Abstract
Inevitably, viruses depend on host factors for their multiplication. Here, we show that hepatitis C virus (HCV) RNA translation and replication depends on Rck/p54, LSm1, and PatL1, which regulate the fate of cellular mRNAs from translation to degradation in the 5′-3′-deadenylation-dependent mRNA decay pathway. The requirement of these proteins for efficient HCV RNA translation was linked to the 5′ and 3′ untranslated regions (UTRs) of the viral genome. Furthermore, LSm1–7 complexes specifically interacted with essential cis-acting HCV RNA elements located in the UTRs. These results bridge HCV life cycle requirements and highly conserved host proteins of cellular mRNA decay. The previously described role of these proteins in the replication of 2 other positive-strand RNA viruses, the plant brome mosaic virus and the bacteriophage Qß, pinpoint a weak spot that may be exploited to generate broad-spectrum antiviral drugs.
Keywords: deadenylation-dependent mRNA decay, HCV, host factors, LSm1–7, Rck/p54
The astonishing diversity in viral life cycles, even inside the same viral group, raises intriguing questions about their origins and evolutionary relationships. Because viruses are obligatory intracellular parasites, they depend on host factors for their multiplication. The requirements for common host factors could provide essential clues about their evolutionary links and would also have important practical implications since these host factors might serve as targets for broad-spectrum antiviral strategies.
The group of positive strand RNA [(+)RNA] viruses encompass over one-third of all virus genera. It includes numerous and serious pathogens, a notable example being the hepatitis C virus (HCV), which is a major cause of chronic liver disease and has chronically infected ≈170 million individuals worldwide. At early times of infection, (+)RNA viral genomes perform 2 essential functions. They act as messengers for translation and as templates for viral replication. Because these 2 functions are mutually exclusive, a key step in the replication of all (+)RNA viruses is the regulated exit of their genomic RNA from the cellular translation machinery to the replication complex, which is always associated to intracellular membranes (1).
The replication of the plant Brome mosaic virus in the yeast Saccharomyces cerevisiae has proven to be a fruitful model system for studying common steps of (+)RNA virus biology in a relatively simple genetic background (2). By using this model system we have shown that the host factors LSm1, LSm6, and LSm7, which are subunits of the heptameric ring LSm1–7, as well as Pat1 and Dhh1 play an essential role in translation and in the translation-replication transit of the BMV genome (3–5). In noninfected cells, these proteins act as activators of decapping in the 5′-3′-deadenylation-dependent mRNA decay pathway (6). Although their precise way of functioning at the molecular level is not totally understood, they have been suggested to determine mRNA fate by facilitating the exit of cellular mRNAs from active translation to a translationally inactive state that allows the assembly of the decapping complex (6–8). These nontranslating mRNAs together with proteins involved in translation repression, mRNA decay, and RNA-mediated silencing accumulate in dynamic cytoplasmic foci referred to as P-bodies (review in refs. 9 and 10). Experiments in yeast indicate that mRNAs targeted to P-bodies can be either decapped and degraded or stored for return to translation (10).
Given the conservation of the 5′-3′-deadenylation-dependent mRNA decay pathway from yeast to humans and the common need of all (+)RNA viruses to regulate the transition of their genomes from active translation to a translationally inactive state to allow replication, an exciting possibility is that the function of Dhh1, LSm1–7, and Pat1 is used not only by BMV to replicate in yeast but also by human viruses to replicate in human cells. By measuring HCV replicon amplification and infectious virus production, we show here that indeed the respective human homologues namely Rck/p54, LSm1–7, and PatL1 (9, 11) are necessary for HCV replication. We also found that they are required for efficient translation of the viral genome and that these requirements are functionally linked to the 5′ and 3′ untranslated regions (UTRs). Furthermore, reconstituted LSm1–7 rings specifically bind to defined sequences in the 5′ and 3′UTRs that are known to play key roles in the regulation of HCV translation and replication. Together this not only demonstrates a conserved utilization of an ancient host cell machinery by the major human pathogen HCV but also opens up perspectives for the development of broadly reactive antiviral drugs.
Results
The Host Factors Rck/p54, LSm1, and PatL1 Promote HCV Replicon Amplification and Infectious HCV Production.
