Summary
How a nucleus is positioned within a highly polarized post-mitotic animal cell is not well understood. In this work, we demonstrate that the Dynactin complex (a regulator of the microtubule motor protein Dynein) is required to maintain the position of the nucleus within post-mitotic Drosophila melanogaster photoreceptor neurons. We show that multiple independent disruptions of Dynactin function cause a relocation of the photoreceptor nucleus toward the brain, and that inhibiting Dynactin causes the photoreceptor to acquire a bipolar appearance with long leading and trailing processes. We find that while the minus-end directed motor Dynein cooperates with Dynactin in positioning the photoreceptor nucleus, the plus-end directed microtubule motor Kinesin acts antagonistically to Dynactin. These data suggest that the maintenance of photoreceptor nuclear position depends on a balance of plus-end and minus-end directed microtubule motor function.
Keywords: Nuclear migration, Dynein, Capping protein, Dynamitin, Kinesin
Introduction
Neurons are highly polarized cells whose cell bodies extend processes specialized for receiving and transmitting information. The location of a neuron’s cell body is defined in large part by the location of its nucleus, and the positioning of a neuron’s cell body with respect to its processes (e.g. axons and dendrites) varies in a neuron-specific way, contributing to the great diversity of neuronal morphologies. Little is known about how the position of the nucleus is maintained as a neuron undergoes the morphological changes accompanying differentiation.
Nuclear positioning makes an important contribution to brain architecture. In insects, for example, neuronal nuclei and neuronal processes are spatially segregated within the brain, with nuclei populating cortical regions while neurites extend into neuropil regions and establish connections (Cajal, 1990; Strausfeld and Meinertzhagen, 1998). While such extreme spatial segregation is not as widely observed in mammals, neuronal nuclear positions are highly stereotyped throughout the mammalian nervous system (Cajal, 1990; Strausfeld and Meinertzhagen, 1998). Genes mutated in several human neurological disorders (including isolated lissencephaly sequence, Miller–Dieker syndrome, and some forms of lissencephaly with cerebellar hypoplasia) have been implicated in nuclear positioning in other systems (Gupta et al., 2002; Olson and Walsh, 2002), although whether defects in the maintenance of nuclear positioning within neurons contribute to these disorders is unknown. Nonetheless, functional studies of these genes in model organisms suggest that the molecular mechanisms that control nuclear positioning may be relevant to human neuronal mispositioning disorders (Morris, 2003; Reinsch and Gonczy, 1998).
Their highly polarized nature and complex morphologies make neurons a favorable system for studying nuclear positioning, yet the mechanisms that maintain nuclear position in postmitotic neurons have not been extensively explored. Both the microtubule cytoskeleton and the actin cytoskeleton have been implicated in positioning nuclei within non-motile animal cells (Starr and Han, 2003). The nucleus is often associated with the focus of microtubule minus ends, and work in non-dividing cultured mammalian cells indicates that the cytoplasmic microtubule network and the minus-end directed microtubule motor Dynein are important for maintaining the focus of microtubule minus ends and nuclear position (Quintyne et al., 1999). In multinucleate Caenorhabditis elegans muscle cells and in Drosophila melanogaster nurse cells, nuclei require anchorage to the actin cytoskeleton to maintain their appropriate positions (Starr and Han, 2003).
In the D. melanogaster compound eye, the precise packing of photoreceptor neurons within each facet of the eye involves the highly stereotyped localization of photoreceptor nuclei. The photoreceptors are generated within a polarized monolayer epithelium (the eye imaginal disc), and the coordinated movements of differentiating photoreceptor nuclei have been described in detail (Tomlinson, 1985). As each photoreceptor differentiates, its nucleus rises toward the apical surface of the eye disc and remains apical while the photoreceptor axon extends toward the basal surface of the eye disc and into the brain. Several mutations that cause photoreceptor nuclei to be displaced toward the brain have been identified and include mutations in genes encoding the Dynactin subunit Glued (Fan and Ready, 1997), the Dynein-associated protein Lis1 (Swan et al., 1999) (the human homolog of which is disrupted in isolated lissencephaly sequence and Miller–Dieker syndrome; Olson and Walsh, 2002; Reiner et al., 1993), the putative microtubule motor regulator Klar (Mosley-Bishop et al., 1999; Welte et al., 1998), and the nuclear lamin Lam DM(0) (Patterson et al., 2004). These studies have demonstrated that the location of the photoreceptor nucleus depends on factors associated with the microtubule cytoskeleton. However, it is essential to determine whether such nuclear relocation reflects nuclear mispositioning within the cell or migration of the entire cell, and whether the defect is simply a secondary consequence of earlier disruptions in mitosis or alterations in the overall apical/basal polarity of the retinal epithelium. The many molecular and genetic tools available in the Drosophila retina facilitate the critical examination of these issues.
The Dynactin complex is an assembly of 11 different subunits that functions as an activator of Dynein (Gill et al., 1991), serving as an adaptor for cargo (Holleran et al., 1996, 2001; Muresan et al., 2001) and enhancing motor processivity (King and Schroer, 2000). The Dynactin subunit Glued couples Dynactin to Dynein by binding to the Dynein intermediate chain (Dic) (Karki and Holzbaur, 1995; Vaughan and Vallee, 1995). Overexpression of a truncated form of Glued that binds to Dic but cannot associate with the rest of the Dynactin complex acts as a powerful inhibitor of Dynein and Dynactin function (Allen et al., 1999; Eaton et al., 2002; Fan and Ready, 1997). Overexpression of the Dynactin subunit Dynamitin disrupts Dynactin complex assembly and also inhibits Dynactin function (Echeverri et al., 1996; Eckley et al., 1999). Biochemical studies have shown that the Dynactin complex also contains Capping Protein (Schafer et al., 1994), a heterodimer composed of the Capping Protein alpha (Cpa) and Capping Protein beta (Cpb) subunits (Cooper et al., 1999). Although best known for capping the barbed ends of filaments of actin, Capping Protein also associates with filaments of the actin-related Arp1 protein, which is a central element of the Dynactin complex (Cooper et al., 1999; Schafer et al., 1996).
In this work we demonstrate, using multiple independent strategies to disrupt Dynactin function, that the Dynactin complex is critical for photoreceptor nuclear positioning and that Dynactin inhibition causes photoreceptor nuclei to leave the retina and move toward the brain. We show that Dynactin acts in postmitotic photoreceptors and that the disruption in nuclear positioning observed reflects the movement of the nucleus within the neuron rather than photoreceptor migration. We isolate loss-of-function mutations in kinesin heavy chain (khc) as strong suppressors of nuclear mispositioning in Glued1 mutants, and we demonstrate that Kinesin antagonizes Glued function in positioning the nuclei of postmitotic photoreceptors, both in the adult eye and in the larval photosensory organ. Our data demonstrate that the maintenance of photoreceptor nuclear position relies on Dynactin activity and suggest that the positioning of photoreceptor nuclei depends on the antagonistic activities of plus-end and minus-end directed microtubule motors.
Materials and methods
Genetics and molecular biology
Unless otherwise indicated, fly stocks were obtained from the Bloomington Stock Center. Glued1 and UAS:GluedDN have been described (Boylan and Hays, 2002; Fan and Ready, 1997; Harte and Kankel, 1982). GluedDN contains the N-terminal 922 amino acids of Glued and behaves similarly to the product of Glued1 (Allen et al., 1999; Eaton et al., 2002; Fan and Ready, 1997). UAS-Dynamitin-GFP flies have been described (Januschke et al., 2002).
cpbF44 was recovered from approximately 4400 lines of EMS-mutagenized FRT40A flies screened for failure to complement cpbM143 lethality. Both cpbM143 and cpbF44 were sequenced and contained a G to A transition introducing a stop codon after amino acid 147 of Cpb. cpbM143 and cpbF44 were independently induced as 14 sequence differences were detected between cpbM143 and cpbF44 within the Cpb transcription unit (flanking the truncation mutation); all cpbF44 polymorphisms were shared with the FRT40A stock used for mutagenesis. cpbM143 was generated in the lab of E. Wieschaus. Df(2L)E.2 was provided by M. Welte. DNA from homozygous Df(2L)E.2 embyros was examined by PCR using multiple primer pairs covering the entire Cpb transcription unit; no Cpb DNA was detected in these animals, suggesting Df(2L)E.2 deletes Cpb. UAS-GlΔ84 was provided by G. Davis, and UAS-Nod:LacZ by S. Thor. Homozygous mutant visual system clones were produced using the eyeless-FLP system (Newsome et al., 2000).
pUAS:Cpb contains a full-length Cpb cDNA (SD07714, Research Genetics) cloned into pUAST (Brand and Perrimon, 1993). pGlass38-1:Gal4 contains 38-1, a pentamer of a 38 bp glass-responsive fragment from the Rh1 enhancer upstream of an hsp70 minimal promoter (Ellis et al., 1993), cloned into pGATb (Brand and Perrimon, 1993). Transgenic flies were created as described (Spradling and Rubin, 1982). pGlass38-1:Gal4 drove expression in the anticipated pattern (Ellis et al., 1993), with expression initiating in photoreceptors seven to eight rows behind the onset of detectable Elav expression. As one row of ommatidia is added every 90 minutes (Wolff and Ready, 1993) and photoreceptor axons reach the brain four to five rows after initiation of Elav expression, the onset of detectable transgene expression lags photoreceptor axon innervation of the target by ≥3 hours. Rescue was obtained by crossing Df(2L)E.2, Bc/+;tubulin:GAL4,UAS:mCD8GFP/+ males to p{w+,UAS:cpb}, cpbM143/SM6:TM6b,Tb virgins. A total of 235 third-instar progeny were scored for UAS:Cpb transgene rescue of cpb lethality. All 37 cpb/Df(2L)E.2 larvae (Bc, non-Tb larvae) were GFP-positive and thus contained both the Gal4 driver and UAS:Cpb; no GFP-negative cpb/Df(2L)E.2 larvae, which did not contain the Gal4 driver, were recovered. Single-cell analysis in Fig. 2 was performed by crossing hsFLP-Actin-FRT-FRT-GAL4,UAS:GFP/Y males to w; c-s or Gl1/TM6b virgins. Progeny were heat-shocked at 38°C for 1 hour each day.
