Abstract
Mechanism-based inhibitors of enzymes, which mimic reactive intermediates in the reaction pathway, have been deployed extensively in the analysis of metabolic pathways and as candidate drugs. The inhibition of cytosine-[C5]-specific DNA methyltransferases (C5 MTases) by oligodeoxynucleotides containing 5-azadeoxycytidine (AzadC) and 5-fluorodeoxycytidine (FdC) provides a well-documented example of mechanism-based inhibition of enzymes central to nucleic acid metabolism. Here, we describe the interaction between the C5 MTase from Haemophilus haemolyticus (M.Hha I) and an oligodeoxynucleotide duplex containing 2-H pyrimidinone, an analogue often referred to as zebularine and known to give rise to high-affinity complexes with MTases. X-ray crystallography has demonstrated the formation of a covalent bond between M.Hha I and the 2-H pyrimidinone-containing oligodeoxynucleotide. This observation enables a comparison between the mechanisms of action of 2-H pyrimidinone with other mechanism-based inhibitors such as FdC. This novel complex provides a molecular explanation for the mechanism of action of the anti-cancer drug zebularine.
Keywords: zebularine, DNA methyltransferase, DNA methylation, M.Hha I, base flipping
Introduction
Modulation of histone modification (acetylation, phosphorylation, and methylation) and DNA methylation are the principal driving forces behind the phenomenon of epigenetics.1–3 While histone modification is restricted to the eukarya, in organisms ranging from bacteriophage to man, differential DNA methylation has been co-opted for the regulation of genetic transactions, including transcription, imprinting, and recombination, and classically provides a barrier to host-specific restriction endonucleases.4–7 Moreover, the recently demonstrated, close molecular similarity between the human DNMT2 protein and M.Hha I,8 a bacterial cytosine-[C5]-specific DNA methyltransferase (C5 MTase) lends strong support to the notion that bacterial DNA MTases represent a generic platform for understanding mechanistic aspects of biological DNA methylation.
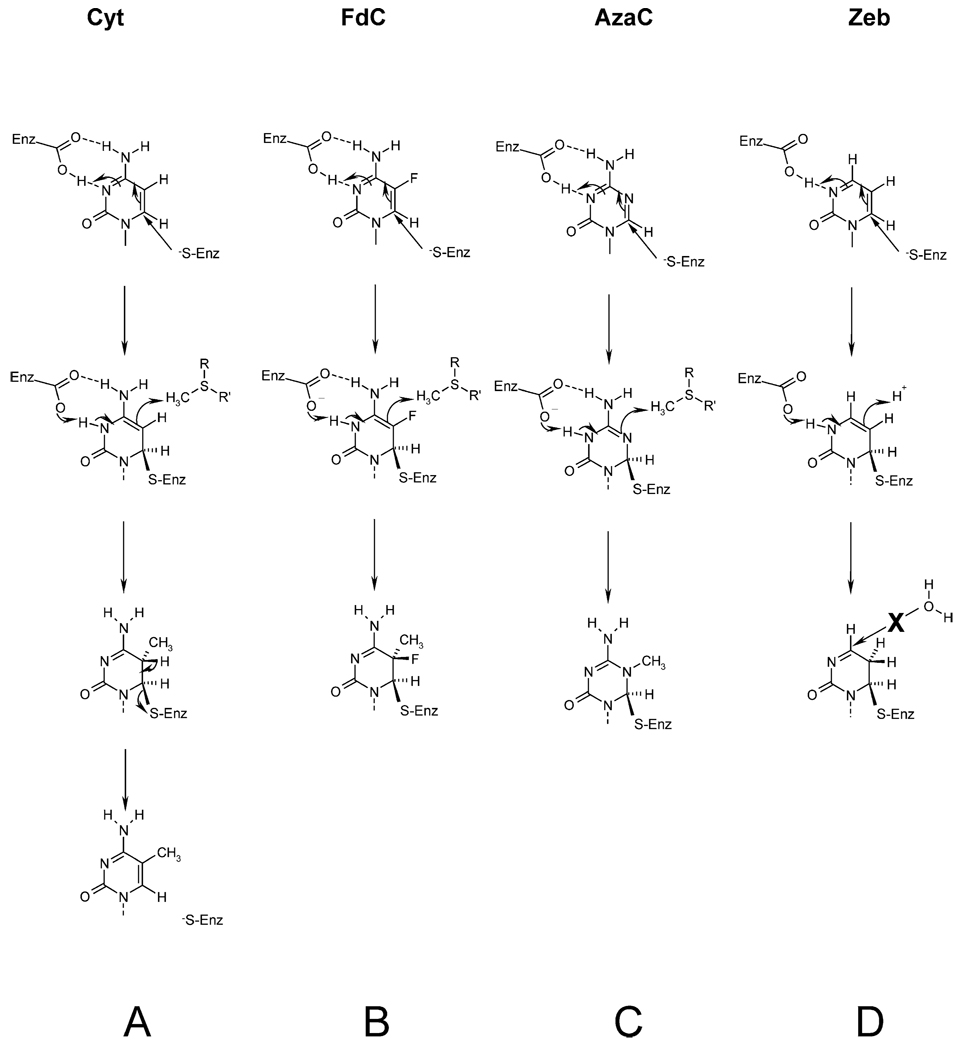
The mechanism of DNA-C5 MTases involves the addition of a protein thiol group (from a cysteine residue in a highly conserved Pro-Cys motif) to the C6 position of the target dC, which activates the carbon atom at the 5 position allowing reaction with S-adenosyl-l-methionine (AdoMet) (Figure 1). Cheng & Roberts9 have recently surveyed the known C5 MTase nucleic acid-related inhibitors (which have been useful in elucidating mechanistic features), including the classical MTase inhibitor 5-azacytidine (AzaC),10 5-fluoro-cytosine (FdC),11 and 5,6-dihydro-5-azacytidine12 as well as compounds such as 4′-thio-2′-deoxycytidine13 in which the sugar moiety is modified. The substitution of the C5 proton by fluorine in FdC has proved to yield an invaluable reagent for studying methyl transfer