Abstract
In Arabidopsis thaliana, flowering-time variation exists among accessions, and the winter-annual (late-flowering without vernalization) versus rapid-cycling (early flowering) growth habit is typically determined by allelic variation at FRIGIDA (FRI) and FLOWERING LOCUS C (FLC). FRI upregulates the expression of FLC, a central floral repressor, to levels that inhibit flowering, resulting in the winter-annual habit. Here, we show that FRI promotes histone H3 lysine-4 trimethylation (H3K4me3) in FLC to upregulate its expression. We identified an Arabidopsis homolog of the human WDR5, namely, WDR5a, which is a conserved core component of the human H3K4 methyltransferase complexes called COMPASS-like. We found that recombinant WDR5a binds H3K4-methylated peptides and that WDR5a also directly interacts with an H3K4 methyltransferase, ARABIDOPSIS TRITHORAX1. FRI mediates WDR5a enrichment at the FLC locus, leading to increased H3K4me3 and FLC upregulation. WDR5a enrichment is not required for elevated H3K4me3 in FLC upon loss of function of an FLC repressor, suggesting that two distinct mechanisms underlie elevated H3K4me3 in FLC. Our findings suggest that FRI is involved in the enrichment of a WDR5a-containing COMPASS-like complex at FLC chromatin that methylates H3K4, leading to FLC upregulation and thus the establishment of the winter-annual growth habit.
INTRODUCTION
The timing of the developmental transition from a vegetative to a reproductive phase (i.e., flowering) is crucial to reproductive success in angiosperms. In a given environment, a plant can respond to environmental signals and integrate the responses with its developmental state to flower at a right time. In Arabidopsis thaliana, naturally occurring flowering-time variation exists among wild accessions, and FRIGIDA (FRI) is a major determinant of natural variation in flowering time (Johanson et al., 2000). The winter-annual (late-flowering without vernalization) versus rapid-cycling (early flowering) growth habit is often determined by allelic variation at FRI and FLOWERING LOCUS C (FLC) (Johanson et al., 2000; Gazzani et al., 2003; Michaels et al., 2003). FRI is a plant-specific protein with coiled-coil domains (Johanson et al., 2000), and FLC is a MADS box transcription factor that quantitatively inhibits the floral transition in Arabidopsis (Michaels and Amasino, 1999; Sheldon et al., 1999). Winter annuals typically have dominant alleles of FRI and FLC, whereas rapid-cycling accessions have either a nonfunctional fri allele or a weak flc allele (Johanson et al., 2000; Gazzani et al., 2003; Werner et al., 2005). The role of FRI is to upregulate FLC expression to levels that inhibit flowering, resulting in the winter-annual growth habit (Johanson et al., 2000).
FLC plays a central role in flowering-time regulation in Arabidopsis. In rapid-cycling accessions that lack FRI, autonomous pathway (AP) genes, such as FCA, FPA, FVE, FY, LUMINIDEPENDENS, and FLOWERING LOCUS D (FLD), constitutively repress FLC expression to promote flowering (Baurle and Dean, 2006). The AP gene-mediated repression can be overcome by a functional FRI, and introgression of FRI into a rapid-cycling accession, such as Columbia (Col), converts it into a winter-annual-like line (Lee et al., 1994). In winter annuals, vernalization (a prolonged cold exposure) overrides FRI function to repress FLC expression, leading to acceleration of flowering after the plants return to warm growth conditions (Michaels and Amasino, 1999; Sheldon et al., 1999).
A number of genes required for FLC expression (or upregulation) have been identified, and these genes can be largely classified into two groups based on their effects on FLC expression. One group consists of general transcriptional regulators that are required for FLC expression in both AP mutants and FRI-containing lines. For instance, mutations in EARLY FLOWERING7 (ELF7) (He et al., 2004), VERNALIZATION INDEPENDENCE5 (Oh et al., 2004), PHOTOPERIOD-INDEPENDENT EARLY FLOWERING1 (PIE1) (Noh and Amasino, 2003), and EARLY FLOWERING IN SHORT DAYS (also known as SDG8) (Kim et al., 2005; Zhao et al., 2005) suppress FLC expression and cause early flowering. In addition to FLC, these genes also regulate other loci; thus, mutations in these genes give rise to pleiotropic phenotypes.
The other group of genes appears to be required specifically for FRI-mediated FLC upregulation, including FRI-LIKE1 (FRL1), FRIGIDA-ESSENTIAL1 (FES1), and SUPPRESSOR OF FRIGIDA4 (SUF4); mutations in these genes strongly suppress FRI-mediated FLC upregulation but only moderately suppress elevated FLC expression in AP mutants (Michaels et al., 2004; Schmitz et al., 2005; Kim et al., 2006; Kim and Michaels, 2006). SUF4 directly interacts with the FLC locus, can also interact with FRI in vitro, and may be involved in the recruitment of FRI to the FLC locus (Kim et al., 2006). However, it remains unclear how FRI activates FLC expression.
Recent studies have shown that chromatin modification plays an important role in modulating FLC expression. For instance, FLC expression requires deposition of the histone variant H2A.Z in FLC chromatin by a PIE1-containing complex whose known components include ACTIN-RELATED PROTEIN6 (also known as SUF3 and ESD1) and SERRATED LEAVES AND EARLY FLOWERING (also known as SWC6) (Choi et al., 2005, 2007; Deal et al., 2005, 2007; Martin-Trillo et al., 2006; March-Diaz et al., 2007). The AP genes FLD, FCA, FPA, and FVE are involved in repressive histone modifications in FLC and repress its expression. FLD, a plant homolog of the human Lysine-Specific Demethylase1 (LSD1) that has been found in histone deacetylase (HDAC) corepressor complexes (Shi et al., 2004; Lee et al., 2006), is involved in H3K4 demethylation and histone deacetylation in FLC (He et al., 2003; Jiang et al., 2007; Liu et al., 2007). Both FCA and FPA, encoding putative RNA-Recognition Motif-type RNA binding proteins, largely act through FLD to repress FLC expression (Liu et al., 2007; Baurle and Dean, 2008). FVE, a retinoblastoma-associated protein, is partly involved in histone deacetylation of FLC chromatin (Ausin et al., 2004). The Polycomb-repressive complex 2 subunit CURLEY LEAF (Schubert et al., 2006) directly interacts with FLC chromatin and mediates deposition of repressive H3 Lys-27 trimethylation in FLC to repress its expression (Jiang et al., 2008). In addition, histone H4 dimethylation at Arg 3 (H4R3) in FLC by Type I and Type II Arg methyltransferases is also associated with FLC repression (Niu et al., 2007; Pei et al., 2007; Wang et al., 2007). Recent studies also reveal that vernalization leads to repressive histone modifications in FLC, such as increased di- and trimethylation of histone H3 at Lys-9 and at Lys-27 and H4R3 dimethylation, to repress FLC expression (reviewed in Sung and Amasino, 2005; Baurle and Dean, 2006; He, 2009).
Histone H3K4 methylation plays an important role in regulating transcription in eukaryotes and is dynamically regulated by H3K4 methyltransferases and demethylases. The ε-amino group of H3K4 residues can be mono-, di-, and trimethylated (Dou et al., 2006; Shilatifard, 2008). H3 lysine-4 trimethylation (H3K4me3) is closely coupled with active gene expression, and the trimethyl H3K4 mark can be recognized by the evolutionarily conserved ATP-dependent chromatin remodeling machines, such as CHD1 and NURF in human, which remodel target gene chromatin leading to transcriptional activation (reviewed in Ruthenburg et al., 2007). In the well-studied Saccharomyces cerevisiae, H3K4 methylation is catalyzed by COMPASS (for Complex Proteins Associated with Set1), which contains an H3K4 methyltransferase known as Set1 (Miller et al., 2001). COMPASS-like complexes have been identified in human, including the hSET1 complex and the MLL1 complex, and are capable of catalyzing H3K4 methylation and activating target gene expression (Shilatifard, 2008). All these complexes contain four evolutionarily conserved core components, namely, a relative of the yeast Set1 and three structural components, including Ash2, RbBP5, and WDR5 (Shilatifard, 2008), and an in vitro reconstituted core complex composed of these four components has H3K4-specific methyltransferase activity (Dou et al., 2006).
It has been shown that levels of H3K4me3 are increased in actively transcribed FLC chromatin (He et al., 2004; Pien et al., 2008). An ELF7-containing complex known as PAF1c is required for FLC upregulation and for the associated H3K4me3 increase in FLC in the FRI background or AP mutants (He et al., 2004; Oh et al., 2004). Furthermore, ATX1, an H3K4 methyltransferase and a homolog of the Drosophila melanogaster TRITHORAX and the yeast Set1 (Alvarez-Venegas et al., 2003), is also required for H3K4me3 in FLC, and the atx1 mutation moderately suppresses FLC expression in the FRI background (Pien et al., 2008). In addition, ATX2 (for ARABIDOPSIS TRITHORAX2), a homolog of ATX1, is also involved in FLC regulation because the atx1 atx2 double mutation strongly suppresses FLC expression in the FRI background (Pien et al., 2008).