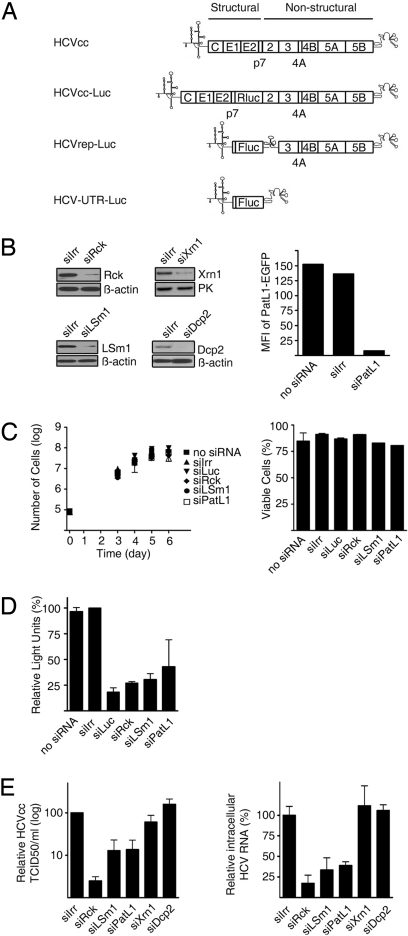
To study whether Rck/p54, LSm1–7, and PatL1 affect HCV replication, we used a gene silencing strategy and used (i) HCV RNA replicons that allow efficient replication but do not result in virus production and (ii) infectious viruses that reproduce the entire virus life cycle (Fig. 1A). The HCV replicons HCVrep-Luc and HCVrep-Neo belong to the 1b genotype and are composed of the HCV 5′UTR, a luciferase reporter or neomycin phosphotransferase selection marker, the internal ribosome entry site (IRES) of the encephalomyocarditis virus (EMCV) followed by the HCV genes for the nonstructural proteins and the HCV 3′UTR. The infectious virus HCVcc has a 2a genotype and was used as such or with a luciferase reporter.
Fig. 1.
Depletion of Rck/p54, LSm1 or PatL1 in hepatoma cell lines impairs HCV replication. (A) Schematic representations of the genomes of HCVcc, HCV Replicons and derivatives used in this study. (B) Huh7-Lunet cells were transfected with siRNA targeting Rck/p54, LSm1, PatL1, Xrn1, Dcp2, or a nontargeting siRNA (siIrr). Immunoblot analyses of Rck/p54, LSm1, Xrn1, Dcp2, β-actin, or pyruvate kinase levels are shown. Because no specific antibody is available for PatL1, to test PatL1 silencing, PatL1-EGFP expression plasmid and siRNAs were cotransfected and fluorescence was analyzed 1 day later by flow cytometry. Values are expressed in mean fluorescence intensity (MFI) (bar graph). Similar silencing results were obtained for Huh7.5 cells. (C) Cell growth of siRNA-transfected cells was followed for 6 days by counting the total number of cells (mean ± SEM; n = 3) (Left). The percentage of viable silenced cells at the day of maximum silencing was measured by propidium iodide staining (mean ± SEM; n = 2) (Right). (D) Huh7-Lunet cells were coelectroporated with the HCVrep-Luc replicon and the siRNAs. The percentage of relative luciferase light units compared with siIrr-transfected cells is shown at the day of most efficient silencing (mean ± SEM; n = 2). (E) Three days after transfection of silenced Huh7.5 cells with HCVcc RNA, the HCVcc infectivity in the supernatant was titrated by a limited dilution assay (Left). The accumulation of intracellular HCVcc mRNA was analyzed by quantitative RT-PCR (Right). Both values were normalized to the amount of transfected RNA (mean ± SEM; n = 3) and are shown relative to siIrr-transfected cells.
We first set up the silencing conditions for the cellular proteins by using specific siRNAs (Fig. 1B). With respect to the LSm1–7 cytoplasmic ring, we focused on the LSm1 subunit that defines the role of the ring in decapping. The other subunits when complexed with LSm8 are also part of a nuclear complex involved in splicing. In all cases, siRNA-mediated silencing resulted in a specific 80–85% reduction of the corresponding proteins when using the nontargeting siRNA siIrr as a reference (Fig. 1B). Importantly, silencing of Rck/p54, LSm1, and PatL1 did not affect cell growth or viability measured by sequential counting of the number of cells, propidium iodide staining or in an ATP assay (Fig. 1 C and D and Fig. S1). In addition, type I-interferons were not induced as judged by lack of MxA protein expression (Fig. S2).
To test whether silencing of any of these factors affects HCV replication, Huh7-Lunet cells were coelectroporated with HCVrep-Luc and a specific siRNA or with siIrr as a negative control. An additional mock-transfected control and a siRNA directed against the replicon-encoded luciferase gene (siLuc) were included. Luciferase values were then measured at times of maximum silencing (Fig. 1D). Down-regulation of Rck/p54, LSm1, and PatL1 resulted in a marked reduction of the luciferase activity by ≈80%, 70%, and 60%, respectively. Similar results were obtained with the HCVrep-Neo replicon (Fig. S3). This reduction was comparable to the 84% reduction observed by directly targeting the HCV replicon with siLuc. With transfection efficiencies of ≈90% and protein knockdowns of 80 to 85%, the values obtained are close to the maximal possible reduction. This strongly suggests that Rck/p54, LSm1, and PatL1 play an important role in HCV replicon amplification. Because the replicon system does not include RNA encapsidation, the observed effects can be explained by defects in HCV RNA translation and/or replication.
