Summary
AdoMet-dependent methylation of histones is part of the “histone code” that can profoundly influence gene expression. We describe the crystal structure of Neurospora DIM-5, a histone H3 lysine 9 methyltranferase (HKMT), determined at 1.98 Å resolution, as well as results of biochemical characterization and site-directed mutagenesis of key residues. This SET domain protein bears no structural similarity to previously characterized AdoMet-dependent methyltransferases but includes notable features such as a triangular Zn3Cys9 zinc cluster in the pre-SET domain and a AdoMet binding site in the SET domain essential for methyl transfer. The structure suggests a mechanism for the methylation reaction and provides the structural basis for functional characterization of the HKMT family and the SET domain.
Introduction
Histones are subject to extensive posttranslational modifications including acetylation, phosphorylation, and methylation, primarily on their N-terminal tails that protrude from the nucleosome. Evidence accumulated over the past few years suggests that such modifications constitute a “histone code” that directs a variety of processes involving chromatin (Jenuwein and Allis, 2001; Strahl and Allis, 2000). Histone methylation represents the most recently recognized component of the histone code. Most histone methylation occurs on lysine, though arginine methylation also occurs on histones H3 and H4 (Ma et al., 2001; Strahl et al., 2001; Wang et al., 2001b). Lysine methylation is highly selective, with the best-characterized sites being K4 and K9 of histone H3. In general, K9 methylation is associated with transcriptionally inactive heterochromatin, while K4 methylation is associated with transcriptionally active euchromatin (Boggs et al., 2002; Litt et al., 2001; Nakayama et al., 2001; Nishioka et al., 2002a). In addition, K9 methylation has been implicated in transcriptional silencing of euchromatic genes such as those involved in cell cycle control (Nielsen et al., 2001; Ogawa et al., 2002), and K4 methylation is involved in silencing of rDNA and telomere sequences in the yeast Saccharomyces cerivisiae (Briggs et al., 2001; Krogan et al., 2002). Methylated K9 of histone H3 is specifically recognized by the chromo domain of heterochromatin protein HP1, which presumably directs the binding of additional proteins involved in the control of chromatin structure and gene expression (Bannister et al., 2001; Jacobs et al., 2001; Lachner et al., 2001).
The discovery that some SET domain proteins are responsible for methylation of lysines in histone tails provided an important advance in our understanding of the workings of the histone code (Rea et al., 2000). The SET domain was originally identified in three Drosophila genes involved in epigenetic processes, Su(var)3–9, En(zeste), and Trithorax (Jenuwein et al., 1998). Mammalian homologs of Drosophila SU(var)3–9 were shown to specifically methylate H3 at lysine 9 (Rea et al., 2000). Soon thereafter, related histone lysine (K) methyltransferases (HKMTs) in various species (see legend of Figure 1) were found to methylate K4, K9, K27, or K36 of H3 and K20 of H4. In addition, K79 of H3 was found to be methylated by a protein containing no SET domain (Feng et al., 2002; Lacoste et al., 2002; Ng et al., 2002; van Leeuwen et al., 2002). The approximately 130 amino acid SET domain is found in a large number of eukaryotic proteins as well as a few bacterial proteins and is not limited to HKMTs. More than 60 SET domain genes have been identified in humans (Pfam database: http://www.sanger.ac.uk/cgi-bin/Pfam/getacc?PF00856), nearly 40 are found in the genome of Arabidopsis thaliana (Baumbusch et al., 2001), and about 10 each are found in Drosophila and the fungi Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Neurosopora crassa. SET proteins can be grouped into families according to the sequences surrounding this distinctive domain (Baumbusch et al., 2001; Kouzarides, 2002). The SUV39 family proteins, which methylate K9 of H3 (O’Carroll et al., 2000; Rea et al., 2000; Schultz et al., 2002; Tamaru and Selker, 2001) or K9 and K27 of H3 (Tachibana et al., 2001) and include the most active HKMTs known to date, contain two cysteine-rich regions flanking the SET domain. These “pre-SET” and “post-SET” domains are required for HKMT activity of SUV39H1 (Rea et al., 2000).
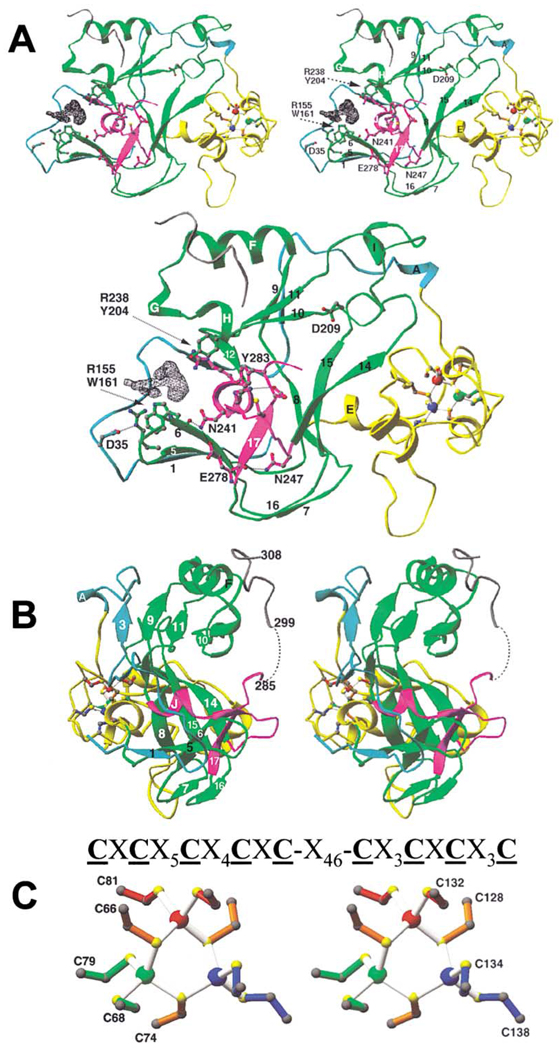
Figure 1. Structure-Based Sequence Alignment of SET Proteins.
The alignment includes (1) all known members of human SUV39 family: SUV39H1 (NP_003164), SUV39H2 (NP_078946), G9a (S30385), Eu-HMT1 (AAM09024), SETDB1 (NP_036564), and CLLL8 (NP_114121); (2) proteins (in bold) that have been shown to have HKMT activity from various species: N. crassa (Nc) DIM-5 (AAL35215), S. pombe (Sp) Clr4 (060016), A. thaliana (At) SUVH4 or KRYPTONITE (AAK28969), S. cerevisiae (Sc) SET1 (P38827) and SET2 (P46995), human SET7 (XP_040150) and PR-SET7 (AAL40879); (3) three bacterial SET proteins: Xylella fastidiosa (Xf) SET (AAF84287), Bradyrhizobium japonicum (Bj) SET (Q9ANB6), and Chlamydophila pneumoniae (Cp) SET (AAD19016); and (4) human EZH2 protein, which appears inactive in vitro (Rea et al., 2000). The residue number and secondary structural elements of DIM-5 (helices A–J and strands 1–17) are shown above the aligned sequences. Dashed lines indicate disordered regions. The color coding is light blue for the N terminus (residues 25–62), yellow for the pre-SET (residues 63–146), green for the SET (residues 147–236 and 248–277), magenta for the signature motifs (SET residues 237–247 and 278–285), and gray for the post-SET (residues 299–308). The amino acids highlighted are invariant (white against black) and conserved (white against gray) among almost all members of the SUV39 family. The number in parentheses indicates the number of amino acids inserted relative to the alignment. The lowercase letters above the sequences indicate the structural/functional role of the corresponding DIM-5 residues: “h” indicates intramolecular hydrophobic interaction, “n” indicates intramolecular nonhydrophobic (polar or charge) interaction, “z” indicates zinc coordination, asterisk indicates structural residue Gly or Pro, and “s” indicates surface-exposed residues potentially important for cofactor or substrate binding or catalysis. The red circles mark the residues that were mutated in this study.
