Abstract
Chromosomal translocation requires formation of paired double strand DNA breaks (DSBs) on heterologous chromosomes. One of the most well characterized oncogenic translocations juxtaposes c-myc and the immunoglobulin heavy chain locus (IgH) and is found in Burkitt’s lymphomas in humans and plasmacytomas in mice. DNA breaks in IgH leading to c-myc/IgH translocations are created by activation induced cytidine deaminase (AID) during antibody class switch recombination or somatic hypermutation. However, the source of DNA breaks at c-myc is not known. Here we provide evidence for the c-myc promoter region being required in targeting AID-mediated DNA damage to produce DSBs in c-myc that lead to c-myc/IgH translocations in primary B lymphocytes. Thus, in addition to producing somatic mutations and DNA breaks in antibody genes, AID is also responsible for the DNA lesions in oncogenes that are required for their translocation.
Introduction
Chromosome translocations have been proposed to require formation of pairs of double-strand DNA breaks (DSBs) and trans-chromosomal ligation of the broken ends (Richardson and Jasin, 2000). Among somatic cells, mature B lymphocytes are particularly prone to DSB formation in Ig loci during antibody gene diversification reactions and to malignant transformation by Ig translocation (Adams et al., 1983; Adams et al., 1982; Adams et al., 1985; Cory et al., 1983; Crews et al., 1982; Dalla-Favera et al., 1983; Erikson et al., 1983; Hamlyn and Rabbitts, 1983; Jankovic et al., 2007; Kuppers and Dalla-Favera, 2001; Leder et al., 1983; Marcu et al., 1983; Stanton et al., 1983; Taub et al., 1982).
Two different reactions diversify antibody genes in mature B cells: class switch recombination (CSR) and somatic hypermutation (SHM). CSR involves deletion of DNA sequences and replacement of one antibody constant region for another while retaining the specificity of the antigen receptor (Chaudhuri et al., 2007; Stavnezer et al., 2008). In contrast, SHM introduces non-templated nucleotide changes in antibody variable region genes, producing variants with differing affinity for antigen (Di Noia and Neuberger, 2007; Peled et al., 2008). Although these reactions are very different, they are both initiated by AID, an enzyme that is expressed primarily in germinal center B cells (Muramatsu et al., 2000; Muramatsu et al., 1999; Revy et al., 2000). The same enzyme is also essential for c-myc/IgH translocations in primary B cells and mouse plasmacytomas (Kovalchuk et al., 2007; Potter, 2003; Ramiro et al., 2006; Ramiro et al., 2004; Takizawa et al., 2008).
AID initiates CSR and SHM by deaminating cytosine residues in single stranded DNA, which is exposed during transcription (Bransteitter et al., 2003; Chaudhuri et al., 2003; Di Noia and Neuberger, 2002; Dickerson et al., 2003; Imai et al., 2003; Petersen-Mahrt et al., 2002; Pham et al., 2003; Rada et al., 2002; Ramiro et al., 2003). The resulting U:G mismatch is recognized by either uracyl DNA glycosylase or mismatch repair proteins and processed by one of several DNA repair pathways to produce point mutations in variable regions or a combination of mutations and DSBs in switch regions (Di Noia and Neuberger, 2007; Peled et al., 2008; Stavnezer et al., 2008). AID introduces multiple DSBs in switch regions during CSR as evidenced by internal deletions (Dudley et al., 2002; Nagaoka et al., 2002; Petersen et al., 2001; Reina-San-Martin et al., 2003) and accumulation of foci of DNA repair factors at the IgH locus preceding CSR (Petersen et al., 2001; Reina-San-Martin et al., 2004). Deletions and duplications are less frequent in Ig variable regions accounting for 5% of all hypermutations (Goossens et al., 1998; Wilson et al., 1998). C-myc translocates to both Ig variable and switch regions and in both cases the translocations are AID dependent (Dorsett et al., 2007; Ramiro et al., 2004).
In addition to targeting Ig loci, AID also produces mutations in a number of other genes including oncogenes (Gordon et al., 2003; Liu et al., 2008; Muschen et al., 2000; Pasqualucci et al., 1998; Pasqualucci et al., 2001; Shen et al., 1998). However, mutations in c-myc are either absent (Pasqualucci et al., 2001; Shen et al., 1998), or occur at a rate that is several orders of magnitude lower than in IgH (Liu et al., 2008). In addition the available sequence analysis shows no evidence of AID dependent deletions or duplications in c-myc or other oncogenes that might suggest AID dependent DSB formation (Gordon et al., 2003; Liu et al., 2008; Mullighan et al., 2007; Muschen et al., 2000; Pasqualucci et al., 1998; Pasqualucci et al., 2001; Shen et al., 1998). Furthermore, AID induced lesions in genes other than antibody genes appear to be repaired by a pathway that is distinct from and less error prone than lesions in IgH (Liu et al., 2008). Indeed, when DSBs have been detected in c-myc in human B cells they have been found to be AID independent (Zan et al., 2003).
One potential source for the DSBs in c-myc is fragile DNA segments that are prone to form non-B-DNA configurations including Z-DNA, triple helical H-DNA, and G-quadruplex DNA (reviewed in (Liu and Levens, 2006; Marcu et al., 1992; Spencer and Groudine, 1991; Wierstra and Alves, 2008)). These sequences are particularly susceptible to AID independent DNA damage and DSB formation during transcription or replication, but also in response to environmental agents and metabolic stress and they have been implicated as a source of the DSBs in c-myc that lead to translocation (reviewed in (Reddy and Vasquez, 2005)).
Here we report on experiments that examine the role of the c-myc promoter and AID in producing the lesions in c-myc that lead to c-myc/IgH translocations in mature B cells in vivo.
Results
c-myc is transcribed from several promoters distributed in a region surrounding the non-coding first exon of the gene (Wierstra and Alves, 2008). To determine whether c-myc transcription is required for translocation to IgH, we used homologous recombination in embryonic stem cells to delete a 2.2 Kbps genomic fragment that includes the region corresponding to c-myc promoters P0, P1 and P2 (Figure 1A, and Supplemental Figure S1). MycΔ/+ mice were born at nearly Mendelian ratio (Figure 1B) and B cell development and activation in response to mitogenic stimuli were unaltered in heterozygous mice (Supplemental Figure S1).
Figure 1. The MycΔ allele is protected from translocating to the IgH.
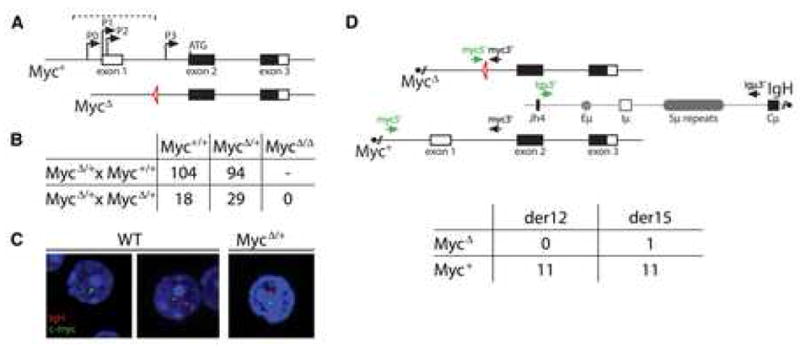
(A) Schematic representation of wild type (Myc+) and mutant (MycΔ)c-myc alleles. The dashed bracket indicates the region deleted and replaced by a loxP site (triangle) in the MycΔ allele. (B) Genotyping summary from the indicated MycΔ/+ cross. (C) RNA-FISH showing monoallelic vs. biallelic c-myc transcription in wild type (WT) vs. MycΔ/+ cells, respectively. (D) Diagram showing the location of the primers used in the PCR assay to detect derivative chromosome 12 (der12, black arrows) and derivative chromosome 15 (der15, green arrows) c-myc/IgH translocations. The table summarizes the number of allele-specific der12 and der15 c-myc/IgH translocations in activated MycΔ/+ B cells, as determined by sequencing. Three independent experiments.