Next we tested whether this role is also detectable with an infectious HCV. At the time of maximal silencing, Huh7.5 cells were transfected with HCVcc RNA. Three days later, cellular supernatants were harvested for titration of infectious particles whereas intracellular HCV RNAs were quantified by quantitative RT-PCR (Fig. 1E). In all cases, HCV production from siRck-, siLsm1- and siPatL1-transfected cells was significantly reduced, the infectious titers being 50-, 10-, and 10-fold lower than in the siIrr control, respectively. Moreover, intracellular HCV RNA levels were also reduced. An inhibition in both, particle production and viral RNA accumulation, is expected for defects in an early step of the viral life cycle such as translation and replication. However, an additional effect on RNA encapsidation, particle morphogenesis or release cannot be excluded. Because all developed systems that allow to study HCV particle production depend on active translation and replication, this possibility was not explored further.
Depletion of the Proteins Dcp2 and Xrn1 Does Not Affect Infectious HCV Production.
In the 5′-3′-deadenylation-dependent mRNA decay pathway, mRNA exit from translation and shortening of the poly(A) tail by deadenylases is followed by decapping via the Dcp1/Dcp2 decapping enzyme and 5′ to 3′ degradation via the exonuclease Xrn1 (6). To test the effect of some late components from this pathway on HCV replication, we selected Xrn1 and Dcp2. Silencing conditions were established, cell toxicity excluded (Fig. 1B and Fig. S1), and the effect on replication of HCVcc was assayed as before (Fig. 1E). No significant differences in the virus titer of the supernatants or in the level of the intracellular HCV RNA between siXrn1-, siDcp2-, or siIrr-transfected cells were observed. These results argue that it is not the decapping and degradation process itself which is important for the HCV life cycle but the proteins acting upstream of it.
Rck/p54, LSm1–7, and PatL1 Affect Translation of the HCV RNA Genome via the 5′ and 3′UTRs.
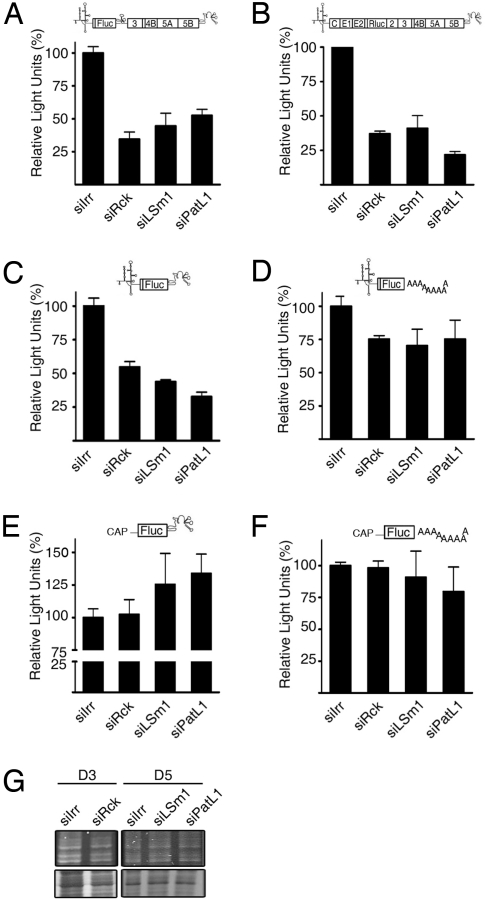
Rck/p54, LSm1 and PatL1 may affect HCV propagation by acting on HCV RNA translation, replication, or both. Most of the HCV proteins required for replication function in cis. As a consequence, one can measure either translation plus replication effects by using a replication-competent HCV derivative as above or only translation effects by using a nonreplicative HCV derivative. To investigate a putative role in translation, we used (i) a HCVrep-Luc replicon and (ii) a derivative of the HCVcc that contains the luciferase ORF fused to the NS2 gene (Fig. 1A). In both cases the NS5B polymerase carries a mutation that inhibits replication and, consequently, any luciferase activity of these derivatives can be attributed solely to translation of the transfected HCV RNA. Rck/p54-, LSm1-, or PatL1-silenced cells were transfected with the corresponding HCV RNAs and luciferase activities were measured 4 h later. When normalized to the abundance of intracellular HCV RNAs, activity reductions by ≈65%, 55%, and 48% were observed with the HCV replicon (Fig. 2A) whereas the reductions with HCVcc were 63%, 59%, and 79% (Fig. 2B), respectively. It is important to note, that the stability of HCV RNA was not significantly affected under these conditions (Fig. S4). By metabolic labeling we could exclude a generalized effect on cellular mRNA translation (Fig. 2G). In addition, translation of a luciferase mRNA flanked by a 5′cap and a 3′poly-(A) tail and with 5′and 3′UTR of nonviral origin was not affected by Rck/p54-, LSm1-, or PatL1- silencing (Fig. 2F). Since the major cis-signals controlling HCV RNA translation and replication are located in the 5′ and 3′UTRs of the HCV genome, we carried out a similar translation analysis with a genotype 1b HCV RNA derivative that contains