As a step to elucidate the mechanism of SET domain HKMTs, we characterized the structure of DIM-5, a K9 histone H3 methyltransferase (MTase) from N. crassa (Tamaru and Selker, 2001). The discovery that this member of the SUV39 family is essential for DNA methylation in vivo revealed a connection between histone methyltion and DNA methylation. This connection has been reinforced by the observation that a HKMT from A. thaliana is also involved in DNA methylation (Jackson et al., 2002).
Results and Discussion
Overall Structure of DIM-5
We used recombinant DIM-5 protein (residues 17 to 318 of Protein Data Bank accession number AF419248) for crystallographic studies (see Experimental Procedures). Electron density maps were calculated using multiwave-length anomalous diffraction data from three intrinsic zinc ions (Table 1). A model of DIM-5 was built and refined to 1.98 Å resolution with a crystallographic R factor of 0.205 and Rfree value of 0.258. The final model includes 1913 protein atoms (with mean B values of 26.9 Å2), 3 zinc ions, and 103 water molecules, with rms deviations of 0.008 Å and 1.5° from ideality for bond lengths and angles, respectively.
Table 1.
Summary of X-Ray Diffraction Data Collection
| Derivative | Native (Zn) | ||
|---|---|---|---|
| Wavelength (Å) | 1.0332 | 1.2834 | 1.2830 |
| Resolution range (Å) | 24.83-1.98 | 24.83-2.3 | 24.83-2.3 |
| Completeness (%)a | 97/95.8 | 98.8/95.3 | 98.8/94.0 |
| R linear (%)a | 0.055/0.276 | 0.063/0.184 | 0.067/0.227 |
| <I/σ(I)> | 15.4 | 18.6 | 18.6 |
| Observed reflections | 112,569 | 77,000 | 78,335 |
| Unique reflections | 20,963 | 13,923 | 14,065 |
| Anomalous zinc sites | 3 | 3 | 3 |
| Overall figure of merit | 0.48 at 2.9 Å resolution | ||
| Overall Z score value | 20.13 at 2.9 Å resolution |
The numerical numbers are given for the whole data set/the highest resolution bin.
Our structural determination on DIM-5 allowed us to perform a structure-guided sequence alignment of SET proteins (Figure 1) that includes human SUV39 family proteins, all verified active HKMTs reported so far, and three bacterial SET proteins. The 318 residue DIM-5 protein is the smallest member of the SUV39 family. It contains four segments: (1) a weakly conserved amino-terminal region (light blue), (2) a pre-SET domain (yellow) containing nine invariant cysteines, (3) the SET region (green) containing signature motifs of NHXCXPN and DY (magenta), and (4) the post-SET region (gray) containing three invariant cysteines. The 9 Cys pre-SET region is unique to the SUV39 family, while the post-SET region is also present in many members of SET1 and SET2 families (Kouzarides, 2002), and even in one bacterial SET protein from Xylella fastidiosa (Figure 1). Two active human HKMTs contain neither pre- nor post-SET regions: SET7 (Wang et al., 2001a) (also called SET9 [Nishioka et al., 2002a]) methylates lysine 4 of histone H3 and SET8 (Fang et al., 2002) (also called PR-SET7 [Nishioka et al., 2002b]) methylates lysine 20 of H4.
The pre-SET residues (yellow) form a 9 Cys cage enclosing a triangular zinc cluster (Figure 2A). The SET residues (green) are folded into six β sheets surrounding the catalytic methyl transfer site (magenta), with a helical cap (αF) above the β sheets. The amino-terminal residues (light blue) appear to be critical to the structural integrity of the molecule: the 38 residue segment extends through nearly the entire back of the molecule in the orientation shown (Figure 2A), providing an edge strand (β1, β2, or β3) to three separate β sheets and a 1 turn helix αA connecting to the pre-SET triangular zinc cage. The overall dimensions of the molecule are 60 × 50 × 30 Å. The triangular zinc cluster and the cofactor binding site are approximately 38 Å apart, located at opposite ends of the molecule along the longest dimension (Figure 2A). A cleft can be seen running across from the cofactor binding site to the zinc cluster (Figure 2B).
Figure 2. DIM-5 Structure.
(A) Front view of ribbons diagram (Carson, 1997) (top, stereo; bottom, mono). The protein is colored according to Figure 1, and the three zinc ions are red, green, and blue balls (as in C). The difference electron density map (black), contoured at 5.5σ above the mean, indicates the presumed cofactor binding site (supported by tests on mutant forms).
(B) Side view. A dashed line indicates the disordered amino acids between strand β17 (magenta) and the post-SET segment (gray).
(C) Stereo diagram of the triangular zinc cluster. Three zinc ions are colored in red, green, and blue, the bridging cysteine residues are colored in orange, and the nonbridging cysteine residues are colored to match their associated zinc ion. The atoms are gray (carbon) and yellow (sulfur). The pre-SET sequence of DIM-5 is shown above. Both Cys-rich segments coordinate the red and blue zinc ions jointly, while the green zinc ion is coordinated solely by the five-Cys segment.
The Pre-SET Domain Forms a Triangular Zinc Cluster
The pre-SET domain contains nine invariant cysteine residues that are grouped into two segments of five and four cysteines separated by various numbers of amino acids (46 in DIM-5). These nine cysteines coordinate three zinc ions to form an equilateral triangular cluster (Figure 2C). Each zinc ion is coordinated by two unique cysteines (six total), and the remaining three cysteine residues (C66, C74, and C128) are each shared by two zinc atoms, thus serving as bridges to complete the tetrahedral coordination of the metal atoms. The distance between zinc atoms is ~3.9 Å, and the Zn-S distance is ~2.3 Å. A similar metal-thiolate cluster can be found in metallothioneins that are involved in zinc metabolism, zinc transfer, and apoptosis (reviewed in Vasak and Hasler, 2000). Methallothioneins often have two metal clusters: a (Me)3Cys9 and a (Me)4Cys11, where Me can be Zn2+, Cd2+, Cu2+, or another heavy metal. The tri-zinc cluster of DIM-5 can be superimposed perfectly upon the (Zn2Cd)Cys9 cluster of rat metallothionein (Robbins et al., 1991) (not shown). The significance of this apparent similarity is unclear.