To examine the transcriptional status of the MycΔ allele we performed RNA fluorescence in situ hybridization (Osborne et al., 2007). Among the Myc+/+ B cells with transcriptionally active alleles (n=168), 76% had one active c-myc allele and 24% had two active alleles. In striking contrast, 97% of the transcriptionally active MycΔ/+ B cells (n=86) had one silent allele, demonstrating that only one of the two alleles was efficiently transcribed in mutant mice (Figure 1C). Consistent with the reported lethality of c-myc deletion (Davis et al., 1993), the MycΔ mutation resulted in intercrosses between MycΔ/+ that failed to produce any MycΔ/Δ homozygous mutants (Figure 1B). We conclude that c-myc transcription is severely impaired by the MycΔ mutation.
To examine the effects of the MycΔ mutation on c-myc/IgH translocations we assayed for these events in heterozygous MycΔ/+ B cells stimulated with LPS and IL-4, using a previously described PCR assay to detect both derivative 12 and 15 chromosomes (Figure 1D and Supplemental Figure S2). Translocations were detected in MycΔ/+ mice, but sequence analysis revealed that 22 of 23 of these were IgH fusions to the wild type Myc+ allele (Figure 1D). Thus, the MycΔ mutation drastically reduces c-myc’s propensity to undergo oncogenic chromosomal translocations to IgH.
c-myc/IgH translocations induced by Cre
The IgH locus on mouse chromosome 12 and the c-myc locus on chromosome 15 co-localize in transcription factories in activated B lymphocytes, and this physical proximity has been proposed to facilitate c-myc/IgH translocations (Osborne et al., 2007; Parada et al., 2004; Roix et al., 2003).
To determine whether the MycΔ allele was accessible for translocation we made use of the phage-derived Cre-loxP recombinase system (Van Duyne, 2001). Cre recognizes 34 nucleotide loxP sites and catalyzes DNA recombination between two such sites. Recombination requires that Cre gain access to the DNA target and the efficiency of the reaction is inversely proportional to the distance between loxP sites (Yu and Bradley, 2001). To test for Cre mediated translocations between the loxP site on the MycΔ allele and IgH we introduced a single loxP site into IgH, upstream of the switch μ region (IgHI/+ mice, Figure 2A and Supplemental Figure S3). As a control for the MycΔ allele we also introduced a single loxP site into the first intron of intact c-myc (MycI/+ mice, Figure 2A and Supplemental Figure S4). IgHI/+ and MycI/+ mice showed normal B lymphocyte development and switching in response to LPS and IL-4 stimulation (Supplemental Figure S3, S4 and data not shown). Compound heterozygous B cells bearing either MycΔ or MycI and IgHI (MycΔ/IgHI and MycI/IgHI respectively) were tested for Cre mediated translocations using a retrovirus expressing Cre. By semi-quantitative analysis, we found that the frequency of c-myc/IgH translocations in MycΔ/IgHI and MycI/IgHI B cells expressing Cre was indistinguishable (0.5×10−5 for der12) (Figure 2B).
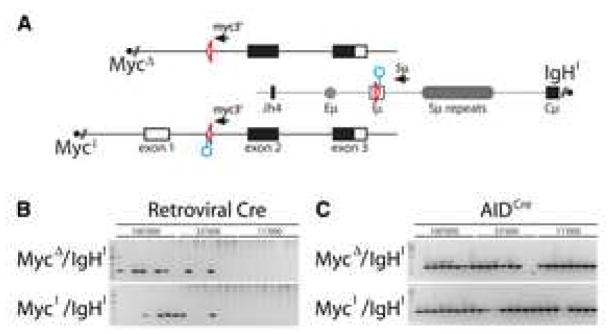
Figure 2. Cre-mediated c-myc/IgH translocations.
(A) Schematic representation of the MycΔ, MycI and IgHI alleles with the PCR primers for detecting der12 c-myc/IgH translocations. Triangles represent loxP sites, circles point to recognition sequences for I-SceI. (B) Translocations by retroviral Cre. Ethidium bromide stained agarose gel with 0.5kb PCR products corresponding to precise loxP-to-loxP c-myc/IgH translocations (as verified by sequencing). B lymphocytes of the indicated genotypes were stimulated with LPS and IL-4 and infected with retroviruses that direct expression of Cre. The number of cells that were assayed in each reaction is indicated. Two independent experiments. (C) Translocations in AIDCre B cells. Agarose gel with 0.5kb PCR products corresponding to precise loxP-to-loxP c-myc/IgH translocations (as verified by sequencing). AIDCre B cells of the indicated genotypes were stimulated with LPS and IL-4. The number of cells assayed in each reaction is indicated. Two independent experiments.
To examine the nature of Cre mediated interchromosomal junctions we sequenced 8 candidate translocations. Irrespective of whether the translocations originated from MycΔ/IgHI or MycI/IgHI B cells, the breakpoints were always precisely at the loxP site. To corroborate these results in the absence of retroviral Cre over-expression, we crossed the MycΔ/IgHI and MycI/IgHI mice to mice where Cre expression is under the control of regulatory elements of aicda (AIDCre/+, Supplemental Figure S5). Cre-mediated translocations were readily detected in both MycΔ/IgHI/AIDCre and MycI/IgHI/AIDCre B lymphocytes upon mitogenic stimulation in vitro and even in Peyer’s Patches in vivo (Figures 2C and Supplemental Figure S6). Thus, Cre rescues translocation of the mutant MycΔ allele in activated B cells in vitro and in vivo. We conclude that both the active (MycI) and silent (MycΔ)alleles reside in Cre-accessible nuclear compartments and remain in sufficient spatial proximity to IgH to permit translocation irrespective of c-myc’s transcriptional status.
DSBs in c-myc catalyzed by I-SceI
AID induced DSBs along the IgH locus can be detected by combining immunocytochemistry to detect accumulation of 53BP1 foci, and fluorescence in situ hybridization to localize IgH (Figure 3 and (Reina-San-Martin et al., 2004)). Background levels of co-localization of 53BP1 foci and IgH were found in AID deficient B cells, and the frequency increased to 42.5% in AID sufficient B cells (Figure 3A and (Reina-San-Martin et al., 2004)). In contrast, when we examined c-myc by the same technique we found no AID dependent increase in 53BP1 foci during CSR (Figure 3A). Retroviral over-expression of AID did however result in DNA damage at c-myc, as determined by increased co-localization of 53BP1 foci (12.2% vs. 4.3% with control, p = 0.007; Figure 3B) and accumulation of mutations in intron 1 (1.2 × 10−4 vs. <0.07×10−4 with control, p = 0.0005; Supplemental Table 1). Thus, our immuno-FISH experiments are in agreement with the sequencing data and fail to show evidence for AID induced DNA damage on c-myc during CSR under physiologic circumstances (Liu et al., 2008; Pasqualucci et al., 2001; Shen et al., 1998). However, c-myc can be an AID target when AID is over-expressed.
Figure 3. I-SceI induced breaks.
