form (32, 57) consisting of the small HoxK subunit with the iron-sulfur clusters and the large HoxG subunit harboring the [NiFe] active site (Fig. 1C). At high redox potential, an EPR signal of Ni(III) was observed. The g values were in agreement with those subsequently assigned to Nir-B (Fig. 1A). The Ni(III) signal of the oxidized MBH was reported to correspond to 0.15 spins per protein, and we found up to 30% Nir-B. In our study we found generally less Nir-B in samples that exhibited a larger fraction of inactive enzyme, as determined by FTIR (see below). Furthermore, a complex and split EPR signal was also found in Ref. 38, which was assigned to a [3Fe4S]+ cluster, coupled to a yet unknown paramagnetic species. This species was reported to have a midpoint potential of +160 mV, where the EPR signal was converted to a narrow line shape, typically representing an uncoupled [3Fe4S]+ cluster. Upon reduction with H2 or dithionite, another EPR signal was observed that was attributed to two coupled [4Fe4S]+ clusters. The redox potential of the transition [4Fe4S]2+/1+ was determined to be −90 mV, which is unusually high for common [4Fe4S] clusters. Our EPR results confirm these findings. The dimeric MBH of R. metallidurans CH34, which is closely related to the R. eutropha MBH, was investigated in a later EPR and redox study (39). In addition to the EPR species found for the R. eutropha MBH another Ni-derived EPR signal, the so called Nia-C was observed, which is known as a catalytic intermediate in anaerobic [NiFe] standard hydrogenases (41, 52). We observed the Nia-C state also in R. eutropha MBH, underlining the close similarity of the hydrogenases of both species.
Our findings for the different paramagnetic states of the redox active centers in R. eutropha MBH and their proposed interactions are compared with those from the earlier studies (38, 39) in supplemental Table S2 and are discussed below.
FTIR Spectroscopy on the Solubilized MBH Dimer
The potential of FTIR spectroscopy for probing redox transitions and the intermolecular interactions within the catalytic center have been demonstrated for the standard [NiFe] hydrogenases from Allochromatium vinosum (14), Desulfovibrio gigas (9, 42), and D. vulgaris (41). The results obtained from these studies guided the assignment of the IR spectra of the R. eutropha MBH presented in this work (Table 2). This approach minimized the uncertainty in the interpretation of spectra caused by unavoidable variations of the sample preparation. Additional information on the assignment of bands was obtained from time-resolved H2 reduction and reoxidation experiments by FTIR (data not shown) and studies of the trimeric MBH complex (see below).
TABLE 2.
Wave-numbers of the CN−and CO stretching modes of theR. eutropha H16 MBH compared with oxygen-sensitive standard [NiFe] hydrogenases (4, 9, 14, 41, 58)
| (Redox) state | Enzyme | CO [cm−1] | CN1 [cm−1] | CN2 [cm−1] |
|---|---|---|---|---|
| Niu-S | R. eutropha H16 MBH | 1943a | 2082 | 2104 |
| A. vinosum MBH | 1950 | 2089 | 2099 | |
| D. gigas | 1950 | 2089 | 2099 | |
| D. vulgaris Miyazaki F | 1958 | 2089 | 2100 | |
| Niu-A | R. eutropha H16 MBH | NDb | ND | ND |
| A. vinosum MBH | 1945 | 2083 | 2093 | |
| D. gigas | 1947 | 2083 | 2093 | |
| D. vulgaris Miyazaki F | 1956 | 2084 | 2094 | |
| Nir-B | R. eutropha H16 MBH | 1948 | 2081 | 2098 |
| A. vinosum MBH | 1944 | 2079 | 2090 | |
| D. gigas | 1946 | 2079 | 2090 | |
| D. vulgaris Miyazaki F | 1955 | 2081 | 2090 | |
| Nir-S | R. eutropha H16 MBH | 1910 | 2055 | 2063 |
| 1936 | 2075 | 2093 | ||
| A. vinosum MBH | 1911 | 2053 | 2067 | |
| 1932 | 2074 | 2086 | ||
| D. gigas | 1914 | 2055 | 2067 | |
| D. vulgaris Miyazaki F | 1922 | 2056 | 2070 | |
| Nia-S | R. eutropha H16 MBH | 1936 | 2075 | 2093 |
| A. vinosum MBH | 1932 | 2074 | 2086 | |
| D. gigas | 1934 | 2075 | 2087 | |
| D. vulgaris Miyazaki F | 1943 | 2075 | 2086 | |
| Nia-C | R. eutropha H16 MBH | 1957 | 2075 | 2097 |
| A. vinosum MBH | 1950 | 2074 | 2087 | |
| D. gigas | 1952 | 2073 | 2086 | |
| D. vulgaris Miyazaki F | 1961 | 2074 | 2085 | |
| Nia-L | R. eutropha H16 MBH | 1899 | 2040 | 2065 |
| A. vinosum MBH | 1898 | 2043 | 2058 | |
| Nia-SR | R. eutropha H16 MBH | 1948 | 2068 | 2087 |
| A. vinosum MBH | 1936 | 2059 | 2073 | |
| D. gigas | 1940 | 2059 | 2073 | |
| D. vulgaris Miyazaki F | 1948 | 2061 | 2074 | |
| Nia-SR′ | R. eutropha H16 MBH | 1926 | 2049 | 2075 |
| A. vinosum MBH | 1921 | 2048 | 2064 | |
| D. gigas | 1923 | — | — | |
| D. vulgaris Miyazaki F | 1933 | — | — | |
| Nia-SR″ | R. eutropha H16 MBH | 1919 | 2046 | 2071 |
| A. vinosum MBH | 1913 | 2043 | 2058 | |
| D. vulgaris Miyazaki F | 1919 | — | — | |
| Niia-S | R. eutropha H16 MBH | 1930 | 2060 | 2076 |
| D. gigas | 1936 | 2063 | 2070 |
a Boldface numbers indicate this work.
b ND, not detected; —, not assigned.