The SET Domain Forms the Active Site
The SET domain resembles a square-sided β barrel topped by a helical cap (αF, αG, αH, and αI). Four β sheets—(1↑ 5↑ 6↓), (7↑ 16↓), (4↓ 14↑ 15↓ 8↑), and (3↑ 9↑ 11↓ 10↑)—form the sides of the barrel and one sheet—(2↓ 12↓)—forms one end (Figure 2A). In the middle of the open end of the barrel is a crossover structure (magenta) formed by threading the β17-loop through an opening formed by a short loop between strands β13 and β14. This brings together the two most-conserved regions of the SET domain: the αJ-β13-loop (N241HXCXPN247) and β17-loop (DY283) (Figure 1). The side chains of these two highly conserved segments are involved in (1) hydrophobic structural packing (I240 of αJ and L279 and F281 of β17), (2) intramolecular side chain-main chain interactions (after a sharp turn at P246, the side chain of N247 interacts with the main chain carbonyl oxygen of E278 and the main chain amide nitrogen of T280), (3) AdoMet binding site and active site formation (R238 and F239 of αJ, N241:E278 pair, H242:D282 pair, and Y283). These invariant residues are clustered together, via pair-wise interactions such as the interactions between N241 and E278 and between H242 and D282, forming an active site in a location immediately next to the AdoMet binding pocket and peptide binding cleft (see below).
Enzymatic Properties of DIM-5
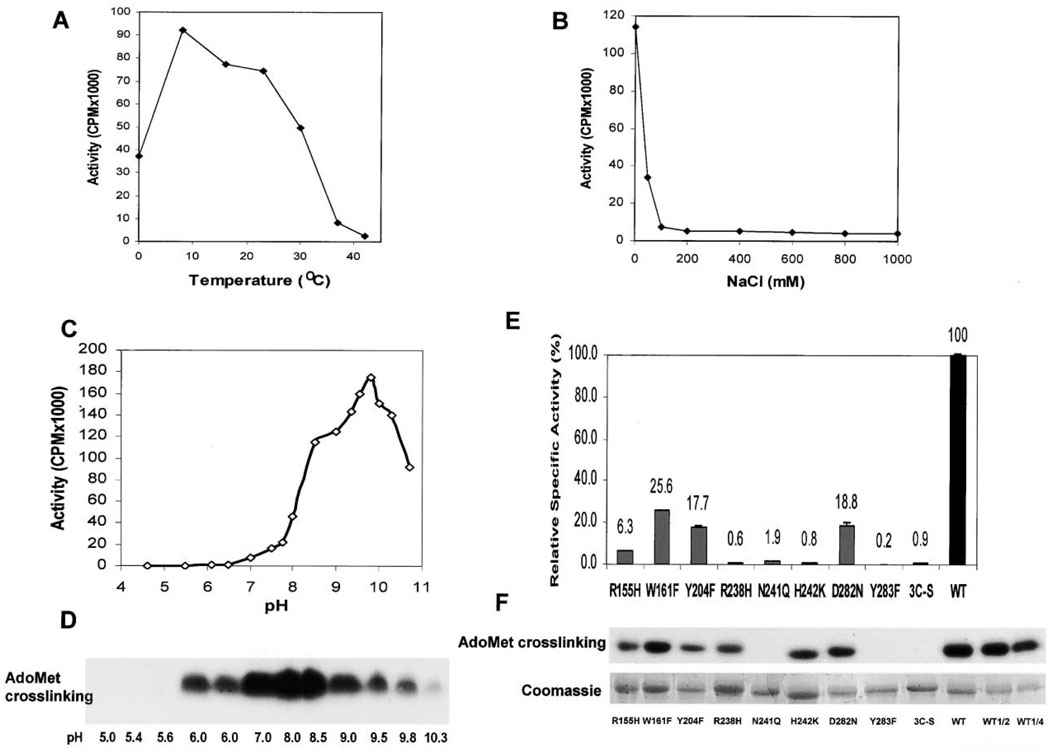
The DIM-5 protein is a very active HKMT in vitro. We noticed several rather unusual properties of DIM-5. (1) Under our laboratory conditions, the enzyme is most active at ~10°C and nearly inactive at 37°C (Figure 3A). (2) DIM-5 is extremely sensitive to salt, e.g., 100 mM NaCl inhibited its activity about 95% (Figure 3B). (3) The enzyme has a high pH optimum. DIM-5 showed maximal activity at ~pH 9.8 (Figure 3C), although it showed strongest crosslinking to AdoMet around pH 8 (Figure 3D). Neither HKMT activity nor AdoMet binding were observed below pH 6.0.
Figure 3. Enzymatic Properties of Recombinant DIM-5.
HKMT activity as functions of (A) temperature, (B) salt concentration, (C) pH, and (D) AdoMet crosslinking as a function of pH. The buffers used were 50 mM Na citrate for pH 5.0–6.0, MES for pH 6.0–6.5, HEPES for pH 7.0–7.5, Tris for pH 8.0–8.5, Bicine for pH 9.0, and glycine for pH 9.35–10.7. To rule out the potential inhibitory effect of Na citrate, both Na citrate and Mes are used for pH 6.0.
(E) Activities of DIM-5 mutants with conservative point mutations. All mutant proteins were expressed to level similar to that of the wild-type, though some were less soluble, and all were monomeric, suggesting that none of the mutations caused gross aggregation of the protein. Various amounts of mutant enzymes were used, the activities were compared to that of serial dilutions of wild-type enzymes purified in the same way, and the specific activity of mutant proteins relative to wild-type was estimated. The activities shown are averages of at least two measurements.
(F) Fluorographic results of an AdoMet crosslinking experiment at pH 8.0 are shown along with results of Coomassie staining to control for the amount of mutant protein tested.
Cofactor Binding Pocket
All known HKMTs use AdoMet as the methyl donor. The most common conformation of AdoMet, or its reaction product AdoHcy, is found in the so-called consensus MTases. These MTases are built around a mixed seven-stranded β sheet, and they include more than 20 structurally characterized MTases acting on carbon, oxygen, or nitrogen atom in DNA, RNA, protein, or small molecule substrates (Cheng and Roberts, 2001). DIM-5 does not share structural similarity to any of these AdoMet-dependent proteins and appears to use a completely different means of interaction with its cofactor.