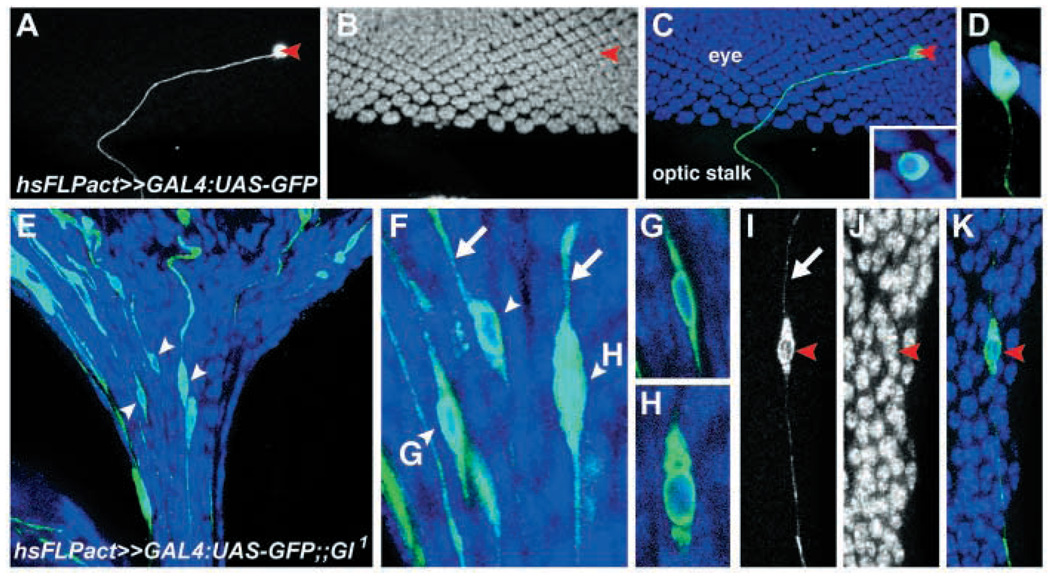
Fig. 2.
Nuclei are mispositioned within Glued mutant photoreceptors. (A–K) All neuronal nuclei in a third instar eye–brain complex are labeled with anti-Elav (blue in C–H,K, white in B,J). Individual cells are labeled with GFP (green in C–H,K, white in A,I) using a heat-shock FLP Act-FRT-FRT-Gal4:UAS-GFP chromosome in a wild-type background (A–D) or a Glued1 mutant animal (E–K). An individual wild-type photoreceptor is marked with GFP in A, and all neuronal nuclei labeled with anti-Elav in B. A and B are merged in C, with a higher magnification view of the marked photoreceptor containing a nucleus in the inset. (D) An individual photoreceptor in another sample is imaged from a side view, with apical at the top. In Glued1 mutants, individual cells in the optic stalk in E (marked with arrowheads) are shown at higher magnification in F. Examples of trailing processes are marked with arrows in F. Cells in F marked with both an arrowhead and a letter are shown at right, with their corresponding letter, at higher magnification. An individual labeled cell in another animal is shown in I, with trailing process indicated by the arrow. I and J are merged in K.
Histology
Third-instar eye–brain complexes were stained as described (Garrity et al., 1999). Primary antibodies were used as indicated: mouse MAb 24B10 anti-Chaoptin (Fujita et al., 1982) (1:200), rat MAb 7E8A10 anti-Elav (O’Neill et al., 1994) (1:50), mouse MAb 40-1a anti-LacZ (1:200) and rabbit anti-PATJ (1:2000). 24B10, 7E8A10, and 40-1a were obtained from the Developmental Studies Hybridoma Bank at the University of Iowa. Secondary antibodies were obtained from Jackson Laboratories and used as described: goat anti-mouse HRP (1:200), goat anti-rat HRP (1:500), goat anti-mouse Cy3 (1:400), goat anti-rabbit FITC (1:200), goat anti-rat Cy5 (1:200), goat anti-mouse FITC (1:200). Confocal images were obtained using a Nikon PCM2000 microscope. SEM was performed as described (Wolff, 2000).
Results
Dynactin is required for proper localization of photoreceptor cell bodies and nuclei
Patterning of the adult compound eye of Drosophila initiates during the third instar phase of larval life, and mutations in the Dynactin subunit Glued strongly disrupt eye development (Fan and Ready, 1997; Harte and Kankel, 1982). Normally the nuclei of differentiating photoreceptors occupy apical regions of the eye disc. In animals heterozygous for the dominant-negative Glued allele Glued1, many photoreceptor nuclei have been shown to accumulate within basal regions of the eye disc (Fan and Ready, 1997). We further characterized the effect of Glued1 on photoreceptor development using an antibody recognizing photoreceptor cell surfaces. In wild type, the region of the differentiating photoreceptor neuron containing the nucleus remained in the retina, while the photoreceptor axon extended through the optic stalk into the brain (Fig. 1A). However, in Glued1 animals, while photoreceptors still extended axons into the brain, the region of the photoreceptor containing the nucleus often appeared to leave the retina and travel through the optic stalk into the brain (Fig. 1B). Staining of photoreceptor nuclei directly demonstrated the movement of photoreceptor nuclei out of the eye disc and into the optic stalk in Glued1 mutants (see below).
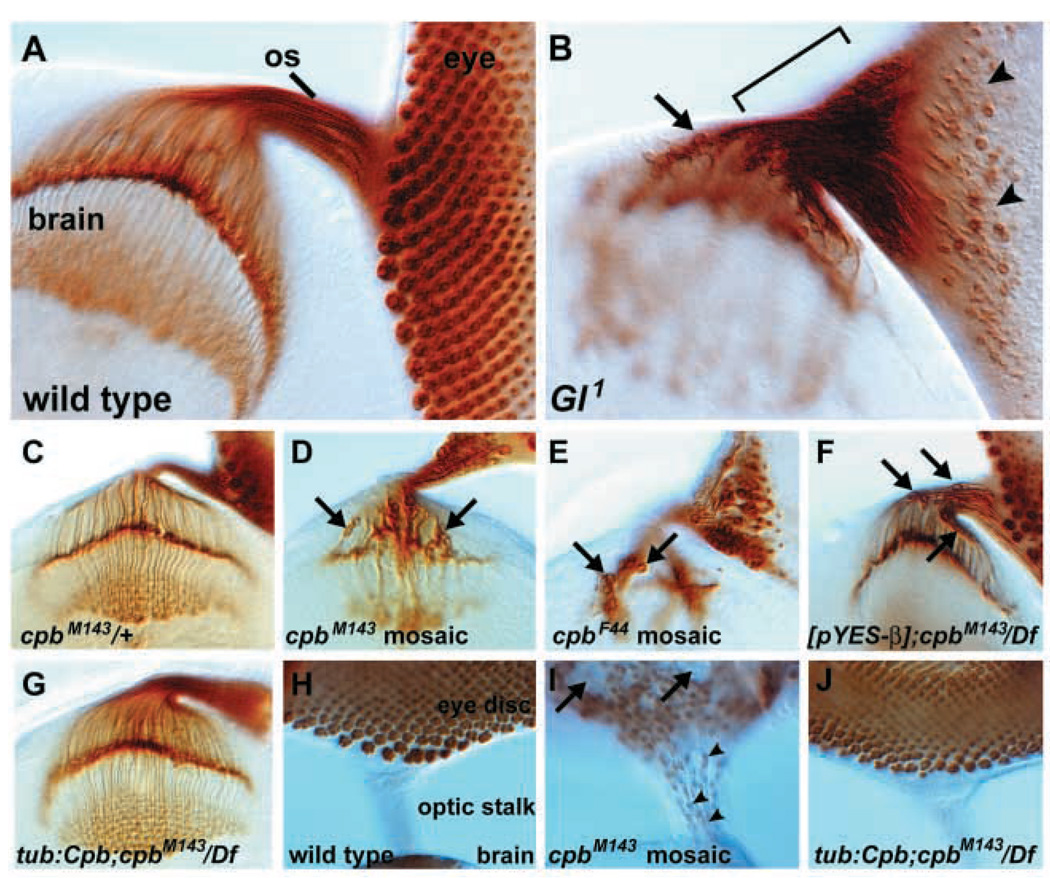
Fig. 1.