Oxidized MBH
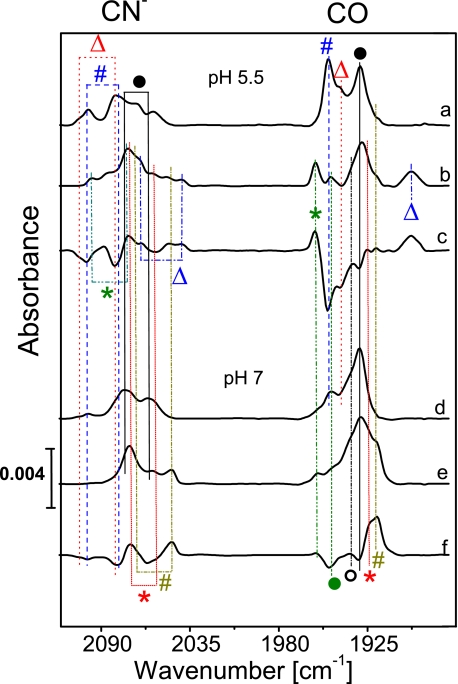
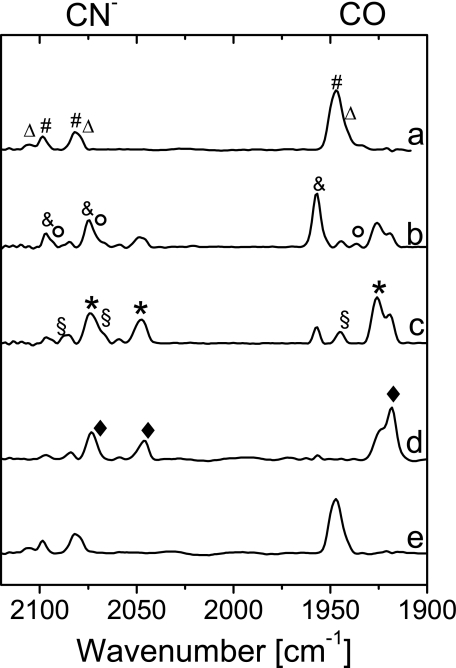
Fig. 4 displays the spectra of the oxidized enzyme recorded at pH 5.5 and 7.0 (traces a and d). The Nir-B state was identified based on the bands observed at 2098 and 2080 cm−1 (CN stretching bands) and 1948 cm−1 (CO stretching) by comparison with the respective bands of anaerobic standard hydrogenases (Table 2). Taking the integrated relative CO band intensities as a direct measure for the relative amounts of nickel states, the Nir-B was estimated to be 10–40% of the total MBH sample, the actual amount being dependent on the pH value and to some degree on the preparation. Generally, an increase of the amount of Nir-B was observed for lower pH values as compared with pH 8.0 used in Ref. 26; however, heterogeneity also increased toward lower pH. At pH 7.0 about 20% Nir-B was observed in the FTIR spectra, which is in agreement with the value estimated from EPR spectroscopy. An additional species with bands at wave-numbers of 1943 (CO) and 2104 cm−1 (CN−) corresponds to a small amount of another oxidized EPR-silent state, which might be either attributed to the so-called Niu-S (unready, silent) state (see Table 2) (14, 41, 42), which shows, in contrast to anaerobic standard [NiFe] hydrogenases, a rather fast H2-mediated re-activation within minutes instead of hours. Alternatively, these bands may originate from another “ready” state, derived from Nir-B, which has been recently observed in the D. vulgaris Miyazaki F enzyme.4 Because of the large number of overlapping bands in the CN stretching region, the second CN stretching mode of this species could not be identified unambiguously for the solubilized MBH dimer (HoxKG, Fig. 1C). However, this CN band position could be resolved in the FTIR spectra of the membrane-attached trimeric MBH (HoxKGZ, see below) and was identified at 2082 cm−1 (Table 2).
FIGURE 4.
FTIR spectra of the R. eutropha MBH, oxidized (as isolated) enzyme (traces a and d), the H2-reduced (1 bar H2) samples (traces b and e), and the corresponding difference spectra (H2-reduced minus oxidized, traces c and f) at different pH values: pH 5. 5, traces a–c; pH 7.0, traces d–f. Definition of the symbols is as follows: Δ (red), Niu/r-S; # (blue), Nir-B; ○, Nir/a-S; ●, Niia-S (only 20% active); Δ (blue), Nia-L; * (green), Nia-C; ● (green), Nia-SR; * (red), Nia-SR′; # (dark yellow), Nia-SR″.
At pH 8.0 only one dominant species was observed in more than 80% of the enzyme preparations (see supplemental Fig. S3). This nickel state showed IR bands at 2076, 2060, and 1930 cm−1 and included only a small fraction of about 15% that could be activated. The major portion, however, did not react with H2 and was therefore attributed to an irreversibly inactive state, designated as Ni(inactive)-S (Niia-S). In this state, solubilized, dimeric MBH could not be activated, even after prolonged incubation with H2. This inactive state is also present in MBH samples at pH 5.5 and pH 7.0, although with smaller amounts ranging between 40 and 60%. A similar inactive state was found in some cases for the D. gigas hydrogenase.5 The Niia-S state was not detectable in the spectra of the HoxKGZ trimer in the cytoplasmic membrane (see below). Therefore, we suggest that Niia-S is a non-native state.
As solubilization of the MBH dimer is associated with a downshift of the optimum pH for H2-oxidizing activity, one may rationalize the higher content of the Nir-B ready state and the lower amount of the inactive Niia-S in the solubilized heterodimer at lower pH values. Even though the Nir-B state was clearly detectable, FTIR signals attributable to the Niu-A state were not observed. This is in agreement with the EPR spectroscopic results.
Reduced MBH
FTIR spectra were recorded for dimeric MBH samples at different pH values (5.5 and 7.0) after incubation under an atmosphere of 1 bar H2 (Fig. 4, traces b and e). At pH 5.5 and a redox potential of −320 mV, IR bands corresponding to Nia-C (2097, 2075, and 1957 cm−1) and Nia-L (2065, 2040, and 1899 cm−1) were observed (Table 2). Contrary to standard [NiFe] hydrogenases Nia-L was found even at ambient temperature (10 °C). The Nia-C and Nia-L state mixture constituted each about 10% of the enzyme, which is in agreement with the data obtained by EPR spectroscopy. Because of the contributions of active Nia-SR and inactive Niia-S (about 50%) states, specifically the spectral cogent CN− stretching region is difficult to disentangle. However, comparison with the spectra of the reduced trimeric, membrane-attached enzyme (see below) allows for an assignment of the bands at 1948, 1926, and 1919 cm−1 in the CO stretching region and at 2068/2087, 2049/2075, and 2046/2071 cm−1 in the CN− stretching region. These frequencies are characteristic for the EPR-silent fully reduced states Nia-SR, Nia-SR′, and Nia-SR″ (Table 2). As described above, the pronounced CO absorption band at 1930 cm−1 did barely change upon incubation with H2 and was therefore assigned to the inactive Ni(inactive)-S state.
At 1 bar H2 and pH 7.0, no Nia-C nor Nia-L but only the fully reduced states Nia-SR, Nia-SR′, Nia-SR″ were observed and assigned on the basis of their characteristic band positions (Fig. 4 and Table 2).
To highlight the fraction of redox active enzyme, the “H2-reduced” minus “oxidized (as isolated)” difference spectra obtained for MBH samples buffered at different pH values are shown Fig. 4 (traces c and f). Bands originating from reduced and oxidized species appear as positive and negative signals, respectively. The difference spectra show that the main redox active species in the solubilized, dimeric MBH was Nir-B in addition to a small amount of “unready” or ready Ni-S state (see above). The large fraction of the EPR-silent Niia-S species, which increased at higher pH values, remained inactive and is therefore not visible in the IR-difference spectra.
The relative contributions of the reduced Nia-SR, Nia-SR′, and Nia-SR″ subspecies depend on the pH (see Fig. 4 and data related to the trimeric species, described below). This behavior has also been observed for the A. vinosum MBH enzyme (14) and suggests the involvement of protonable groups in the equilibria between these states.