Although increased H3K4me3 has been shown to be associated with FLC chromatin in the FRI background, this increase is also associated with elevated FLC expression in AP mutants (He et al., 2004; Kim et al., 2005). Hence, the role of FRI in the H3K4 methylation of FLC chromatin is yet to be determined. In addition, little is known on how H3K4me3 is deposited at FLC and other loci. Furthermore, although recent studies have shown that the AP genes, such as FLD and FVE, are involved in H3K4 demethylation and deacetylation of FLC chromatin, it is essentially unknown how FRI overcomes these repressive modifications to upregulate FLC expression.
Here, we show that WDR5a, a homolog of a structural component of the human COMPASS-like complexes, binds histone H3 tails and also interacts with the ATX1 H3K4 methyltransferase. FRI mediates WDR5a enrichment at the FLC locus, resulting in an increase in H3K4me3, which leads to FLC upregulation to inhibit flowering. Furthermore, we found that FRI does not disrupt the recruitment of an FLC repressor, FLD, to the FLC locus, but may compromise FLD-mediated H3K4 demethylation to overcome the FLC repression mediated by the AP genes FLD, FCA, and FPA.
RESULTS
WDR5a, an Arabidopsis Homolog of the Human WDR5, Represses the Floral Transition
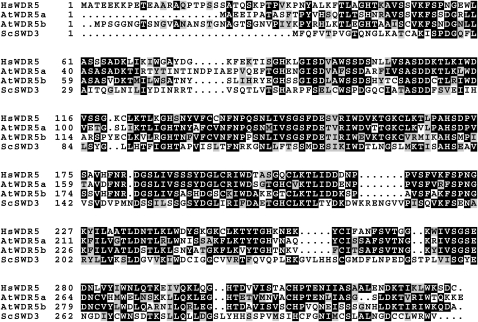
In an effort to identify the Arabidopsis homologs of core components of the human COMPASS-like complexes, we found that there are two Arabidopsis homologs of the human WDR5, namely, At WDR5a and At WDR5b (Figure 1). The amino acid sequence identity between WDR5a and the human WDR5 over the entire WDR5a is 63%, and the identity between WDR5b and the human WDR5 over the entire WDR5b is 58%.
Figure 1.
Amino Acid Sequence Alignment of Arabidopsis WDR5a (At WDR5a) and WDR5b (At WDR5b) with S. cerevisiae SWD3 (Sc SWD3) and Homo sapiens WDR5 (Hs WDR5).
Numbers refer to amino acid residues. Identical residues are shaded black, and similar residues are shaded gray.
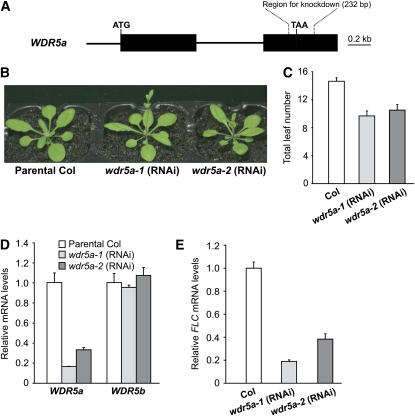
We sought to address biological functions of these two genes. First, we identified a loss-of-function mutant of WDR5b with a T-DNA insertion in its coding region; however, no obvious phenotypes were observed in wdr5b mutants (see Supplemental Figure 1 online). As no wdr5a mutants were identified, a double-stranded RNA interference (RNAi) approach using a WDR5a-specific fragment with no homology to WDR5b (Figure 2A), was employed to specifically knock down WDR5a expression. Two independent homozygous transgenic lines with a single T-DNA locus, wdr5a-1 (RNAi) and wdr5a-2 (RNAi), were created. These lines grown in long days developed normally except that they flowered earlier than the parental Col (Figures 2B and 2C). We further quantified transcript levels of WDR5a and WDR5b in these two lines and found that WDR5a expression was greatly reduced, whereas levels of WDR5b transcripts remained unchanged in both lines compared with Col (Figure 2D), indicating that the RNAi specifically knocks down only WDR5a expression. In addition, we characterized another seven independent wdr5a (RNAi) lines and found that all of them flowered earlier than Col (see Supplemental Table 1 online). Together, these data show that WDR5a represses the floral transition.
Figure 2.
WDR5a Represses the Floral Transition in Arabidopsis.
(A) WDR5a gene structure. Exons are represented by filled boxes; the start and stop codons are marked as ATG and TAA, respectively. The 232-bp region used to knock down WDR5a expression is indicated by broken lines.
(B) wdr5a (RNAi) lines grown in long days. Col is the parental accession used in the RNAi-mediated WDR5a suppression.
(C) Flowering times of wdr5a (RNAi) lines grown in long days. The total number of primary rosette and cauline leaves at flowering was counted, and for each line, 16 plants were scored. The values shown are means ± sd.
(D) Relative mRNA levels of WDR5a and WDR5b in seedlings of wdr5a (RNAi) lines quantified by real-time PCR. Relative expression to parental Col is presented, with sd for three quantitative PCR replicates ([D] and [E]).
(E) Relative FLC mRNA levels in seedlings of wdr5a (RNAi) lines quantified by real-time PCR.
[See online article for color version of this figure.]
WDR5a Promotes the Expression of FLC and an FLC Homolog
FLC is a central floral repressor in Arabidopsis. We examined whether WDR5a promotes FLC expression to repress flowering (note that FLC is expressed at a low level in Col, a rapid-cycling accession) and found that FLC transcript levels in wdr5a (RNAi) lines were strongly reduced compared with Col (Figure 2E). Hence, WDR5a indeed upregulates FLC expression to delay flowering. Recent studies have shown that FLC homologs, including FLOWERING LOCUS M (FLM) (Scortecci et al., 2001), MADS BOX AFFECTING FLOWERING2 (MAF2), and MAF4 (Ratcliffe et al., 2003; Gu et al., 2009), moderately repress Arabidopsis flowering. We examined the expression of these three genes in wdr5a-1 and -2 (RNAi) lines and found that MAF4 transcript levels were reduced, whereas FLM and MAF2 were expressed in both lines at levels similar to those in Col (see Supplemental Figure 2 online). Thus, WDR5a upregulates the expression of the floral repressors FLC and MAF4. Interestingly, these two genes are still expressed at very low levels in both RNAi lines, which most likely is due to low residual levels of WDR5a.
As described earlier, recent studies have identified a number of genes required for FLC expression. We examined the expression of a few of these FLC regulators in wdr5a (RNAi) lines, including ATX1, PIE1, ELF7, FRL1, FES1, and SUF4. None of these genes was affected by WDR5a knockdown (see Supplemental Figure 3A online), indicating that WDR5a may directly promote FLC expression.
WDR5a Knockdown Specifically Suppresses FRI-Mediated FLC Upregulation but Not FLC Activation upon Loss of FLD Activity
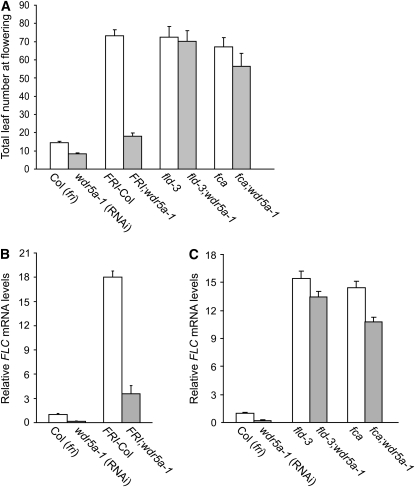
FLC is upregulated by FRI and repressed by the AP genes; thus, either the presence of a functional FRI or a mutation in an AP gene causes delayed flowering due to elevated FLC expression. To evaluate the genetic interaction of FRI with WDR5a suppression, a functional FRI from FRI-Col (Lee et al., 1994) was introduced into wdr5a-1 and -2 (RNAi) lines. The late-flowering phenotypes conferred by FRI were strongly suppressed by WDR5a knockdown (Figure 3A; see Supplemental Figure 4 online). To evaluate the effect of WDR5a knockdown on FLC expression in the fld mutant, fld was introduced into the wdr5a (RNAi) lines. Interestingly, the late-flowering phenotypes of fld were not suppressed by WDR5a knockdown (Figure 3A; see Supplemental Figures 3B and 4 online). Recently, it has been shown that another AP gene FCA mainly acts through FLD to repress FLC expression (Liu et al., 2007). We introduced fca into the wdr5a-1 line and found that the late-flowering phenotypes of fca mutants were only slightly suppressed by WDR5a knockdown (Figure 3A).
Figure 3.
Effect of WDR5a Knockdown on FLC-Dependent Late Flowering.
(A) Flowering times of the indicated genotypes grown in long days. The total number of primary rosette and cauline leaves at flowering was scored, and 10 to 15 plants were counted for each line. The values shown are means ± sd.