Dynactin is required to position photoreceptor cell bodies and nuclei in the developing third instar eye disc. Photoreceptor cell membranes are stained with anti-Chaoptin in A–G. (A) In wild type, photoreceptor cell bodies (as defined in this figure by the position of the nucleus) are precisely arranged in clusters in the apical region of the eye disc and project axons through the optic stalk (os) into the brain's optic lobe. (B) In Glued1 mutants, many photoreceptor neuron cell bodies leave the apical region of the eye disc (arrowheads) and travel into the optic stalk (bracket) and brain (arrow). (C) Heterozygous cpbM143 animals have a wild-type photoreceptor axon projection pattern, with photoreceptor cell bodies positioned in the eye disc. (D) In heterozygous cpbM143 animals with homozygous cpbM143/cpbM143 patches in the visual system, many photoreceptor cell bodies leave the eye disc and enter the brain (arrows). (E) An independently generated cpb allele, cpbF44, also causes photoreceptor cell bodies to enter the brain (arrows) in eye clones. (F) Photoreceptor mispositioning (arrows) is also observed in cpbM143/Df(2L)E.2 animals rescued from early lethality by expression of Cpb from a genomic transgene [pYES-β] (see text for details). (G) cpbM143/Df(2L)E.2 animals rescued by ubiquitous expression of a wild-type Cpb cDNA have normal photoreceptor positioning. Photoreceptor nuclei are stained with anti-Elav in H–J. (H) In wild type, photoreceptor nuclei remain in the eye disc and do not enter the optic stalk. (I) Photoreceptor nuclei are mispositioned in cpbM143 mosaic eye discs, with patches of eye tissue missing nuclei (arrows) and Elav-staining nuclei found in the optic stalk (arrowheads). (J) cpbM143/Df(2L)E.2 animals rescued by ubiquitous expression of a Cpb cDNA have normal photoreceptor nuclear positioning.
To further establish that Glued1 defects reflected disruptions in Dynactin function, we used two other approaches to disrupt the Dynactin complex. As described below, we overexpressed Drosophila Dynamitin, which also inhibits Dynactin function in flies (Duncan and Warrior, 2002; Januschke et al., 2002), in photoreceptor neurons. We also examined loss-of-function mutations in the Dynactin subunit Cpb by generating animals whose visual systems contained homozygous mutant clones of the cpb strong loss-of-function mutation cpbM143. In these cpbM143 mosaic animals, the nuclear regions of many photoreceptors were observed in the optic stalk and brain (Fig. 1C,D).
To confirm that the cpbM143 mutant photoreceptor defect was due to a loss of cpb function, we isolated an additional strong loss-of-function cpb allele, cpbF44, from an EMS mutagenesis and obtained a chromosomal deficiency uncovering the cpb locus, Df(2L)E.2 (see Materials and methods for details). When animals contained homozygous mutant clones of cpbF44 cells or homozygous mutant clones of Df(2L)E.2, a similar movement of photoreceptor nuclear regions toward the brain was observed (Fig. 1E and data not shown). cpb/Df(2L)E.2 animals did not survive to third instar, preventing the classic genetic demonstration that these cpb alleles behaved as strong loss-of-function mutations. Fortunately, we found that the [pYES-β] genomic transgene, which contains the CPB coding region (Hopmann et al., 1996), was able to rescue the lethality of cpb/Df(2L)E.2 animals, but did not rescue the previously described cpb bristle defect (Hopmann et al., 1996). This suggested that [pYES-β] was a partially functional rescue construct that could be used to examine the visual systems of otherwise cpb/Df(2L)E.2 animals. We found that [pYES-β];cpbM143/Df(2L)E.2 animals displayed a photoreceptor defect similar to that of other cpb mutants, consistent with nuclear mispositioning resulting from the loss of cpb function (Fig. 1F). We further confirmed that the defect was due to the loss of cpb function by successfully rescuing the cpbM143/Df(2L)E.2 photoreceptor defects (as well as the cpb bristle defects) by expression of a wild-type Cpb cDNA under the control of a heterologous promoter (Fig. 1G and data not shown). Staining of photoreceptor nuclei directly demonstrated the movement of photoreceptor nuclei out of the eye disc and into the optic stalk in cpb mutants (Fig. 1H–J).
The bifunctional nature of Cpb, which associates with filaments of actin as well as filaments of Arp1, means that loss of Cpb also increases filamentous actin levels (Hopmann and Miller, 2003). Nonetheless, previous studies have shown that increases in filamentous actin alone, such as those observed in hypomorphic cpb alleles or in actup mutants, do not cause photoreceptor nuclear mispositioning (Benlali et al., 2000; Hopmann and Miller, 2003). Together with the Glued1 and Dynamitin data, the cpb observations yield a consistent picture that alterations in Dynactin subunits cause mispositioning of photoreceptor cell bodies and nuclei, and indicate that Dynactin, and not just the Glued subunit, has an important role in photoreceptor development.
Dynactin is required for maintenance of nuclear positioning within postmitotic photoreceptors
The mispositioning of photoreceptor nuclei in Dynactin mutants raised the question of whether these disruptions reflect altered positioning of the nucleus within the photoreceptor or simply migration of the entire photoreceptor. To address this question, single photoreceptors were labeled in wild type and in Glued1 mutants. Wild-type photoreceptors exhibit a highly polarized morphology in which the region of the photoreceptor containing the nucleus lies in the apical region of the eye disc and an axon extends basally into the brain (Fig. 2A–D). Glued1 mutant photoreceptors whose nuclei have entered the optic stalk had highly altered morphologies, with both leading and trailing processes extending from the regions of the cell where the misplaced nucleus was located (Fig. 2E–K). We quantified leading and trailing processes of misplaced Glued1 photoreceptors, considering only those with no other labeled cells or processes nearby. Of these 13 neurons, 12 had clearly detectable leading and trailing processes. The leading process (axon) extended into the target region and the trailing process extended back into the eye disc. These data demonstrate that inhibition of Dynactin function dramatically alters the position of the nucleus within the photoreceptor.
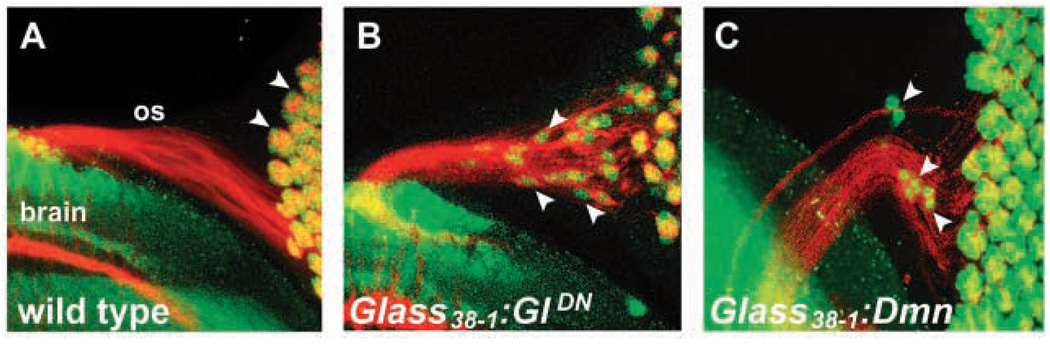
The Dynactin complex also controls the pattern of mitoses within the Drosophila retina (Fan and Ready, 1997). To determine whether nuclear mispositioning is a secondary consequence of the earlier mitotic requirement for Dynactin, we examined the effects of specifically inhibiting the Dynactin complex in postmitotic photoreceptors. Conditional inhibition of Dynactin function can be achieved through inducible expression of a truncated, dominant-negative form Glued (GluedDN) that resembles the protein product of Glued1 (Allen et al., 1999; Fan and Ready, 1997). GluedDN was expressed under the control of the postmitotic photoreceptor-specific Glass 38-1 promoter, which initiates expression in the photoreceptors only after their axons have entered the brain (see Materials and methods). Expression of GluedDN under the control of Glass 38-1 caused photoreceptor nuclei to move into the optic stalk (Fig. 3A,B). Overexpression of Dynamitin under the control of Glass 38-1 caused similar photoreceptor nuclear positioning defects (Fig. 3C). These data demonstrate that Dynactin is required postmitotically in photoreceptors to maintain nuclear position and that the disruptions in nuclear positioning observed are not simply a secondary consequence of mitotic defects.
Fig. 3.
Dynactin is required postmitotically to maintain photoreceptor nuclear positioning. Third instar eye–brain complexes are stained with anti-Chaoptin (red) and anti-Elav (green). No photoreceptor nuclei (arrowheads) are seen in the optic stalk in wild type (A). Expression of dominant-negative Glued or overexpression of Dynamitin in differentiated photoreceptors under the control of Glass38-1 causes nuclei to leave the eye disc and enter the optic stalk (B,C, arrowheads).