EPR Spectroscopic Characterization of the MBH Protein as a Constituent of the Cytoplasmic Membrane
Preparations of the solubilized dimeric R. eutropha MBH contained a large protein fraction (up to 80%) that did not react with hydrogen, hence being catalytically inactive. Also the active fraction itself showed peculiar properties such as the appearance of Nia-L in nonilluminated samples. These findings indicate structural and functional rearrangements because of the detachment of the enzyme from the membrane. Thus, the spectroscopic studies were expanded to entire membrane fragments containing the native trimeric form of the R. eutropha MBH.
MBH at Positive Redox Potentials
Fig. 5 shows the EPR spectra of the R. eutropha cytoplasmic membrane. The spectrum of the oxidized (as isolated) membrane at 20 K and pH 7.0 (trace b) exhibited a complex line shape with a superposition of several paramagnetic species. In the air-saturated membrane sample at a redox potential of approximately +290 mV, the prominent contribution was represented by the complex broad and split EPR signal (Fig. 5, labeled by ▿), which results from the coupling of a [3Fe4S]+ complex with another paramagnetic center. The broad signal component is not present in the spectra of periplasmic anaerobic standard hydrogenases, such as that of D. vulgaris Miyazaki F (see Fig. 5, trace a). To test whether the complex spectrum is because of MBH-derived paramagnetic centers, membrane preparations of the R. eutropha mutant strain HF359, which carries a deletion in the MBH large subunit gene hoxG, have been investigated by EPR spectroscopy (supplemental Fig. S2). It has been shown previously that this strain does not possess HoxG as well as the mature subunit HoxK in the membrane (46). The spectral features attributable to the coupled [3Fe4S]+ cluster were only visible in MBH-containing wild-type membranes but were absent in the ΔhoxG mutant. Only a weak signal of negligible intensity was found at g = 2, the origin of which is unknown (supplemental Fig. S2).
FIGURE 5.
EPR spectra of R. eutropha MBH attached to the membrane (traces b–f) and oxidized (as isolated) D. vulgaris Miyazaki F hydrogenase (trace a). The enhanced (trace a) shows signals from superimposed gx and gy components of Niu-A and Nir-B. Experimental conditions are as follows: 1 milliwatt microwave power; 1 mT modulation amplitude, 12.5 kHz modulation frequency. Trace b, EPR spectra of oxidized (as isolated or reoxidized) MBH from a preparation of the cytoplasmic membrane at T = 20 K (redox potential +290 mV) showing signals from Nir-B and split signal from [3Fe4S]+. Trace c, oxidized (as isolated) from cytoplasmic membrane preparations at T = 80 K. Trace d, MBH, partially reduced with 5 mm β-mercaptoethanol (+40 mV), recorded at T = 20 K (the complex split [3Fe4S] signal has disappeared, and a narrow signal arises at a redox potential of +40 mV), Enhanced trace, Nir-B. Trace e, MBH partially reduced with 5 mm β-mercaptoethanol (+40 mV), recorded at T = 80 K; the dashed line represents the simulated spectrum of Nir-B. Trace f, H2-reduced samples from cytoplasmic membrane preparation (redox potential −390 mV). Experimental conditions for spectra (traces b–f) are as follows: 10 milliwatt microwave power; microwave frequency 9.56 GHz; 1 mT modulation amplitude, 12.5 kHz modulation frequency.
The membrane-attached enzyme also showed signals attributable to a Ni(III) center. The respective gx and gy values (2.30 and 2.17) agree with the Nir-B state observed in standard hydrogenases (51–54). The gz component is superimposed by the complex split signal of the [3Fe4S] cluster. Compared with the EPR signal of [3Fe4S]1+, the relative intensity of the double-integrated Ni(III) signal is ∼40%. Because FTIR spectroscopy indicates >80% Nir-B in these samples (see below), we conclude that approximately two spins per protein contribute to the split [3Fe4S]+ signal (see also results from dimeric MBH).
At 80 K, and a redox potential of +290 mV, the EPR signals attributed to Nir-B are no longer visible (Fig. 5, trace c). This indicates that at redox potentials, at which the complex [3Fe4S]+ signal is visible, the relaxation behavior of Nir-B in the R. eutropha hydrogenase is different from that observed in standard [NiFe] hydrogenases. Thus, both the Nir-B and the [3Fe-4S]+ center seem to be magnetically coupled to another paramagnetic center.
Upon mild reduction of the as-isolated membrane sample with 5 mm β-mercaptoethanol, the redox potential decreased to +40 mV resulting in a transformation of the complex broad and split EPR signal into a narrow signal, as it is usually observed for an uncoupled [3Fe4S]+ cluster (Fig. 5, trace d). This narrow signal at g = 2.0 is very similar to that found for the [3Fe4S]+ center in the D. vulgaris Miyazaki F standard [NiFe] hydrogenase (Fig. 5, trace a).
At 20 K and +40 mV the spectrum of Nir-B showed a broadening of each g component (Fig. 5, trace d). However, at higher temperatures (80 K) the EPR signals from Nir-B were strong and now well resolved, and the spectrum could be readily simulated, yielding all three g values (Table 1 and Fig. 5, trace e). This result indicates that the magnetic coupling to another paramagnetic state is no longer present at +40 mV. The absence of the paramagnetic species inducing a splitting of the [3Fe4S]+ signal at +290 mV also changed the relaxation behavior of the Nir-B in such a way that it can be observed at 80 K, a temperature where the EPR signal of the uncoupled [3Fe4S]+ cluster is no longer present.
Comparison of the second integrals of the broad and narrow EPR signals derived from the [3Fe4S]+ cluster at +290 and +40 mV showed that the intensity of the narrow signal was less than 50% of the broad and split signal. This is consistent with one spin per protein for the [3Fe4S]+ cluster. The total integral of the broad and split signal observed at a redox potential of +290 mV versus standard hydrogen electrode in the membrane corresponds to two spins per protein indicating the occurrence of an additional yet unknown high potential paramagnetic center coupling to [3Fe4S]1+.
It is important to note that signals attributable to the Niu-A state were not observed under any conditions. Traces of Niu-A below the detection limit (<2%) cannot be excluded.
Reduced MBH
To analyze the catalytically relevant reduced states of the MBH, the membrane samples were incubated under anaerobic conditions at pH 7.0 in the presence of 1 bar H2. The Nir-B and the [3Fe4S]+ signals disappeared, and weak broad EPR spectral features emerged in the low field range between 3100 and 3300 G, where the signals of the reduced Nia-C and Nia-L states are expected (Fig. 5, trace f). However, these broad features could also reflect superpositions of signals from other membrane proteins harboring paramagnetic metal species. On the high field side, between 3350 and 3700 G, a broad EPR spectrum emerged, displaying more than three spectral components in the g value range from 2.0 to 1.82. These signals cannot be attributed to only one reduced [4Fe4S]+ species. For anaerobic standard hydrogenases, two coupled [4Fe4S]+ centers have been proposed. For R. eutropha MBH, the signals may be attributed to a single [4Fe4S]+ center, which is coupled to the reduced [3Fe4S]0 cluster with a spin S = 2. This is supported by the relative quantification of the double-integrated signals of the broad [4Fe4S]+ signal in H2-reduced MBH compared with the split [3Fe4S]+ signal in oxidized samples (Fig. 5, trace b), which was attributed to two coupled paramagnetic species, each having a spin S = 1/2. The calculation revealed only one-half of the intensity for the [4Fe4S]+ signals in H2-reduced samples indicating that only one spin per protein (S = 1/2) is present, i.e. only one of the two [4Fe4S]+ clusters is reduced.