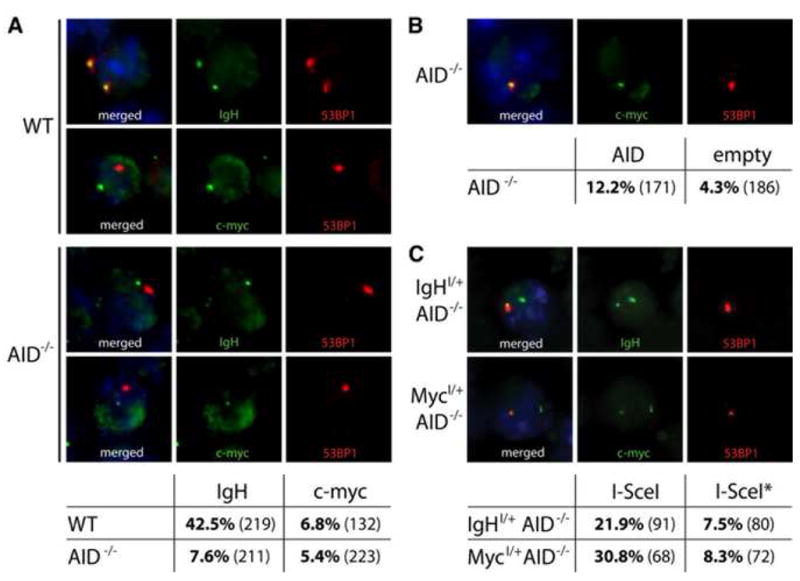
(A) Immuno-FISH analysis of 53BP1 DNA damage foci at IgH or c-myc loci in activated B cells. Representative images and table showing co-localization frequencies as percentage of cells analyzed in LPS and IL-4 stimulated wild type (WT) or AID−/− B lymphocytes. The number of cells analyzed is shown in parentheses. Three independent experiments. (B) Immuno-FISH analysis of AID induced 53BP1 DNA damage foci in AID−/− B cells infected with AID or empty virus control. Representative images and table showing co-localization frequencies at c-myc (p = 0.007 with Student’s T-Test). (C) Immuno-FISH analysis of I-SceI induced 53BP1 DNA damage foci in IgHI/+AID−/− (top) or MycI/+AID−/− B cells (bottom). Representative images and table showing co-localization frequencies at IgH or c-myc (as indicated) in I-SceI or I-SceI* expressing B lymphocytes.
To examine the consequence of DSB formation at c-myc, we used the yeast derived I-SceI endonuclease, which recognizes an 18 bps site that is otherwise absent from the mouse genome (reviewed in (Stoddard, 2005)). DNA breaks were created in the presence or absence of AID using I-SceI to cut at cognate target sites introduced into the MycI and IgHI alleles (Figure 2A and Supplemental Figures S3 and S4). Similar to AID induced DNA damage, retroviral expression of I-SceI in activated MycI/+AID−/− or IgHI/+AID−/− B lymphocytes produced frequent DSBs in c-myc or IgH respectively. As determined by immuno-FISH, 30.8% of MycI/+ B cells expressing I-SceI showed co-localization of c-myc and 53BP1 and 21.9% of IgHI/+ B cells exhibited c-myc/53BP1 co-localization (Figure 3C). Control B lymphocytes infected with a catalytically inactive form of I-SceI (I-SceI*) showed background levels of DNA damage at c-myc or IgH (Figure 3C). We conclude that I-SceI efficiently targets MycI/+ and IgHI/+ in activated B cells.
c-myc/IgH translocations catalyzed by I-SceI
To determine whether DSBs produced by I-SceI can serve as substrates for c-myc/IgH translocation we assayed for these events in LPS and IL-4 stimulated MycI/+IgHI/+AID−/−B cells expressing I-SceI or I-SceI* (Figure 4). Whereas we found no translocation with the retrovirus alone or I-SceI*, I-SceI expression produced der15 translocations at a rate of 10×10−5 (Figures 4B, 6, Supplemental Figures S7, S10 and data not shown; Note: while we assayed both der12 and der15 translocations, the close proximity of the binding sites of the der12 primers to the I-SceI sequences and processing of the DNA ends, see below, appears to result in the underestimation of their frequency). Thus, DSBs introduced by I-SceI in c-myc and IgH result in c-myc/IgH transchromosomal rearrangements.
Figure 4. I-SceI induced translocations in AID deficient B lymphocytes.
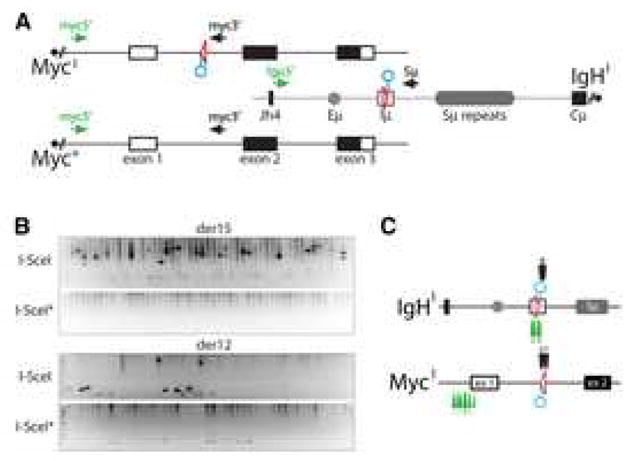
(A) Schematic representation of the MycI, Myc+ and IgHI alleles with the PCR primers for detecting der12 and der15 c-myc/IgH translocations. Circles point to the position of the I-SceI sites. (B) I-SceI rescues translocations in the absence of AID. Representative agarose gels with PCR products corresponding to c-myc/IgH translocations (as verified by sequencing and/or Southern Blot, see Supplemental Figure S7). MycI/+IgHI/+AID−/− B cells were stimulated with LPS and IL-4 and infected with I-SceI or I-SceI* control retroviruses. 100,000 cells were assayed each in lane. Three independent experiments. (C) Map of translocation breakpoints from I-SceI expressing MycI/+IgHI/+AID−/− B cells (see also Supplemental Table 2). Arrows point to c-myc/IgH translocation breakpoints for der12 (black) or der15 (green).
Figure 6. Translocations induced by AID, I-SceI or Cre in AID−/− cells.
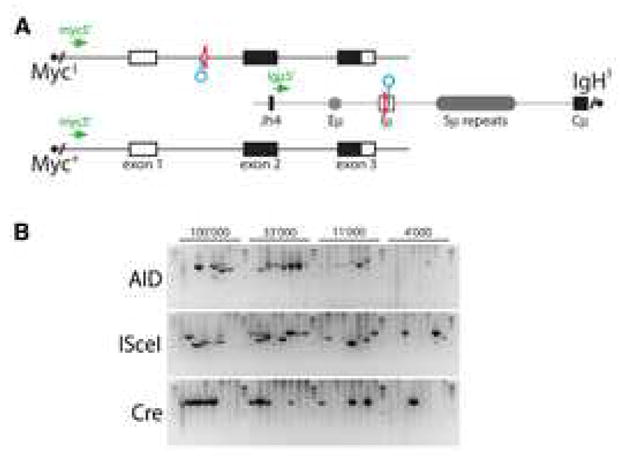
(A) Schematic representation of the MycI, Myc+ and IgHI alleles with the PCR primers for detecting der15 c-myc/IgH translocations. Triangles represent loxP sites, circles point to recognition sequences for I-SceI. (B) Representative agarose gels with PCR products corresponding to c-myc/IgH translocations (as verified by sequencing and/or Southern Blot, see Supplemental Figure S10). MycI/+ IgHI/+ AID−/− B cells were stimulated with LPS and IL-4 and infected with AID, I-SceI, or Cre, as indicated. DNA equivalents for the indicated number of cells were assayed in each reaction. Two independent experiments.