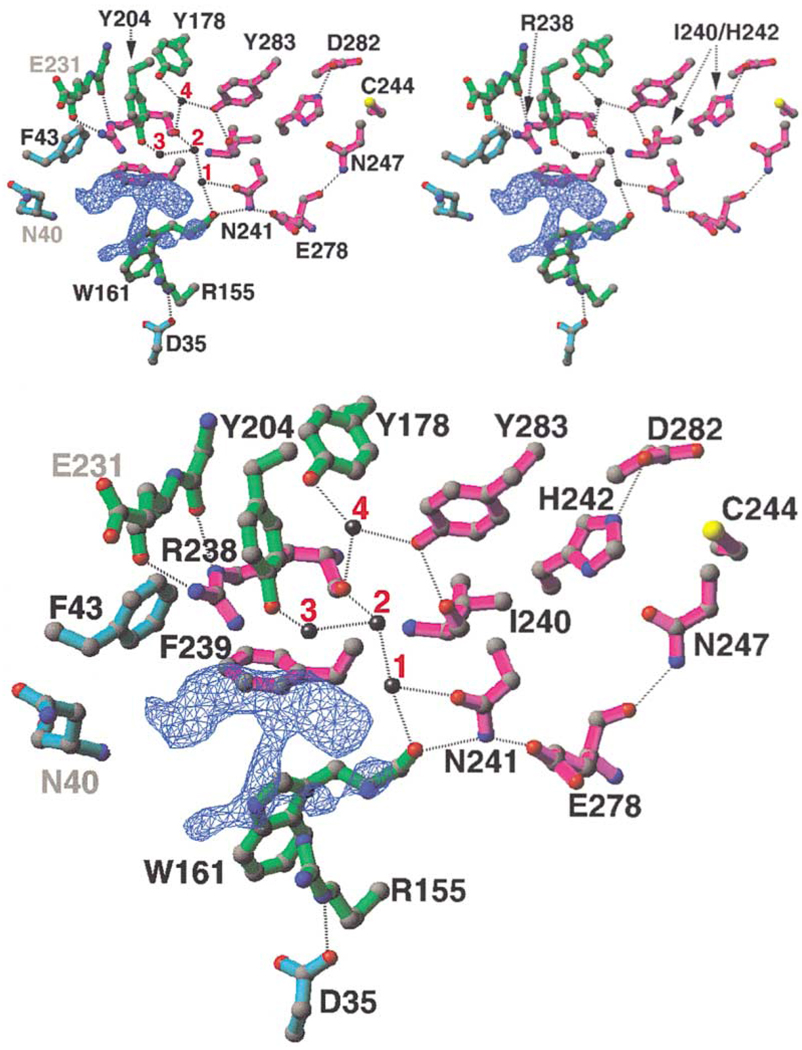
A difference electron density is observed in an open pocket on one end of the DIM-5 molecule opposite from the triangular zinc cluster (Figure 2A and Figure 4). We interpret this density as the cofactor product, AdoHcy, which was present during crystal growth (see Experimental Procedures). Although part of the AdoHcy can be fit into the density (not shown), it is difficult to fit the entire molecule, particularly because there is no recognizable density for the adenine ring of AdoHcy. This could potentially reflect flexibility of the cofactor bound to DIM-5. Unlike the “consensus” MTases where AdoMet/AdoHcy binds in a relatively closed pocket with hydrophobic stacking on both sides of adenine ring (Fauman et al., 1999), the density we observe is located in an open pocket, sitting above the antiparallel strands β5 and β6 and against the short helix αJ (Figure 2A). This environment may contribute to its flexibility or allow multiple conformations in the absence of substrate. The flexibility may also result from low pH during crystallization (pH 5.4–5.6), a condition in which no UV crosslinking of AdoMet to the protein was observed (Figure 3D). At low pH the adenine ring might not interact stably enough with DIM-5 to be crosslinked to the protein or observed in the structure.
Figure 4. The Cofactor Binding and Active Site.
Close-up view of the proposed cofactor binding site and the adjacent active site (top, stereo; bottom, mono). The difference electron density map (blue) is contoured at 5.5σ; the water molecules are numbered 1–4. Dashed lines indicate the hydrogen bonds. The residues are colored as in Figure 1. The water at site 2 is hydrogen bonded to the main chain carbonyl oxygen atom of R238 and to the water molecules at sites 1 and 3, which in turn interacts with the side chain carbonyl oxygen of N241 and the side chain hydroxyl oxygen of Y204, respectively.
The significance of this density is further enhanced by the highly conserved residues with which it is surrounded. Two conserved arginines (R155 of β5 and R238 of αJ) and three aromatic residues (W161, Y204, and F239) directly contact the density (Figure 4). The side chains of these two arginines are locked in place by other conserved residues: the guanidino group of R155 is parallel to the plane of the W161 indole ring and ion pairs with D35; and the guanidino group of R238 is surrounded by three aromatic rings, F43, F239, and Y204, and its two terminal nitrogen atoms (N∊ and Nη2) form hydrogen bonds to the main chain carbonyl oxygen atoms of G230 and E231, respectively (Figure 4).
We made conservative substitutions for several of the residues surrounding this density: R155H, W161F, Y204F, and R238H (see Experimental Procedures). The enzymatic activities of all the mutants were reduced ranging from a 75% reduction (W161F) to nearly inactive (R238H) (Figure 3E). The ability of these mutants to bind AdoMet, as measured by crosslinking, was also reduced but not abolished (Figure 3F). It appears that the reduced AdoMet binding alone could account for the reduction in HKMT activity for the R155H, W161F, and Y204F mutations. The R238H mutation, however, caused a much greater reduction in HKMT activity than in AdoMet binding, suggesting that R238 may also play roles in other aspects of catalysis (see below). In SUV39H1 and SUV39H2, a histidine is in the position of R238 in DIM-5; changing this histidine to an arginine resulted in at least 20-fold increase of activity in SUV39H1 (Rea et al., 2000), consistent with the greatly reduced activity in the converse R238H mutants of DIM-5.
Putative Peptide Binding Cleft
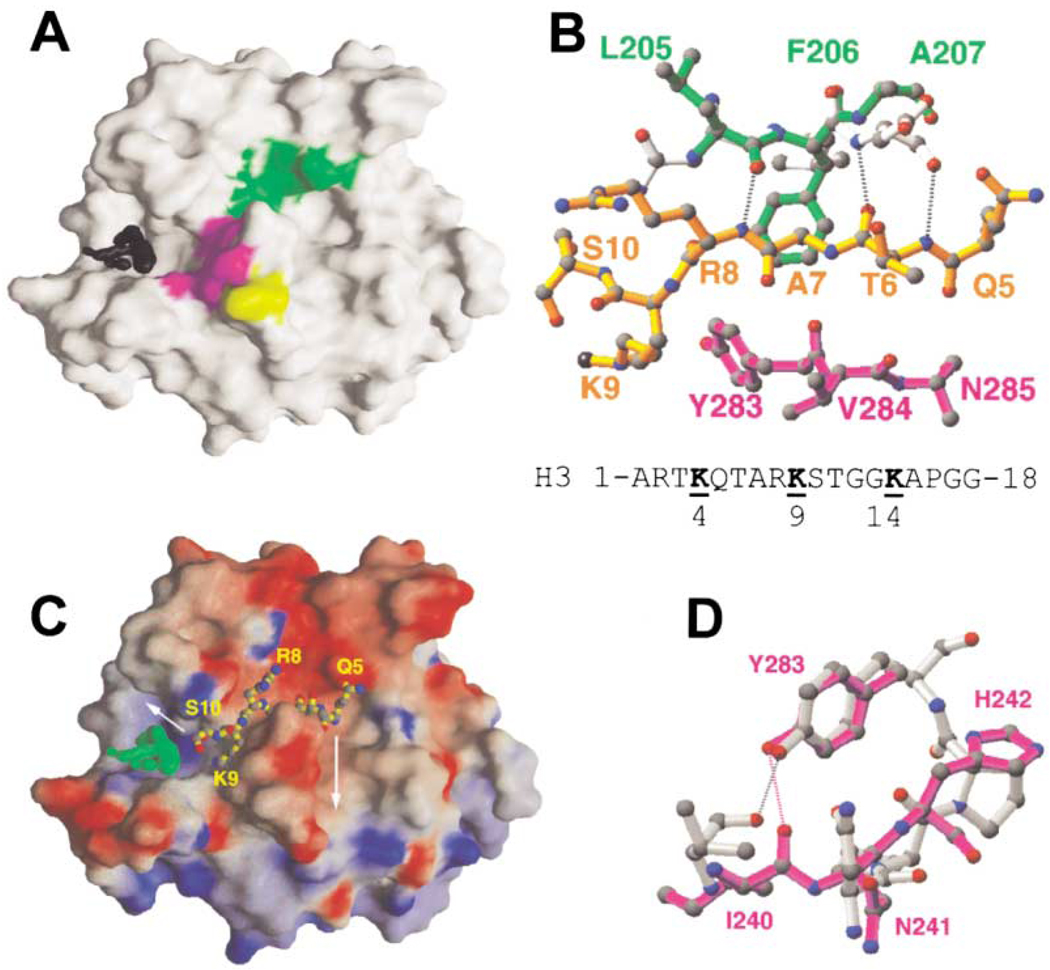
The cleft along the surface emanating from the presumed cofactor binding site is the likely binding site for the substrate polypeptide (Figure 2B). One side of this cleft is formed by strand β10 (green in Figure 5A)—the outermost strand of the β sheet (3↑ 9↑ 11↓ 10↑)—and the other side is formed by the loop after strand β17, which is the beginning of the disordered carboxy-terminal residues (286–299).