Re-oxidation of the membrane samples under air led to the same EPR signals of Nir-B and the [3Fe4S]+ center as observed previously in the untreated, as-isolated MBH (Fig. 5, trace b). This observation clearly indicates that the H2-mediated reduction is completely reversible when the MBH is associated with the cytoplasmic membrane.
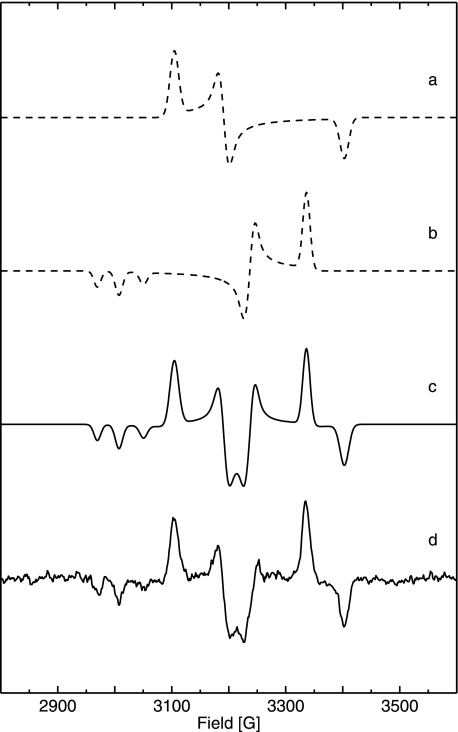
Reduction with 1 bar H2 at pH 5.5 led to the same broad EPR signals of reduced [4Fe4S]+ on the high field side as at pH 7.0 (Fig. 5, trace f). At higher temperature (80 K) the background signals from paramagnetic metal centers became weaker, and a well resolved EPR spectrum of Nia-C appeared. Upon illumination by white light for 25 min at 80 K, the Nia-C was converted completely to Nia-L, which appeared in three sub-species, with slightly different gx values (Table 1 and Fig. 6). The experimental “dark-minus-light” difference spectrum is shown together with its simulation in Fig. 6, trace b. Simulated spectra of the pure Nia-C and Nia-L states, the latter with negative intensity, are given in Fig. 6 (traces a and c). The integrated intensities of the Nia-C and Nia-L spectra are identical within the experimental error showing the complete conversion from Nia-C to Nia-L. The respective g values are very similar to those from standard hydrogenases (Table 1). When the membrane sample was kept in the dark for 15 min at 200 K, the Nia-L state was completely back-converted to the Nia-C state. Hence, the [NiFe] active site of the trimeric MBH shows the same light-induced Nia-C/Nia-L inter-conversion as reported for anaerobic standard [NiFe] hydrogenases (20, 60).
FIGURE 6.
Experimental and simulated EPR spectra of reduced, membrane-attached R. eutropha MBH pH 5. 5 at T = 80 K (for g values see Table 1). Trace a, simulation of Nia-C spectrum. Trace b, simulation of the spectrum of superimposed Nia-L states (inverted intensities) showing three different gx values. Trace c, sum of simulated Nia-C and Nia-L (inverted intensities) spectra. Trace d, experimental EPR difference spectrum (dark minus light) of H2-reduced cytoplasmic membrane at T = 80 K, Experimental conditions are as follows: 10 milliwatt microwave power; microwave frequency 9.56 GHz; 1 mT modulation amplitude, 12.5 kHz modulation frequency.
FTIR Spectroscopic Characterization of the MBH Protein as a Constituent of the Cytoplasmic Membrane
Oxidized MBH
The FTIR spectra of oxidized, membrane-attached MBH at pH 7.0 and at potentials between +250 and +300 mV displayed the characteristic band signature of an almost pure (up to 90%) Nir-B state (Fig. 7, trace a). The characteristic CN− stretching modes at 2098 and 2081 cm−1 and a CO stretching at 1948 cm−1 were in agreement with those of the Nir-B state of standard [NiFe] hydrogenases from D. gigas and D. vulgaris Miyazaki F (Table 2) (21, 41, 42). Only relatively small contributions (less than 20%) to the spectrum might originate from an EPR-silent oxidized form, which was also observed to a larger extent in the isolated MBH dimer. Signals attributable to the Niu-A state, if present at all, did not exceed the detection limit (<2%).
FIGURE 7.
FTIR spectra of the membrane-attached R. eutropha MBH from cytoplasmic membrane preparations as follows: oxidized (as isolated, +250 to +300 mV) at pH 7.0 (trace a), reduced at pH 5.5 with 0.05 bar H2 in N2 (−280 mV) (trace b), reduced at pH 5.5 with 1 bar H2 in N2 (−320 mV) (trace c), reduced at pH 7.0 with 1 bar H2 in N2 (−410 mV) (trace d), and re-oxidized with air (+250 to +300 mV) (trace e). Definition of the symbols is as follows: Δ, Niu/r-S; #, Nir-B; ○, Nir/a-S; &, Nia-C; §, Nia-SR; *, Nia-SR′; ♦, Nia-SR″.
Reduced MBH
Upon exposure to 1 bar H2 at pH 7.0, which results in a redox potential of −410 mV, the bands of the Nir-B state disappeared, and new bands were observed at lower wave numbers. According to previous data on standard [NiFe] hydrogenases (21), these bands are assigned to the fully reduced Nia-SR states. The most prominent stretching vibrations are located at 1926 and 1919 cm−1 in the CO region and some broader CN− stretchings at 2071 and 2046 cm−1 (Fig. 7, trace d). These band pairs can be attributed to the Nia-SR′ and Nia-SR″ sub-states. Their band positions and relative intensities slightly change as a function of the pH. Re-oxidation of the reduced membrane sample in air or argon resulted in an FTIR spectrum identical to that obtained prior to H2 reduction, irrespective of the pH value (Fig. 7, trace e). These findings clearly show that almost 100% of the membrane-attached MBH is reversibly redox-active.
Fig. 7 (traces b and c) illustrates the FTIR spectra of membrane samples at pH 5.5 reduced by applying different concentrations of H2 in a nitrogen atmosphere. The spectrum of the aerobic as-isolated sample shows predominantly Nir-B and a small amount of another oxidized EPR-silent state, e.g. Niu-S, as observed at pH 7.0 (data not shown). Reduction with H2 at 0.05 bar leads to the formation of a considerable amount of Nia-C (trace b), which was not observed at pH 7.0. Only small amounts of the Nia-SR′ and SR″ states were observed, probably because of the more positive redox potential (approximately −280 mV at pH 5.5). The observation of Nia-C with a characteristic CO stretching at 1957 cm−1 and corresponding CN stretching frequencies at 2075 and 2097 cm−1 is in agreement with the EPR spectra obtained from the same material at pH 5.5. Additionally, small amounts of the (Nir-S) Nia-S were found corresponding to the positions of the CO band at 1936 cm−1 and the CN− bands at 2075 and 2093 cm−1. This transitional state was also detectable in a time-resolved gas-exchange experiment (data not shown).