reactions (Figure 1). Indeed, Chen et al.14 incorporated FdC into a specific duplex and were able to trap M.Hae III in a covalent complex through the conserved Pro-Cys motif. Subsequently, the crystallization of a covalent ternary complex between M.Hha I, AdoMet, and an FdC oligonucleotide led to the discovery of the phenomenon of base flipping and provided a structural basis of this mechanism-based inhibition.15 Earlier suggestions that the catalytic mechanism of DNA methyltransfer involves transient disruption of the DNA duplex proved well founded.16 – 18 However, while the presence of a fluorine atom at the C5 position of the target base renders covalent attack irreversible, it does not significantly impair or stimulate initial complex formation or base flipping. In a similar manner, replacement of C5 by a nitrogen atom in AzaC does not influence initial binding events; rather, nucleophilic attack is facilitated at the C6 position19 and methyl transfer, although possible, is substantially retarded.20
Figure 1.
The reaction pathway of C5 MTases in the presence and in the absence of mechanism-based inhibitors. (a) The reaction pathway for all C5 MTases involves the transfer of the labile methyl group from S-adenosyl-l-methionine (AdoMet) to the 5 position of the cytosine ring, proceeds through a covalent intermediate at position C6.14 The nucleophilic attack upon the C6 position of cytosine drives the subsequent acquisition of the labile methyl group from AdoMet. (Note, the protonation status of Glu119 in M.Hha I36). (b) The inhibition by FdC. Following covalent complex formation and methyl transfer, the analogue remains bound to the active-site Cys, since abstraction of F cannot be achieved. (c) The inhibition by AzaC. Following covalent complex formation at a C6 with enhanced reactivity, slow methyl transfer may take place, but there is no H at C5 to abstract and the covalent complex persists. (d) The inhibition by zebularine. Following covalent complex formation at a C6 with enhanced reactivity as with AzaC, facilitated deamination at C4 cannot proceed,33 since the amino moiety is absent from the analogue. Note that the water molecule nearest to the C4 atom is 3.6 Å away and the water molecule nearest to the C5 atom is 3.3 Å away.
The covalent attachment of a C5 MTase to its recognition sequences will presumably lead to persistent but aberrant nucleoprotein complexes throughout the genome.21,22 This leads to a cumulative depletion of the enzyme from the nuclear pool, leading to the net demethylation of the genome: the repair of damage as a consequence of nucleoprotein adduct formation subsequently takes place and may be, in part, error-prone.22
The observation that oligodeoxynucleotide duplexes containing zebularine at the target dC form high-affinity, SDS-resistant complexes with M.Msp I23,24 and M.Hga I-225 suggests the zebularine-containing DNA could be an effective inhibitor.