Photoreceptor nuclear movement occurs without disruption of apical/basal polarity
The displacement of photoreceptor nuclei from apical regions of the eye disc toward more basal regions could reflect an overall disruption in apical/basal polarity of the eye disc. The apical/basal polarity of developing photoreceptors was assessed by examining the distribution of the Drosophila β-catenin Armadillo and the PDZ-domain-containing protein PATJ (Pielage et al., 2003). Armadillo localizes to the zonula adherens separating the apical and basolateral membrane domains of developing photoreceptors (Pellikka et al., 2002), while PATJ localizes to the apical membrane domain (Izaddoost et al., 2002). In wild-type eye discs, Armadillo is concentrated just beneath the apical tips of the developing photoreceptors (Fig. 4A). In Glued1 animals Armadillo was still present near apical regions of the eye disc, even in areas completely devoid of apical photoreceptor nuclei (Fig. 4B). Thus, this marker of apical/basal polarity was retained even when photoreceptor nuclei moved basally. Similar results were obtained when Glued1 mutants were visualized in cross-section using both Armadillo and PATJ. Apical localization of PATJ and Armadillo were observed in Glued1 and the relative apical/basal ordering of these markers was maintained (Fig. 4C,D). These data suggest that the alterations in photoreceptor morphology are not caused by a loss of apical/basal polarity within the developing photoreceptors.
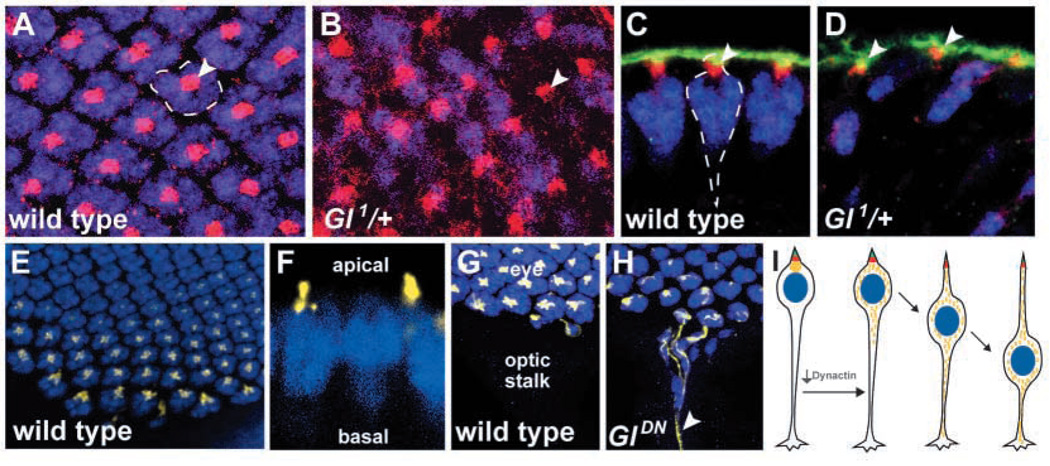
Fig. 4.
Apical markers are not disrupted, but Nod:LacZ is mislocalized, in Glued mutants. Third instar eye discs are stained with anti-Elav (blue), anti-Armadillo (red), and anti-PATJ (green) (A–D). Views from the apical surface (A,B) show evenly spaced apical markers in wild type (A), with each ommatidial cluster (dashed outline) centered under a concentration of Armadillo (arrowhead). Armadillo staining is largely normal in Glued1 (B), despite the presence of ommatidia devoid of nuclei (arrowhead). Side views show that in wild type (C), each ommatidium (dashed line) has a distinct apical clustering of Armadillo and PATJ (arrowhead). Glued animals appear to retain apical markers (arrowheads) even when photoreceptor nuclei are mispositioned (D). Nod:LacZ (yellow) expressed in postmitotic photoreceptor neurons under the control of the Glass38-1 promotor localizes apical to photoreceptor nuclei (anti-Elav, blue) in the most mature photoreceptor neurons (E, apical surface, and F, side view) and is not found in axons or in the optic stalk (G). When dominant-negative Glued is expressed in photoreceptor neurons using the Glass38-1 promoter, Nod:LacZ staining is distributed throughout the axons of the most mature cells (H, arrowhead). In (I), consequences of Dynactin disruption are summarized, synthesizing the data obtained in Fig 2–Fig 4. Inhibition of Dynactin function in the postmitotic neuron causes the photoreceptor nucleus (blue) to be displaced toward the axon terminal. Despite nuclear movement, a trailing process remains and apical markers (PATJ in green, Armadillo in red) are retained. Nod: LacZ (yellow), however, becomes mislocalized from its wild-type apical position and enters the axon.
Glued maintains microtubule cytoskeleton organization
Dynactin has important functions in the organization of the microtubule cytoskeleton in many systems. The microtubule cytoskeleton of developing photoreceptors is highly polarized, with microtubule minus ends concentrated apical to the nucleus as detected using antisera recognizing gamma-tubulin (Swan et al., 1999). A similar apical focus is observed when using the fusion protein Nod:LacZ (Fig. 4E,F), which often co-localizes with microtubule minus ends (Clark et al., 1997). The relatively ubiquitous expression of gamma-tubulin in the retina complicated the analysis of gamma-tubulin localization when retinal patterning was disrupted (J.L.W. and P.A.G., unpublished). Therefore, we examined the effect of Glued on factors associated with the microtubule cytoskeleton by expressing Nod:LacZ specifically in postmitotic photoreceptors. In animals expressing GluedDN in postmitotic photoreceptors as well as in Glued1 mutants, Nod:LacZ was no longer exclusively concentrated in apical regions of photoreceptors, but rather spread into the photoreceptor axons (Fig. 4H, data not shown). Thus, while the overall apical/basal polarity of the photoreceptors was not disrupted in Glued mutants, the spatial organization of the microtubule cytoskeleton-associated factor Nod:LacZ was affected.
Glued cooperates with Dynein in photoreceptor nuclear positioning but is antagonized by Kinesin
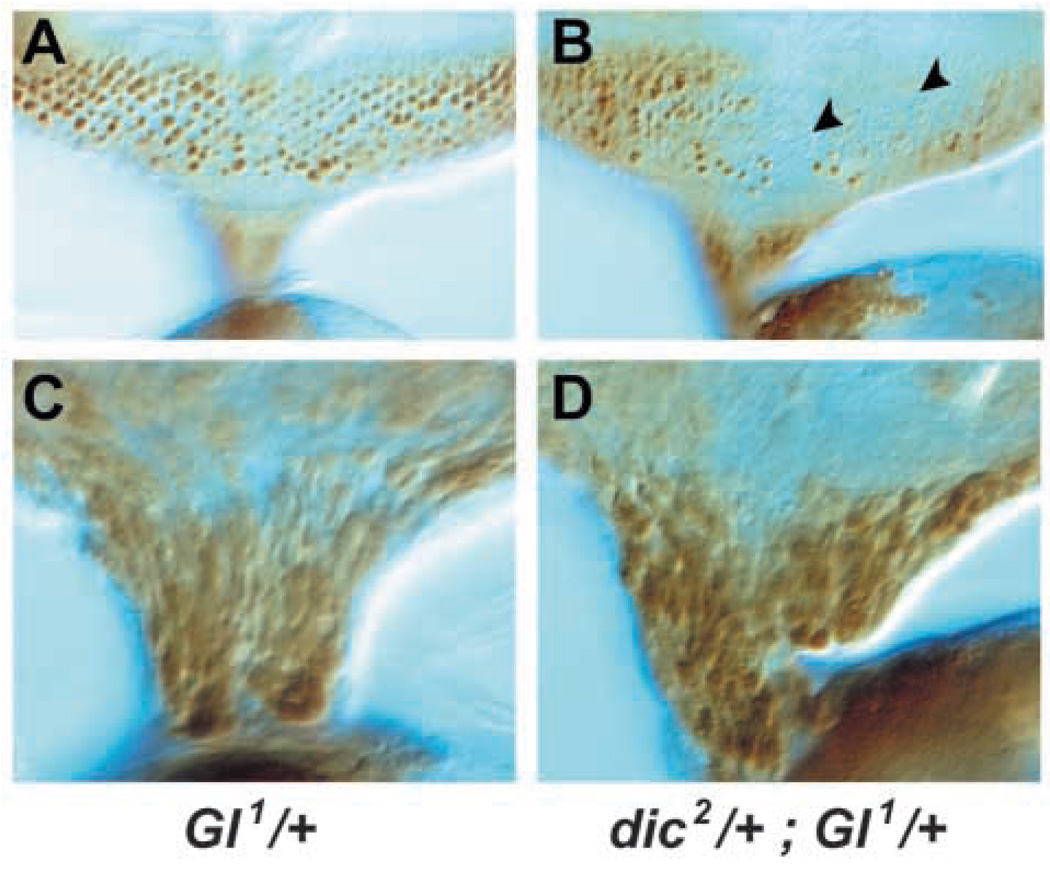
Dynactin activates the microtubule motor Dynein, and strong loss-of-function mutations in dynein intermediate chain (dic) are dominant enhancers of the rough eye phenotype of Glued1 mutants (Boylan and Hays, 2002). As Dynein and Dynactin may play multiple roles together during eye development, we examined the effect of a reduction in dic gene dosage upon photoreceptor nuclear positioning in Glued1 animals. A twofold reduction in dic gene dosage caused a further decrease in the number of photoreceptor nuclei in apical regions of Glued1 mutant eye discs (Fig. 5A,B). This did not reflect a simple reduction in the number of photoreceptors generated, as large numbers of photoreceptor nuclei were crowded at the base of the eye disc and entered the optic stalk in both animals (Fig. 5C,D). Thus, a larger fraction of photoreceptor nuclei left apical positions when the level of dic gene activity was reduced, consistent with Dynein and Dynactin acting together in this process.