(B) Relative FLC mRNA levels in seedlings of FRI-Col and FRI;wdr5a-1 quantified by real-time PCR. Relative expression to Col is presented, with sd for three quantitative PCR replicates ([B] and [C]).
(C) Relative FLC mRNA levels in seedlings of fld, fld;wdr5a-1, fca, and fca;wdr5a-1 quantified by real-time PCR.
Consistent with the flowering phenotypes, elevated FLC expression in fld mutants was only slightly reduced, whereas the transcript levels of FLC in the FRI background were greatly reduced by WDR5a knockdown (Figures 3B and 3C). In addition, FLC expression in fca mutants was slightly suppressed by WDR5a knockdown (Figure 3C). To rule out the possibility of that WDR5a knockdown might suppress FRI expression, we examined FRI expression in FRI;wdr5a-1 and found that it was not affected by WDR5a knockdown (see Supplemental Figure 3C online). Together, these data show that WDR5a knockdown specifically suppresses FRI-mediated FLC upregulation, but not elevated FLC expression in fld or fca mutants. Thus, WDR5a is required for FLC upregulation by FRI.
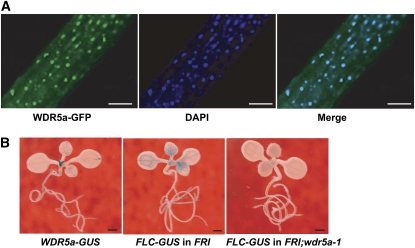
WDR5a Is Localized in the Nucleus and Preferentially Expressed in Shoot and Root Apical Regions and Vasculature
WDR5a, like the human WDR5, may act to activate target-gene expression. Consistent with its role as a transcriptional activator, the WDR5a fusion protein with green fluorescent protein (GFP) was specifically localized to the nucleus (Figure 4A). To examine the spatial expression pattern of WDR5a, we fused the 5′ promoter plus part of the coding region of WDR5a with the reporter gene GUS (for β-GLUCURONIDASE). WDR5a was preferentially expressed in shoot and root apical regions in seedlings, which are enriched with dividing cells, and was also expressed in vasculature (Figure 4B). This pattern is nearly identical to that of FLC-GUS (He et al., 2003) in the FRI background (Figure 4B). WDR5a knockdown nearly eliminated FLC-GUS expression in shoot apical regions and leaf vasculature in the FRI background (Figure 4B), consistent with the notion that WDR5a functions as an activator that mediates FLC upregulation by FRI.
Figure 4.
Cellular Localization of WDR5a and the Expression Pattern of WDR5a.
(A) Nuclear localization of the WDR5a-GFP fusion protein in roots of transgenic Arabidopsis seedlings. The blue 4',6-diamidino-2-phenylindole (DAPI) staining indicates nuclei. WDR5a-GFP and DAPI fluorescence was imaged using a laser scanning confocal microscope. Bars = 50 μm.
(B) Spatial expression patterns of the GUS reporter gene translationally fused to FLC or WDR5a in seedlings. The lines containing FLC-GUS carry a null flc allele. Bars = 1.0 mm.
Recombinant WDR5a Binds K4-Methylated Histone H3 Peptides
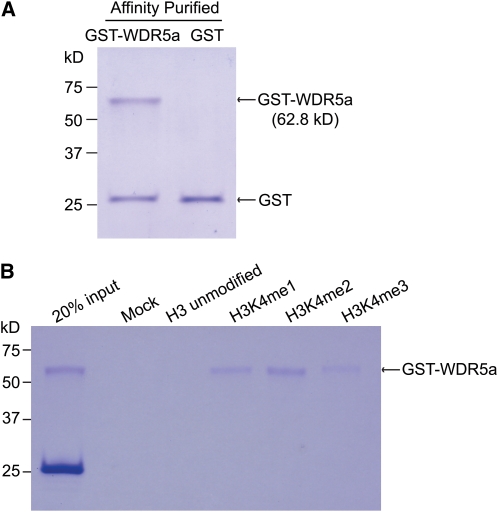
The human WDR5 recognizes and binds to the histone H3 N-terminal tail (Wysocka et al., 2005; Ruthenburg et al., 2006) and presents the H3K4 side chain for processive methylation: from unmodified K4 to mono- to di- to trimethylated form as WDR5 knockdown causes a strong reduction in mono-, di-, and trimethyl H3K4 in human cells (Wysocka et al., 2005; Dou et al., 2006; Ruthenburg et al., 2006). It was of interest to determine whether WDR5a may directly interact with the K4-methylated H3 tails. First, glutathione S-transferase (GST)-tagged WDR5a was expressed in Escherichia coli and purified by affinity purification (Figure 5A). Next, we performed H3 peptide pull down assays using GST-WDR5a. Like the human WDR5 (Wysocka et al., 2005), the WDR5a fusion protein was enriched in the K4-mono-, di-, or trimethylated peptide pulldown with a stronger association with the K4-dimethylated H3 peptides (Figure 5B). Thus, WDR5a can recognize and bind K4-methylated H3 tails.
Figure 5.
Histone H3 Peptide Pull-Down Assays Using a Recombinant WDR5a.
(A) Affinity-purified GST and GST-WDR5a fusion protein from E. coli. Proteins were analyzed by SDS-PAGE and Coomassie blue staining. The lower band in the left lane is a degradation product of GST-WDR5a. Molecular mass markers are indicated on the left.
(B) Peptide pull-down assays with GST-WDR5a and H3 peptides. A mixture of ∼10-fold excess of GST with GST-WDR5a was incubated with 5.0 μg of each peptide. Proteins bound to the peptide resins were eluted and analyzed by SDS-PAGE and Coomassie blue staining. The mock is a control without any peptides.
FRI Mediates WDR5a Enrichment at the FLC Locus
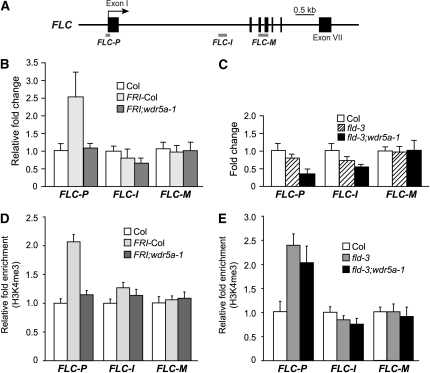
To investigate whether WDR5a bound to FLC chromatin, we performed chromatin immunoprecipitation (ChIP) using the human anti-WDR5 antibody, which recognizes both WDR5a and WDR5b (see Supplemental Figure 5 online). Previously, it has been shown that in actively transcribed FLC chromatin, levels of H3K4me3 increase in the region around the transcription start site (TSS) (FLC-P; as shown in Figure 6A) but not in the 3′ region of Intron I of FLC (FLC-I) or the middle of FLC (FLC-M) (He et al., 2004; Saleh et al., 2008a). We first quantified amounts of the immunoprecipitated FLC fragments from Col and wdr5a-1 (RNAi) seedlings and found that WDR5a knockdown led to a reduction in WDR5a binding to the FLC-P region, whereas the amounts of FLC-I and FLC-M fragments in wdr5a-1 were similar to those in Col (see Supplemental Figure 6 online). These data suggest that WDR5a directly interacts with the FLC locus.
Figure 6.
Role of WDR5a in FLC activation.
(A) Schematic structure of genomic FLC and the regions examined by ChIP. The arrow indicates the transcription start site; filled boxes represent exons.
(B) WDR5a enrichment at the FLC locus in presence of FRI. The amounts of FLC fragments immunoprecipitated from seedlings were quantified by real-time PCR and subsequently normalized to an internal control (TUBLIN2 [TUB2]). The fold enrichments of FRI-Col and FRI;wdr5a-1 over Col are shown. Data in the graphs are average values from two ChIP experiments (each quantified in triplicate), and error bars represent sd ([B] to [E]).
(C) Loss of FLD function does not cause WDR5a enrichment at the FLC locus.
(D) Relative levels of trimethyl H3K4 in FLC chromatin in Col, FRI-Col, and FRI;wdr5a-1 seedlings determined by real-time quantitative PCR. The amounts of DNA fragments after ChIP were quantified and subsequently normalized to an internal control (TUB2). The fold changes of FRI-Col and FRI;wdr5a-1 over Col at the indicated regions are shown.
(E) Relative levels of trimethyl H3K4 in FLC chromatin in Col, fld-3, and fld-3;wdr5a-1 seedlings. The fold changes of fld-3 and fld-3;wdr5a-1 over Col at the indicated regions are shown.
Second, we investigated whether a functional FRI would mediate WDR5a enrichment at FLC using ChIP. Indeed, we found that WDR5a was enriched in the region around TSS (FLC-P) but not in FLC-I or FLC-M in the presence of FRI (Figure 6B). Furthermore, WDR5a knockdown eliminates the WDR5a enrichment at FLC-P (Figure 6B). Together, these data show that FRI mediates WDR5a enrichment at the FLC-P region, consistent with the H3K4me3 enrichment in this region in the presence of FRI (He et al., 2004) (also see Figure 6D).