Here, we describe the structure of a complex between the bacterial DNA MTase M.Hha I and an oligodeoxynucleotide duplex containing zebularine incorporated at the position normally occupied by the base targeted for methylation. The enzyme forms a covalent complex in the absence of methyl transfer from AdoMet, unlike that formed between a duplex similarly substituted with FdC. We present a generalized framework for the inhibitory properties of this and the other known C5 MTase inhibitors based on facilitated flipping and electrostatic properties of the flipped nucleotide.
Results
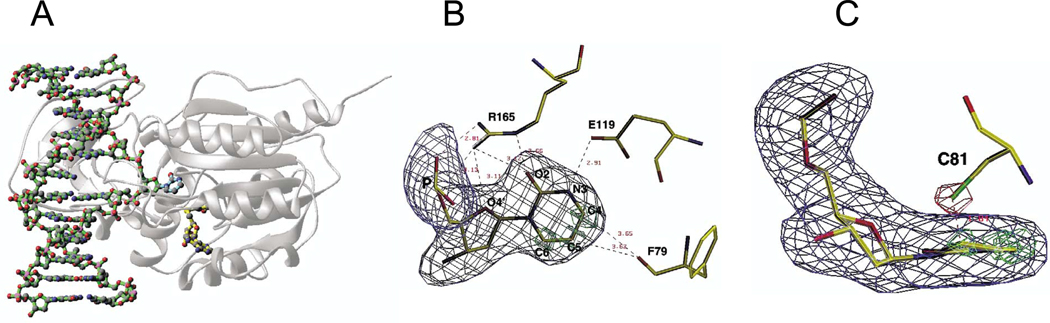
The structure of M.Hha I in a ternary complex with AdoHcy and a 13-mer non-palindromic DNA duplex containing a 5′-GZGC-3′-5′GCGC-3′ with zebularine as the target nucleotide on one strand was determined by X-ray crystallography. The target zebularine was flipped out of the DNA helix (Figure 2(a)), while the dG on the complementary strand remained stacked within the DNA helix. The absence of the amino (NH2) group of the zebularine was confirmed by the (Fo − Fc′ αc) electron density map, in which the zebularine was omitted from the structure factor calculation (Figure 2(b)). The zebularine–DNA enzyme complex is highly similar to that formed through FdC (see Figure 10 of Kumar et al.13) within the estimated coordinate error (0.3 Å ). The truncation at the N4 (NH2 group) position leads to the loss of two enzyme–DNA hydrogen bonds.
Figure 2.
Structure of M.Hha I–AdoHcy–DNA containing zebularine. (a) The structural impasse between the proposed mechanism (outlined in Figure 1) and the barrier to substrate access in duplex DNA was overcome elegantly by the phenomenon of protein-induced base flipping.15 Once the target base (zebularine here) is released from the constraints of the Watson–Crick base-pair, conventional active-site chemistry is facilitated. (b) Zebularine difference electron density maps (Fo − Fc, αc) superimposed on the refined coordinates with carbon atoms being yellow, oxygen atoms red, nitrogen atom blue, and sulfur atom green, respectively. The blue electron density map contoured at 5.0σ was computed with the zebularine moiety omitted from the atomic model. The green electron density maps contoured above 5.5σ were calculated with the C4, C5, and C6 atoms of zebularine omitted from the atomic model, respectively. The zebularine is constrained in the plane of the ring by a highly conserved network of hydrogen bonds (via E119 and R165) and van der Waals interactions between the main-chain C=O group of F79 and C4 and C5 atoms. (c) A view perpendicular to (b), looking edge-on at the flipped zebularine molecule. A covalent bond is observed between C6 of the zebularine ring and an invariant thiolate side-chain C81, approaching the C6 perpendicular to the ring. The red electron density map contoured above 10.0σ was calculated with the sulfur atom of C81 omitted from the atomic model.