At a redox potential of −320 mV, which was achieved in the presence of 1 bar H2 at pH 5.5, the relative amount of Nia-C is considerably lower (Fig. 7, trace c). Interestingly, all three sub-states of the fully reduced form are visible, i.e. Nia-SR, Nia-SR′, and Nia-SR″. The two CO bands assigned to the Nia-SR′ and SR″ states show different relative intensities as compared with the spectrum taken at pH 7.0. Also, the two dominant bands of the CN− stretching modes are slightly shifted (2049 and 2075 cm−1) and broadened and show different relative intensities compared with the spectrum at pH 7 (2046 and 2071 cm−1). This comparison allows for a more reliable assignment of these bands to Nia-SR′ (1926, 2049, and 2075 cm−1) and Nia-SR″ (1919, 2046, and 2071 cm−1) (Table 2). Furthermore, Nia-SR is detectable with bands at 1946 (CO), 2068 (CN−), and 2087 (CN−) cm−1.
DISCUSSION
The heterodimeric membrane-bound hydrogenase of R. eutropha H16 is composed of a large subunit containing the [NiFe] active site and a small subunit presumably harboring one [3Fe4S] and two [4Fe4S] clusters. This basic composition, including the specific cofactors, is similar to that of the well studied anaerobic standard [NiFe] hydrogenases. By applying FTIR spectroscopy and chemical quantification of the cyanide content, we present evidence that the catalytic center of the R. eutropha MBH is equipped with one CO and two CN− ligands (see also Ref. 26).
In its native environment, the MBH has a trimeric structure, i.e. it is tightly bound to its primary electron acceptor, a membrane-integral cytochrome b. The treatment with detergent results in the detachment of MBH dimer from the membrane. The resulting solubilized MBH dimer has been repeatedly demonstrated to be suitable for electrochemical and electrocatalytic studies (11, 26). Because protein film voltammetry solely probes the electroactive protein, the inactive protein fraction does not influence the results in those experiments. The present EPR and FTIR results reveal that the solubilized dimeric MBH contains considerable amounts of the irreversibly inactive Niia-S state, even after fast and mild purification by affinity chromatography. Nevertheless, the catalytic site active fraction of the solubilized MBH dimer exhibited the redox states known from oxygen-sensitive standard [NiFe] hydrogenases. Notably, the Niia-S state within the MBH dimer could be detected and quantified only by FTIR spectroscopy, because this particular state is EPR-silent. In EPR spectroscopic experiments, the heterogeneity of the solubilized enzyme samples just results in a lower net yield of EPR-visible signals. Therefore, we are not able to draw conclusions about the state composition of the catalytic site from the earlier studies on MBH dimer preparations, where FTIR spectroscopy was not applied (38, 39).
Because the spectroscopic experiments of solubilized dimeric MBH were hampered by the inactive enzyme fraction, a new approach focused on the preparation of an MBH sample in its native trimeric state. Thus, we performed FTIR and EPR studies on isolated cytoplasmic membrane fractions containing overproduced trimeric MBH, including the b-type cytochrome. To our knowledge, this is the first spectroscopic study on a membrane-bound [NiFe] hydrogenase in its native environment. Only the trimeric MBH preparation was found to be 100% reducible by H2 and re-oxidizable to full extent with argon or air.
The integrated spectroscopic analysis by EPR and FTIR revealed that almost all functional [NiFe] states, which have been characterized in the standard [NiFe] hydrogenases of anaerobic bacteria, were also found in the MBH albeit with one remarkable exception. There was no indication for the presence of the oxygen-inactivated Niu-A state under any conditions. Niu-A represents a state in which an oxygen species, presumably a peroxo ligand (16, 17), blocks the active site. The resulting unready, inactive enzyme requires a long term reactivation with H2 to acquire a catalytically active conformation. Two other hydrogenases in R. eutropha, the soluble, hexameric NAD+-reducing hydrogenase and the cytoplasmic H2-sensing hydrogenase, share the exceptional ability of being quickly reactivated after exposure to oxygen, and for none of them have Niu-A signals been detected (25, 61).
The prevention of forming the Niu-A state appears to be a prerequisite for H2 catalysis in the presence of O2 and represents a fundamental feature of the so-called oxygen-tolerant [NiFe] hydrogenases. This conclusion is strongly supported by the spectroscopic analysis of the O2-tolerant hydrogenases from R. metallidurans CH34 (39), A. aeolicus (62), and A. ferrooxidans (51), all of which do not exhibit the Niu-A state.
In view of the close structural similarity of the R. eutropha MBH and other O2-tolerant membrane-bound hydrogenases from R. metallidurans and A. aeolicus with anaerobic standard [NiFe] hydrogenases, the following question arises. What is the structural and/or functional basis for the observed O2 tolerance?
FTIR spectroscopy on the different redox states of the [NiFe] active site shows that the trimeric membrane-attached oxidized form of the MBH mainly resides in the Nir-B state with a minor contribution of an unready and/or ready EPR silent state (Niu/r′-S). Both states were found to react completely with H2 and exhibit similar EPR and FTIR parameters as observed for anaerobic standard [NiFe] hydrogenases (21).4 Upon reduction with H2, the reduced Nia-C and Nia-L states, the fully reduced Nia-SR species as well as reduced [4Fe4S]+ clusters were identified. During reduction and re-oxidation of the samples, the Nir-S/Nia-S states also were observed as intermediates. The redox-dependent stretching mode frequencies of the CO and CN− ligands were in good agreement to those observed in standard [NiFe] hydrogenases. Only minor shifts of ±5 cm−1 were noted. A larger shift of up to +10 cm−1 was observed only for one CN− stretching mode, indicating differences in the protein environment of one of the CN ligands. However, the generally small frequency deviations compared with the respective IR bands in standard [NiFe] hydrogenases allowed the identification of all functional redox states. This observation is in accordance with the notion that, apart from Niu-A, the active site of the trimeric MBH is very similar to that of standard hydrogenases.
In addition to the lack of Niu-A, another feature of the trimeric MBH was intriguing, namely the complex and split EPR signal attributed to the [3Fe4S]+ cluster. A similar complex EPR spectrum of the [3Fe4S]+ cluster has been observed previously for the solubilized membrane-bound hydrogenases of R. metallidurans (39) and A. aeolicus (62) as well as for the enzyme of A. vinosum (7, 63–65). In all three cases this signal has been attributed to a [3Fe-4S]+ cluster magnetically coupled to another paramagnetic center (for an overview see supplemental Table S2). In the case of the A. vinosum MBH, it was proposed that the additional paramagnetic center could be either the Ni(III) in the Nir-B state of the active site (7, 63–65), or a cofactor Xox that is only paramagnetic in its oxidized form and mediates a magnetic coupling between the Nir-B and the [3Fe4S]+ cluster. This Xox center was proposed to be either an additional Fe3+ species located between the [NiFe] site and the medial cluster, i.e. close to the proximal Fe-S cluster (7, 64, 66) or, alternatively, the proximal [4Fe4S] cluster itself in a high potential [4Fe4S]3+ form (65).