only the HCV 5′UTR followed by a luciferase ORF and the HCV 3′UTR (Fig. 1A). The luciferase values were comparable to the ones obtained with the complete replicon (Fig. 2C). To test whether the observed translation inhibition depended on the HCV 5′UTR, HCV 3′UTR or both, we generated luciferase-reporter derivatives in which either the HCV 3′UTR was exhanged by a 3′poly(A)-tail or the HCV 5′UTR by a capped, unrelated 5′UTR. Silencing of Rck/p54, LSm1, and PatL1 had no significant effect on the translation of any of these RNAs (Fig. 2 D and E). In addition, EMCV-IRES mediated translation was also not significantly inhibited by silencing (Fig. S5). This suggests that HCV RNA translation specifically depends on Rck/p54, LSm1, and PatL1, and that this dependence is linked to the presence of both UTRs.
Fig. 2.
Rck/p54, LSm1 and PatL1 silencing influences HCV RNA translation. Huh7-Lunet and Huh7.5 cells were transfected with siRNAs targeting Rck/p54, LSm1, PatL1, or a nontargeting siRNA, siIrr. The silenced cells were further transfected with (A) a nonreplicating bicistronic Luciferase replicon (HCVrep-Luc-GND), (B) a nonreplicating Luciferase-HCVcc (HCVcc-Luc-GNN), (C) a derivative (HCV-UTRs-Luc) containing the HCV 5′ and 3′UTRs from genotype 1b flanking the firefly luciferase ORF, (D) a derivative from HCV-UTRs-Luc in which the HCV 3′ UTR was exchanged by a poly(A) tail, (E) a derivative from HCV-UTRs-Luc in which the HCV 5′ UTR was exchanged by capped, nonviral 5′UTR, and (F) a derivative [CAP-Luc-Poly(A)] containing the 5‘capped, nonviral 5′UTR followed by the firefly luciferase ORF and a poly(A) tail. The luciferase activity was measured 4 h after transfection and normalized to the respective intracellular RNA levels measured by quantitative RT-PCR (mean ± SEM; n = 3). (G) To examine the influence of Rck/p54-, LSm1- and PatL1-silencing on the synthesis of cellular proteins, silenced cells were labeled with [35S]methionine for 30 min, separated on a denaturating polyacrylamide gel and visualized by autoradiography (Lower). Gels were coomassie-stained to visualize protein-loading (Upper).
Reconstituted LSm1–7 Rings Bind Directly and Specifically to Translation/Replication Regulatory Signals in the HCV 5′ and 3′UTRs.
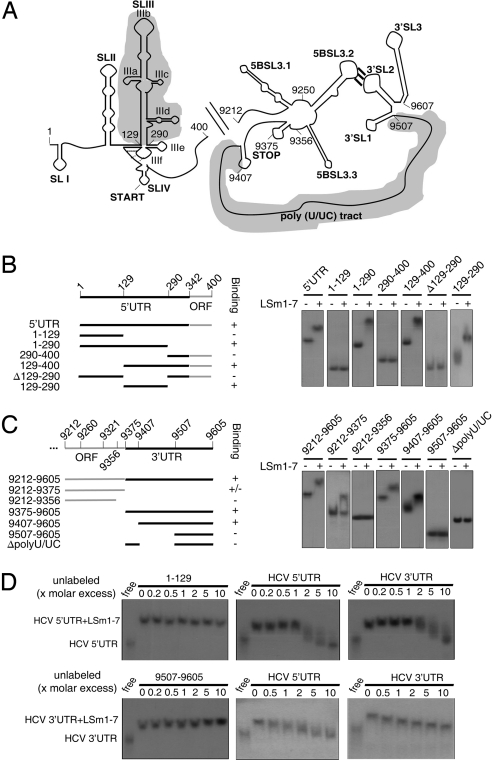
At least 2 possible models can be considered by which Rck/p54, LSm1, and PatL1 can act on the HCV life cycle. First, silencing of these proteins may alter the host physiology thereby exerting a nonspecific effect on HCV replication. The toxicity tests performed in Rck-, LSm1-, and PatL1-silenced cells, however, render this possibility unlikely. Alternatively, these proteins may have a direct and specific effect on the virus and hence directly interact with viral RNA or proteins. In yeast cells, the corresponding proteins Dhh1, Pat1, and the LSm1–7 ring have been shown to interact in vivo (6), and there is evidence of a direct interaction of the LSm1–7 ring with deadenylated cellular mRNAs (8, 12). Considering a direct interaction model, it seemed possible that the LSm1–7 ring could interact with the 5′ and 3′UTRs of HCV since they are essential regions in the regulation of viral translation and replication (13), and our translation results suggested a functional link to these sequences. To examine this possibility, we reconstituted functional human LSm1–7 rings according to a recently reported strategy (14) (Fig. S6), and performed electromobility shift assays with HCV RNA fragments (Fig. 3). Incubation of the LSm1–7 rings with the corresponding 32P-labeled transcripts demonstrated strong binding to both UTR regions reflected by a complete band shift (Fig. 3 B and C). This binding was specific because addition of excess unlabeled 5′ or 3′UTR sequences resulted in binding competition (Fig. 3D), whereas addition of excess unlabeled nonbinding HCV RNA sequences did not.