Fig. 5.
Glued nuclear mispositioning is enhanced by dynein intermediate chain reduction. Third instar eye discs were stained with anti-Elav. (A,B) Apical surface of eye disc. Glued1 mutants show small areas devoid of apical photoreceptor nuclei (A), while dic2/+; Glued1/+ animals (B) have much larger areas devoid of nuclei (arrowheads). (C,D) Basal surface of eye disc and optic stalk. The greater absence of photoreceptor neuron nuclei in apical regions of the eye disc in dic2/+; Glued1/+ animals is not simply due to an absence of photoreceptor neurons, as large numbers of photoreceptor nuclei are clustered at the base of the optic stalk in both Glued1 (C) and dic2/+; Glued1/+ animals (D).
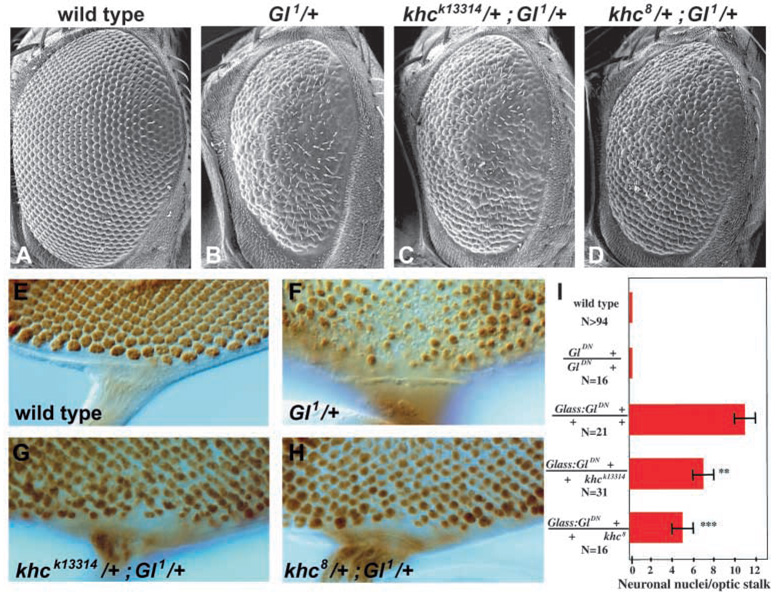
To identify additional factors that interact with Dynactin to control nuclear positioning, a genetic screen was performed to identify genes that dominantly enhanced or suppressed the Glued1 external eye phenotype. From a collection of approximately 1800 stocks containing transposon-induced lethal mutations, several stocks were identified that had no dominant effect on eye development in a wild-type background, but were dominant enhancers or suppressors of Glued1. Two dominant suppressors of Glued1, khck13219 and khck13314, were alleles of kinesin heavy chain (khc), which encodes a subunit of the plus-end directed microtubule motor kinesin (Fig. 6A–C). The interaction with Glued1 was further confirmed using the null allele khc8 (Fig. 6C). Examination of developing eye discs demonstrated that a twofold reduction of khc gene dosage greatly increased the number of photoreceptor nuclei present in apical regions of Glued1 mutant eye discs (Fig. 6E–H). This suggested that khc acts antagonistically to Glued in photoreceptor nuclear positioning.
Fig. 6.
Glued nuclear mispositioning is suppressed by kinesin heavy chain reduction. Scanning electron micrographs of adult eyes (A–D). (A) Wild type. (B) Gl1/+ animals have smaller eyes with disorganized ommatidia. (C,D) Reduction of kinesin heavy chain gene dosage partially suppresses Gl1/+ eye defect. Apical regions of third instar eye discs in which photoreceptor nuclei are stained with anti-Elav (E–H). (I) Suppression of the Glass-GluedDN phenotype quantified by counting the number of Elav-positive nuclei in the optic stalks of animals with 15 to 22 rows of photoreceptor development. The average for each genotype was 19 rows of development. Error bars are s.e.m. and asterisks denote P-value of unpaired t-test (**P<0.01, ***P<0.001).
To determine whether khc mutations interacted with Glued1 in postmitotic photoreceptors, khc gene dosage was reduced in animals expressing dominant-negative Glued under the control of the postmitotic Glass38-1 promoter (Fig. 6I). Wild-type animals (n>50 hemispheres) or animals containing the dominant-negative Glued transgene without the Glass 38-1 promoter (n>20) never contained photoreceptor nuclei within their optic stalks. Glass38-1:GluedDN animals contained an average of 11±1 photoreceptor nuclei within the optic stalk (± s.e.m., n=21). However, Glass38-1:GluedDN animals heterozygous for either khck13314 or khc8 showed a significant reduction in the number of photoreceptor nuclei in the optic stalk (7±1, n=31, unpaired t-test P<0.01 for khck13314/+ and 5±1, n=16, P<0.001 for khc8/+). Thus, a twofold reduction in khc gene dosage suppressed the effects of postmitotic expression of dominant-negative Glued, consistent with Glued and khc acting antagonistically within differentiated photoreceptors to regulate nuclear positioning.
Glued and Kinesin Heavy Chain also act antagonistically in positioning Bolwig photoreceptor nuclei
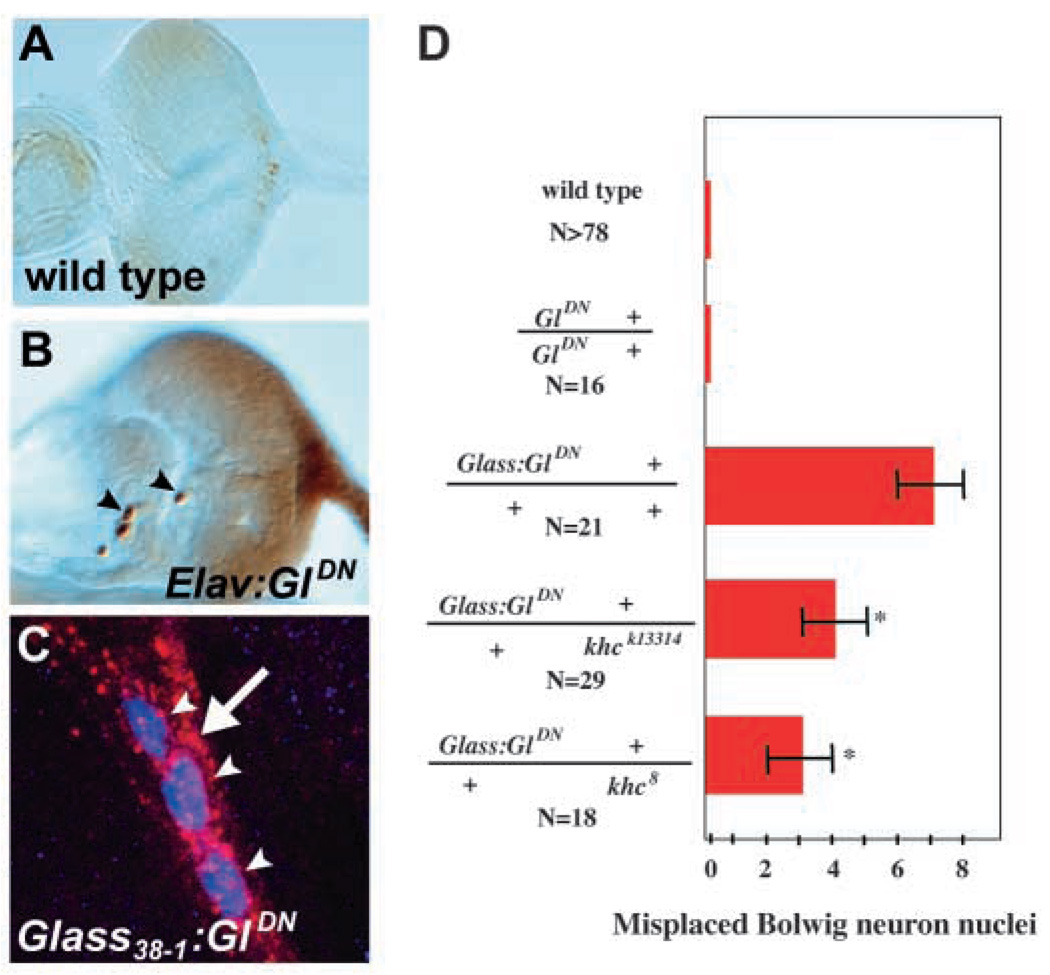
We examined the interaction between Glued and khc in other photoreceptors by examining the Bolwig organ, a cluster of 12 photosensitive neurons that differentiate during embryonic development and extend axons into the brain (Schmucker et al., 1997). By second and third instar larval stages, Bolwig photoreceptor nuclei are located near the anterior tip of the larva and their axons extend over the eye/antennal disc into the brain, a distance of >200 µm. In wild-type second instar animals, photoreceptor neuron differentiation has not yet begun in the eye disc and no neuronal nuclei are present there (Fig. 7A). However, when GluedDN was expressed in postmitotic Bolwig photoreceptors, their nuclei appeared on the surface of the eye/antennal disc (Fig. 7B,C). Thus, as in the photoreceptors of the adult eye, expression of GluedDN in Bolwig photoreceptors caused their nuclei to be positioned closer to their axon termini; in many cases, the Bolwig nuclei were over 150 µm closer than normal to their axon terminals in the brain.