Translocation breakpoints were found precisely at the I-SceI site and also at distances of up to 2178 bps from the site of the DSB (Figure 4C, and Supplemental Table 2). The average distance of the breakpoint position from the I-SceI site on c-myc was 50 bps for der12 and 1744 bps for der15, and for IgH it was 39 bps for der12 and 91 bps (Supplemental Table 2). Out of 21 translocation junctions, 19 involved 1–7 bps of microhomology (average microhomology of 2.9 bps) and 2 of them included insertions (Figure 5 and Supplemental Table 2). We conclude that repair of I-SceI breaks frequently involves processing of the ends and microhomology mediated joining.
Figure 5. I-SceI induced translocations in AID sufficient B lymphocytes.
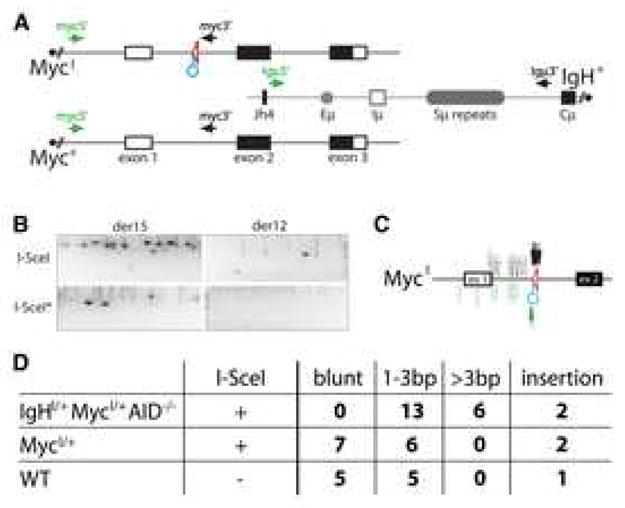
(A) Schematic representation of the MycI, Myc+ and IgH+ alleles with the PCR primers for detecting der12 and der15 c-myc/IgH translocations. Circle points to recognition sequence for I-SceI. (B) I-SceI directed translocations in the presence of AID. Representative agarose gels with PCR products corresponding to c-myc/IgH translocations (as verified by sequencing and/or Southern Blot, see Supplemental Figure S8). MycI/+ B cells were stimulated with LPS and IL-4 and infected with I-SceI (three independent experiments) or I-SceI* control. 100,000 cells were assayed in each lane. (C) Map of translocation breakpoints from stimulated MycI/+ B cells infected with I-SceI (see also Supplemental Table 3). Filled arrows point to c-myc/IgH translocation breakpoints from der12 (black) or der15 (green). Empty arrows indicate the distribution of breakpoints obtained in the absence of I-SceI. (D) Table shows summary of the extent of junctional microhomology from c-myc/IgH translocations (see Supplemental Tables 2, 3, and 4).
Source of DSB in c-myc during c-myc/IgH translocation
To determine if I-SceI breaks can be joined to AID induced lesions we examined MycI/+ B cells infected with I-SceI or I-SceI* encoding retroviruses. Baseline levels of AID mediated translocation were enhanced by I-SceI cutting in MycI/+ B cells (for der15 I-SceI*: 0.2×10−5 vs. I-SceI: 0.8×10−5, p = 0.0002; Figure 5B and Supplemental Figure S8). Consistent with this enhancement, 15 of 17 translocations produced in MycI/+ B cells infected with I-SceI were MycI to IgH translocations, while only 2 involved the wild type Myc+ allele (Supplemental Table 3). Whereas the breakpoints for c-myc/IgH translocations cloned from wild type control B cells were spread over 1.4 Kbps surrounding the first exon of c-myc, 10/15 I-SceI induced MycI breakpoints were found precisely at the I-SceI site and the remainder mapped within 74 bps of the site (Figure 5C and Supplemental Table 3). In contrast to the breakpoints produced by joining two I-SceI sites in the absence of AID (Figure 4C, Supplemental Table 2), 7 of the junctions between an I-SceI DSB in c-myc and an AID lesion in IgH were blunt, 2 had an insertion, and the remaining 6 showed 1–3 bps of microhomology (overall average microhomology: 0.8 bps; Figure 5D, Supplemental Table 2). A similar relationship between direct versus microhomology mediated joins is found in c-myc/IgH translocations in wild type B cells (Figure 5D and Supplemental Table 4) and in physiological CSR, where 30–60% of the joints are blunt and the remainder use 1–4 bps of microhomology (reviewed in (Chaudhuri et al., 2007; Stavnezer et al., 2008)).
Whereas I-SceI induced DSBs in c-myc enhanced translocation frequency (Figure 5B), we found no difference in translocation frequency between IgHI/+ B cells infected with I-SceI or I-SceI* (for der15 0.14×10−5 vs. 0.2×10−5 respectively; Supplemental Figure S9). The breakpoint was at the I-SceI cognate site in 2 out of 4 translocations involving the IgHI allele, confirming that similar to MycI, IgHI was also substrate for I-SceI cleavage and recombination (Supplemental Figure S9). Therefore, although I-SceI generates DSBs at c-myc and IgH with comparable efficiency (Figure 3C), it enhances the frequency of c-myc/IgH translocation in MycI/+ but not IgHI/+ B cells. We conclude that I-SceI induced breaks in c-myc or IgH can be joined to AID produced breaks in IgH or c-myc, respectively, to create c-myc/IgH translocations. In addition, we find that DSBs in c-myc but not IgH are limiting for translocation since a single I-SceI break in c-myc but not IgH enhances translocation.
To determine whether the source of the DSB alters the frequency of translocation we compared the incidence of c-myc/IgH transchromosomal rearrangements in MycI/+IgHI/+AID−/− B cells infected with AID or I-SceI or Cre expressing retroviruses. We found that the rate of translocation was similar for all three DNA damage-inducing enzymes (for der15 5×10−5 for AID, 10×10−5 for I-SceI and 3×10−5 for Cre, Figure 6 and Supplemental Figure S10). We conclude that the rate of c-myc/IgH translocation is similar irrespective of whether the DSBs are induced by AID, I-SceI or Cre.
To determine whether AID is essential to create the breaks in c-myc that lead to c-myc/IgH translocation we induced DSBs in the IgH locus of IgHI/+AID−/− or the c-myc locus of MycI/+AID−/− B cells using I-SceI. In contrast to IgHI/+MycI/+AID−/− B cells (Figure 4), and to AID sufficient MycI/+ and IgHI/+ B cells (Figure 5 and Supplemental Figure S9), I-SceI induced DSBs in either c-myc or IgH alone, in MycI/+AID−/− or IgHI/+AID−/− B cells respectively, failed to produce c-myc/IgH translocations (Figure 7). Thus, when I-SceI alone or I-SceI and AID coordinately generate DSBs on both chromosomes they result in c-myc/IgH translocations. In contrast, in the absence of AID, I-SceI-induced DSBs generated solely at IgH or c-myc do not produce translocations. We conclude that AID is essential for the DSBs in both of the chromosomes involved in c-myc/IgH translocation.
Figure 7. AID is essential for the lesion in c-myc that leads to c-myc/IgH translocation.