Fig. 3.
Reconstituted LSm1–7 rings bind to specific HCV 5′ and 3′UTR regions. (A) Schematic representation of the secondary structures of the HCV 5′ and 3′ ends. Upstream of the 3′UTR, the NS5B coding sequence containing an RNA cruciform structure is shown. This structure includes the 5BSL3.2 loop. Its long-range interaction with the 3′SL3 loop in the 3′Xtail of the 3′UTR is essential for replication. Shadowed regions highlight the binding sites of the LSm1–7 rings. (B and C, Left) The constructs used in the electromobility shift assays are shown. The numbers refer to the nucleotide positions in the genome of the HCV Con1 strain. (B and C, Right) Radiolabeled, gel-purified RNA transcripts were incubated with the reconstituted LSm1–7 rings. After complex formation, products were separated on a nondenaturating polyacrylamide gel and visualized by autoradiography. (D) Labeled HCV 5′ and 3′UTR RNAs (HCV sequences 1–400 and 9,375–9,605, respectively) were incubated with reconstituted LSm1–7 rings in the presence of increasing amounts of unlabeled HCV 5′ and 3′UTR transcripts. As noncompeting controls, unlabeled RNAs negative for LSm1–7 ring binding (HCV sequences 1–129 and 9,507–9,605) were used. After complex formation, products were treated as in B and C.
To identify the viral RNA motifs involved in the interactions with the LSm1–7 rings, we systematically deleted domains of defined RNA structure and function from the HCV 5′ and 3′UTRs respectively. The HCV 5′UTR contains the 4 stem loop structures SLI to SLIV (Fig. 3A). SLI is required in replication but is dispensable for translation. SLII, SLIII and SLIV form the internal ribosomal entry site. From these, SLIII is proposed to interact with the 40S ribosomal subunits thus playing a key role in translation initiation. The SLII and SLIII stem loops function in replication as well (13, 15). Electromobility shift analysis showed that the SLIII was both necessary and sufficient for binding of the LSm1–7 ring to the 5′UTR region (Fig. 3B) as evidenced by binding to this RNA motif (positions 129–290) and by loss of binding upon removal of it.
The HCV 3′UTR consists of a variable region, a poly (U/UC) tract and a highly conserved terminal region termed 3′X tail that consists of 3 stem loop structures (13). Importantly, the 3′UTR is not only required for replication but also for efficient translation (16). The electromobility shift analysis revealed a robust area of binding corresponding to the poly(U/UC) tract. Binding to the LSm1–7 ring was lost when this area was deleted (transcripts Δ polyU/UC) and gained when it was added (transcripts 9,507–9,605 and 9,407–9,605). The length and sequence composition of this region has recently been shown to have an important function in HCV RNA replication and the binding of host factors that could regulate this function has been suggested (17). An additional weak binding was observed to the NS5B coding sequence that includes a RNA cruciform structure (transcript 9,212–9,375). This binding was lost when the transcript was reduced to the positions 9,212–9,356. Because this remaining HCV sequence still contained the cruciform structure, either this was not the target of binding or the complete sequence might be required for proper folding to allow binding. Thus, in summary, the LSm1–7 ring binds robustly and directly to 2 important motifs in the 5′ and 3′UTR regions that are involved in the regulation of translation and replication of HCV.