Figure 5. Putative Peptide Binding Cleft.
(A) Front view of GRASP surface (Nicholls et al., 1991). The difference electron density map (black) is contoured at 5.5σ. Strand β10 is green, N241, H242, and Y283 are magenta, and C244 is yellow.
(B) Superimposition of Drosophila HP1 β strand (light gray) (Jacobs and Khorasanizadeh, 2002; PDBcode 1KNA) and DIM-5 strand β10 (green). Dashed lines indicate the hydrogen bonds between HP1 and H3 peptide (yellow). The DIM-5 residues on the other side of the HP1 peptide are colored magenta. The dimethylated (methyl groups in black) target nitrogen atom occupies water site 2 (see Figure 4). The sequence of histone H3 peptide is shown at the bottom; both K4 and K14 are five residues away from K9.
(C) The docked H3 peptide lies in the putative peptide binding cleft. The cleft extends in both directions following turns as indicated. Surface charge distribution is displayed as blue for positive, red for negative, and white for neutral.
(D) Superimposition of active site NPPY residues of TaqI DNA-adenine amino MTase (gray) (Goedecke et al., 2001; PDB code 1G38) and the proposed DIM-5 active site residues N241, H242, and Y283 (magenta). The Tyr in both cases is hydrogen bonded to a main chain amide nitrogen atom (dashed bonds).
Structural studies have shown that heterochromatin protein HP1 binds to a methylated histone H3 peptide by inserting it as an antiparallel β strand between two HP1 strands, forming a hybrid three-stranded β sheet (Jacobs and Khorasanizadeh, 2002; Nielsen et al., 2002). Encouraged by the fact that one side of the DIM-5 cleft is a strand (β10), we superimposed the HP1 β strand (Drosophila HP1 residues 60–62) onto DIM-5 strand β10 (residue 205–207) (Figure 5B). The superimposition placed the H3 peptide (e.g., Q5-S10 as observed in HP1) in the DIM-5 cleft (Figure 5C) and residues Y283-V284-N285 following strand β17 on the other side of the peptide (Figure 5B). An induced-fit mechanism is used in HP1, in which the amino-terminal tail of the free HP1 adopts a β strand-like conformation upon interacting with the H3 peptide (Nielsen et al., 2002). In a similar way, binding of the H3 peptide may induce residues Y283-V284-N285 of DIM-5 and subsequent disordered residues to adopt a more stable β strand conformation that interacts with the peptide to form a hybrid sheet.
Target Lysine Binding Site
The most interesting result of the docking experiment is the placement of the target K9 immediately next to the presumed cofactor binding site (Figure 5C) with the target nitrogen atom occupying the position of a water molecule (site 2 in Figure 4). We propose that water site 2 is the likely active site of DIM-5, where the terminal amino group (NH3) of the substrate lysine would form a hydrogen bond with main chain carbonyl oxygen atom of R238. Many highly conserved residues, mainly from the two signature motifs (magenta), surround this site. Side chains of N241, H242, Y283, and Y204 form an inner circle immediately around site 2 (Figure 4). Residues E278, D282, and Y178 form an outer circle via interactions with the inner-circle residues: E278 interacts with N241, D282 interacts with H242, and Y178 interacts with Y283 via a water molecule (site 4) (Figure 4). Unlike protein arginine MTases or small molecule glycine N-MTase, which uses acidic residue(s) to neutralize the positive charge on the substrate amino group (Fu et al., 1996; Zhang et al., 2000), no acidic residue is immediately present in the proposed active site of DIM-5. Nevertheless, the combination of the negative dipole moment at the carboxyl end of helix αJ (R238 and F239), the negatively polarized main chain carbonyl oxygen atoms (I240 and W161), the side chain hydroxyl oxygen atoms of Y178, Y204, and Y283, and the asparagine oxygen atom of N241 might increase the nitrogen electron density enough to allow a nucleophilic attack on the AdoMet methylsulfonium group. The proton elimination step in conjunction with the methyl transfer is likely accomplished through a charge relay system involving H242 and D282, much as in protein arginine MTases (Zhang et al., 2000).
One observation consistent with this mechanism is the unusually high optimal pH (~10) of DIM-5 (Figure 3C), despite the fact that AdoMet binding is much more favorable in solutions of lower pH (Figure 3D). At pH 10, the amino group of target lysine (with a typical pKa value of 10) may be partially neutralized and the conserved tyrosines Y283, Y204, and Y178 (also with typical pKa values of 10) near the active site may be deprotonated; both deprotonations would facilitate methyl transfers.
The importance of the proposed active site residues is supported by site-directed mutagenesis experiments. Conservative changes at three residues (N241Q, H242K, and Y283F) immediately surrounding water site 2 essentially abolished HKMT activity (Figure 3E). Y283F has the lowest residual activity, suggesting that the hydroxyl group of Y283 is critical; it is hydrogen bonded to the backbone amide nitrogen of I240 and immediately adjacent to water site 2. Y283 is also one of the most conserved residues of the SET domain, being invariant in most of the SET-containing proteins in the Pfam database. Mutations of the two residues proposed to be involved in proton elimination, H242K and D282N, abolished and reduced HKMT activity, respectively. As expected, both mutants retained AdoMet crosslinking, though at reduced levels (Figure 3F). The complete loss of AdoMet crosslinking in N241Q and Y283F mutant proteins is somewhat unexpected. For N241Q, perhaps the longer glutamine side chain prevents the two hydrogen bonds forming between the side chain amino group of N241 and both the backbone carbonyl of W161 and the side chain of E278 (Figure 4). Interrupting the W161-N241-E278 interactions probably disrupts local structure, having a more deleterious effect than the replacement of side chain in the W161F mutant. It is also possible that both N241 and Y283 interact with the adenine ring of AdoMet, which is likely involved in the UV crosslinking, although not observed in the crystal.
The presumptive active site of DIM-5 is reminiscent of the consensus NPPY motif involved in the amino-methylation of adenine or cytosine in DNA (Blumenthal and Cheng, 2001; Goedecke et al., 2001; Gong et al., 1997) and of the glutamine in peptide release factor (Heurgue-Hamard et al., 2002; Nakahigashi et al., 2002). Remarkably, the invariant N241 and Y283 of DIM-5 are superimposable onto the first and the last amino acids of NPPY in TaqI DNA adenine MTase (Figure 5D). This suggests a potential similarity in the catalytic mechanism between histone lysine MTases and DNA amino-MTases. In the latter case, the amino group (NH2) that becomes methylated is positioned for an in-line attack on AdoMet by hydrogen bonding to the backbone carbonyl connecting the two inflexible prolines (Goedecke et al., 2001). The equivalent backbone carbonyl in DIM-5 is probably that of R238. Since this particular carbonyl needs to be relatively immobile to hinder the free rotation of the amino group bound at site 2, the great reduction of HKMT activity in R238H mutant could be the result of a small change or flexibility in the position of the backbone carbonyl oxygen atom that fails to interact properly with the target amino group.