Third, we examined whether in fld mutants WDR5a binding to FLC chromatin was also increased but found that WDR5a was not enriched in FLC-P, FLC-I, or FLC-M in fld relative to wild-type Col (Figure 6C). Hence, WDR5a enrichment is not associated with elevated FLC expression upon loss of FLD activity. Interestingly, WDR5a knockdown led to a moderate reduction in WDR5a binding to FLC-P in fld;wdr5a-1 relative to fld and Col (Figure 6C), which may contribute to a slight reduction in FLC expression in fld mutants as shown in Figure 3C.
WDR5a Enrichment at FLC Is Required Specifically for Elevated H3K4me3 in the Presence of FRI
To investigate the effect of WDR5a knockdown on the H3K4me3 state in FLC in the FRI background, we performed ChIP using anti-H3K4me3. Consistent with the previous findings (He et al., 2004), H3K4me3 was enriched in the region around TSS in the FRI background. Furthermore, we found that WDR5a knockdown eliminated this H3K4me3 enrichment in FLC-P (Figure 6D); hence, this enrichment mediated by a functional FRI is WDR5a dependent.
We further examined the H3K4me3 state of FLC chromatin in fld mutants upon WDR5a knockdown. Consistent with our previous findings (He et al., 2004; Jiang et al., 2007), H3K4me3 was enriched in the FLC-P region but not in FLC-I in fld relative to Col (Figure 6E). Recent studies show that dimethylated H3K4 is enriched in the middle of FLC (FLC-M) upon loss of FLD activity (Liu et al., 2007). Interestingly, the levels of H3K4me3 in FLC-M were not increased in fld relative to Col (Figure 6E).
Next, we compared the levels of H3K4me3 in FLC-P in fld and fld;wdr5a-1 seedlings. In contrast with the strong reduction in H3K4me3 in the FRI background upon WDR5 knockdown (Figure 6D), the level of H3K4me3 in FLC-P in fld;wdr5a-1 was close to that in fld (Figure 6E); hence, WDR5a suppression has little effect on H3K4me3 in FLC chromatin upon loss of FLD activity. Together, these data show that WDR5a enrichment at FLC chromatin is required specifically for the H3K4me3 increase in the presence of FRI.
WDR5a Interacts with the ATX1 Histone H3K4 Methyltransferase
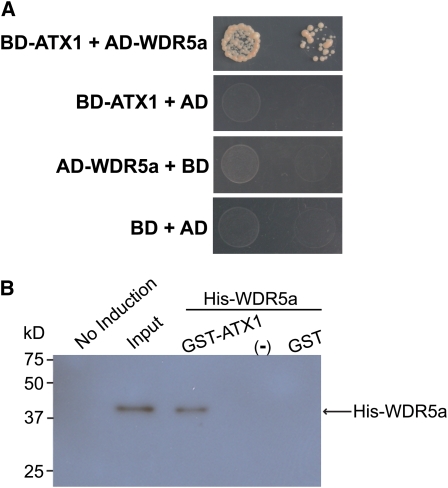
As noted above, WDR5a enrichment at FLC is required for FRI-mediated increase of H3K4me3 in FLC chromatin. WDR5a is expected to act in the context of an H3K4 methyltransferase complex to promote FLC expression. Recent studies show that ATX1, an H3K4 methyltransferase (Alvarez-Venegas et al., 2003), is required for H3K4me3 in FLC (Pien et al., 2008). We sought to address whether WDR5a and ATX1 act as part of a complex that catalyzes H3K4me3. First, yeast two-hybrid assays were performed using the full-length WDR5a and ATX1 proteins; we found that these two proteins interacted strongly in yeast (Figure 7A). Next, we performed GST-ATX1 pull-down experiments using protein extracts from E. coli expressing GST-ATX1 or recombinant WDR5a (see Supplemental Figure 7 online) and found that GST-ATX1, but not GST, could effectively bind WDR5a (Figure 7B). Together, these data suggest that WDR5a and ATX1 may act in a complex to mediate H3K4 methylation.
Figure 7.
WDR5a Interacts with the ATX1 Histone H3K4 Methyltransferase.
(A) Interaction of WDR5a with ATX1 in yeast. Full-length WDR5a and ATX1 proteins were fused to GAL4 activation (AD) and DNA binding domains (BD), respectively. Yeast strains harboring these fusion constructs and/or empty vectors, as indicated, were grown on a selective medium lacking histidine and adenine.
(B) GST-ATX1 pull-down assays. An equal amount of the bacteria extract containing GST or GST-ATX1, or noninduced E. coli extract indicated as −, was incubated with the extract containing His-WDR5a. Proteins were recovered using glutathione-linked resins and analyzed by immunoblotting with anti-WDR5. Input corresponds to ∼3% of the His-WDR5a extract used in the pull-down assays.
Effect of FRI and Loss of FLD Function on FLC Upregulation
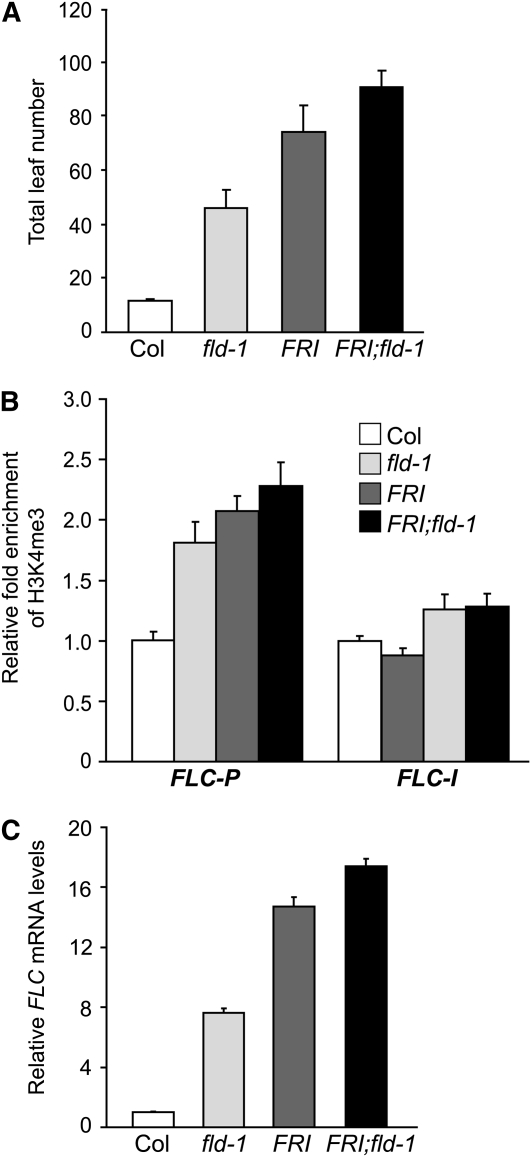
FRI can override FLD-mediated FLC repression to upregulate FLC expression in winter annuals. We sought to address how FRI might interfere with FLD function. We first investigated the genetic interaction of FRI with fld. A functional FRI was introduced into an fld mutant. FRI;fld flowered later than either fld or FRI-Col (Figure 8A). Furthermore, we examined the H3K4me3 state of FLC chromatin in FRI-Col, fld, and FRI;fld seedlings using ChIP. The levels of H3K4me3 in FRI;fld were higher than those in either FRI-Col or fld (Figure 8B). In addition, we examined FLC expression in these lines and found that the transcript levels of FLC in FRI;fld were higher than those in either FRI-Col or fld (Figure 8C), consistent with the H3K4me3 levels in these lines. Hence, the effect of FRI and loss of FLD function on the H3K4me3 state and FLC upregulation appears partially additive.
Figure 8.
Effect of FRI and Loss of FLD Activity on H3K4me3 in FLC Chromatin and FLC Upregulation.
(A) Flowering times of Col, fld-1 (a weak allele), FRI-Col, and FRI;fld-1 grown in long days. The total number of primary rosette and cauline leaves at flowering was scored, and 13 to 15 plants for each line were counted. The values shown are means ± sd.
(B) Relative levels of trimethyl H3K4 in FLC chromatin in Col, fld-1, FRI-Col, and FRI;fld-1seedlings. The fold changes of the indicated genotypes over Col at the indicated regions are shown (Col and FRI-Col as described in Figure 6D). Data in the graphs are average values from two ChIP experiments (each quantified in triplicate), and error bars represent sd. The examined regions are as illustrated in Figure 6A. (C) Relative FLC mRNA levels in seedlings of Col, fld-1, FRI-Col, and FRI;fld-1quantified by real-time PCR. Relative expression to Col is presented, with sd for three quantitative PCR replicates.