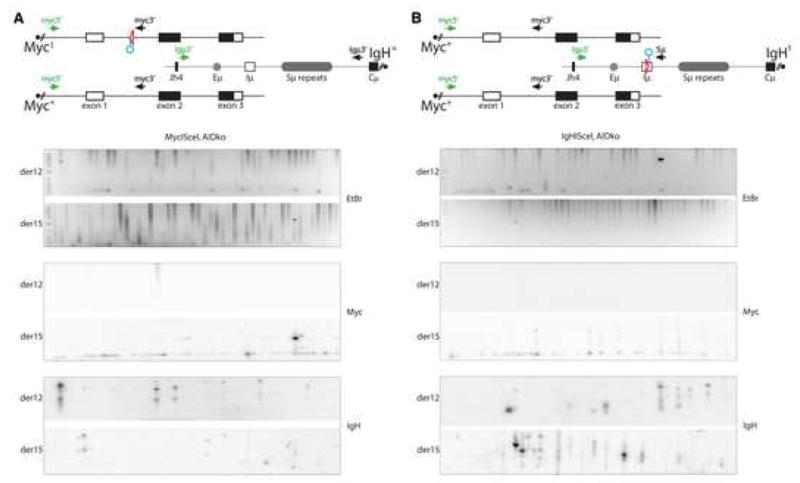
(A) Top: schematic representation of the MycI, Myc+ and IgH+ alleles with the PCR primers for detecting der12 and der15 c-myc/IgH translocations. Circle indicates I-SceI site. Bottom: a representative ethidium bromide (EtBr) stained agarose gel was also Southern blotted and probed for c-myc and IgH, as indicated, to verify translocations. MycI/+AID−/− B cells were stimulated with LPS and IL-4 and infected with an I-SceI encoding retrovirus. 100,000 cells were assayed in each lane. No translocations were identified out of 3.4×107 cells analyzed. Three independent experiments. (B) Top: schematic representation of the Myc+ and IgHI alleles with the PCR primers for detecting der12 and der15 c-myc/IgH translocations. Circle indicates I-SceI site. Bottom: a representative ethidium bromide (EtBr) stained agarose gel was also Southern blotted and probed for c-myc and IgH, as indicated to verify translocations. IgHI/+ AID−/− B cells were stimulated with LPS and IL-4 and infected with I-SceI. 100,000 cells were assayed in each lane. No translocations were identified in 6.9×107 cells. Seven independent experiments.
Discussion
DSBs are dangerous lesions because they can lead to genomic instability and oncogenic chromosomal translocations. Although DSBs rarely accumulate in somatic cells, they are obligate intermediates in CSR, a reaction that involves introduction of multiple targeted DSBs in the IgH locus in mature B lymphocytes. The genome is normally protected from AID dependent IgH translocations by ATM-, p53- and p19Arf-dependent pathways that detect and signal DNA damage and oncogenic stress (Ramiro et al., 2006). Nevertheless, these protective mechanisms sometimes fail, as evidenced by involvement of the IgH locus in nearly all cancer-associated chromosomal translocations in mature B cell lymphomas and multiple myeloma (Kuppers and Dalla-Favera, 2001).
AID creates the lesions in the IgH locus that lead to translocations (Franco et al., 2006; Petersen et al., 2001; Ramiro et al., 2006; Ramiro et al., 2004). However, DSBs in IgH alone would not be sufficient for translocation because aberrant joining of heterologous chromosomes requires formation of paired DNA breaks (Richardson and Jasin, 2000), and the source of DNA damage in the oncogenes that are translocated in mature B cell malignancies has not been determined (Kuppers and Dalla-Favera, 2001).
In Burkitt’s lymphoma and in murine plasmacytoma, IgH is translocated to c-myc leading to deregulated expression of c-myc and malignant transformation (Adams et al., 1983; Adams et al., 1982; Adams et al., 1985; Crews et al., 1982; Dalla-Favera et al., 1983; Erikson et al., 1983; Hamlyn and Rabbitts, 1983; Marcu et al., 1983; Taub et al., 1982). Recurrent translocation between c-myc and IgH has been attributed in part to proximity of the two loci in B cells due to the dynamic relocation of the two genes to transcription factories following B cell activation (Osborne et al., 2007; Roix et al., 2003). Consistent with a role for transcription in facilitating c-myc/IgH translocation our experiments show that the DNA region containing the c-myc promoters is required for efficient translocation in stimulated B cells. However, this effect does not appear to be due to transcriptionally regulated proximity of the two genes or accessibility to recombinase as indicated by equivalent levels of Cre mediated translocation between IgHI and transcriptionally crippled MycΔ or active MycI alleles. In light of the current discussion about “breakage first” or “contact first” models of chromosomal translocations, our data does not speak directly to the issue of physical proximity, but suggest that proximity is not limiting. Although loss of the c-myc promoter region interferes with transcription and c-myc/IgH translocation, it does not interfere with c-myc’s ability to come into contact with IgH or with its accessibility to Cre.
Instead, it appears that the c-myc promoter region is essential to recruit AID to initiate DSB formation. This is consistent with the finding that Ig transcriptional regulators including promoters, enhancers and the E-box motif CAGGTG (Michael et al., 2003) are involved in targeting AID to Igs (Di Noia and Neuberger, 2007; Peled et al., 2008; Stavnezer et al., 2008). Although the elements required for bringing AID to Ig or c-myc DNA have not yet been precisely defined, in vitro experiments with artificial substrates and in E. coli show that transcription is essential because it exposes single stranded DNA which is the substrate for AID (Chaudhuri et al., 2003; Dickerson et al., 2003; Pham et al., 2003; Ramiro et al., 2003). Furthermore, the formation of R-loops during transcription has been proposed to play a role in targeting AID (Yu et al., 2003). Thus, our observations suggest a transcriptional requirement for AID targeting to oncogenes, but the mechanism by which transcription targets AID to any gene remains to be determined.
Although increased AID expression was found to increase c-myc/IgH translocation, this effect could have been due to higher levels of DNA breaks on c-myc or IgH or both (Dorsett et al., 2008; Ramiro et al., 2006; Takizawa et al., 2008). Previous work suggested that DSBs in c-myc are AID independent and might be due to increased susceptibility of fragile non-B-DNA to environmental agents like reactive oxygen intermediates or transcriptional or replication-linked DNA damage (Belotserkovskii et al., 2007; Boles and Hogan, 1987; Lieber et al., 2003; Wang and Xu, 2004; Yang and Hurley, 2006). This idea was supported by the finding of AID independent DSBs in c-myc in B cell lymphomas (Zan et al., 2003), the relative rarity of somatic mutations in c-myc (Liu et al., 2008; Pasqualucci et al., 2001; Shen et al., 1998), and the lack of deletions or duplications in c-myc that would mark DSB formation (Gordon et al., 2003; Liu et al., 2008; Mullighan et al., 2007; Muschen et al., 2000; Pasqualucci et al., 1998; Pasqualucci et al., 2001; Shen et al., 1998). Despite these findings, our experiments show that AID can indeed induce lesions and mutations at c-myc, and demonstrate that AID is essential for the lesions in c-myc that result in c-myc/IgH translocation. Nevertheless, the absence of easily detectable DNA damage without AID over-expression suggests that formation of DNA lesions on c-myc may be one of the limiting factors in c-myc/IgH translocation under physiologic conditions. Consistent with this notion, we find that increasing the frequency of DSB formation by the I-SceI endonuclease at c-myc but not IgH enhances translocation. Since I-SceI targets c-myc and IgH with similar efficiency, we can also conclude that AID-induced DSB formation at c-myc is lower than at IgH. Although the DNA lesions introduced by either I-SceI or AID alone produced c-myc/IgH translocation, the extent of DNA processing and the structure of the intermolecular joints differed. The most extensive resection of DNA ends was found at c-myc when I-SceI mediated translocations in the absence of AID. Why the same resection was not detected at IgH under the same conditions is unclear, but the data is consistent with the existence of locus specific DNA damage processing factors (Liu et al., 2008). Whereas I-SceI induced breaks were resolved by a mechanism biased toward junctional microhomology reminiscent of alternative NHEJ (Richardson and Jasin, 2000), AID breaks produced a combination of blunt and short microhomology joints similar to those found in class switch recombination (Stavnezer et al., 2008). The difference in trans-chromosomal joints between I-SceI and AID may be explained by recruitment of a distinct set of repair enzymes to the two types of DNA damage. In the case of I-SceI a DSB is introduced directly by the yeast endonuclease and must be recognized by the DNA damage recognition and repair system before being processed and joined. In contrast, AID induced U:G mismatches are recognized by mismatch and base excision repair enzymes before recruitment of additional repair factors that then process the lesions to produce DSBs. Thus, the difference in the joints produced by I-SceI breaks and AID lesions may in part be due to the distinct mechanisms that recognize the initial lesion and produce the DSB.