Fig. 7.
Glued function is required to position Bolwig organ nuclei, where it is antagonized by kinesin heavy chain. Wild-type second instar eye-antennal discs stained with anti-Elav show no neuronal nuclei in the eye-antennal disc along the path of the Bolwig nerve (A), while animals expressing dominant-negative Glued in postmitotic Bolwig photoreceptor neurons, using either the Elav promoter (B) or Glass38-1 (C), have neuronal nuclei (arrowheads in B,C) along the path of the Bolwig nerve (in C, anti-Chaoptin in red, anti-Elav in blue, arrow indicates Bolwig nerve). Misplaced Bolwig nuclei in animals that express dominant-negative Glued under Glass38-1 control were quantified in D. The number of misplaced Bolwig neuron nuclei was reduced when animals were heterozygous for loss-of-function mutations in khc. *P<0.05.
The interaction between Glued and khc in Bolwig photoreceptors was assessed by counting the number of Bolwig nuclei on the surface of the eye/antennal disc (Fig. 7D). While wild-type and UAS:GluedDN animals had no neuronal nuclei in this region, Glass38-1:GluedDN animals contained 7±1 (n=21). A reduction of khc gene dosage in Glass38-1:GluedDN; khck13314/+ and Glass38-1:GluedDN; khc8/+ animals significantly reduced this to 4±1 (n=29, P<0.05) and 3±1 (n=16, P<0.05), respectively. These data further support the functional antagonism of Glued and khc in photoreceptor nuclear positioning.
Discussion
Although the proper positioning of neuronal cell bodies and the nuclei they contain is a central feature of brain morphogenesis, relatively little is known about how the position of a nucleus is maintained within a postmitotic neuron. Here we have examined the function of Dynactin in maintenance of nuclear positioning in postmitotic photoreceptor neurons. We saw that nuclear positioning shows impressive plasticity, as disruption of Dynactin function after photoreceptor axons had extended into their target region in the brain caused the nucleus to move away from the neuron’s apical tip and toward the growth cone, giving the photoreceptor a ‘bipolar’ morphology (compared with its normally ‘unipolar’ morphology). Interestingly, we found that reductions in Kinesin partially compensated for the effects of reduced Dynactin activity. Taken together, these data establish an essential role for Dynactin in the morphological organization of postmitotic photoreceptors and suggest that a balance of plus- and minus-end directed microtubule motor activity could influence the position of the nucleus.
Glued acts in postmitotic photoreceptors to control nuclear positioning
Establishing and maintaining appropriate nuclear position is a general challenge for eukaryotic cells and the mechanisms that control nuclear positioning vary with cell type and developmental stage (Morris, 2003; Starr and Han, 2003). In neurons, the position of the nucleus is initially established at the end of the precursor cell’s mitosis and changes as the neuron migrates into position and acquires its differentiated morphology. Thus, nuclear positioning is a dynamic process integrated into the differentiation program of a neuron.
Several alternative models have been proposed for the role of the Dynactin subunit Glued in neuronal positioning in the fly eye (Fan and Ready, 1997). As Glued1 mutations affect both mitosis and nuclear positioning in the eye, it has been difficult to assess whether Glued activity is required specifically in postmitotic neurons. In fact, broad expression of the cell-cycle inhibitor p21 behind the morphogenetic furrow (the region in which photoreceptors differentiate) partially suppressed the Glued1 nuclear positioning defect, suggesting that disruptions in Dynactin might lead to a nuclear positioning defect by simply disrupting the coordination of cell-cycle progression and nuclear movement (Fan and Ready, 1997). Our results demonstrate that Dynactin activity is required within the postmitotic photoreceptor to regulate nuclear positioning. We also see nuclear mispositioning when Glued function is inhibited in postmitotic photoreceptors of the Bolwig organ, indicating that this function is not specific for photoreceptors generated in the eye disc.
Our analysis has focused on the positioning of the nucleus within the photoreceptor neuron. It is interesting to consider whether other constituents of the cell body are similarly mispositioned when Dynactin function is disrupted. Our analysis of single Glued1 photoreceptors indicates that mispositioned nuclei are surrounded by a concentration of other cellular material, as evidenced by the accumulation of CD8:GFP (a transmembrane protein associated with cell surfaces as well as secretory vesicles) around the nuclei in Fig. 2H,I. Thus it is possible that not only nuclei, but also other elements of the cell body, are mispositioned in these animals.
Kinesin exerts an antagonistic influence on photoreceptor nuclear positioning
Our finding that Glued collaborates with Dynein in photoreceptor neuron nuclear positioning raises the question of whether other motor proteins contribute to this process. From a screen for genes that promote or antagonize Glued function in the retina, we identified loss-of-function alleles of kinesin heavy chain (khc) and demonstrated that a reduction in khc dosage reduced the amount of photoreceptor nuclear mispositioning observed in Glued1 animals. These data suggest that nuclear mispositioning does not result simply from the poisoning of axonal transport, as a decrease in khc function exacerbates the axonal transport defects of Glued1 animals (Martin et al., 1999). Furthermore, the observation of a Glued/khc interaction in postmitotic photoreceptors of the adult eye and the larval Bolwig organ indicates that the interplay between Glued and Kinesin occurs within the differentiating photoreceptor. Taken together, our data suggest that the two may normally play antagonistic roles in positioning the photoreceptor nucleus. The fact that strong photoreceptor nuclear mispositioning is not observed in animals containing homozygous mutant clones of khc tissue in the retina (Brendza et al., 2000) (J.L.W. and P.A.G., unpublished) is perhaps not surprising, as the nucleus normally resides near the apical surface of the retina and adjacent to the focus of microtubule minus ends, leaving little room for further apical movement. While the microtubule motor proteins Dynein and Kinesin are important for nuclear positioning in many cell types (Cottingham and Hoyt, 1997; DeZwaan et al., 1997; Duncan and Warrior, 2002; Januschke et al., 2002; Morris, 2003; Reinsch and Gonczy, 1998; Requena et al., 2001), a role for microtubule motors in nuclear positioning in postmitotic neurons has not been previously established.
Roles of Dynein, Dynactin and Kinesin in photoreceptor nuclear positioning
Dynein and Dynactin control a number of cellular processes through their effects on the structure of the microtubule cytoskeleton and through the transportation of cargo along microtubules. In particular, Dynein and Dynactin regulate nuclear positioning in many dividing and migrating eukaryotic cells (Morris, 2003; Reinsch and Gonczy, 1998). How do Dynein and Dynactin control photoreceptor nuclear position and how might Kinesin exert an antagonistic influence? One possibility is that the photoreceptor nucleus may be a cargo moved directly by the Dynein/Dynactin complex. The proximity of the photoreceptor nucleus to the focus of microtubule minus ends in wild-type animals would be consistent with Dynein and Dynactin working to move the nucleus toward the focus of microtubule minus ends, while the antagonistic interaction between Dynactin and Kinesin could reflect the direct coupling of the nucleus to both minus-end and plus-end directed motors. Thus, the position of the photoreceptor nucleus would reflect the relative balance of opposing motor activities, with Dynein predominating under normal circumstances in the photoreceptors. Such coupling to opposite-polarity microtubule motors has been implicated in the movement of other organelles, such as mitochondria and lipid droplets (Gross, 2003). This scenario would be consistent with the movement of the photoreceptor nucleus away from the focus of microtubule minus ends in animals mutant for klar, a gene implicated in the coordination of plus- and minus-end directed motors attached to lipid droplets in the Drosophila embryo (Mosley-Bishop et al., 1999; Welte et al., 1998). Although the mechanism by which Klar may regulate microtubule motors is unknown, klar genetically interacts with the nuclear lamin Lam DM(0) (Patterson et al., 2004), raising the possibility that Klar could be involved in the coordination of Dynein and Kinesin motors associated with the photoreceptor nuclear envelope.