FRI Does Not Disrupt the Recruitment of FLD to FLC Chromatin but May Compromise FLD Function
Although it has been shown that FLD is an FLC repressor, it was unknown whether FLD acted directly on the FLC locus or indirectly. To examine the interaction of FLD with the FLC locus, we performed ChIP assays using a transgenic line expressing a functional myc-tagged FLD (Figure 9A). FLD was enriched in the region around TSS of FLC but not in the 3′ region of Intron I or the middle of FLC (Figure 9B). Thus, FLD interacts with FLC chromatin to mediate H3K4 demethylation in FLC.
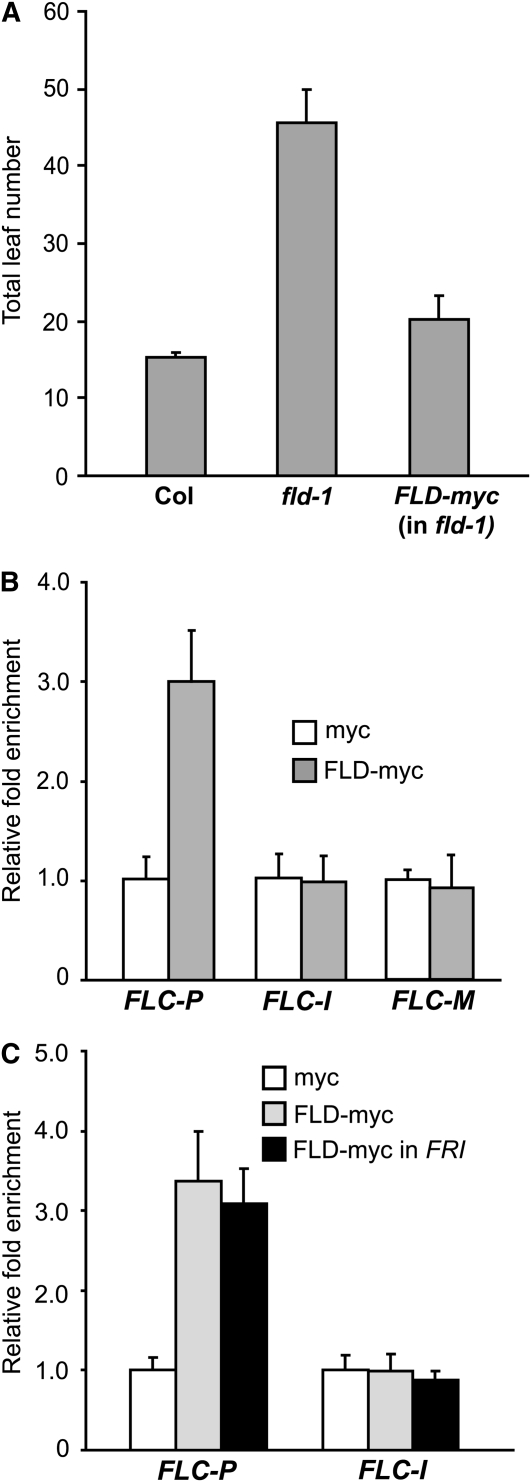
Figure 9.
Recruitment of FLD to the FLC Locus.
(A) FLD-myc rescues the late-flowering phenotypes of fld-1 mutants grown in long days. The total number of primary rosette and cauline leaves at flowering was scored, and 7 to 10 plants were counted for each line.
(B) Recruitment of FLD to the FLC locus. The amounts of FLC fragments immunoprecipitated from seedlings of an fld-1 mutant line expressing a functional FLD-myc (labeled as FLD-myc) and a transgenic Col line expressing 35S Promoter-myc (labeled as myc) were quantified by real-time PCR and subsequently normalized to an internal control (TUB2). The fold enrichments of FLD-myc over the control (myc) are shown. Data in the graphs are average values from two ChIP experiments (each quantified in triplicate), and error bars are sd ([B] and [C]). The examined regions are as illustrated in Figure 6A.
(C) Recruitment of FLD to the FLC locus in the presence of FRI. The amounts of DNA fragments immunoprecipitated from seedlings of myc, FLD-myc, and a FRI/-;fld-1 line expressing a functional FLD-myc (labeled as FLD-myc in FRI) were quantified by real-time PCR and subsequently normalized to TUB2. The fold enrichments of the indicated genotypes over the control (myc) are shown.
Next, we investigated the effect of FRI on FLD binding to FLC chromatin and, surprisingly, found that in the presence of FRI, FLD still bound to FLC chromatin and that levels of FLD recruited to the FLC locus were similar with and without FRI (Figure 9C). Thus, FRI does not disrupt the recruitment of FLD to FLC chromatin. The FLD protein at FLC is at least partially functional because loss of FLD activity gives rise to a moderate increase of H3K4me3 in FLC in the FRI background (Figure 8B). Interestingly, FLD function appears compromised in the presences of FRI as indicated by the less than predicted fully additive effect of FRI and fld on H3K4me3 in FLC.
DISCUSSION
In this study, we show that FRI mediates the enrichment of WDR5a at FLC chromatin and that WDR5a can bind the K4-methylated H3 tails and directly interact with the ATX1 H3K4 methyltransferase. The enrichment of WDR5a causes elevated H3K4me3 in FLC and FLC upregulation. In rapid-cycling accessions that lack FRI, an AP component, FLD, is recruited to the FLC locus to mediate H3K4 demethylation, and loss of FLD activity causes an increase in H3K4me3 that does not require WDR5a enrichment at FLC chromatin. FRI does not disrupt the recruitment of FLD to the FLC locus but may compromise FLD function. Our findings suggest that FRI is involved in the enrichment of a WDR5a-containing COMPASS-like complex at the FLC locus that methylates H3K4, leading to FLC upregulation and thus the establishment of winter-annual growth habit.
Two Distinct Mechanisms Underlie Elevated H3K4me3 in FLC Chromatin
Previously, it has been shown that elevated H3K4me3 in FLC chromatin is associated with FLC upregulation in the FRI background or AP mutants, such as fld and fca, and requires the PAF1c complex (He et al., 2004; Kim et al., 2005); however, the underlying mechanisms are unclear. The findings in this study suggest that there are two distinct mechanisms by which H3K4me3 can be elevated at the FLC locus. In the first mechanism, FRI mediates WDR5a enrichment followed by H3K4me3 enrichment at the FLC locus, particularly in the region around the transcription start site, part of which is required for elevated FLC expression because an 80-bp deletion in the 5′ untranslated region of the FLC-P region suppresses FLC expression (He et al., 2004). The H3K4me3 enrichment in the presence of FRI requires WDR5a. In the second mechanism, loss of FLD activity disrupts H3K4 demethylation, resulting in H3K4me3 enrichment in FLC chromatin. This does not require WDR5a enrichment at the FLC locus. Therefore, both WDR5a enrichment-dependent H3K4 methylation and disruption of H3K4 demethylation can cause elevated H3K4me3 in FLC chromatin.
FLD-Mediated H3K4 Demethylation in FLC Chromatin
FLD is a plant homolog of the human LSD1 that has been found in HDAC corepressor complexes and demethylates mono- and dimethyl H3K4 (Shi et al., 2004; Lee et al., 2006). Upon loss of FLD activity, H3K4me3, and to a lesser degree, H3K4 dimethylation in FLC chromatin increase (He et al., 2004; Jiang et al., 2007; Liu et al., 2007). The increase in H3K4me3 may be due to an increased availability of dimethyl H3K4 to an H3K4-methyltransferase complex (also see discussion below); on the other hand, the reduction of H3K4me3 in the presence of FLD may be attributed to a decrease in the dimethyl K4 level.
There are several lines of evidence to suggest that FLD functions as an H3K4 demethylase. First, a point mutation that changes a Pro into a Leu in the conserved FAD (for flavin adenine dinucleotide; a cofactor for catalysis) binding subdomain in the FLD protein eliminates its function (Chen et al., 2006; Liu et al., 2007). In addition, we noticed that the purified FLD protein from E. coli, like the purified LSD1, was light yellow, which is characteristics of FAD binding proteins (Shi et al., 2004). Second, recent studies show that LSD1 LIKE1 (LDL1), another Arabidopsis homolog of LSD1, specifically demethylates H3K4 and that LDL1 mediates H3K4 demethylation in FLC and acts in partial redundancy with FLD to repress FLC expression (Jiang et al., 2007; Spedaletti et al., 2008). Taken together, these findings suggest that FLD and LDL1 function as H3K4 demethylases to repress FLC repression.
A Putative WDR5a-Containing H3K4 Methyltransferase Complex Involved in FRI-Mediated H3K4me3 in FLC
The yeast COMPASS complex and the human COMPASS-like complexes all contain four evolutionarily conserved core components (Shilatifard, 2008). Besides WDR5a and WDR5b, Arabidopsis also has homologs of the other three core components of the COMPASS-like complexes, including seven homologs of the yeast Set1 H3K4 methyltransferase (two of these homologs are ATX1 and ATX2) (Alvarez-Venegas et al., 2003; Springer et al., 2003), a single homolog of RbBP5, and a single homolog of Ash2 (data not shown), suggesting that COMPASS-like complexes are evolutionarily conserved in Arabidopsis.