The target zebularine is rotated out of the helix and into the catalytic pocket of the enzyme, as seen in each of the previously solved DNA-M. Hha I-cofactor ternary structures. This dramatic structural change, first observed for M.Hha I,15 forms the basis of enzymatic sequence-specific methylation of cytosine bases in duplex DNA. Subsequently, the crystal structures of a growing number of nucleic acid-modifying enzymes have revealed that the flipping of a target base from the DNA duplex pervades enzymatic DNA modification.7
The introduction of zebularine did not distort the structure of the complex grossly. However, the introduction of the zebularine molecule results in a covalent bond distance of 1.84 Å between the C6 atom and sulfur atom of the nucleophile, Cys81 (Figure 2(c)). In conjunction with the reactive state of Cys81, the distance between O∈1 of Glu119 and N3 of zebularine is 2.91 Å, where the N3 atom is proposed to be protonated during the reaction and forms a hydrogen bond with the acidic side-chain (Figure 2(b)). The covalent bond distance between Cys81 and C6, and the hydrogen bond distance between Glu119 and N3 are consistent with the suggestion that the N3 protonation and the sulfhydryl attack on C6 are concerted events. In addition, the side-chain of Arg165 has extensive interactions with three oxygen atoms: the terminal Nη1 (NH2 group) forms a charge–charge interaction with the phosphate O1p (2.8 Å ), Nη2 forms a hydrogen bond with the deoxyribose O4′ (3.1 Å ), and both Nη2 and N∈ form hydrogen bonds with O2 (3.1 Å and 2.7 Å, respectively) (Figure 2(b)). The net positive charge carried by the Arg is balanced mainly by the negative charge of the phosphate group. The O4′ oxygen atom has been calculated to carry a total charge of only −0.16e,26 suggesting that it is a weak hydrogen bond acceptor. The O2 atom has been calculated to have a net negative charge of −0.51e that rendered it a good acceptor for hydrogen bonds.
The crystallographic data indicate that the zebularine moiety alone, when incorporated into DNA, is sufficient to produce strong inhibition of M.Hha I, since it occupies the active site of the enzyme with formation of a covalent bond between the sulfur atom of Cys81 and C6 of the base. Moreover, the covalent complex was formed in the presence of AdoHcy, which precluded the need for methyl transfer for its formation. Consistent with the structural data, molecular orbital calculations indicate that, like dC and FdC, C6 of zebularine is still a potential site for nucleophilic attack (Figure 3). Covalent adduct formation is the natural course of the enzyme-catalyzed reaction, and the pyrimidinone is more reactive than the pyrimidine.23 One would expect an enzyme capable of adding a thiol group across the 5,6 double bond of a pyrimidine to do the same with the more reactive pyrimidinone. The only thing that could prevent this is if the pyrimidinone binds to the enzyme in a manner substantially different from that of the pyrimidine; but the X-ray structure shows that this is not the case.
Figure 3.
Molecular orbital calculations. Ab initio geometries were calculated with the program SPARTAN (Wave-function, Irvine, CA) as described.37 1-Methylcytosine (instead of the sugar ring at position 1) was used to model the flipped base. Dark blue indicates a high value for the orbital, and the red indicates a low value, while green/yellow is in between. High values for the lowest unoccupied molecular orbital (LUMO) are seen over C6 and C4 for both 1-methylcytosine and N3 protonated 1-methylcytosine in all three cases, indicating that these are potential sites for nucleophilic attack. The 1-methyl-6-sulfhydryl-enol derivative of cytosine was used to model the intermediate formed by the enzyme after nucleophilic attack at C6. High values of the highest occupied molecular orbital (HOMO) are confined to C5 in cytosine and FdC, where the highly reactive orbital at C5 is then poised for attack on the methyl group of AdoMet, but between C4 and C5 in 2-H pyrimidinone. This is consistent with the observation that zebularine does not seem to be methylated at all.38
However, what remains unexplained is why the bond is permanent, i.e. why the β-elimination to reverse adduct formation does not take place. In the case of AzaC, there is no proton available for extraction, while for FdC an F is more difficult to abstract than a proton.11 However, if the pyrimidinone analogue acts as a mimic of an intermediate in the minor deaminative pathway catalyzed by C5 MTases,19,20 (see Discussion) then the pyrimidinone does not fall on the major catalytic pathway, but represents a stable end-point in the absence of the 4-amino moiety, in a manner similar to that of the inhibitory zebularine complex with cytidine deaminase,27,28 but without the formation of a 3,4-hydrate (see Figure 1(d)).
Discussion
The proposed increase in the reactivity of C6 in zebularine24 is coupled to the reduction in the barrier to base flipping as observed for the inhibitory duplexes containing an abasic site or a mismatch at the target dC.29 – 31 This increase in reactivity at C6 in zebularine-substituted DNA has been substantiated by the finding that replacement of the catalytic Cys in M.MspI by either Thr or Ser, but not by Tyr or any other side-chain, leads to the formation of high-affinity, SDS-resistant complexes with DNA.24 Hurd et al.24 argued that the observed formation of denaturant-resistant complexes between the Thr and Ser mutants is consistent with increased reactivity at the C6 position, as has been observed between such mutants and AzaC-substituted DNA.19 These experiments indicate that the activation energy barrier to nucleophilic attack at the C6 position of the flipped base is diminished considerably in both zebularine and AzaC-containing DNA. Increased reactivity of 5-methyl-2-pyrimidinone (the 5-methyl derivative of zebularine) to nucleophilic attack at C6 has been demonstrated,32 consistent with the idea that the C6 position in zebularine, and its derivatives, is more reactive than the corresponding location in pyrimidines.