Mono-, di-, and trimethylated lysines have been observed in histones (Duerre and Chakrabarty, 1975) but very little information is available about the methylation status of individual residues. Nevertheless, we have found that DIM-5 efficiently methylates dimethylated lysine 9 of histone H3 peptide, and DIM-5 is capable of adding 1–3 methyl groups to K9 of histone H3 peptide (unpublished data of H.T., X.Z., D. McMillen, J. Nakayama, P. Singh, D. Allis, S. Grewal, X.C., and E.U.S.). We propose that water sites 1 and 3 (Figure 4), which hydrogen bonded to site 2, may accommodate the methyl group(s) on mono- and dimethylated lysine substrates. These additional interactions may help position the nitrogen atom and enhance its reactivity.
The Post-SET Domain
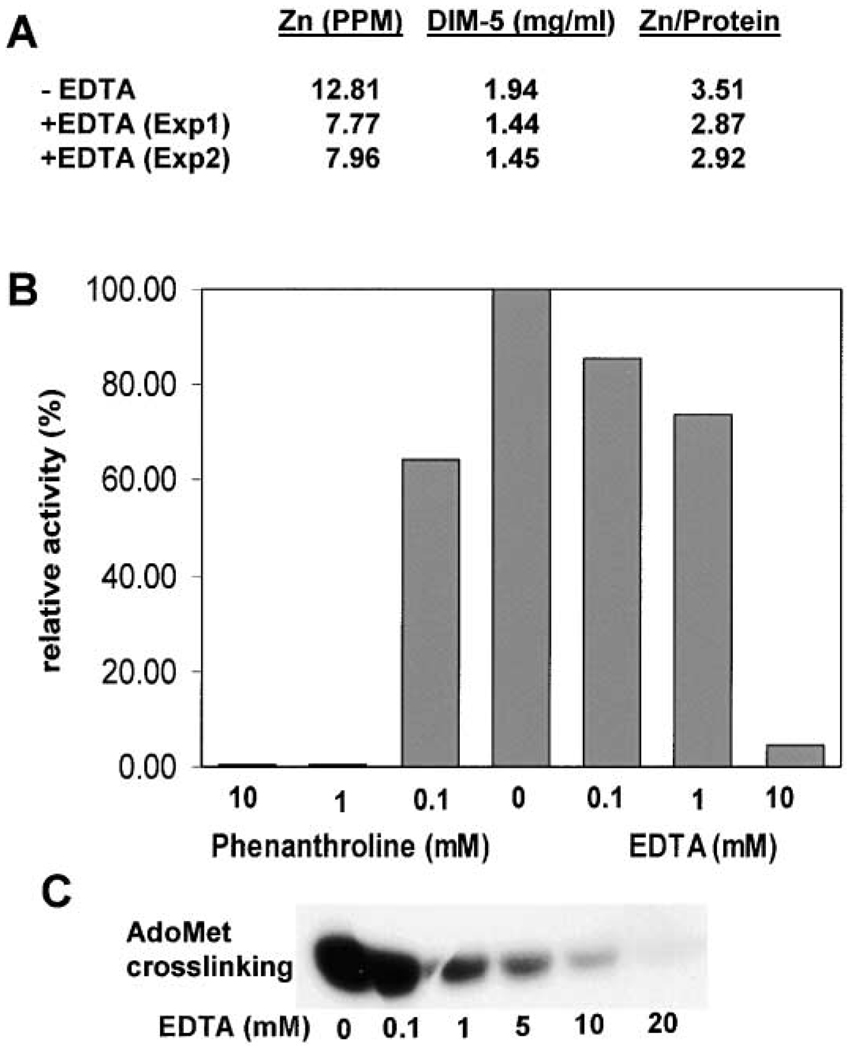
The C terminus, including the post-SET region, is mostly disordered in the crystal except for the segment between residues 299 and 308 (gray in Figures 2A and 2B). This 10 residue segment, identified through M303 in selenomethionine-substituted DIM-5 protein (see Experimental Procedures), was stabilized in the interface between two crystallographic-related molecules. We hypothesize that this segment (along with the adjacent disordered residues) will adopt a different structure upon binding to substrate. The post-SET region contains three conserved cysteine residues that appear to be essential for HKMT activity in the SUV39 family. Changing all three cysteines to serines (3C-S) abolished DIM-5 activity (Figure 3E), as did a Cys to Tyr substitution at C1279 in SETDB1 (Schultz et al., 2002), which corresponds to C306 of DIM-5. While the exact role of the three post-SET cysteines cannot be determined from the current structure, one intriguing possibility is that, when coupled with the fourth cysteine from the loop formed by the signature motif N241HXCXPN247 (C244 in PerDIM-5), these form an additional metal binding site. Several observations are consistent with this hypothesis. (1) Of the more than 50 SET protein sequences that we have examined to date, there appears to be an absolute correlation between the presence of the post-SET and a cysteine corresponding to C244 of DIM-5 (see Figure 1 for examples). In addition, replacing the Cys corresponding to C244 with alanine in SUV39H1 or SETDB1 abolished HKMT activity (Rea et al., 2000; Schultz et al., 2002). (2) The total zinc content of DIM-5 protein is 3.51 (Figure 6A), indicating that more than three zinc ions are present. (3) Incubation of metal chelators, phenanthroline or EDTA, with DIM-5 protein inhibited its activity and significantly reduced AdoMet binding (Figures 6B and 6C). Interestingly, even when EDTA completely abolished DIM-5 activity, the protein still retained approximately three (2.9) zinc ions (Figure 6A). As the triangular zinc cluster is quite stable, it is conceivable that the chelated zinc was coordinated by the three post-SET cysteines and C244 (yellow in Figure 5A), which is near the active site. (4) Like the metal chelators, simultaneous mutation of the three cysteines (3C-S) also caused a complete loss of DIM-5 activity and AdoMet crosslinking (Figures 3E and 3F), consistent with the idea that the post-SET cysteines are involved in AdoMet binding. Perhaps the observed disorder of the post-SET is partly, or fully, responsible for the poor density of the AdoHcy in the current structure.
Figure 6. Metal Chelators Inhibit DIM-5 Activity.
(A) Analysis of zinc content of DIM-5 with and without EDTA treatment. DIM-5 protein was incubated with 20 mM EDTA for 2 days, at which time HKMT activity was no longer detectable. To remove zinc bound to EDTA, the protein was either dialyzed (Exp1) or subjected to gel filtration chromatography (Exp2) against 20 mM glycine (pH 9.8), 5% glycerol, 0.5 mM DTT, and 1 mM EDTA.
(B) Purified DIM-5 protein (1 mg/ml in 20 mM glycine [pH 9.8], 5% glycerol) was incubated with various concentration of 1,10-phenanthroline or EDTA for 18 hr at 4°C. The enzyme was diluted 80-fold and assayed for HKMT activity under standard conditions, except that no DTT was present.
(C) Fluorographic results of AdoMet crosslinking in the presence of EDTA.