Alternatively, Dynein and Dynactin could also play more indirect roles in photoreceptor nuclear positioning. For example, in non-motile interphase mammalian tissue culture cells, Dynactin co-localizes with the focus of microtubule minus ends and Dynactin disruption defocuses these minus ends (Quintyne et al., 1999). Since the photoreceptor nucleus normally lies adjacent to the focus of microtubule minus ends, it is possible that nuclear movement could then be a secondary consequence of microtubule minus-end redistribution. Such redistribution could potentially be dependent upon Kinesin activity. In C. elegans embryos, zyg-12 is required for close association of the nucleus with the focus of microtubule minus ends and the ZYG-12 protein may act as a physical link between Dynein and the nuclear envelope (Malone et al., 2003). However, no functional equivalent of ZYG-12 has been identified in Drosophila. While ZYG-12 has homology to the Hook family of proteins, analysis of Drosophila hook indicates that it is involved in regulating secretory and endocytic pathways rather than photoreceptor nuclear localization (Walenta et al., 2001). In a similar model, Dynein and Dynactin could also control the apical/basal positioning of the focus of microtubule minus ends. In Saccharomyces cerevisiae, Dynein associated with the cell cortex is postulated to control the movement of microtubules along the interior surface of the cell (Lee et al., 2003). In photoreceptors, association of Dynein with the apical cortex of the cell might act similarly to move microtubule minus ends toward the apical tip of the photoreceptor.
To begin to test the effect of Dynactin inhibition on factors associated with the microtubule cytoskeleton, we have examined the distribution of the fusion protein, Nod:LacZ, which colocalizes with microtubule minus ends in wild-type animals. We see a strong delocalization of Nod:LacZ in Glued mutants (Fig. 4) and in cpb mutants (J.L.W. and P.A.G., unpublished). The movement of Nod:LacZ into the axon would be consistent with a defocusing of microtubule minus ends and even alterations in the overall polarity of the microtubule cytoskeleton. Such microtubule disorganization would cause Nod:LacZ to no longer travel to a single destination in Glued1 mutants. However, an alternative explanation is that Dynactin is required for the movement of Nod:LacZ to microtubule minus ends. In this scenario, Nod:LacZ would not necessarily be localized at minus ends and thus no longer serve as an effective microtubule minus-end marker in Glued1 mutants. It is interesting to note that despite the strong effects of reducing the gene dosage of dic and khc on nuclear mispositioning in GluedDN mutants, we did not see detectable effects of reducing dic or khc gene dosage on Nod:LacZ distribution in GluedDN animals (J.L.W. and P.A.G., unpublished). Thus the redistribution of Nod:LacZ may be unrelated to the mispositioning of the photoreceptor nucleus, although only a dramatic alteration in the distribution of Nod:LacZ would be detected in our assay. A more detailed analysis of microtubule organization in photoreceptors with disruptions in Dynactin functions awaits the development of additional tools.
How the distinct regions of a neuron (the axons, dendrites and nucleus-containing cell body) are properly positioned is a central question in neuronal cell biology, about which little is known. Here we have shown that Dynein and Dynactin play a major role in maintaining the position of the nucleus within a postmitotic photoreceptor neuron, and that Kinesin can antagonize this function. It will be of interest to determine whether Dynactin may be directly involved in coupling the apical/basal polarity of the photoreceptor neuron to the polarity of the microtubule cytoskeleton; for example, through association with factors involved in apical/basal polarization of the photoreceptor. Another key issue for the future is to determine whether these effects of Dynein, Dynactin and Kinesin on photoreceptor nuclear migration reflect their association with the photoreceptor nucleus and/or the effects of these complexes on the microtubule cytoskeleton more generally.
Acknowledgments
We thank Hugo Bellen, Graeme Davis, Tom Hays, Roberta Hopmann, Katherine Miller, William Saxton, Stefan Thor, Michael Welte, Kevin Cook and the Bloomington Stock Center, and the Developmental Studies Hybridoma Bank for reagents, Nicki Watson for SEM assistance, Tim Tayler and Frank Miskevich for microscopy assistance, and Garrity lab members and Frank Gertler for discussions. We thank Linda Huang, Mark Rosenzweig, Frank Solomon, and an anonymous reviewer for comments on the manuscript. This work was supported by grants to P.A.G. from the National Eye Institute, the Raymond and Beverly Sackler Foundation, and the McKnight Foundation. J.L.W. was supported by an NIH Predoctoral Training Grant and A.C. by an undergraduate research grant to MIT from the Howard Hughes Medical Institute.
References
- Allen MJ, Shan X, Caruccio P, Froggett SJ, Moffat KG, Murphey RK. Targeted expression of truncated glued disrupts giant fiber synapse formation in Drosophila. J. Neurosci. 1999;19:9374–9384. doi: 10.1523/JNEUROSCI.19-21-09374.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlali A, Draskovic I, Hazelett DJ, Treisman JE. act up controls actin polymerization to alter cell shape and restrict Hedgehog signaling in the Drosophila eye disc. Cell. 2000;101:271–281. doi: 10.1016/s0092-8674(00)80837-5. [DOI] [PubMed] [Google Scholar]
- Boylan KL, Hays TS. The gene for the intermediate chain subunit of cytoplasmic dynein is essential in Drosophila. Genetics. 2002;162:1211–1220. doi: 10.1093/genetics/162.3.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brendza RP, Sheehan KB, Turner FR, Saxton WM. Clonal tests of conventional kinesin function during cell proliferation and differentiation. Mol. Biol. Cell. 2000;11:1329–1343. doi: 10.1091/mbc.11.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajal SR. New Ideas on the Structure of the Nervous System in Man and Vertebrates. Cambridge, MA: MIT Press; 1990. [Google Scholar]
- Clark IE, Jan LY, Jan YN. Reciprocal localization of Nod and kinesin function proteins indicates microtubule polarity in the Drosophila oocyte, epithelium, neuron, and muscle. Development. 1997;124:461–470. doi: 10.1242/dev.124.2.461. [DOI] [PubMed] [Google Scholar]
- Cooper JA, Hart MC, Karpova TS, Schafer DA. Capping protein. In: Kreis T, Vale R, editors. Guidebook to the Cytoskeletal and Motor Proteins. New York: Oxford University Press; 1999. pp. 62–64. [Google Scholar]
- Cottingham FR, Hoyt MA. Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J. Cell Biol. 1997;138:1041–1053. doi: 10.1083/jcb.138.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZwaan TM, Ellingson E, Pellman D, Roof DM. Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration. J. Cell Biol. 1997;138:1023–1040. doi: 10.1083/jcb.138.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan JE, Warrior R. The cytoplasmic dynein and kinesin motors have interdependent roles in patterning the Drosophila oocyte. Curr. Biol. 2002;12:1982–1991. doi: 10.1016/s0960-9822(02)01303-9. [DOI] [PubMed] [Google Scholar]
- Eaton BA, Fetter RD, Davis GW. Dynactin is necessary for synapse stabilization. Neuron. 2002;34:729–741. doi: 10.1016/s0896-6273(02)00721-3. [DOI] [PubMed] [Google Scholar]
- Echeverri CJ, Paschal BM, Vaughan KT, Vallee RB. Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J. Cell Biol. 1996;132:617–633. doi: 10.1083/jcb.132.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckley DM, Gill SR, Melkonian KA, Bingham JB, Goodson HV, Heuser JE, Schroer TA. Analysis of dynactin subcomplexes reveals a novel actin-related protein associated with the arp1 minifilament pointed end. J. Cell Biol. 1999;147:307–320. doi: 10.1083/jcb.147.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MC, O’Neill EM, Rubin GM. Expression of Drosophila glass protein and evidence for negative regulation of its activity in non-neuronal cells by another DNA-binding protein. Development. 1993;119:855–865. doi: 10.1242/dev.119.3.855. [DOI] [PubMed] [Google Scholar]
- Fan SS, Ready DF. Glued participates in distinct microtubule-based activities in Drosophila eye development. Development. 1997;124:1497–1507. doi: 10.1242/dev.124.8.1497. [DOI] [PubMed] [Google Scholar]
- Fujita SC, Zipursky SL, Benzer S, Ferrus A, Shotwell SL. Monoclonal antibodies against the Drosophila nervous system. Proc. Natl. Acad. Sci. USA. 1982;79:7929–7933. doi: 10.1073/pnas.79.24.7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity PA, Lee CH, Salecker I, Robertson HC, Desai CJ, Zinn K, Zipursky SL. Retinal axon target selection in Drosophila is regulated by a receptor protein tyrosine phosphatase. Neuron. 1999;22:707–717. doi: 10.1016/s0896-6273(00)80730-8. [DOI] [PubMed] [Google Scholar]