It has been demonstrated that C5 MTases, in addition to their primary function in transmethylation, promote the deamination of flipped dC duplex DNA.19,20 This side-reaction proceeds either through a route that involves the formation of a Cys-dC covalent adduct19 or by a mechanism that is independent of thiolate attack at the C6 position of the flipped base.32 Whichever mechanism applies to the deamination step, it is clear that base flipping is likely to contribute to enzyme-induced deamination. C5 MTase-mediated deamination of cytosine is similar in principle to the highly efficient reaction catalyzed by cytidine deaminases.27 Indeed, it is this enzyme that forms the primary target for zebularine as determined by a series of biochemical and structural studies.27,28 Moreover, it has been suggested that a combined administration of zebularine and AzadC could be more potent than the MTase inhibitor alone.33 This would result primarily from a steady-state increase in the intracellular concentration of the methylation inhibitor, as cytidine deaminase is effectively blocked by zebularine.33 Therefore, it seems plausible to propose that the potency of zebularine as an inhibitor of DNA methylation arises as an inhibitor of methyl transfer itself, and as a suicide inhibitor in the DNA cytosine deamination pathway catalyzed by C5 MTases.
In support of this hypothesis is the observation that zebularine-modified DNA cannot be methylated by the C5 MTases M.Msp I and M.Hha I even after prolonged exposure to AdoMet (P.J.H. & D.P.H., unpublished results), in contrast to AzaC-substituted DNA.20 The latter could arise from the inappropriate juxtaposition of the C5 atom in zebularine-substituted DNA or, more likely (since the two carbon atoms appear to be superimposable structurally), by virtue of altered chemistry at the C6 position in a manner similar to that observed for cytidine deaminases.27,28 This is consistent with the molecular orbital calculations that show that C5 of zebularine is not active (Figure 3); therefore, as shown empirically, reaction with AdoMet does not take place (or is very slow).
The complex between M.Hha I and zebularine-substituted DNA, reveals that the outcome of sequence-specific recognition is the formation of a covalent complex similar in structure to that observed with a related DNA duplex containing FdC. However, the combination of two less hydrogen bonds in the recognition site together with the enhanced reactivity of the C6 position make zebularine a potent DNA MTase inhibitor.
Materials and Methods
Oligodeoxynucleotide synthesis and purification
Oligondeoxynucleotides were prepared as described23,34 with minor modifications. Oligodeoxy-nucleotides were synthesized on a 1 µmol scale ( × 3) using NH3 labile (FOD) deoxynucleoside phosphoramidites. The phosphoramidite of 5′-dimethoxytrityl-2-pyrimidinone-1-β-d-2′-deoxyribofuranoside was dissolved in anhydrous acetonitrile at the usual concentration of 0.1 M. All syntheses were performed trityl-on. After synthesis, deblocking was performed at room temperature for four to five hours in 35% (v/v) aqueous NH3.
Oligodeoxynucleotides were initially purified trityl-on using reverse-phase HPLC as described.23,34 After detriritylation of the product oligodeoxynucleotide with aqueous acetic acid, a further round of reverse-phase (RP) HPLC was undertaken. Following purification, the products were further analyzed by ion-pair (IP) RP HPLC using a DNAsep® column (Transgenomic, San Jose). The major oligodeoxynucleotide fragments were collected and subjected to further chemical and mass spectrometry analysis.
After resuspension in water, 1 µl of the oligodeoxy-nucleotide (100 µM) was mixed with 10 µl of matrix (25 mg of 3-hydoxypicolinic acid dissolved in 300 µl of acetonitrile, 700 µl of water) and 2 µl spotted onto the target for matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) analysis using a Micromass TOF2E mass spectrometer. Difficulties are often encountered during the synthesis and purification of pyrimidinone-containing oligodeoxynucleotides.34 Primarily, these arise owing to the reactivity of d4HC. Following detritylation, many impurities were visible during HPLC analysis, and further mass analysis was essential in order to isolate and purify the pyrimidinone-containing oligodeoxy-nucleotide. Two major species were isolated and subjected to MALDI-TOF MS, the results of this indicated the presence of the oligodeoxynucleotide TGTCAGd4HCGCATGG (expected m/z, 3975.65 calculated m/z 3973.9). However the major product gave a calculated m/z 3990.7. The mass analysis is consistent with the major product resulting from the addition of the d4HC with NH3 (expected m/z 3992.65). The NH3 addition across the 5,6 double bond of the pyrimidinone, with the formation of a saturated double bond and the NH2 adduct, is consistent with the mass analysis. Further confirmation was shown with the incubation of the pyrimidinone-oligodeoxynucleotide with NH3, which resulted in a shift in retention time to the predicted NH2 adduct.