Although the precise mechanism by which AID introduces lesions in DNA is still debated, AID is thought to produce U:G mismatches in DNA by cytidine deamination (Di Noia and Neuberger, 2007; Peled et al., 2008; Stavnezer et al., 2008). In B cells undergoing SHM or CSR, uracyl is detected and removed from antibody genes by error prone mechanisms leading to mutations or DSBs. Other genes, including oncogenes, appear to repair such lesions in a relatively error free manner, which may explain the nearly undetectable level of AID mediated mutation in genes like c-myc (Supplemental Table 1 and (Liu et al., 2008; Pasqualucci et al., 2001; Shen et al., 1998)). Our experiments reveal that like the error prone repair pathways that process the lesions at the IgH locus, the relatively error free repair of AID dependent lesions in c-myc can also lead to oncogene translocation.
Experimental Procedures
Mice
MycΔ/+, IgHI/+, MycI/+, and AIDCre/+ mice were generated by homologous recombination in C57BL/6 albino ES cells. Details of the targeting vectors, screening by Southern Blot and genotyping PCR are provided in the legends to the Supplemental Figures. AID−/− mice (Muramatsu et al., 2000) were backcrossed to C57BL/6 for at least 10 generations. All experiments were in agreement with protocols approved by the Rockefeller University and NIH Institutional Animal Care and Use Committee.
Retroviruses
Cre, I-SceI, I-SceI*, and AID were PCR-amplified and cloned into the pMX-IRES-GFP plasmid. In I-SceI*, D44A and D145A substitutions to inactivate the catalytic sites were introduced by PCR (Stoddard, 2005).
B cell cultures and retroviral transduction
Resting B lymphocytes were isolated from mouse spleens by immunomagnetic depletion with anti-CD43 MicroBeads (Miltenyi Biotech), and cultured at 0.5×106 cells/ml in R10 medium: RPMI supplemented with L-glutamine, sodium pyruvate, antibiotic/antimycotic, 50μM 2-mercaptoethanol (all from GIBCO-BRL), and 10% fetal calf serum (Hyclone). B cells were stimulated in the presence of 25 μg/ml lipopolysaccharide (LPS, E. coli serotype 0111:B4, Sigma) and 5 ng/ml mouse recombinant IL-4 (Sigma), and analyzed after 96 hours. For the analysis of translocations in MycΔ/+ mice (Figure 1D) cultures were supplemented with 50ng/ml mouse recombinant BAFF (R&D Systems) to boost the translocation frequency. For infection experiments, retroviral supernatants were prepared by co-transfecting BOSC23 cells with pCL-Eco and pMX-IRES-GFP-derived plasmids (encoding for Cre, I-SceI, I-SceI*, or AID) using Fugene 6, 72 hours before infection. At 20 and 44 hours of lymphocyte culture, retroviral supernatants were added and B cells spinoculated at 1150g for 1.5 hours in the presence of 10μg/ml polybrene. After 4 hours at 37C, supernatants were replaced with LPS and IL4 supplemented R10. B cells were analyzed at 96 hours from the beginning of their culture.
Flow cytometry and cell sorting
For FACS analysis, spleen cell suspensions were stained with fluorochrome-conjugated anti-CD19, anti-CD3, anti-IgM, anti-IgD, and anti-IgG1 (Pharmingen). CFSE-labeling for cell division was at 37C for 10 min in 5μM carboxyfluorescein succinimidyl esther. In all experiments, retrovirally infected GFP+ cells were sorted to >95% purity with a FACSVantage SE or FACSAria instruments (Becton Dickinson).
RNA fluorescence in situ hybridization (RNA FISH)
Stimulated splenic B cells were fixed on slides and analyzed by RNA FISH as described (Gribnau et al., 1998; Gribnau et al., 2000; Chakalova et al., 2004).
Immuno-FISH
Immunocytochemistry staining followed by fluorescence in situ hybridization (Immuno-FISH) was as described (Petersen et al., 2001; Difilippantonio et al., 2000).
PCR assay for translocations
PCR reactions were performed on genomic DNA from 105 cells, unless otherwise indicated. Using Expand Long Template PCR System nested reactions were performed with the following primers: for derivative chromosome 12 translocations first round, 5-TGAGGACCAGAGAGGGATAAAAGAGAA-3 and 5-GGGGAGGGGGTGTCAAATAATAAGA-3; for derivative chromosome 12 translocations second round, 5-CACCCTGCTATTTCCTTGTTGCTAC-3 and 5-GACACCTCCCTTCTACACTCTAAACCG-3; for derivative chromosome 15 translocations first round, 5-ACTATGCTATGGACTACTGGGGTCAAG-3 and 5-GTGAAAACCGACTGTGGCCCTGGAA-3; for derivative chromosome 15 translocations second round, 5-CCTCAGTCACCGTCTCCTCAGGTA-3 and 5-GTGGAGGTGTATGGGGTGTAGAC-3; for derivative chromosome 12 translocations (upstream of switch μ) first round, 5-GGCAACTTCAAATTCATTAAACCACAT-3 and 5-GGGGAGGGGGTGTCAAATAATAAGA-3; for derivative chromosome 12 translocations (upstream of switch μ) second round, 5-AAATGTGAGTGACCCAGACA ACG-3 and 5-GACACCTCCCTTCTACACTCTAAACCG-3; PCR conditions were according to manufacturer instructions with 29 cycles for the first round (10 cycles at 92C, 10 s; 60C, 30 s; and 68C, 7 min; followed by 19 cycles at 92C, 15 s; 60C, 30 s; and 68C, 7 min with 20 s of additional extension time per cycle) and 25 cycles for the second round (10 cycles at 92C, 10 s; 60C, 30 s; and 68C, 4 min; followed by 15 cycles at 92C, 15 s; 60C, 30 s; and 68C, 4 min with 20 s of additional extension time per cycle). PCR products were gel-purified and sequenced.
Southern Blotting
Southern blot analysis was performed as described (Ramiro et al., 2006). Oligoprobes were: for der15 c-myc 5-GCCGCCACTTTACTGGACTGCGCAGG-3; for der15 IgH 5-GAGGGAGCCGGCTGAGAGAAGTTGGG-3; for der12 c-myc 5-GCAGCGATTCAGCACTGGGTGCAGG-3; for der12 IgH (3′ of switch μ) 5-CCTGGTATACAGGACGAAACTGCAGCAG-3; for der12 IgH (5′ of switch μ) 5-TTCAGTCATTGCTTTAGGGGGAG-3. P value was calculated using the 2-Tail Fisher’s exact test.
Mutational analysis
Primers 5-TGGTCTTTCCCTGTGTTCTTTCTG-3 and 5-GACACCTCCCTTCTACACTCTAAACCG-3 were used to amplify a portion of c-myc intron 1 with Pfu polymerase and 25 cycles of amplification (95C, 10 s; 58C, 30 s; and 72C, 90 s), prior to cloning in TOPO vector and sequencing.