Based on the present data only the two latter interpretations are plausible explanations for the unusual EPR signal of the [3Fe4S]+ cluster in the R. eutropha MBH. A direct coupling between the Nir-B and the medial [3Fe4S]+ cluster would require an exchange of the cluster positions resulting in a proximal [3Fe4S]+ cluster and a medial [4Fe4S] cluster (39). Such a cluster exchange would explain the split signal of the oxidized [3Fe4S]1+. However, the amino acid composition at the binding sites of the Fe-S clusters does not support this hypothesis, because the medial Fe-S cluster is coordinated by only three cysteine ligands, which is not sufficient for the coordination in a [4Fe4S] center (see supplemental Fig. S4). Moreover, we observed that in β-mercaptoethanol-treated samples of trimeric MBH (redox potential +40 mV), the split [3Fe4S]+ EPR signal is converted completely into the narrow signal of an uncoupled [3Fe4S]+. This conversion had no major effect on the Nir-B signal (except for a modified relaxation behavior) indicating that the [3Fe4S]+ split EPR signal does not result from a direct magnetic coupling between [3Fe4S]+ and Nir-B. This is supported by the observation that the FTIR bands related to the Nir-B state did not change upon mild reduction by β-mercaptoethanol (data not shown).
The [3Fe4S]+ EPR spectra of R. eutropha H16 MBH are similar to those reported for the purified dimeric MBH of the closely related strain R. metallidurans CH34 that displayed the complex EPR signal only at high redox potentials (+323 mV) (39). This indicates that the splitting of the EPR signal is associated with the oxidation of the postulated cofactor X (Xox). Such a cofactor is in full agreement with the observation that the R. eutropha H16 MBH exhibits the complex [3Fe4S]+ EPR split signal at a redox potential of +290 mV, which transforms to a narrow signal at lower redox potential of +40 mV (Fig. 4). The latter signal is generally indicative for an uncoupled [3Fe4S]+ center (7, 62, 64, 66).
Notably, the relaxation behavior of Nir-B in R. eutropha MBH was found to be different from that of standard hydrogenases. At high redox potential (+290 mV), the Nir-B signals vanished at temperatures above 60 K. This demonstrates that the proposed high potential paramagnetic species that causes the split EPR signal of [3Fe4S]+ leads, by weak magnetic interaction, also to enhanced spin relaxation of Nir-B. This indicates a close proximity to the medial [3Fe4S] cluster and also a proximity to the active site nickel. The proposed high potential species is therefore consistent with the presence of an additional iron close to the position of the proximal [4Fe4S] cluster or, alternatively, with the presence of an oxidized high potential [4Fe4S]3+ at the proximal position.
The presence of an additional metal ion at medium distance to the [Ni-Fe] active site is known for some [NiFe] hydrogenases, whose structures have been solved. In case of Desulfomicrobium baculatum (67) and Desulfovibrio desulfuricans (58) iron and magnesium were identified, respectively. Such an additional metal species (as Xox) has not yet been found in the R. eutropha H16 MBH.
Further indication for an additional high potential paramagnetic species with S = 1/2 is based on the integral signal intensity of the complex split [3Fe-4S]+ EPR signal, corresponding to more than one spin per protein (supplemental Table S1), whereas the narrow [3Fe-4S]+ EPR signal corresponds to one spin per protein.
Significant differences as compared with anaerobic standard hydrogenases were also found in the EPR spectra of the reduced [NiFe] and [4Fe4S] states. In the case of anaerobic standard [NiFe] hydrogenases, a strong coupling is observed between Ni-C and the reduced proximal [4Fe4S] cluster. The temperature-dependent EPR spectra of the MBH, incubated with H2 showed, however, no indication for a magnetic coupling between Nia-C/Nia-L and reduced [4Fe4S]. This finding indicates that the electron, which is released by the heterolytic cleavage of H2 resulting in the Nia-C state, is transferred all the way to the terminal [4Fe4S] cluster of the MBH. This cluster, however, is too far away from the catalytic site to induce coupled EPR signals of Nia-C/Nia-L. Moreover, relative spin quantification of the MBH for the reduced signal showed only 50% of the intensity as compared with the split signal indicating only one reduced [4Fe4S] cluster per protein after incubation with 1 bar H2. This again indicates a unique redox behavior of the proximal Fe-S cluster in R. eutropha MBH.
The peculiar spectroscopic features of the R. eutropha MBH point to a modification at or near the proximal Fe-S cluster, whereas the active [NiFe] site seems to have a highly similar structure as compared with anaerobic standard [NiFe] hydrogenases. This is supported by the results of a recent biochemical and electrochemical investigation of the MBH (34). The exchange of amino acids close to the active site, which are unique in the MBH and closely related O2-tolerant hydrogenases, revealed mutant proteins with a significantly lowered Ki toward oxygen. However, these MBH variants still showed a considerable O2 tolerance, which was still orders of magnitude higher that that of O2-sensitive standard [NiFe] hydrogenases (34). These results indicate that the reason for O2 tolerance may not be intimately related to the unique amino acid composition in the first coordination shell of the MBH active site. The oxygen tolerance could be explained by an alteration of the proximal Fe-S cluster being functional in preventing the formation of the oxygen-inhibited Niu-A state. Remarkably, there are two additional cysteine residues in close vicinity to the proximal [4Fe4S] cluster that are absent in oxygen-sensitive standard [NiFe] hydrogenases (see supplemental Fig. S4). These two additional cysteines may play a crucial role in the unusual redox potential dependent splitting of the [3Fe4S]+ EPR signal and might provide additional binding sites for an unusual proximal cluster, of higher complexity than a standard-type cubane. Interestingly, these two additional cysteines are only conserved in other membrane-bound hydrogenases, like those from R. metallidurans (39) and A. aeolicus (62), which have been shown to be or are proposed to be O2-tolerant (supplemental Fig. S4).
Further studies, including the construction of R. eutropha MBH mutant proteins with altered coordination of the proximal and medial iron-sulfur clusters, are envisaged to explore their detailed structure and their potential role in the unusual oxygen tolerance of the membrane-bound hydrogenases from aerobic H2-oxidizing bacteria.
Supplementary Material
Acknowledgments
We are grateful to Wolfgang Lubitz and Maria Pandelia (Max Plank Institute) for providing a hydrogenase sample from D. vulgaris Miyazaki F and for helpful discussions. Robert Bittl is gratefully acknowledged for helpful discussions. Metal analyses have been performed with the help of Silke Leimkühler and Meina Neumann using ICP-OES (University of Potsdam).
This work was supported by Deutsche Forsch ungs ge mein schaft Grants SFB 498 TP C1, TP C9 and by Cluster of Excellence “Unifying Concepts in Catalysis,” and by the Federal Ministry of Education and Research (BMBF, project “Bio-H2”).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S4.