Crystallography
The structure of M.Hha I in a ternary complex with AdoHcy and a short DNA duplex containing zebularine, referred to as the Z13 structure, was determined by X-ray crystallography. M.Hha I used for crystallization was purified as described and dialyzed exhaustively to remove endogenously bound AdoMet. Two 13-mer oligonucleotides were annealed to form a 12 base-pair asymmetric duplex with 5′ single thymine overhangs:
5′-TCCATGCGCTGAC—3′
3′--GGTACGZGACTGT-5′
where Z = zebularine. Crystals of the Z13 complex were grown by mixing the M.Hha I–AdoHcy–DNA complex, with an equal volume of 20% (w/v) polyethylene glycol 8000, 100 mM calcium acetate, 50 mM sodium cacodylate (pH 6.5), and equilibrating the mixture against 1 ml of the latter solution at 16 °C. The Z13 crystals formed in the same space group, R32, as in the previously reported ternary structures with cell dimensions a = b = 96.19 Å, and c = 315.75 Å. One Z13 complex was present per asymmetric unit. X-ray diffraction intensities were measured from a single frozen crystal, at 100 K, at a wavelength of 0.979 Å at beamline X8C of the National Synchrotron Light Source, Brookhaven National Laboratory.
The structure was solved by the difference Fourier method using the previously solved 9MHT (PDB) ternary structure (M.Hha I with AdoHcy and a DNA identical in sequence with that used here but containing an abasic site in place of zebularine) as the initial model. The model was refined against diffraction data measured from 39.7 to 2.50 Å (98.1% complete and 19,591 reflections) using X-PLOR.35 After several rounds of least-squares refinement, manual model building, and placement of well-ordered solvent molecules (interpreted as water) by examination of difference electron density, the crystallographic R-factor ( = ΣlFo − Fcl/ ΣFo, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively) and R-free (10% of total reflection) were reduced to 17.7% and 24.3%, respectively. During the refinement, the sulfur atom of Cys81 was omitted and its position was determined by difference electron density. The distance between the Cys81 sulfur atom and the zebularine C6 atom revealed a covalent bond distance that was included in the last round of refinement. The final model includes 2591 protein atoms (with mean B values of 16.5 Å 2), 517 DNA atoms, 26 AdoHcy atoms and 246 water molecules, with r.m.s. deviations of 0.01 Å and 1.5° from ideality for bond lengths and angles, respectively.
Acknowledgments
We thank Susan Sunay and Aiping Dong for technical assistance during protein purification and crystallization. The study was supported, in part, by the US National Institutes of Health GM49245 to X.C. and through BBSRC and Wellcome studentships to P.J.H. and M.J.D., respectively: the Krebs Institute is a BBSRC designated centre for Biomolecular research. Work in the laboratory of B.A.C. was supported by the UK BBSRC, the EU and the Wellcome Trust.
Abbreviations
- C5 MTase
cytosine-[C5]-specific DNA methyltransferase
- AzadC
5-azadeoxycytidine
- FdC
5-fluorodeoxycytidine
- M.Hha I
C5 MTase from Haemophilus haemolyticus
- AdoMet
S-adenosyl-l-methionine
- AdoHcy
S-adenosyl-l-homo-cysteine
Footnotes
Protein Data Bank accession code
The coordinates have been deposited with the RCSB Protein Data Bank with accession code 1M0E.
Contributor Information
X. Cheng, Email: xcheng@emory.edu.
D.P. Hornby, Email: d.hornby@sheffield.ac.uk.