Discussion
Our data demonstrate that HCV translation and replication decreases when the levels of Rck/p54, LSm1–7, and PatL1 are down-regulated. This was observed for the HCV genotype 1b and 2a for which replicons and viral derivatives were readily available. Most strikingly, the respective homologs play a similar role in BMV translation and replication in yeast, and Hfq, a homolog of LSm1 in bacteria, is required for the replication of the (+)RNA bacteriophage Qß (18). Furthermore, efficient retrotransposition of the yeast retrovirus-like elements Ty-1 and Ty-3 also depends on Dhh1, Lsm1, and Pat1 (19). One common theme for (+)RNA viruses and retroviruses is that both need to regulate the transition of the genomic RNA from translation to encapsidation (20). Thus, the dependence of at least some members of both virus groups on particularly those host proteins that are involved in the transit of cellular mRNAs from translation to another fate suggests that they have hijacked this function for their own benefit. Interestingly, Rck/p54, LSm1–7, and PatL1 are core components of P-bodies. These foci are sites where nontranslating mRNAs accumulate for different fates such as degradation, storage or returning to translation. Whether P-body formation itself is required for the HCV life cycle is an interesting issue (19) yet to be resolved in subsequent studies.
The function of LSm1–7 rings as activators of decapping of cellular mRNAs seems to involve their binding to short oligo (A) tracts at the 3′end of deadenylated cellular mRNAs. This binding then inhibits trimming of the 3′end while simultaneously promotes decapping and subsequent 5′ to 3′ degradation (8, 12, 21). However, the role of LSm1–7 rings on virus life cycles may be different because viral RNAs have different requirements for their eventual fates. In case of HCV RNA, the LSm1–7 rings are required for efficient translation. This function might be mediated by the direct interaction of the LSm1–7 rings with sequences in both, the 5′ and 3′UTR regions (Fig. 3). These interactions could facilitate rearrangements in the viral RNP structure and composition, recruiting proteins such as Rck/p54 and PatL1 from the cellular mRNA repression/decay machinery and, instead of promoting decay, might promote HCV RNA translation and subsequent transfer to replication. This view is consistent with a recent proposal made for the regulation of mRNAs generated by poxviridae. Viruses of this family generate viral mRNAs with an additional oligo(A) tract located at their 5′ends. Bergman and colleagues have shown that binding of LSm1–7 rings to such a tract at the 5′end of reporter RNAs does not result in mRNA decay but rather in RNA stabilization through inhibition of decapping and degradation (22). This effect was proposed to be mediated by the simultaneous binding of Lsm1–7 rings to the 5′ and the 3′ends.
Silencing of Rck/p54, LSm1, or PatL1 affects HCV RNA translation and intracellular HCV RNA accumulation. This may be explained by an effect solely at the translation level or by an independent effect on both translation and replication as observed in the BMV model. Such an apparently antagonistic function, to promote both translation and exit from translation, is not without precedent as cellular proteins acting in 2 antagonistic processes such as translation initiation and translation repression have been described (23). An advantage of using a single complex for opposing outcomes seems to be the possibility of responding rapidly to different cellular requirements. A similar advantage might apply for the regulation of the viral life cycle.
In conclusion, the functional conservation of cellular and viral regulatory circuits across kingdoms and virus groups mark a weak spot that can be exploited for the generation of broad-spectrum antiviral drugs. Our observation that the individual, transient knock-down of Rck/p54, LSm1–7, and PatL1 proteins in human cells is not toxic and the fact that the respective yeast knockout strains are viable, stress the feasibility of such an approach for the future.
Materials and Methods
Plasmids, siRNA, and Antibodies.
We present the plasmids, siRNA and the antibodies used in this study in SI Text and in Table S1 and Table S2.
In Vitro Transcription and Capping Reaction.
In vitro transcripts of HCVcc, HCV replicons, and Luciferase reporter derivatives were performed by using RNAMaxx High Yield Transcription Kit (Stratagene) or MEGAScript Kit (Ambion) with T3 or T7 polymerase according to the manufacturer's instructions. After the in vitro transcription capped RNA was generated using the ScriptCap m7G Capping System (Epicentre Biotechnologies). Transcripts used in electromobility shift assays were in vitro transcribed using T7 and SP6 polymerases (Fermentas GmbH) and labeled with [α-32P]UTP.
Cell Culture, RNA Transfections, and Knockdown of Host Factors by RNAi.
The Huh7.5 and Huh7-Lunet cells, subclones of the hepatoma cell line Huh7, were described (24, 25). Two different RNA transfection protocols were used, transfection by lipofectamine 2000 (Invitrogen) and electroporation (26). For silencing, 50 nM siRNA was optimal for Lipofectamine 2000 and 1 μM or 4 μM siRNA for electroporation. In all cases siRNA transfection efficiencies were determined 4 h after transfection using fluorescence-labeled siRNA and cytometry. The knockdown of Rck/p54, Xrn1, and Dcp2 required 1 siRNA transfection and efficient silencing was achieved 3 to 4 days after. The knockdown of Lsm1 required 2 to 3 successive transfections and efficient silencing was achieved 6 to 7 days after the initial transfection. A similar procedure was used for the transient knockdown of PatL1. The viability of the silenced cells was assessed by quantification of propidium iodide (PI) (MBL International,), by measurement of intracellular ATP-levels using CellTiterGlo (Promega) or by growth rate, counting cells up to 6 days after transfection. The ATP assay (Fig. S1) was used to analyze cell viability of the lipofectamine-transfected cells whereas growth rate and propidium iodide incorporation was used to analyze viability of the electroporated cells (Fig. 1C).