W. Lubitz and M. Pandelia, personal communication.
A. L. De Lacey, personal communication.
- MBH
- membrane-bound hydrogenase
- FTIR
- Fourier transform infrared
- ICP-OES
- inductive coupled plasma optical emission spectroscopy
- mT
- millitesla.
REFERENCES
- 1.Cammack R., Frey M., Robson R. (eds) ( 2001) Hydrogen As a Fuel: Learning from Nature, Taylor and Francis Ltd., London [Google Scholar]
- 2.Volbeda A., Charon M. H., Piras C., Hatchikian E. C., Frey M., Fontecilla-Camps J. C. ( 1995) Nature 373, 580– 587 [DOI] [PubMed] [Google Scholar]
- 3.Higuchi Y., Yagi T., Yasuoka N. ( 1997) Structure 5, 1671– 1680 [DOI] [PubMed] [Google Scholar]
- 4.Peters J. W., Lanzilotta W. N., Lemon B. J., Seefeldt L. C. ( 1998) Science 282, 1853– 1858 [DOI] [PubMed] [Google Scholar]
- 5.Nicolet Y., Piras C., Legrand P., Hatchikian C. E., Fontecilla-Camps J. C. ( 1999) Struct. Fold. Design 7, 13– 23 [DOI] [PubMed] [Google Scholar]
- 6.Pilak O., Mamat B., Vogt S., Hagemeier C. H., Thauer R. K., Shima S., Vonrhein C., Warkentin E., Ermler U. ( 2006) J. Mol. Biol. 358, 798– 809 [DOI] [PubMed] [Google Scholar]
- 7.Surerus K. K., Chen M., van der Zwaan J. W., Rusnak F. M., Kolk M., Duin E. C., Albracht S. P., Münck E. ( 1994) Biochemistry 33, 4980– 4993 [DOI] [PubMed] [Google Scholar]
- 8.Albracht S. P. ( 1994) Biochim. Biophys. Acta 1188, 167– 204 [DOI] [PubMed] [Google Scholar]
- 9.Volbeda A., Garcia E., Piras C., de Lacey A. L., Fernandez V. M., Hatchikian E. C., Frey M., Fontecilla-Camps J. C. ( 1996) J. Am. Chem. Soc. 118, 12989– 12996 [Google Scholar]
- 10.Pierik A. J., Roseboom W., Happe R. P., Bagley K. A., Albracht S. P. ( 1999) J. Biol. Chem. 274, 3331– 3337 [DOI] [PubMed] [Google Scholar]
- 11.Vincent K. A., Parkin A., Lenz O., Albracht S. P., Fontecilla-Camps J. C., Cammack R., Friedrich B., Armstrong F. A. ( 2005) J. Am. Chem. Soc. 127, 18179– 18189 [DOI] [PubMed] [Google Scholar]
- 12.Adams M. W. ( 1990) Biochim. Biophys. Acta 1020, 115– 145 [DOI] [PubMed] [Google Scholar]
- 13.Cammack R., Fernandez V. M., Schneider K. ( 1986) Biochimie 68, 85– 91 [DOI] [PubMed] [Google Scholar]
- 14.Bleijlevens B., van Broekhuizen F. A., De Lacey A. L., Roseboom W., Fernandez V. M., Albracht S. P. ( 2004) J. Biol. Inorg. Chem. 9, 743– 752 [DOI] [PubMed] [Google Scholar]
- 15.Lamle S. E., Albracht S. P., Armstrong F. A. ( 2004) J. Am Chem. Soc. 126, 14899– 14909 [DOI] [PubMed] [Google Scholar]
- 16.Ogata H., Hirota S., Nakahara A., Komori H., Shibata N., Kato T., Kano K., Higuchi Y. ( 2005) Structure 13, 1635– 1642 [DOI] [PubMed] [Google Scholar]
- 17.Volbeda A., Martin L., Cavazza C., Matho M., Faber B. W., Roseboom W., Albracht S. P., Garcin E., Rousset M., Fontecilla-Camps J. C. ( 2005) J. Biol. Inorg. Chem. 10, 239– 249 [DOI] [PubMed] [Google Scholar]
- 18.Lubitz W., Reijerse E., van Gastel M. ( 2007) Chem. Rev. 107, 4331– 4365 [DOI] [PubMed] [Google Scholar]
- 19.Fernandez V. M., Hatchikian E. C., Cammack R. ( 1985) Biochim. Biophys. Acta 832, 69– 79 [Google Scholar]
- 20.Brecht M., van Gastel M., Buhrke T., Friedrich B., Lubitz W. ( 2003) J. Am. Chem. Soc. 125, 13075– 13083 [DOI] [PubMed] [Google Scholar]
- 21.Kurkin S., George S. J., Thorneley R. N., Albracht S. P. ( 2004) Biochemistry 43, 6820– 6831 [DOI] [PubMed] [Google Scholar]
- 22.Aragno M., Schlegel H. G. ( 1992) in The Prokaryotes ( Balows H. G., Trüper H. G., Dworkin M., Harder W., Schleifer K. H. eds) pp. 344– 384, Springer-Verlag Inc., New York [Google Scholar]
- 23.Friedrich B., Schwartz E. ( 1993) Annu. Rev. Microbiol. 47, 351– 383 [DOI] [PubMed] [Google Scholar]
- 24.Vignais P. M., Billoud B. ( 2007) Chem. Rev. 107, 4206– 4272 [DOI] [PubMed] [Google Scholar]
- 25.Burgdorf T., Löscher S., Liebisch P., Van der Linden E., Galander M., Lendzian F., Meyer-Klaucke W., Albracht S. P., Friedrich B., Dau H., Haumann M. ( 2005) J. Am. Chem. Soc. 127, 576– 592 [DOI] [PubMed] [Google Scholar]
- 26.Vincent K. A., Cracknell J. A., Lenz O., Zebger I., Friedrich B., Armstrong F. A. ( 2005) Proc. Natl. Acad. Sci. U. S. A. 102, 16951– 16954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buhrke T., Lenz O., Krauss N., Friedrich B. ( 2005) J. Biol. Chem. 280, 23791– 23796 [DOI] [PubMed] [Google Scholar]
- 28.Van der Linden E., Burgdorf T., Bernhard M., Bleijlevens B., Friedrich B., Albracht S. P. ( 2004) J. Biol. Inorg. Chem. 9, 616– 626 [DOI] [PubMed] [Google Scholar]
- 29.Happe R. P., Roseboom W., Egert G., Friedrich C. G., Massanz C., Friedrich B., Albracht S. P. ( 2000) FEBS Lett. 466, 259– 263 [DOI] [PubMed] [Google Scholar]
- 30.Duché O., Elsen S., Cournac L., Colbeau A. ( 2005) FEBS J. 272, 3899– 3908 [DOI] [PubMed] [Google Scholar]
- 31.Bernhard M., Benelli B., Hochkoeppler A., Zannoni D., Friedrich B. ( 1997) Eur. J. Biochem. 248, 179– 186 [DOI] [PubMed] [Google Scholar]
- 32.Schink B., Schlegel H. G. ( 1979) Biochim. Biophys. Acta 567, 315– 324 [DOI] [PubMed] [Google Scholar]