References
- 1.Bestor TH. Gene silencing. Methylation meets acetylation. Nature. 1998;393:311–312. doi: 10.1038/30613. [DOI] [PubMed] [Google Scholar]
- 2.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 3.Bestor TH. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000;16:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 4.Redaschi N, Bickle TA. DNA restriction and modification systems. In: Neidhart FC, Curtiss R III, Ingraham JL, Brooks Low KB, Magasanik B, Reznikoff WS, Riley R, Schaecter M, Umbarger HE, editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd edit. ASM Press: Washington, DC; 1996. pp. 773–781. [Google Scholar]
- 5.Bestor TH, Verdine GL. DNA Methyltransferases. Curr. Opin. Cell. Biol. 1994;6:380–389. doi: 10.1016/0955-0674(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 6.Rountree MR, Bachman KE, Herman JG, Baylin SB. DNA methylation, chromatin inheritance, and cancer. Oncogene. 2001;20:3156–3165. doi: 10.1038/sj.onc.1204339. [DOI] [PubMed] [Google Scholar]
- 7.Cheng X. Structure and function of DNA methyltransferases. Annu. Rev. Biophys. Biomol. Struct. 1995;24:293–318. doi: 10.1146/annurev.bb.24.060195.001453. [DOI] [PubMed] [Google Scholar]
- 8.Dong A, Yoder JA, Zhang X, Zhou L, Bestor TH, Cheng X. Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA. Nucl. Acids Res. 2001;29:439–448. doi: 10.1093/nar/29.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng X, Roberts RJ. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucl. Acids Res. 2001;29:3784–3795. doi: 10.1093/nar/29.18.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 11.Santi DV, Garrett CE, Barr PJ. On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell. 1983;33:9–10. doi: 10.1016/0092-8674(83)90327-6. [DOI] [PubMed] [Google Scholar]
- 12.Sheikhnejad G, Brank A, Christman JK, Goddard A, Alvarez E, Ford H, Jr, et al. Mechanism of inhibition of DNA (cytosine C5)-methyltransferases by oligodeoxyribonucleotides containing 5,6-dihydro-5-AzaCytosine. J. Mol. Biol. 1999;285:2021–2034. doi: 10.1006/jmbi.1998.2426. [DOI] [PubMed] [Google Scholar]
- 13.Kumar S, Horton JR, Jones GD, Walker RT, Roberts RJ, Cheng X. DNA containing 4′-thio-2′-deoxycytidine inhibits methylation by HhaI methyltransferase. Nucl. Acids Res. 1997;25:2773–2783. doi: 10.1093/nar/25.14.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen L, MacMillan AM, Chang W, Ezaz-Nikpay K, Lane WS, Verdine GL. Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry. 1991;30:11018–11025. doi: 10.1021/bi00110a002. [DOI] [PubMed] [Google Scholar]
- 15.Klimasauskas S, Kumar S, Roberts RJ, Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 16.Wu JC, Santi DV. Kinetic and catalytic mechanism of HhaI methyltransferase. J. Biol. Chem. 1987;262:4778–4786. [PubMed] [Google Scholar]
- 17.Baker DJ, Kan JLC, Smith SS. Recognition of structural perturbations in DNA by human DNA(cytosine-5)methyltransferase. Gene. 1988;74:207–210. doi: 10.1016/0378-1119(88)90288-0. [DOI] [PubMed] [Google Scholar]
- 18.Smith SS, Kan JLC, Baker DJ, Kaplan BE, Dembek P. Recognition of unusual DNA structures by human DNA(cytosine-5)methyltransferase. J. Mol. Biol. 1991;217:39–51. doi: 10.1016/0022-2836(91)90609-a. [DOI] [PubMed] [Google Scholar]
- 19.Gabbara S, Sheluho D, Bhagwat AS. Cytosine methyltransferase from Escherichia coli in which active site cysteine is replaced with serine is partially active. Biochemistry. 1995;34:8914–8923. doi: 10.1021/bi00027a044. [DOI] [PubMed] [Google Scholar]