Supplementary Material
Acknowledgments
All members of the Nussenzweig lab for discussions. Dr. Barry Stoddard (Fred Hutchinson Cancer Research Center, Seattle) for suggestions on catalytically inactive I-SceI*. The Rockefeller University Gene Targeting Facility, Klara Velinzon and Tamara Shengelia for FACSorting, Matthias Muellenbeck and Nancy Wong for technical assistance, and David Bosque and Tom Eisenreich for help in managing the mouse colonies. The work was supported in part by NIH grants to M.C.N. The A.N. lab was supported by the Intramural research Program of the NIH, National Cancer Institute, Center for Cancer Research. D.F.R. is a Fellow of the Leukemia and Lymphoma Society and M.C.N. is a HHMI Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams JM, Gerondakis S, Webb E, Corcoran LM, Cory S. Cellular myc oncogene is altered by chromosome translocation to an immunoglobulin locus in murine plasmacytomas and is rearranged similarly in human Burkitt lymphomas. Proc Natl Acad Sci U S A. 1983;80:1982–1986. doi: 10.1073/pnas.80.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams JM, Gerondakis S, Webb E, Mitchell J, Bernard O, Cory S. Transcriptionally active DNA region that rearranges frequently in murine lymphoid tumors. Proc Natl Acad Sci U S A. 1982;79:6966–6970. doi: 10.1073/pnas.79.22.6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- Belotserkovskii BP, De Silva E, Tornaletti S, Wang G, Vasquez KM, Hanawalt PC. A triplex-forming sequence from the human c-MYC promoter interferes with DNA transcription. J Biol Chem. 2007;282:32433–32441. doi: 10.1074/jbc.M704618200. [DOI] [PubMed] [Google Scholar]
- Boles TC, Hogan ME. DNA structure equilibria in the human c-myc gene. Biochemistry. 1987;26:367–376. doi: 10.1021/bi00376a006. [DOI] [PubMed] [Google Scholar]
- Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci U S A. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakalova L, Carter D, Fraser P. RNA fluorescence in situ hybridization tagging and recovery of associated proteins to analyze in vivo chromatin interactions. Methods Enzymol. 2004;375:479–493. doi: 10.1016/s0076-6879(03)75029-0. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Basu U, Zarrin A, Yan C, Franco S, Perlot T, Vuong B, Wang J, Phan RT, Datta A, et al. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv Immunol. 2007;94:157–214. doi: 10.1016/S0065-2776(06)94006-1. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- Cory S, Adams JM, Gerondakis SD, Miller JF, Gamble J, Wiener F, Spira J, Francke U. Fusion of DNA region to murine immunoglobulin heavy chain locus corresponds to plasmacytoma-associated chromosome translocation. EMBO J. 1983;2:213–216. doi: 10.1002/j.1460-2075.1983.tb01407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews S, Barth R, Hood L, Prehn J, Calame K. Mouse c-myc oncogene is located on chromosome 15 and translocated to chromosome 12 in plasmacytomas. Science. 1982;218:1319–1321. doi: 10.1126/science.7146913. [DOI] [PubMed] [Google Scholar]
- Dalla-Favera R, Martinotti S, Gallo RC, Erikson J, Croce CM. Translocation and rearrangements of the c-myc oncogene locus in human undifferentiated B-cell lymphomas. Science. 1983;219:963–967. doi: 10.1126/science.6401867. [DOI] [PubMed] [Google Scholar]
- Davis AC, Wims M, Spotts GD, Hann SR, Bradley A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 1993;7:671–682. doi: 10.1101/gad.7.4.671. [DOI] [PubMed] [Google Scholar]
- Di Noia J, Neuberger MS. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002;419:43–48. doi: 10.1038/nature00981. [DOI] [PubMed] [Google Scholar]
- Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- Dickerson SK, Market E, Besmer E, Papavasiliou FN. AID mediates hypermutation by deaminating single stranded DNA. J Exp Med. 2003;197:1291–1296. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio MJ, Zhu J, Chen HT, Meffre E, Nussenzweig MC, Max EE, Ried T, Nussenzweig A. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature. 2000;404:510–514. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett Y, McBride KM, Jankovic M, Gazumyan A, Thai TH, Robbiani DF, Di Virgilio M, San-Martin BR, Heidkamp G, Schwickert TA, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett Y, Robbiani DF, Jankovic M, Reina-San-Martin B, Eisenreich TR, Nussenzweig MC. A role for AID in chromosome translocations between c-myc and the IgH variable region. J Exp Med. 2007;204:2225–2232. doi: 10.1084/jem.20070884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley DD, Manis JP, Zarrin AA, Kaylor L, Tian M, Alt FW. Internal IgH class switch region deletions are position-independent and enhanced by AID expression. Proc Natl Acad Sci U S A. 2002;99:9984–9989. doi: 10.1073/pnas.152333499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erikson J, ar-Rushdi A, Drwinga HL, Nowell PC, Croce CM. Transcriptional activation of the translocated c-myc oncogene in burkitt lymphoma. Proc Natl Acad Sci U S A. 1983;80:820–824. doi: 10.1073/pnas.80.3.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco S, Alt FW, Manis JP. Pathways that suppress programmed DNA breaks from progressing to chromosomal breaks and translocations. DNA Repair (Amst) 2006;5:1030–1041. doi: 10.1016/j.dnarep.2006.05.024. [DOI] [PubMed] [Google Scholar]
- Goossens T, Klein U, Kuppers R. Frequent occurrence of deletions and duplications during somatic hypermutation: implications for oncogene translocations and heavy chain disease. Proc Natl Acad Sci U S A. 1998;95:2463–2468. doi: 10.1073/pnas.95.5.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon MS, Kanegai CM, Doerr JR, Wall R. Somatic hypermutation of the B cell receptor genes B29 (Igbeta, CD79b) and mb1 (Igalpha, CD79a) Proc Natl Acad Sci U S A. 2003;100:4126–4131. doi: 10.1073/pnas.0735266100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribnau J, de Boer E, Trimborn T, Wijgerde M, Milot E, Grosveld F, Fraser P. Chromatin interaction mechanism of transcriptional control in vivo. EMBO J. 1998;17:6020–6027. doi: 10.1093/emboj/17.20.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribnau J, Diderich K, Pruzina S, Calzolari R, Fraser P. Intergenic transcription and developmental remodeling of chromatin subdomains in the human beta-globin locus. Mol Cell. 2000;5:377–386. doi: 10.1016/s1097-2765(00)80432-3. [DOI] [PubMed] [Google Scholar]
- Hamlyn PH, Rabbitts TH. Translocation joins c-myc and immunoglobulin gamma 1 genes in a Burkitt lymphoma revealing a third exon in the c-myc oncogene. Nature. 1983;304:135–139. doi: 10.1038/304135a0. [DOI] [PubMed] [Google Scholar]
- Imai K, Slupphaug G, Lee WI, Revy P, Nonoyama S, Catalan N, Yel L, Forveille M, Kavli B, Krokan HE, et al. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat Immunol. 2003;4:1023–1028. doi: 10.1038/ni974. [DOI] [PubMed] [Google Scholar]
- Jankovic M, Nussenzweig A, Nussenzweig MC. Antigen receptor diversification and chromosome translocations. Nat Immunol. 2007;8:801–808. doi: 10.1038/ni1498. [DOI] [PubMed] [Google Scholar]
- Kovalchuk AL, duBois W, Mushinski E, McNeil NE, Hirt C, Qi CF, Li Z, Janz S, Honjo T, Muramatsu M, et al. AID-deficient Bcl-xL transgenic mice develop delayed atypical plasma cell tumors with unusual Ig/Myc chromosomal rearrangements. J Exp Med. 2007;204:2989–3001. doi: 10.1084/jem.20070882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppers R, Dalla-Favera R. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene. 2001;20:5580–5594. doi: 10.1038/sj.onc.1204640. [DOI] [PubMed] [Google Scholar]