Recent studies show that both ATX1 and ATX2 are involved in FLC regulation (Pien et al., 2008). ATX1, an H3K4 methyltransferase, is directly involved in H3K4me3 in FLC chromatin and required for FLC upregulation in the presence of FRI (Pien et al., 2008), whereas the biochemical role of ATX2 in FLC regulation is unclear, as its biochemical function is distinct from ATX1 (Saleh et al., 2008b). We have found that WDR5a interacts with ATX1, suggesting that these proteins may act in a complex. In addition, the spatial expression patterns of WDR5a, ATX1, and FLC overlap. It has been shown that like FLC, the GUS reporter driven by the ATX1 promoter, can be readily detected in vasculature and shoot and root apical regions in seedlings (Saleh et al., 2008b). We have found that like FLC, WDR5a is preferentially expressed in vasculature and shoot and root apical regions in seedlings. Together, these findings suggest that ATX1 is part of a WDR5a-containing complex that is enriched at the FLC locus by a functional FRI to methylate H3K4, leading to FLC activation.
The human WDR5 is an essential structural component of the COMPASS-like H3K4 methyltransferase complexes. It recognizes and binds H3 tails as revealed by peptide pulldown and peptide binding assays and functions to present H3K4 for methylation by an H3K4 methyltransferase (Wysocka et al., 2005; Dou et al., 2006; Ruthenburg et al., 2006). We have found that WDR5a can bind to K4-methylated H3 peptides with a stronger association with H3K4me2. Although the recombinant WDR5a was not enriched in the K4-unmodified H3 peptide pulldown (Figure 5B), it is likely that WDR5a may recognize and bind unmodified H3K4 peptides at a low affinity. Given the close homology and functional similarity of WDR5a and the human WDR5, it is likely that WDR5a may function to present the H3K4 side chain for methylation in target loci.
H3K4 Methylation in fld;wdr5a (RNAi) Mutants
Levels of methylated H3K4 are dynamically regulated by H3K4 methyltransferases and demethylases. Prior to being demethylated, the K4 residues must be methylated first; hence, although disruption of H3K4 demethylation at the FLC locus upon loss of FLD activity causes an increase in methylated K4 residues, it is expected that at least a low level of H3K4 methylation would still be required for the increase in H3K4me3 in fld mutants. Interestingly, knockdown or suppression of WDR5a-dependent H3K4 methylation in FLC has little effect on the H3K4 trimethylation of FLC chromatin in fld mutants. One possibility is that the elevated H3K4 trimethylation upon loss of FLD activity is WDR5a independent. A second possibility is that in the event of WDR5a knockdown, the remaining WDR5a proteins (see Supplemental Figure 5 online) may act in the context of an ATX1-containing COMPASS-like complex to processively convert mono- and dimethyl H3K4 to trimethyl H3K4 in fld mutants. As noted above, loss of FLD activity is expected to give rise to an increase in both mono- and dimethyl H3K4, and the increased substrate availability to the H3K4 methyltransferase complex at the FLC locus in fld mutants may compensate for the reduced complex levels upon WDR5a knockdown.
FRI-Mediated WDR5a Enrichment at the FLC Locus
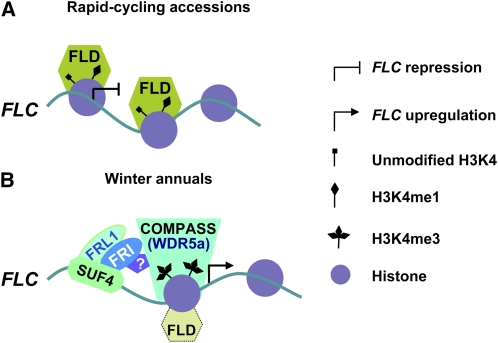
Although both FRI and loss of FLD activity lead to elevated H3K4me3 in FLC, we have found that WDR5a is enriched at the FLC locus only in the presence of FRI. As noted above, WDR5a and ATX1 may act as part of a COMPASS-like complex to methylate H3K4 in FLC. Now the question is how a functional FRI mediates the enrichment of a WDR5a-containing complex at the FLC locus. Recent studies show that SUF4, a zinc finger protein, directly binds the FLC promoter and that SUF4 also can interact with FRI and an FRI homolog, FRL1, and may be involved in the recruitment of FRI to the FLC locus (Kim et al., 2006; Kim and Michaels, 2006). As illustrated in Figure 10, we speculate that the FRI proteins localized in the FLC locus via SUF4 may promote the recruitment of a WDR5a- and ATX1-containing COMPASS-like complex to this locus, leading to elevated H3K4me3 and thus FLC upregulation.
Figure 10.
Model for Regulation of FLC Expression by FRI and FLD.
(A) In rapid-cycling accessions, FLD mediates repressive histone modifications to repress FLC expression and thus accelerate flowering. FLD (or a putative FLD-containing HDAC corepressor complex) is recruited to the FLC locus and mediates demethylation of mono- and dimethyl H3K4 and deacetylation of core histone tails to establish a repressive chromatin environment.
(B) FRI mediates WDR5a enrichment followed by elevated H3K4me3 in FLC to upregulate FLC expression resulting in the winter-annual growth habit. In winter annuals, SUF4 recognizes and binds to the FLC promoter and subsequently recruits FRI and FRL1 to the FLC locus. FRI may promote the recruitment of a WDR5a-containing COMPASS-like H3K4 methyltransferase complex to FLC chromatin to catalyze H3K4 methylation and may also cause a partial disruption of the function of FLD or an FLD complex, resulting in FLC upregulation. FRL1 and/or other cofactors may be involved in FRI-mediated enrichment of a WDR5a-containing complex at FLC.
[See online article for color version of this figure.]
As noted earlier, FRI can override FLD to upregulate FLC expression in winter annuals. We have found that FRI does not disrupt the recruitment of FLD (or FLD-containing corepressor complex) to FLC chromatin but may partially disrupt FLD function. Because the AP genes FCA and FPA mainly function through FLD to repress FLC expression (Liu et al., 2007; Baurle and Dean, 2008), a functional FRI may also disrupt the FLC repression mediated by these two genes. Taken together, our study suggests that FRI may play a dual role in the H3K4 trimethylation of FLC chromatin, namely, enriching an H3K4 methyltransferase complex and compromising H3K4 demethylation at FLC, to upregulate FLC expression and thus establish the winter-annual growth habit.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana fld-1, fld-3 (He et al., 2003), fca-9 (Baurle and Dean, 2008), and FRI-Col (Lee et al., 1994) were described previously. The wdr5b mutant was isolated from the Versailles transformant collection (Samson et al., 2002). Plants were grown under cool white fluorescent light in long days (16 h light /8 h night) at ∼22°C.
RNA Analysis
Total RNAs from aerial parts of ∼10-d-old seedlings grown in long days were extracted as described previously (Jiang et al., 2007). cDNAs were reverse transcribed from total RNAs with Moloney murine leukemia virus reverse transcriptase (Promega). Real-time quantitative PCR was performed on an ABI Prism 7900HT sequence detection system using SYBR Green PCR master mix as described previously (Jiang et al., 2007). Each sample was quantified in triplicate and normalized using TUB2 (At_5g62690) as the endogenous control. Primers used for the amplification of FLC, FLM, MAF2, MAF4, TUB2, and ACT2 have been described previously (Jiang et al., 2007; Gu et al., 2009), and the primers used for WDR5a and WDR5b amplification are specified in Supplemental Table 2 online.
Knockdown of WDR5a Expression via Double-Stranded RNAi
A 232-bp WDR5a-specific fragment (from +957 to +1188 of WDR5a cDNA; the transcription start site is set as +1) was used to create a hairpin RNA by the AGRICOLA consortium (Hilson et al., 2004); the resulting binary plasmid was introduced into Agrobacterium tumefaciens strain GV3101 carrying pMP90 and pSOUP helper plasmids through electroporation and subsequently was introduced into Col by the floral dip method (Clough and Bent, 1998).
Plasmid Construction
To construct the WDR5a-GFP plasmid, the entire coding sequence of WDR5a except the stop codon (1.0 kb) was inserted between the 35S promoter and GFP in the pMDC85 vector (Curtis and Grossniklaus, 2003) via Gateway technology (Invitrogen); the coding sequence is in-frame with the downstream GFP reporter gene. To construct the WDR5a-GUS plasmid, a 1.75-kb WDR5a genomic fragment (from −1617 to +150; A of the start codon as +1) including a 1.6-kb 5′ promoter plus a 0.15-kb genomic coding sequence was inserted into the pBGWFS7 vector (Karimi et al., 2005) via Gateway technology; the genomic coding sequence is in-frame with the downstream GUS reporter gene. To construct the FLD-myc plasmid, a 4424-bp FLD (from −1900 to +2524; A of the start codon as +1) genomic fragment including a 1.9-kb 5′ promoter plus the 2.5-kb genomic coding sequence was inserted into a vector derived from pC-TAPa (Rubio et al., 2005) in which a stop codon has been introduced in between the coding sequences for the 9x c-myc tag and the IgG binding domain; FLD is in-frame with the downstream 9x c-myc.