- 20.Gabbara S, Bhagwat AS. The mechanism of inhibition of DNA (cytosine-5-)-methyltransferases by 5-azacytosine is likely to involve methyltransfer to the inhibitor. Biochem. J. 1995;307:87–92. doi: 10.1042/bj3070087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santi DV, Norment A, Garrett CE. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-AzaCytosine. Proc. Natl Acad. Sci. USA. 1984;81:6993–6997. doi: 10.1073/pnas.81.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson-Grusby L, Laird PW, Magge SN, Moeller BJ, Jaenisch R. Mutagenicity of 5-aza-2′-deoxycytidine is mediated by the mammalian DNA methyltransferase. Proc. Natl Acad. Sci. USA. 1997;94:4681–4685. doi: 10.1073/pnas.94.9.4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor C, Ford K, Connolly BA, Hornby DP. Determination of the order of substrate addition to MspI DNA methyltransferase using a novel mechanism-based inhibitor. Biochem. J. 1993;291:493–504. doi: 10.1042/bj2910493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurd PJ, Whitmarsh AJ, Baldwin GS, Kelly SM, Waltho JP, Price NC, et al. Mechanism-based inhibition of C5-cytosine DNA methyltransferases by 2-H pyrimidinone. J. Mol. Biol. 1999;286:389–401. doi: 10.1006/jmbi.1998.2491. [DOI] [PubMed] [Google Scholar]
- 25.Baldwin GS, Kelly SM, Price NC, Wilson GW, Connolly BA, Artymiuk PJ, Hornby DP. Ligand-induced conformational states of the cytosine-specific DNA methyltransferase M.HgaI-2. J. Mol. Biol. 1994;235:545–553. doi: 10.1006/jmbi.1994.1012. [DOI] [PubMed] [Google Scholar]
- 26.Saenger W. Principles of Nucleic Acid Structure. New York: Springer; 1984. [Google Scholar]
- 27.Frick L, Yang C, Marquez VE, Wolfenden R. Binding of pyrimidin-2-one ribonucleoside by cytidine deaminase as the transition state analogue 3,4 dihydrouridine and the contribution of the 4-hydroxyl group to its binding affinity. Biochemistry. 1989;28:9423–9430. doi: 10.1021/bi00450a027. [DOI] [PubMed] [Google Scholar]
- 28.Betts L, Xiang S, Short SA, Wolfenden R, Carter CW., Jr Cytidine deaminase. The 2.3 Å crystal structure of an enzyme: transition state analogue complex. J. Mol. Biol. 1994;235:635–656. doi: 10.1006/jmbi.1994.1018. [DOI] [PubMed] [Google Scholar]
- 29.Smith SS, Hardy TA, Baker DJ. Human DNA (cytosine-5) methyltransferase selectively methylates duplex DNA containing mispairs. Nucl. Acids Res. 1987;15:6899–6916. doi: 10.1093/nar/15.17.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klimasauskas S, Roberts RJ. M.HhaI binds tightly to substrates containing mismatches at the target base. Nucl. Acids Res. 1995;23:1388–1395. doi: 10.1093/nar/23.8.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang AS, Shen JC, Zingg JM, Mi S, Jones PA. HhaI and HpaII DNA methyltransferases bind DNA mismatches, methylate uracil and block DNA repair. Nucl. Acids Res. 1995;23:1380–1387. doi: 10.1093/nar/23.8.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharath A, Weinhold E, Bhagwat AS. Reviving a dead enzyme. Cytosine deamination promoted by an inactive DNA methyltransferase and an S-adenosylmethionine analogue. Biochemistry. 2000;39:14611–14616. doi: 10.1021/bi001610e. [DOI] [PubMed] [Google Scholar]
- 33.Jeong LS, Buenger G, McCormack J, Cooney DA, Hao Z, Marquez VE. Carbocyclic analogues of the potent cytidine deaminase inhibitor 1-(β-d-ribofuranosyl)-1,2-dihydropyrimidin-2-one (zebularine) J. Med. Chem. 1998;41:2572–2578. doi: 10.1021/jm980111x. [DOI] [PubMed] [Google Scholar]
- 34.Connolly BA, Newman PC. Synthesis and properties of oligonucleotides containing 4-thiothymidine, 5-methyl-2-pyrimidinone-1-β-d(2′-deoxyriboside) and 2-thiothymidine. Nucl. Acids Res. 1989;17:4957–4974. doi: 10.1093/nar/17.13.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brünger AT. X-PLOR. A system for X-ray crystallography and NMR, version 3.1. New Haven, CT: Yale University; 1992. [Google Scholar]
- 36.O’Gara M, Klimasauskas S, Roberts RJ, Cheng X. Enzymatic C5-cytosine methylation of DNA: mechanistic implications of new crystal structures for HhaI methyltransferase-DNA-AdoHcy complexes. J. Mol. Biol. 1996;261:634–645. doi: 10.1006/jmbi.1996.0489. [DOI] [PubMed] [Google Scholar]
- 37.Smith SS, Niu L, Baker DJ, Wendel JA, Kane SE, Joy DS. Nucleoprotein-based nanoscale assembly. Proc. Natl Acad. Sci. USA. 1997;94:2162–2167. doi: 10.1073/pnas.94.6.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hurd PJ. PhD thesis. UK: University of Sheffield; 1996. A study of the catalytic properties of M.Msp I DNA methyltransferase. [Google Scholar]