Conclusions
We determined the crystal structure of a histone H3 lysine 9 MTase, DIM-5 from N. crassa, and carried out mutational and biochemical studies to illuminate the mechanism of this enzyme. We found that the highly conserved residues of the pre-SET region form a triangular zinc cluster, Zn3Cys9, and that residues in the SET domain are essential for the cofactor binding and methyl transfer. The SET domain also has a cleft that is the likely binding site for the methylatable amino-terminal tail of histone H3. The post-SET region may also contribute to cofactor binding and catalysis by forming another zinc binding site in conjunction with a conserved cysteine near the active site. These results provide insight into a common fold and the catalytic mechanism for the SUV39 family histone H3 lysine 9 MTases. Finally, this work provides an example of completely unrelated, structurally distinct proteins that carry out a common function, in this case AdoMet-dependent methyl transfer.
Experimental Procedures
Protein Expression and Purification
N. crassa DIM-5 protein was expressed as a GST fusion. A segment of the wild-type dim-5 ORF, including amino acid residues 17–318, was amplified from pGEX-5X-3/DIM-5 (Tamaru and Selker, 2001) and subcloned between the BamHI and EcoRI sites in pGEX2T (Amersham-Pharmacia), yielding pXC379. E. coli strain BL21(DE3) Codon plus RIL (Stratagene) carrying pXC379 was grown in LB medium supplemented with 10 µM ZnSO4 at 37°C to OD600 = 0.5, shifted to 22°C, and induced with 0.4mMIPTG overnight at 22°C. The proteins were purified using Glutathione-Sepharose 4B (Amersham-Pharmacia), UnoQ6 (Bio-Rad), and Superdex 75 columns (Amersham-Pharmacia). The GST tag was cleaved by applying thrombin to fusion proteins bound to the Glutathione-Sepharose column, leaving five additional residues (GSHMG) in front of amino acid 17 of DIM-5. All purification buffers contained 1 mM DTT and no EDTA. The protein was stored in the Superdex 75 column buffer containing 20 mM glycine (pH 9.8), 150 mM NaCl, 1 mM DTT, and 5% glycerol. Se-containing DIM-5 (with five methionines) was expressed in a methionine auxotroph strain (B834) grown in the presence of Se-methionine, and the protein was purified similarly to the native protein.
Methyl Transfer Activity Assay
The activity was assayed in a 20 µl reaction containing 50 mM glycine (pH 9.8), 2 mM DTT, 40–80 µM unlabeled AdoMet (Sigma), 0.5 µCi [methyl-3H]AdoMet (78 Ci/mmol, NEN NET155H), 0.25–0.5 µg of DIM-5 protein, and 2–5 µg histones (calf thymus histones Sigma H4524, Roche 223565, or recombinant chicken erythrocyte histones, a gift from Dr. V. Ramakrishnan). The reaction was incubated at room temperature for 10–15 min and methylation was analyzed either by SDS-PAGE and fluorography or by precipitation with 20% TCA, filtration (Milipore GF/F filter), washing, and liquid scintillation counting. Under these conditions, DIM-5 activity was linearly related to reaction time and amount of enzyme and AdoMet and histone were saturating. For some reason, the relatively crude Sigma H4524 histone preparations generally gave 2- to 4-fold higher incorporation than either the Roche preparations or the recombinant histones.
AdoMet Binding Assay by UV Crosslinking
Twenty microliters of purified DIM-5 protein (2–5 µg) was incubated with 0.5 µCi of [methyl-3H]AdoMet (78 Ci/mmol, NEN NET155H) overnight at 4°C. Samples were added to a 96-well plate on ice and placed 8 cm from an inverted UV transilluminator (VWR, 302 nm) for 1 hr. The protein was then separated by SDS-PAGE, stained with Coomassie, and subjected to fluorography.
Mutagenesis
Amino acid replacements of DIM-5 to yield R155H, W161F, Y204F, manuR238H, N241Q, H242K, D282K, and Y283F were made using Quik-Change site-directed mutagenesis protocol (Stratagene) using pXC379 and primer pairs to generate CAC, TTC, TTC, CAC, CAG, AAA, AAC, and TTC codons in place of AGG, TGG, TAC, AGG, AAC, CAC, GAC, and TAT codons, respectively. The DIM-5 mutant 3C to 3S, in which all three invariant cysteines in the post-SET region are replaced by serines, was generated by PCR using a mutagenic 3′ primer. All mutants were sequenced to verify the presence of the intended mutation and the absence of additional mutations. The only exception is the Y204F mutant, which carries an additional Asp substitution (A24D) in the N-terminal region that was not observed in the structure. Mutant proteins, along with wild-type, were purified from 100–200 ml of induced cultures. A disposable column containing 0.5 ml of Glutathione-Sepharose 4B (Amersham-Pharmacia) was used for each mutant. The mutant proteins were separated from GST by on-column thrombin cleavage and then used for enzymatic assay (using calf thymus histones Sigma H4524 as substrate), AdoMet binding by crosslinking analysis, and analytical gel filtration chromatography for native protein size determination.
Zinc Content Analysis
One sample of untreated and two samples of EDTA-treated DIM-5 protein (about 2 ml of 2 mg/ml each) was analyzed for the presence of 20 elements on a Thermo Jarrell-Ash Enviro 36 ICAP analyzer at the Chemical Analysis Laboratory of the University of Georgia at Athens. In order to calculate the molar ratio of Zn to protein, the precise concentration of the untreated DIM-5 protein was determined by amino acid analysis (averaging two independent measurements) performed at the Keck Facilities at Yale University. The extinction coefficient (29,559 M−1cm−1) derived from the amino acid analysis was used to estimate the protein concentration of the EDTA-treated samples.
Crystallography
Purified DIM-5 protein was concentrated to about 10–15 mg/ml in 20 mM glycine (pH 9.8), 150 mM NaCl, 1 mM DTT, 5% glycerol, and 600 µM AdoHcy. Crystals were obtained using the hanging drop method, with mother liquor containing 1.1–1.2 M ammonium sulfate and 100mM Na citrate (pH 5.4–5.6) at 16°C. Crystals belong to space group P212121 with cell dimensions of 36.73 × 81.56×101.27 Å. Each asymmetric unit contains one molecule. Complete data sets were collected from a native crystal near the Zn absorption edge (Table 1) and a SeMet-incorporated crystal at both Se and Zn absorption edges (not shown). The data were processed using the HKL package (Otwinowski and Minor, 1997). SOLVE (Terwilliger and Berendzen, 1999) first revealed the positions of three zinc atoms and RESOLVE (Terwilliger, 2000) was then used to modify the electron density map. The modified map was of good quality at 2.9 Å resolution to place amino acids of DIM-5 into the recognizable densities using O (Jones and Kjeldgard, 1997). In parallel, SOLVE determined the positions of five selenium atoms: two of them (SeMet 233 and 248) were confirmed by Zn-phased map, and three of them (SeMet 75, 85, and 303) served as markers in the primary sequence during tracing. The resultant model was refined against the data collected at wavelength of 1.0332 Å in the resolution range of 24.8–1.98 Å, using the X-PLOR program suite (Brünger, 1992). Three segments of DIM-5 were not observed in the final model: the N-terminal 8 residues (17–24) (these may not be present in the native DIM-5 protein as there is an in-frame splicing site immediately after these residues); residues 89–99 of the pre-SET domain (these are deleted in many of the SUV39 proteins) (see Figure 1); and the majority of the C-terminal 34 amino acids (the C terminus is also highly variable in length and sequence among SET proteins except for the three-Cys post-SET region). Among the nonglycine and nonproline residues, 86% are in most favored and 14% in additional allowed regions of a Ramachandran plot (Laskowski, 1993).