Peptide Pull-Down Assay
GST-WDR5a and GST were overexpressed in Escherichia coli and affinity-purified using glutathione-linked resins according to the manufacturer's instructions (Sigma-Aldrich). Peptide pulldown was performed as described by Wysocka et al. (2005). Briefly, a mixture of ∼10-fold excess of GST (40 μg) with ∼4.0-μg GST-WDR5a (estimated) was first precleared with avidin beads (Sigma-Aldrich); subsequently, 5.0-μg H3 peptides with unmodified, mono-, di-, or trimethylated K4, conjugated with biotin (Millipore), were incubated with the avidin beads and the protein mixture; beads were washed six times and subsequently the proteins bound to the beads were eluted by boiling using a 2× SDS-PAGE loading buffer. The mock is a control in which the protein mixture was incubated with the avidin beads alone. Elutes were analyzed by SDS-PAGE and Coomassie Brilliant Blue staining.
Protein Pull-Down Assay
The pulldowns were performed as described by Calonje et al. (2008). Briefly, after isopropyl-β-d-thiogalactopyranoside induction, E. coli (BL21 DE3) cells expressing GST-ATX1, His-WDR5a, or GST were harvested by centrifugation, resuspended in 1.0-mL binding buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Triton X-100, 10% glycerol, 1 mM phenylmethylsulphonyl fluoride, and 1× Roche proteinase inhibitors), and sonicated. The protein extracts were centrifuged and an equal amount of the extract (300 μL for each extract) containing GST or GST-ATX1, or noninduced E. coli extract, was mixed with the His-WDR5a extract with rotating for 4 h at 4°C; subsequently, 40 μL of glutathione-linked resins (Sigma-Aldrich) was added into the mixture and incubated with rotating for another 2 h at 4°C. The beads were washed four times with the binding buffer. Proteins were eluted and further analyzed by immunoblotting using an antibody raised against the human WDR5 (Abcam).
ChIP and Real-Time Quantitative PCR Assay
The ChIP experiments were performed as described previously using 10-d-old seedlings (Johnson et al., 2002). Rabbit polyclonal anti-trimethyl-histone H3 (Lys 4) (Abcam), anti-WDR5 (Abcam), and anti-c-Myc (Lab Vision) were used in the immunoprecipitation experiments. Rabbit IgG (Millipore) was used as a negative control in each ChIP experiment to check the background levels of DNA fragments. The amounts of the immunoprecipitated genomic DNA were quantified by real-time PCR. Quantitative measurements of FLC-P, FLC-I, and FLC-M fragments were performed on an ABI Prism 7900HT sequence detection system using SYBR Green PCR master mix. Primers used to amplify FLC-P and TUB2 have been described previously (Jiang et al., 2007), and primers used to amplify FLC-I and FLC-M are specified in Supplemental Table 2 online. Each of the immunoprecipitations was repeated independently once. The relative fold changes are the average of six measurements of two ChIP experiments (each quantified in triplicate), and error bars are standard deviations of the six quantitative PCR assays.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At_3g49660 (WDR5a) and At_4g02730 (WDR5b).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Analyses of a Loss-of-Function wdr5b Mutant.
Supplemental Figure 2. Relative mRNA Levels of FLM, MAF2, and MAF4 in Seedlings of wdr5a (RNAi) Lines Quantified by Real-Time PCR.
Supplemental Figure 3. Analysis of Expression of the Indicated Genes upon WDR5a Knockdown.
Supplemental Figure 4. Effect of WDR5a Knockdown on the Late-Flowering Phenotypes of FRI-Col and fld Grown in LDs.
Supplemental Figure 5. Immunoblot Analysis of WDR5a in the Indicated Lines Using an Antibody Raised against the Human WDR5.
Supplemental Figure 6. WDR5a Knockdown Leads to Decreased Binding of WDR5a to FLC Chromatin.
Supplemental Figure 7. SDS-PAGE Analysis of Total Protein Extracts from E. coli Harboring GST-ATX1, His-WDR5a, or GST Plasmids.
Supplemental Table 1. Total Leaf Number at Flowering of wdr5a (RNAi) Lines Grown in Long Days.
Supplemental Table 2. Sequences of Primers Used in RT-PCR and ChIP-PCR Experiments.
Supplementary Material
Acknowledgments
We thank Toshiro Ito and R. Jose Dinney for critically reading this manuscript, Wannian Yang and Jiafu Jiang for experimental assistance, and the AGRICOLA consortium for providing the double-stranded RNAi plasmid targeting WDR5a. This work was supported by grants from the Singapore Ministry of Education (AcRF Tier 2; T207B3105) and the National University of Singapore (AcRF Tier 1; R-154-000-294-112) and by the Temasek Life Sciences Laboratory to Y.H.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Yuehui He (dbshy@nus.edu.sg).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Alvarez-Venegas, R., Pien, S., Sadder, M., Witmer, X., Grossniklaus, U., and Avramova, Z. (2003). ATX-1, an Arabidopsis homolog of TRITHORAX, activates flower homeotic genes. Curr. Biol. 13 627–637. [DOI] [PubMed] [Google Scholar]
- Ausin, I., Alonso-Blanco, C., Jarillo, J.A., Ruiz-Garcia, L., and Martinez-Zapater, J.M. (2004). Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat. Genet. 36 162–166. [DOI] [PubMed] [Google Scholar]
- Baurle, I., and Dean, C. (2006). The timing of developmental transitions in plants. Cell 125 655–664. [DOI] [PubMed] [Google Scholar]
- Baurle, I., and Dean, C. (2008). Differential interactions of the autonomous pathway RRM proteins and chromatin regulators in the silencing of Arabidopsis targets. PLoS ONE 3 e2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calonje, M., Sanchez, R., Chen, L., and Sung, Z.R. (2008). EMBRYONIC FLOWER 1 participates in polycomb group-mediated AG gene silencing in Arabidopsis. Plant Cell 20 277–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Yang, Y., Wang, F., Wan, K., Yamane, K., Zhang, Y., and Lei, M. (2006). Crystal structure of human histone Lysine-Specific Demethylase 1 (LSD1). Proc. Natl. Acad. Sci. USA 103 13956–13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, K., Kim, S., Kim, S.Y., Kim, M., Hyun, Y., Lee, H., Choe, S., Kim, S.G., Michaels, S., and Lee, I. (2005). SUPPRESSOR OF FRIGIDA3 encodes a nuclear ACTIN-RELATED PROTEIN6 required for floral repression in Arabidopsis. Plant Cell 17 2647–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, K., Park, C., Lee, J., Oh, M., Noh, B., and Lee, I. (2007). Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 134 1931–1941. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Curtis, M.D., and Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal, R.B., Kandasamy, M.K., McKinney, E.C., and Meagher, R.B. (2005). The nuclear actin-related protein ARP6 is a pleiotropic developmental regulator required for the maintenance of FLOWERING LOCUS C expression and repression of flowering in Arabidopsis. Plant Cell 17 2633–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal, R.B., Topp, C.N., McKinney, E.C., and Meagher, R.B. (2007). Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell 19 74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou, Y., Milne, T.A., Ruthenburg, A.J., Lee, S., Lee, J.W., Verdine, G.L., Allis, C.D., and Roeder, R.G. (2006). Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 13 713–719. [DOI] [PubMed] [Google Scholar]
- Gazzani, S., Gendall, A.R., Lister, C., and Dean, C. (2003). Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol. 132 1107–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, X., Jiang, D., Wang, Y., Bachmair, A., and He, Y. (2009). Repression of the floral transition via histone H2B monoubiquitination. Plant J. 57 522–533. [DOI] [PubMed] [Google Scholar]
- He, Y., Doyle, M.R., and Amasino, R.M. (2004). PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 18 2774–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y., Michaels, S.D., and Amasino, R.M. (2003). Regulation of flowering time by histone acetylation in Arabidopsis. Science 302 1751–1754. [DOI] [PubMed] [Google Scholar]
- He, Y. (2009). Control of the transition to flowering by chromatin modifications. Mol. Plant 2 http://dx.doi.org/10.1093/mp/ssp1005. [DOI] [PubMed]
- Hilson, P., Allemeersch, J., Altmann, T., Aubourg, S., Avon, A., Beynon, J., Bhalerao, R.P., Bitton, F., Caboche, M., Cannoot, B., Chardakov, V., and Cognet-Holliger, C. (2004). Versatile gene-specific sequence tags for Arabidopsis functional genomics: Transcript profiling and reverse genetics applications. Genome Res. 14 2176–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, D., Yang, W., He, Y., and Amasino, R.M. (2007). Arabidopsis relatives of the human Lysine-Specific Demethylase 1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell 19 2975–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, D., Wang, Y., Wang, Y., and He, Y. (2008). Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS ONE 3 e3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson, U., West, J., Lister, C., Michaels, S., Amasino, R., and Dean, C. (2000). Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290 344–347. [DOI] [PubMed] [Google Scholar]