HCV-Replication Assays.
Huh7-Lunet or Huh7 cells were coelectroporated with 10 μg yeast RNA, 1 μg of the corresponding replicon, and siRNA. For HCVrep-Luc, 1 μM of the corresponding siRNAs was used whereas for HCVrep-Neo, either 1 or 4 μM of siRNAs was used (27). Replication was measured either in colony-formation assays (HCVrep-Neo) or by quantification of intracellular replicon-encoded Luciferase (HCVrep-Luc and HCVrep-Luc-GND) as described (26, 28, 29).
To investigate the effect of the knockdown of the analyzed proteins on HCVcc replication, silenced cells were transfected by Lipofectamine with HCVcc RNA at the time of most efficient silencing. To maintain the protein knockdown of LSm1 and PatL1, an additional transfection with siRNA was required 24 h later. Intracellular HCVcc RNA levels and infectious HCVcc particles in the supernatant of transfected cells were quantified at various time points up to 72 h after transfection. The obtained values were standardized to the amount of transfected RNA quantified 4 h after transfection to equalize transfection efficiencies.
HCV-Translation Assays.
Analysis of HCV translation was performed with different luciferase constructs. After transfection by Lipofectamine of the respective RNAs, luciferase activities were measured 4 h later and normalized to the total amount of protein. Then this value was corrected by the amount of the HCV RNA that was obtained by qRT-PCR using specific Taq-Man primers and probes (Table S3) after normalization to internal 18S RNA.
Titration of Infectious HCVcc Particles and RNA Quantification.
Titration of infectious particles in the supernatant of HCVcc RNA-transfected cells was performed as described in ref. 28. For RNA quantification, 40 ng total RNA were reverse transcribed using random primers and SuperScript III according to manufacturer's recommendations (Invitrogen). The cDNA was amplified with specific primers and probes (listed in Table S3) using an ABI Prism 900HT sequence detection system (Applied Biosystems). The amplifications were standardized to an internal 18S control (ABI Taqman HS99999901 s1*; Applied Biosystems) using a relative quantification analysis from the SDS 2.3 software (Applied Biosystems).
Electromobility Shift Assays.
Expression, purification of individual LSm proteins and reconstitution of complexes were performed as described in ref. 14. Three hundred cpm of gel-purified in vitro transcribed HCV-RNAs were incubated with 10 pmol LSm protein heptameric complexes in a buffer containing 20 mM Hepes-NaOH, pH 7.5, 200 mM NaCl, 2 mM MgCl2, 0.1 U/μL RNasin, and 0.1 μg/μL yeast tRNA at 30 °C for 1 h. Samples were loaded on prerun 5% native polyacrylamide gels, and run at 4 °C for 2 h and 30 mA. Gels were autoradiographed on maximum sensitivity films (KODAK Biomax MS). For assays that included RNA competition, increasing amounts of RNA competitor were added to the reactions (Fig. 3D). The assays for binding to Xenopus U1 and U6 snRNAs were performed as described (14).
Supplementary Material
Acknowledgments.
We thank Apath LLC, R. Bartenschlager (Univ. of Heidelberg, Germany), O. Haller (Univ. of Freiburg, Germany), C. Kambach (Paur Scherrer Institut, Villigen, Switzerland), M. Kiledjian (Rutgers Univ., Piscataway, NJ), J. Lykke-Andersen (Univ. of Colorado, Boulder, CO), I.W. Mattaj (European Molecular Biology Laboratory, Heidelberg), F. Gebauer (Centre de Regulació Genòmica, Barcelona), S. Bradrick (Duke Univ. Medical Center, Durham, NC), and M. Niepmann (Justus-Liebig-Univ., Giessen, Germany) for reagents; and B. Lindenbach for critically reading the manuscript and for helpful discussions. This work was supported by Spanish Ministerio de Educación y Ciencia Grants BFU2004–00654 and BFU2007–66933/BMC, the Deutsche Forschungsgemeinschaft Me 1061/4 as part of the clinical research network KFO 129, SFB581, and FOR-855.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906413106/DCSupplemental.