Acknowledgments
We thank Dr. V. Ramakrishnan (Cambridge) for recombinant histone, Ms. Fang Fang Yin for making 3C-S mutant, and Drs. D. Reines, P.W. Wade, and R.M. Blumenthal for critical comments on the manuscript. These studies were supported in part by U.S. Public Health Services grants GM49245 and GM61355 (to X.Z. and X.C.) and GM35690 (to E.U.S). X-ray data were collected at beamline 17-ID in the facilities of IMCA-CAT at the Advanced Photon Source. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract number W-31-109-Eng-38.
Footnotes
Accession Numbers
The coordinates for DIM-5 can be found in the Protein Data Bank with the accession number 1ML9.
References
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, Schulz I, Reuter G, Aalen RB. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 2001;29:4319–4333. doi: 10.1093/nar/29.21.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal RM, Cheng X. A Taq attack displaces bases. Nat. Struct. Biol. 2001;8:101–103. doi: 10.1038/84072. [DOI] [PubMed] [Google Scholar]
- Boggs BA, Cheung P, Heard E, Spector DL, Chinault AC, Allis CD. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat. Genet. 2002;30:73–76. doi: 10.1038/ng787. [DOI] [PubMed] [Google Scholar]
- Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT. X-PLOR. A System for X-Ray Crystallography and NMR, 3.1 edn. New Haven, CT: Yale University; 1992. [Google Scholar]
- Carson M. Ribbons. Methods Enzymol. 1997;227:493–505. [PubMed] [Google Scholar]
- Cheng X, Roberts RJ. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 2001;29:3784–3795. doi: 10.1093/nar/29.18.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerre JA, Chakrabarty S. Methylated basic amino acid composition of histones from the various organs from the rat. J. Biol. Chem. 1975;250:8457–8461. [PubMed] [Google Scholar]
- Fang J, Feng Q, Ketel CS, Wang H, Cao R, Xia L, Erdjument-Bromage H, Tempst P, Simon JA, Zhang Y. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr. Biol. 2002;12:1086–1099. doi: 10.1016/s0960-9822(02)00924-7. [DOI] [PubMed] [Google Scholar]
- Fauman EB, Blumenthal RM, Cheng X. Structure and evolution of Adomet-dependent methyltransferases. In: Cheng X, Blumenthal RM, editors. S-Aden-osylmethionine-Dependent Methyltransferases: Structures and Functions. River Edge, NJ: World Scientific; 1999. pp. 1–38. [Google Scholar]
- Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- Fu Z, Hu Y, Konishi K, Takata Y, Ogawa H, Gomi T, Fujioka M, Takusagawa F. Crystal structure of glycine N-methyltransferase from rat liver. Biochemistry. 1996;35:11985–11993. doi: 10.1021/bi961068n. [DOI] [PubMed] [Google Scholar]
- Goedecke K, Pignot M, Goody RS, Scheidig AJ, Weinhold E. Structure of the N6-adenine DNA methyltransferase M.TaqI in complex with DNA and a cofactor analog. Nat. Struct. Biol. 2001;8:121–125. doi: 10.1038/84104. [DOI] [PubMed] [Google Scholar]
- Gong W, O’Gara M, Blumenthal RM, Cheng X. Structure of PvuII DNA-(cytosine N4) methyltransferase, an example of domain permutation and protein fold assignment. Nucleic Acids Res. 1997;25:2702–2715. doi: 10.1093/nar/25.14.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurgue-Hamard V, Champ S, Engstrom A, Ehrenberg M, Buckingham RH. The hemK gene in Escherichia coli encodes the N(5)-glutamine methyltransferase that modifies peptide release factors. EMBO J. 2002;21:769–778. doi: 10.1093/emboj/21.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Taverna SD, Zhang Y, Briggs SD, Li J, Eissenberg JC, Allis CD, Khorasanizadeh S. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 2001;20:5232–5241. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Laible G, Dorn R, Reuter G. SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell. Mol. Life Sci. 1998;54:80–93. doi: 10.1007/s000180050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Kjeldgard M. Electron-density map interpretation. Methods Enzymol. 1997;277:173–208. doi: 10.1016/s0076-6879(97)77012-5. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A. COMPASS, a histone H3 (lysine 4) methyltransferase required for telomeric silencing of gene expression. J. Biol. Chem. 2002;277:10753–10755. doi: 10.1074/jbc.C200023200. [DOI] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Lacoste N, Utley RT, Hunter J, Poirier GG, Cote J. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J. Biol. Chem. 2002;277:30421–30424. doi: 10.1074/jbc.C200366200. [DOI] [PubMed] [Google Scholar]
- Laskowski RA. PROCHECK: a program to check the stereo-chemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science. 2001;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- Ma H, Baumann CT, Li H, Strahl BD, Rice R, Jelinek MA, Aswad DW, Allis CD, Hager GL, Stallcup MR. Hormone-dependent, CARM1-directed, arginine-specific methylation of histone H3 on a steroid-regulated promoter. Curr. Biol. 2001;11:1981–1985. doi: 10.1016/s0960-9822(01)00600-5. [DOI] [PubMed] [Google Scholar]
- Nakahigashi K, Kubo N, Narita S, Shimaoka T, Goto S, Oshima T, Mori H, Maeda M, Wada C, Inokuchi H. HemK, a class of protein methyl transferase with similarity to DNA methyl transferases, methylates polypeptide chain release factors, and hemK knockout induces defects in translational termination. Proc. Natl. Acad. Sci. USA. 2002;99:1473–1478. doi: 10.1073/pnas.032488499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O’Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, Kouzarides T. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P, Reinberg D. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 2002a;16:479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R, et al. PR-Set7 Is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell. 2002b;9:1201–1213. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- O’Carroll D, Scherthan H, Peters AH, Opravil S, Haynes AR, Laible G, Rea S, Schmid M, Lebersorger A, Jerratsch M, et al. Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol. Cell. Biol. 2000;20:9423–9433. doi: 10.1128/mcb.20.24.9423-9433.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F-and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Robbins AH, McRee DE, Williamson M, Collett SA, Xuong NH, Furey WF, Wang BC, Stout CD. Refined crystal structure of Cd, Zn metallothionein at 2.0 Å resolution. J. Mol. Biol. 1991;221:1269–1293. [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Briggs SD, Brame CJ, Caldwell JA, Koh SS, Ma H, Cook RG, Shabanowitz J, Hunt DF, Stallcup MR, Allis CD. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr. Biol. 2001;11:996–1000. doi: 10.1016/s0960-9822(01)00294-9. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr. D. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- Vasak M, Hasler DW. Metallothioneins: new functional and structural insights. Curr. Opin. Chem. Biol. 2000;4:177–183. doi: 10.1016/s1367-5931(00)00082-x. [DOI] [PubMed] [Google Scholar]
- Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell. 2001a;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P, Zhang Y. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001b;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhou L, Cheng X. Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. EMBO J. 2000;19:3509–3519. doi: 10.1093/emboj/19.14.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]