- Johnson, L., Cao, X., and Jacobsen, S. (2002). Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 12 1360–1367. [DOI] [PubMed] [Google Scholar]
- Karimi, M., De Meyer, B., and Hilson, P. (2005). Modular cloning in plant cells. Trends Plant Sci. 10 103–105. [DOI] [PubMed] [Google Scholar]
- Kim, S., Choi, K., Park, C., Hwang, H.J., and Lee, I. (2006). SUPPRESSOR OF FRIGIDA 4, encoding a C2H2-type zinc finger protein, represses flowering by transcriptional activation of Arabidopsis FLOWERING LOCUS C. Plant Cell 18 2985–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.Y., He, Y., Jacob, Y., Noh, Y.S., Michaels, S., and Amasino, R. (2005). Establishment of the vernalization-responsive, winter-annual habit in Arabidopsis requires a putative histone H3 methyl transferase. Plant Cell 17 3301–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.Y., and Michaels, S.D. (2006). SUPPRESSOR OF FRI 4 encodes a nuclear-localized protein that is required for delayed flowering in winter-annual Arabidopsis. Development 133 4699–4707. [DOI] [PubMed] [Google Scholar]
- Lee, I., Michaels, S.D., Masshardt, A.S., and Amasino, R.M. (1994). The late-flowering phenotype of FRIGIDA and LUMINIDEPENDENS is suppressed in the Landsberg erecta strain of Arabidopsis. Plant J. 6 903–909. [Google Scholar]
- Lee, M.G., Wynder, C., Bochar, D.A., Hakimi, M.A., Cooch, N., and Shiekhattar, R. (2006). Functional interplay between histone demethylase and deacetylase enzymes. Mol. Cell. Biol. 26 6395–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F., Quesada, V., Crevillen, P., Baurle, I., Swiezewski, S., and Dean, C. (2007). The Arabidopsis RNA-binding protein FCA requires a Lysine-Specific Demethylase 1 homolog to downregulate FLC. Mol. Cell 28 398–407. [DOI] [PubMed] [Google Scholar]
- March-Diaz, R., Garcia-Dominguez, M., Florencio, F.J., and Reyes, J.C. (2007). SEF, a new protein required for flowering repression in Arabidopsis, interacts with PIE1 and ARP6. Plant Physiol. 143 893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Trillo, M., Lazaro, A., Poethig, R.S., Gomez-Mena, C., Pineiro, M.A., Martinez-Zapater, J.M., and Jarillo, J.A. (2006). EARLY IN SHORT DAYS 1 (ESD1) encodes ACTIN-RELATED PROTEIN 6 (AtARP6), a putative component of chromatin remodelling complexes that positively regulates FLC accumulation in Arabidopsis. Development 133 1241–1252. [DOI] [PubMed] [Google Scholar]
- Michaels, S., and Amasino, R. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S.D., Bezerra, I.C., and Amasino, R.M. (2004). FRIGIDA-related genes are required for the winter-annual habit in Arabidopsis. Proc. Natl. Acad. Sci. USA 101 3281–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S.D., He, Y., Scortecci, K.C., and Amasino, R.M. (2003). Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc. Natl. Acad. Sci. USA 100 10102–10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, T., Krogan, N.J., Dover, J., Erdjument-Bromage, H., Tempst, P., Johnston, M., Greenblatt, J.F., and Shilatifard, A. (2001). COMPASS: A complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. USA 98 12902–12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu, L., Lu, F., Pei, Y., Liu, C., and Cao, X. (2007). Regulation of flowering time by the protein arginine methyltransferase AtPRMT10. EMBO Rep. 8 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh, Y.S., and Amasino, R.M. (2003). PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell 15 1671–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, S., Zhang, H., Ludwig, P., and van Nocker, S. (2004). A mechanism related to the yeast transcriptional regulator Paf1c is required for expression of the Arabidopsis FLC/MAF MADS box gene family. Plant Cell 16 2940–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei, Y., Niu, L., Lu, F., Liu, C., Zhai, J., Kong, X., and Cao, X. (2007). Mutations in the Type II protein arginine methyltransferase AtPRMT5 result in pleiotropic developmental defects in Arabidopsis. Plant Physiol. 144 1913–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pien, S., Fleury, D., Mylne, J.S., Crevillen, P., Inze, D., Avramova, Z., Dean, C., and Grossniklaus, U. (2008). ARABIDOPSIS TRITHORAX 1 dynamically regulates FLOWERING LOCUS C activation via histone H3 lysine-4 trimethylation. Plant Cell 20 580–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe, O.J., Kumimoto, R.W., Wong, B.J., and Riechmann, J.L. (2003). Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15 1159–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio, V., Shen, Y., Saijo, Y., Liu, Y., Gusmaroli, G., Dinesh-Kumar, S.P., and Deng, X.W. (2005). An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 41 767–778. [DOI] [PubMed] [Google Scholar]
- Ruthenburg, A.J., Allis, C.D., and Wysocka, J. (2007). Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell 25 15–30. [DOI] [PubMed] [Google Scholar]
- Ruthenburg, A.J., Wang, W., Graybosch, D.M., Li, H., Allis, C.D., Patel, D.J., and Verdine, G.L. (2006). Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat. Struct. Mol. Biol. 13 704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh, A., Alvarez-Venegas, R., and Avramova, Z. (2008. a). Dynamic and stable histone H3 methylation patterns at the Arabidopsis FLC and AP1 loci. Gene 423 43–47. [DOI] [PubMed] [Google Scholar]
- Saleh, A., Alvarez-Venegas, R., Yilmaz, M., Le, O., Hou, G., Sadder, M., Al-Abdallat, A., Xia, Y., Lu, G., Ladunga, I., and Avramova, Z. (2008. b). The highly similar Arabidopsis homologs of TRITHORAX ATX1 and ATX2 encode proteins with divergent biochemical functions. Plant Cell 20 568–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson, F., Brunaud, V., Balzergue, S., Dubreucq, B., Lepiniec, L., Pelletier, G., Caboche, M., and Lecharny, A. (2002). FLAGdb/FST: A database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res. 30 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz, R.J., Hong, L., Michaels, S., and Amasino, R.M. (2005). FRIGIDA-ESSENTIAL 1 interacts genetically with FRIGIDA and FRIGIDA-LIKE 1 to promote the winter-annual habit of Arabidopsis thaliana. Development 132 5471–5478. [DOI] [PubMed] [Google Scholar]
- Schubert, D., Primavesi, L., Bishopp, A., Roberts, G., Doonan, J., Jenuwein, T., and Goodrich, J. (2006). Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 25 4638–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scortecci, K.C., Michaels, S.D., and Amasino, R.M. (2001). Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J. 26 229–236. [DOI] [PubMed] [Google Scholar]
- Sheldon, C.C., Burn, J.E., Perez, P.P., Metzger, J., Edwards, J.A., Peacock, W.J., and Dennis, E.S. (1999). The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y., Lan, F., Matson, C., Mulligan, P., Whetstine, J.R., Cole, P.A., Casero, R.A., and Shi, Y. (2004). Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119 941–953. [DOI] [PubMed] [Google Scholar]
- Shilatifard, A. (2008). Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr. Opin. Cell Biol. 20 341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spedaletti, V., Polticelli, F., Capodaglio, V., Schinina, M.E., Stano, P., Federico, R., and Tavladoraki, P. (2008). Characterization of a lysine-specific histone demethylase from Arabidopsis thaliana. Biochemistry 47 4936–4947. [DOI] [PubMed] [Google Scholar]
- Springer, N.M., Napoli, C.A., Selinger, D.A., Pandey, R., Cone, K.C., Chandler, V.L., Kaeppler, H.F., and Kaeppler, S.M. (2003). Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots. Plant Physiol. 132 907–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, S., and Amasino, R.M. (2005). Remembering winter: Toward a molecular understanding of vernalization. Annu. Rev. Plant Biol. 56 491–508. [DOI] [PubMed] [Google Scholar]
- Wang, X., Zhang, Y., Ma, Q., Zhang, Z., Xue, Y., Bao, S., and Chong, K. (2007). SKB1-mediated symmetric dimethylation of histone H4R3 controls flowering time in Arabidopsis. EMBO J. 26 1934–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, J.D., Borevitz, J.O., Uhlenhaut, N.H., Ecker, J.R., Chory, J., and Weigel, D. (2005). FRIGIDA-independent variation in flowering time of natural Arabidopsis thaliana accessions. Genetics 170 1197–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka, J., Swigut, T., Milne, T.A., Dou, Y., Zhang, X., Burlingame, A.L., Roeder, R.G., Brivanlou, A.H., and Allis, C.D. (2005). WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121 859–872. [DOI] [PubMed] [Google Scholar]
- Zhao, Z., Yu, Y., Meyer, D., Wu, C., and Shen, W.H. (2005). Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat. Cell Biol. 7 1256–1260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.