References
- 1.Gamarnik AV, Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alves-Rodrigues I, Galao RP, Meyerhans A, Diez J. Saccharomyces cerevisiae: A useful model host to study fundamental biology of viral replication. Virus Res. 2006;120:49–56. doi: 10.1016/j.virusres.2005.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diez J, Ishikawa M, Kaido M, Ahlquist P. Identification and characterization of a host protein required for efficient template selection in viral RNA replication. Proc Natl Acad Sci USA. 2000;97:3913–3918. doi: 10.1073/pnas.080072997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mas A, et al. Host deadenylation-dependent mRNA decapping factors are required for a key step in brome mosaic virus RNA replication. J Virol. 2006;80:246–251. doi: 10.1128/JVI.80.1.246-251.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noueiry AO, et al. Yeast Lsm1p–7p/Pat1p deadenylation-dependent mRNA-decapping factors are required for brome mosaic virus genomic RNA translation. Mol Cell Biol. 2003;23:4094–4106. doi: 10.1128/MCB.23.12.4094-4106.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coller J, Parker R. Eukaryotic mRNA decapping. Annu Rev Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 7.Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tharun S, Parker R. Targeting an mRNA for decapping: Displacement of translation factors and association of the Lsm1p–7p complex on deadenylated yeast mRNAs. Mol Cell. 2001;8:1075–1083. doi: 10.1016/s1097-2765(01)00395-1. [DOI] [PubMed] [Google Scholar]
- 9.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 11.Scheller N, et al. Identification of PatL1, a human homolog to yeast P body component Pat1. Biochim Biophys Acta. 2007;1773:1786–1792. doi: 10.1016/j.bbamcr.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 12.Chowdhury A, Mukhopadhyay J, Tharun S. The decapping activator Lsm1p–7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. Rna. 2007;13:998–1016. doi: 10.1261/rna.502507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tellinghuisen TL, et al. Studying hepatitis C virus: Making the best of a bad virus. J Virol. 2007;81:8853–8867. doi: 10.1128/JVI.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaric B, et al. Reconstitution of two recombinant LSm protein complexes reveals aspects of their architecture, assembly, and function. J Biol Chem. 2005;280:16066–16075. doi: 10.1074/jbc.M414481200. [DOI] [PubMed] [Google Scholar]
- 15.Isken O, et al. Nuclear factors are involved in hepatitis C virus RNA replication. RNA. 2007;13:1675–1692. doi: 10.1261/rna.594207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song Y, et al. The hepatitis C virus RNA 3′-untranslated region strongly enhances translation directed by the internal ribosome entry site. J Virol. 2006;80:11579–11588. doi: 10.1128/JVI.00675-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.You S, Rice CM. 3′ RNA elements in hepatitis C virus replication: Kissing partners and long poly(U) J Virol. 2008;82:184–195. doi: 10.1128/JVI.01796-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franze de Fernandez MT, Eoyang L, August JT. Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature. 1968;219:588. doi: 10.1038/219588a0. [DOI] [PubMed] [Google Scholar]
- 19.Beckham CJ, Parker R. P bodies, stress granules, and viral life cycles. Cell Host Microbe. 2008;3:206–212. doi: 10.1016/j.chom.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahlquist P. Parallels among positive-strand RNA viruses, reverse-transcribing viruses, and double-stranded RNA viruses. Nat Rev Microbiol. 2006;4:371–382. doi: 10.1038/nrmicro1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He W, Parker R. The yeast cytoplasmic LsmI/Pat1p complex protects mRNA 3′ termini from partial degradation. Genetics. 2001;158:1445–1455. doi: 10.1093/genetics/158.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bergman N, et al. Lsm proteins bind and stabilize RNAs containing 5′ poly(A) tracts. Nat Struct Mol Biol. 2007;14:824–831. doi: 10.1038/nsmb1287. [DOI] [PubMed] [Google Scholar]
- 23.Abaza I, Gebauer F. Trading translation with RNA-binding proteins. RNA. 2008;14:404–409. doi: 10.1261/rna.848208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quinkert D, Bartenschlager R, Lohmann V. Quantitative analysis of the hepatitis C virus replication complex. J Virol. 2005;79:13594–13605. doi: 10.1128/JVI.79.21.13594-13605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lohmann V, Korner F, Dobierzewska A, Bartenschlager R. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J Virol. 2001;75:1437–1449. doi: 10.1128/JVI.75.3.1437-1449.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson JA, Richardson CD. Hepatitis C virus replicons escape RNA interference induced by a short interfering RNA directed against the NS5b coding region. J Virol. 2005;79:7050–7058. doi: 10.1128/JVI.79.11.7050-7058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindenbach BD, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 29.Lohmann V, et al. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J Virol. 2003;77:3007–3019. doi: 10.1128/JVI.77.5.3007-3019.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.