- 33.Kortlüke C., Horstmann K., Schwartz E., Rohde M., Binsack R., Friedrich B. ( 1992) J. Bacteriol. 174, 6277– 6289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ludwig M., Cracknell J. A., Vincent K. A., Armstrong F. A., Lenz O. ( 2009) J. Biol. Chem. 284, 465– 477 [DOI] [PubMed] [Google Scholar]
- 35.De Lacey A. L., Fernandez V. M., Rousset M., Cammack R. ( 2007) Chem. Rev. 107, 4304– 4330 [DOI] [PubMed] [Google Scholar]
- 36.Vincent K. A., Cracknell J. A., Clark J. R., Ludwig M., Lenz O., Friedrich B., Armstrong F. A. ( 2006) Chem. Commun. 28, 5033– 5035 [DOI] [PubMed] [Google Scholar]
- 37.Ihara M., Nishihara H., Yoon K. S., Lenz O., Friedrich B., Nakamoto H., Kojima K., Honma D., Kamachi T., Okura I. ( 2006) Photochem. Photobiol. 82, 676– 682 [DOI] [PubMed] [Google Scholar]
- 38.Schneider K., Patil D. S., Cammack R. ( 1983) Biochim. Biophys. Acta 748, 353– 361 [Google Scholar]
- 39.Knüttel K., Schneider K., Erkens A., Plass W., Müller A., Bill E., Trautwein A. X. ( 1994) Bull. Pol. Acad. Sci. Chem. 42, 495– 511 [Google Scholar]
- 40.Bagley K. A., Duin E. C., Roseboom W., Albracht S. P., Woodruff W. H. ( 1995) Biochemistry 34, 5527– 5535 [DOI] [PubMed] [Google Scholar]
- 41.Fichtner C., Laurich C., Bothe E., Lubitz W. ( 2006) Biochemistry 45, 9706– 9716 [DOI] [PubMed] [Google Scholar]
- 42.De Lacey A. L., Hatchikian E. C., Volbeda A., Frey M., Fontecilla-Camps J. C., Fernandez V. M. ( 1997) J. Am. Chem. Soc. 119, 7181– 7189 [Google Scholar]
- 43.Lenz O., Gleiche A., Strack A., Friedrich B. ( 2005) J. Bacteriol. 187, 6590– 6595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Witholt B., Boekhout M., Brock M., Kingma J., van Heerikhuizen H., Leij L. D. ( 1976) Anal. Biochem. 74, 160– 170 [DOI] [PubMed] [Google Scholar]
- 45.Schwartz E., Gerischer U., Friedrich B. ( 1998) J. Bacteriol. 180, 3197– 3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schubert T., Lenz O., Krause E., Volkmer R., Friedrich B. ( 2007) Mol. Microbiol. 66, 453– 467 [DOI] [PubMed] [Google Scholar]
- 47.Bradford M. M. ( 1976) Anal. Biochem. 72, 248– 254 [DOI] [PubMed] [Google Scholar]
- 48.Laemmli U. K. ( 1970) Nature 227, 680– 685 [DOI] [PubMed] [Google Scholar]
- 49.Stesmans A., Vangorp G. ( 1989) Rev. Sci. Instrum. 60, 2949– 2952 [Google Scholar]
- 50.Stoll S., Schweiger A. ( 2006) J. Magn. Res. 178, 42– 55 [DOI] [PubMed] [Google Scholar]
- 51.Schröder O., Bleijlevens B., de Jongh T. E., Chen Z., Li T., Fischer J., Förster J., Friedrich C. G., Bagley K. A., Albracht S. P., Lubitz W. ( 2007) J. Biol. Inorg. Chem. 12, 212– 233 [DOI] [PubMed] [Google Scholar]
- 52.Foerster S., Stein M., Brecht M., Ogata H., Higuchi Y., Lubitz W. ( 2003) J. Am. Chem. Soc. 125, 83– 93 [DOI] [PubMed] [Google Scholar]
- 53.Trofanchuk O., Stein M., Gessner C., Lendzian F., Higuchi Y., Lubitz W. ( 2000) J. Biol. Inorg. Chem. 5, 36– 44 [DOI] [PubMed] [Google Scholar]
- 54.van Gastel M., Stein M., Brecht M., Schröder O., Lendzian F., Bittl R., Ogata H., Higuchi Y., Lubitz W. ( 2006) J. Biol. Inorg. Chem. 11, 41– 51 [DOI] [PubMed] [Google Scholar]
- 55.Foerster S., van Gastel M., Brecht M., Lubitz W. ( 2005) J. Biol. Inorg. Chem. 10, 51– 62 [DOI] [PubMed] [Google Scholar]
- 56.Dole F., Medina M., More C., Cammack R., Bertrand P., Guigliarelli B. ( 1996) Biochemistry 35, 16399– 16406 [DOI] [PubMed] [Google Scholar]
- 57.Podzuweit H. G., Schneider K., Knüttel H. ( 1987) Biochim. Biophys. Acta 905, 435– 446 [DOI] [PubMed] [Google Scholar]
- 58.Rousset M., Montet Y., Guigliarelli B., Forget N., Asso M., Bertrand P., Fontecilla-Camps J. C., Hatchikian E. C. ( 1998) Proc. Natl. Acad. Sci. U. S. A. 95, 11625– 11630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Agrawal A. G., van Gastel M., Gärtner W., Lubitz W. ( 2006) J. Phys. Chem. B. 110, 8142– 8150 [DOI] [PubMed] [Google Scholar]
- 60.Fichtner C., van Gastel M., Lubitz W. ( 2003) Phys. Chem. Chem. Phys. 5, 5507– 5513 [Google Scholar]
- 61.Buhrke T., Löscher S., Lenz O., Schlodder E., Zebger I., Andersen L. K., Hildebrandt P., Meyer-Klaucke W., Dau H., Friedrich B., Haumann M. ( 2005) J. Biol. Chem. 280, 19488– 19495 [DOI] [PubMed] [Google Scholar]
- 62.Brugna-Guiral M., Tron P., Nitschke W., Stetter K. O., Burlat B., Guigliarelli B., Bruschi M., Giudici-Orticoni M. T. ( 2003) Extremophiles 7, 145– 157 [DOI] [PubMed] [Google Scholar]
- 63.Albracht S. P., Albrechtellmer K. J., Schmedding D. J., Slater E. C. ( 1982) Biochim. Biophys. Acta 681, 330– 334 [Google Scholar]
- 64.Albracht S. P., Vanderzwaan J. W., Fontijn R. D. ( 1984) Biochim. Biophys. Acta 766, 245– 258 [Google Scholar]
- 65.Albracht S. P., Kalkman M. L., Slater E. C. ( 1983) Biochim. Biophys. Acta 724, 309– 316 [Google Scholar]
- 66.Coremans J. M., van der Zwaan J. W., Albracht S. P. ( 1992) Biochim. Biophys. Acta 1119, 157– 168 [DOI] [PubMed] [Google Scholar]
- 67.Garcin E., Vernede X., Hatchikian E. C., Volbeda A., Frey M., Fontecilla-Camps J. C. ( 1999) Structure 7, 557– 566 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.