- Leder P, Battey J, Lenoir G, Moulding C, Murphy W, Potter H, Stewart T, Taub R. Translocations among antibody genes in human cancer. Science. 1983;222:765–771. doi: 10.1126/science.6356357. [DOI] [PubMed] [Google Scholar]
- Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- Liu J, Levens D. Making myc. Curr Top Microbiol Immunol. 2006;302:1–32. doi: 10.1007/3-540-32952-8_1. [DOI] [PubMed] [Google Scholar]
- Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- Marcu KB, Bossone SA, Patel AJ. myc function and regulation. Annu Rev Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- Marcu KB, Harris LJ, Stanton LW, Erikson J, Watt R, Croce CM. Transcriptionally active c-myc oncogene is contained within NIARD, a DNA sequence associated with chromosome translocations in B-cell neoplasia. Proc Natl Acad Sci U S A. 1983;80:519–523. doi: 10.1073/pnas.80.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael N, Shen HM, Longerich S, Kim N, Longacre A, Storb U. The E box motif CAGGTG enhances somatic hypermutation without enhancing transcription. Immunity. 2003;19:235–242. doi: 10.1016/s1074-7613(03)00204-8. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- Muschen M, Re D, Jungnickel B, Diehl V, Rajewsky K, Kuppers R. Somatic mutation of the CD95 gene in human B cells as a side-effect of the germinal center reaction. J Exp Med. 2000;192:1833–1840. doi: 10.1084/jem.192.12.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka H, Muramatsu M, Yamamura N, Kinoshita K, Honjo T. Activation-induced deaminase (AID)-directed hypermutation in the immunoglobulin Smu region: implication of AID involvement in a common step of class switch recombination and somatic hypermutation. J Exp Med. 2002;195:529–534. doi: 10.1084/jem.20012144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Mitchell JA, Horton A, Wood AL, Bolland DJ, Corcoran AE, Fraser P. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5:e192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada LA, Sotiriou S, Misteli T. Spatial genome organization. Exp Cell Res. 2004;296:64–70. doi: 10.1016/j.yexcr.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Migliazza A, Fracchiolla N, William C, Neri A, Baldini L, Chaganti RS, Klein U, Kuppers R, Rajewsky K, et al. BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc Natl Acad Sci U S A. 1998;95:11816–11821. doi: 10.1073/pnas.95.20.11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Neumeister P, Goossens T, Nanjangud G, Chaganti RS, Kuppers R, Dalla-Favera R. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- Peled JU, Kuang FL, Iglesias-Ussel MD, Roa S, Kalis SL, Goodman MF, Scharff MD. The Biochemistry of Somatic Hypermutation. Annu Rev Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- Petersen S, Casellas R, Reina-San-Martin B, Chen HT, Difilippantonio MJ, Wilson PC, Hanitsch L, Celeste A, Muramatsu M, Pilch DR, et al. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414:660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- Potter M. Neoplastic development in plasma cells. Immunol Rev. 2003;194:177–195. doi: 10.1034/j.1600-065x.2003.00061.x. [DOI] [PubMed] [Google Scholar]
- Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, Neuberger MS. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Callen E, Difilippantonio S, Chen HT, McBride KM, Eisenreich TR, Chen J, Dickins RA, Lowe SW, et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006;440:105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Eisenreich T, Difilippantonio S, Chen-Kiang S, Muramatsu M, Honjo T, Nussenzweig A, Nussenzweig MC. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Ramiro AR, Stavropoulos P, Jankovic M, Nussenzweig MC. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat Immunol. 2003;4:452–456. doi: 10.1038/ni920. [DOI] [PubMed] [Google Scholar]
- Reddy MC, Vasquez KM. Repair of genome destabilizing lesions. Radiat Res. 2005;164:345–356. doi: 10.1667/rr3419.1. [DOI] [PubMed] [Google Scholar]
- Reina-San-Martin B, Chen HT, Nussenzweig A, Nussenzweig MC. ATM is required for efficient recombination between immunoglobulin switch regions. J Exp Med. 2004;200:1103–1110. doi: 10.1084/jem.20041162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-San-Martin B, Difilippantonio S, Hanitsch L, Masilamani RF, Nussenzweig A, Nussenzweig MC. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J Exp Med. 2003;197:1767–1778. doi: 10.1084/jem.20030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405:697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat Genet. 2003;34:287–291. doi: 10.1038/ng1177. [DOI] [PubMed] [Google Scholar]
- Shen HM, Peters A, Baron B, Zhu X, Storb U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 1998;280:1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- Spencer CA, Groudine M. Control of c-myc regulation in normal and neoplastic cells. Adv Cancer Res. 1991;56:1–48. doi: 10.1016/s0065-230x(08)60476-5. [DOI] [PubMed] [Google Scholar]
- Stanton LW, Watt R, Marcu KB. Translocation, breakage and truncated transcripts of c-myc oncogene in murine plasmacytomas. Nature. 1983;303:401–406. doi: 10.1038/303401a0. [DOI] [PubMed] [Google Scholar]
- Stavnezer J, Guikema JE, Schrader CE. Mechanism and Regulation of Class Switch Recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard BL. Homing endonuclease structure and function. Q Rev Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- Takizawa M, Tolarova H, Li Z, Dubois W, Lim S, Callen E, Franco S, Mosaico M, Feigenbaum L, Alt FW, et al. AID expression levels determine the extent of cMyc oncogenic translocations and the incidence of B cell tumor development. J Exp Med. 2008 doi: 10.1084/jem.20081007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taub R, Kirsch I, Morton C, Lenoir G, Swan D, Tronick S, Aaronson S, Leder P. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc Natl Acad Sci U S A. 1982;79:7837–7841. doi: 10.1073/pnas.79.24.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duyne GD. A structural view of cre-loxp site-specific recombination. Annu Rev Biophys Biomol Struct. 2001;30:87–104. doi: 10.1146/annurev.biophys.30.1.87. [DOI] [PubMed] [Google Scholar]
- Wang G, Xu XS. Peptide nucleic acid (PNA) binding-mediated gene regulation. Cell Res. 2004;14:111–116. doi: 10.1038/sj.cr.7290209. [DOI] [PubMed] [Google Scholar]
- Wierstra I, Alves J. The c-myc promoter: still MysterY and challenge. Adv Cancer Res. 2008;99:113–333. doi: 10.1016/S0065-230X(07)99004-1. [DOI] [PubMed] [Google Scholar]
- Wilson PC, de Bouteiller O, Liu YJ, Potter K, Banchereau J, Capra JD, Pascual V. Somatic hypermutation introduces insertions and deletions into immunoglobulin V genes. J Exp Med. 1998;187:59–70. doi: 10.1084/jem.187.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Hurley LH. Structure of the biologically relevant G-quadruplex in the c-MYC promoter. Nucleosides Nucleotides Nucleic Acids. 2006;25:951–968. doi: 10.1080/15257770600809913. [DOI] [PubMed] [Google Scholar]
- Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- Yu Y, Bradley A. Engineering chromosomal rearrangements in mice. Nat Rev Genet. 2001;2:780–790. doi: 10.1038/35093564. [DOI] [PubMed] [Google Scholar]
- Zan H, Wu X, Komori A, Holloman WK, Casali P. AID-dependent generation of resected double-strand DNA breaks and recruitment of Rad52/Rad51 in somatic hypermutation. Immunity. 2003;18:727–738. doi: 10.1016/s1074-7613(03)00151-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.