- Gill SR, Schroer TA, Szilak I, Steuer ER, Sheetz MP, Cleveland DW. Dynactin, a conserved, ubiquitously expressed component of an activator of vesicle motility mediated by cytoplasmic dynein. J. Cell Biol. 1991;115:1639–1650. doi: 10.1083/jcb.115.6.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross SP. Dynactin: coordinating motors with opposite inclinations. Curr. Biol. 2003;13:R320–R322. [PubMed] [Google Scholar]
- Gupta A, Tsai LH, Wynshaw-Boris A. Life is a journey: a genetic look at neocortical development. Nat. Rev. Genet. 2002;3:342–355. doi: 10.1038/nrg799. [DOI] [PubMed] [Google Scholar]
- Harte PJ, Kankel DR. Genetic analysis of mutations at the Glued locus and interacting loci in Drosophila melanogaster. Genetics. 1982;101:477–501. doi: 10.1093/genetics/101.3-4.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleran EA, Ligon LA, Tokito M, Stankewich MC, Morrow JS, Holzbaur EL. beta III spectrin binds to the Arp1 subunit of dynactin. J. Biol. Chem. 2001;276:36598–36605. doi: 10.1074/jbc.M104838200. [DOI] [PubMed] [Google Scholar]
- Holleran EA, Tokito MK, Karki S, Holzbaur EL. Centractin (ARP1) associates with spectrin revealing a potential mechanism to link dynactin to intracellular organelles. J. Cell Biol. 1996;135:1815–1829. doi: 10.1083/jcb.135.6.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopmann R, Miller KG. A balance of capping protein and profilin functions is required to regulate actin polymerization in Drosophila bristle. Mol. Biol. Cell. 2003;14:118–128. doi: 10.1091/mbc.E02-05-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopmann R, Cooper JA, Miller KG. Actin organization, bristle morphology, and viability are affected by actin capping protein mutations in Drosophila. J. Cell Biol. 1996;133:1293–1305. doi: 10.1083/jcb.133.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaddoost S, Nam SC, Bhat MA, Bellen HJ, Choi KW. Drosophila Crumbs is a positional cue in photoreceptor adherens junctions and rhabdomeres. Nature. 2002;416:178–183. doi: 10.1038/nature720. [DOI] [PubMed] [Google Scholar]
- Januschke J, Gervais L, Dass S, Kaltschmidt JA, Lopez-Schier H, St Johnston D, Brand AH, Roth S, Guichet A. Polar transport in the Drosophila oocyte requires Dynein and Kinesin I cooperation. Curr. Biol. 2002;12:1971–1981. doi: 10.1016/s0960-9822(02)01302-7. [DOI] [PubMed] [Google Scholar]
- Karki S, Holzbaur EL. Affinity chromatography demonstrates a direct binding between cytoplasmic dynein and the dynactin complex. J. Biol. Chem. 1995;270:28806–28811. doi: 10.1074/jbc.270.48.28806. [DOI] [PubMed] [Google Scholar]
- King SJ, Schroer TA. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat. Cell Biol. 2000;2:20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
- Lee WL, Oberle JR, Cooper JA. The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J. Cell Biol. 2003;160:355–364. doi: 10.1083/jcb.200209022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, White JG. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–836. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- Martin M, Iyadurai SJ, Gassman A, Gindhart JG, Hays TS, Jr, Saxton WM. Cytoplasmic dynein, the dynactin complex, and kinesin are interdependent and essential for fast axonal transport. Mol. Biol. Cell. 1999;10:3717–3728. doi: 10.1091/mbc.10.11.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris NR. Nuclear positioning: the means is at the ends. Curr. Opin. Cell Biol. 2003;15:54–59. doi: 10.1016/s0955-0674(02)00004-2. [DOI] [PubMed] [Google Scholar]
- Mosley-Bishop KL, Li Q, Patterson L, Fischer JA. Molecular analysis of the klarsicht gene and its role in nuclear migration within differentiating cells of the Drosophila eye. Curr. Biol. 1999;9:1211–1220. doi: 10.1016/s0960-9822(99)80501-6. [DOI] [PubMed] [Google Scholar]
- Muresan V, Stankewich MC, Steffen W, Morrow JS, Holzbaur EL, Schnapp BJ. Dynactin-dependent, dynein-driven vesicle transport in the absence of membrane proteins: a role for spectrin and acidic phospholipids. Mol. Cell. 2001;7:173–183. doi: 10.1016/s1097-2765(01)00165-4. [DOI] [PubMed] [Google Scholar]
- Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- O’Neill EM, Rebay I, Tjian R, Rubin GM. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78:137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- Olson EC, Walsh CA. Smooth, rough and upside-down neocortical development. Curr. Opin. Genet. Dev. 2002;12:320–327. doi: 10.1016/s0959-437x(02)00305-2. [DOI] [PubMed] [Google Scholar]
- Patterson K, Molofsky AB, Robinson C, Acosta S, Cater C, Fischer JA. The functions of Klarsicht and nuclear lamin in developmentally regulated nuclear migrations of photoreceptor cells in the Drosophila eye. Mol. Biol. Cell. 2004;15:600–610. doi: 10.1091/mbc.E03-06-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellikka M, Tanentzapf G, Pinto M, Smith C, McGlade CJ, Ready DF, Tepass U. Crumbs, the Drosophila homologue of human CRB1/RP12, is essential for photoreceptor morphogenesis. Nature. 2002;416:143–149. doi: 10.1038/nature721. [DOI] [PubMed] [Google Scholar]
- Pielage J, Stork T, Bunse I, Klambt C. The Drosophila cell survival gene discs lost encodes a cytoplasmic Codanin-1-like protein, not a homolog of tight junction PDZ protein Patj. Dev. Cell. 2003;5:841–851. doi: 10.1016/s1534-5807(03)00358-7. [DOI] [PubMed] [Google Scholar]
- Quintyne NJ, Gill SR, Eckley DM, Crego CL, Compton DA, Schroer TA. Dynactin is required for microtubule anchoring at centrosomes. J. Cell Biol. 1999;147:321–334. doi: 10.1083/jcb.147.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns WB, Caskey CT, Ledbetter DH. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature. 1993;364:717–721. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- Reinsch S, Gonczy P. Mechanisms of nuclear positioning. J. Cell Sci. 1998;111:2283–2295. doi: 10.1242/jcs.111.16.2283. [DOI] [PubMed] [Google Scholar]
- Requena N, Alberti-Segui C, Winzenburg E, Horn C, Schliwa M, Philippsen P, Liese R, Fischer R. Genetic evidence for a microtubule-destabilizing effect of conventional kinesin and analysis of its consequences for the control of nuclear distribution in Aspergillus nidulans. Mol. Microbiol. 2001;42:121–132. doi: 10.1046/j.1365-2958.2001.02609.x. [DOI] [PubMed] [Google Scholar]
- Schafer DA, Gill SR, Cooper JA, Heuser JE, Schroer TA. Ultrastructural analysis of the dynactin complex: an actin-related protein is a component of a filament that resembles F-actin. J. Cell Biol. 1994;126:403–412. doi: 10.1083/jcb.126.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DA, Jennings PB, Cooper JA. Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J. Cell Biol. 1996;135:169–179. doi: 10.1083/jcb.135.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker D, Jackle H, Gaul U. Genetic analysis of the larval optic nerve projection in Drosophila. Development. 1997;124:937–948. doi: 10.1242/dev.124.5.937. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Rubin GM. Transposition of cloned P elements into Drosophila germline chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- Starr DA, Han M. ANChors away: an actin based mechanism of nuclear positioning. J. Cell Sci. 2003;116:211–216. doi: 10.1242/jcs.00248. [DOI] [PubMed] [Google Scholar]
- Strausfeld N, Meinertzhagen IA. The insect neruon: types, morphologies, fine structure, and relationship to the architectonics of the insect nervous system. In: Harrison FW, Locke M, editors. Microscopic Anatomy of Invertebrates. Vol. 11(B) New York: Wiley and Sons; 1998. pp. 487–538. [Google Scholar]
- Swan A, Nguyen T, Suter B. Drosophila Lissencephaly-1 functions with Bic-D and dynein in oocyte determination and nuclear positioning. Nat. Cell Biol. 1999;1:444–449. doi: 10.1038/15680. [DOI] [PubMed] [Google Scholar]
- Tomlinson A. The cellular dynamics of pattern formation in the eye of Drosophila. J. Embryol. Exp. Morphol. 1985;89:313–331. [PubMed] [Google Scholar]
- Vaughan KT, Vallee RB. Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150Glued. J. Cell Biol. 1995;131:1507–1516. doi: 10.1083/jcb.131.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walenta JH, Didier AJ, Liu X, Kramer H. The Golgi-associated hook3 protein is a member of a novel family of microtubule-binding proteins. J. Cell Biol. 2001;152:923–934. doi: 10.1083/jcb.152.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte MA, Gross SP, Postner M, Block SM, Wieschaus EF. Developmental regulation of vesicle transport in Drosophila embryos: forces and kinetics. Cell. 1998;92:547–557. doi: 10.1016/s0092-8674(00)80947-2. [DOI] [PubMed] [Google Scholar]
- Wolff T. Histological techniques for the Drosophila eye. Part II: Adult. In: Sullivan W, Ashburner M, Hawley RS, editors. Drosophila Protocols. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 224–243. [Google Scholar]
- Wolff T, Ready DF. Pattern formation in the Drosophila retina. In: Bate M, Martinez-Arias A, editors. The development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1993. pp. 1277–1325. [Google Scholar]