Abstract
Approximately 1% of the open reading frames in the human genome encode proteins that function as DNA or RNA helicases. These enzymes act in all aspects of nucleic acid metabolism where the complementary strands of DNA:DNA or DNA:RNA duplexes require to be transiently opened. However, they perform wider roles in nucleic acid metabolism due to their ability to couple the energy derived from hydrolysis of ATP to their unidirectional translocation along strands of DNA∕RNA. In this way, helicases can displace proteins from DNA∕RNA, drive the migration of DNA junctions (such as the Holliday junction recombination intermediate), or generate superhelical tension in nucleic acid duplexes. Here, we review a subgroup of DNA helicase enzymes, the RecQ family, that has attracted considerable interest in recent years due to their role not only in suppression of genome instability, but also in the avoidance of human disease. We focus particularly on the protein structural motifs and the multiple assembly states that characterize RecQ helicases and discuss novel biophysical techniques to study the different RecQ structures present in solution. We also speculate on the roles of the different domains and oligomeric forms in defining which DNA structures will represent substrates for RecQ helicase-mediated transactions.
The transient separation of the complementary strands of nucleic acid duplexes is a fundamental step in most DNA and RNA metabolic processes. Helicases are a seemingly ever expanding family of enzymes that catalyze DNA strand separation in a reaction coupled to the binding and hydrolysis of nucleotide triphosphate (NTP) (Matson et al., 1994; Singleton and Wigley, 2002; von Hippel, 2004). A detailed description of the different superfamilies of helicases and their properties is outside the scope of this article, and we refer readers to recent reviews (Lohman et al., 2008; Singleton et al., 2007; von Hippel, 2004). RecQ enzymes are a subfamily of helicases that play an essential role in the maintenance of genome stability by acting at the interface between DNA replication, recombination, and repair (Bachrati and Hickson, 2008; Bohr, 2008; Hickson, 2003). Mutations in the genes of three human RecQ family members are linked to defined genetic disorders associated with genomic instability, cancer predisposition, and features of premature aging; namely, Bloom’s syndrome (BLM gene mutations), Werner’s syndrome (WRN gene mutations), and Rothmund–Thomson syndrome, RAPADILINO and Baller-Gerold syndrome (all caused by mutation of RECQ4) (Ellis et al., 1995; Kitao et al., 1999; Siitonen et al., 2003; Van Maldergem et al., 2006; Yu et al., 1996). Hence, understanding how these enzymes function and regulate their DNA processing activities is important if we are to shed light on the mechanisms that control the integrity of our genome and prevent cancer and aging.
Since the discovery of the first RecQ enzyme from E. coli made by Nakayama and colleagues over 20 years ago (Nakayama et al., 1984), numerous other RecQ family helicase genes have been found in all kingdoms of life. Unicellular organisms, such as bacteria and yeasts only have one or two RecQ helicase genes per species, while higher eukaryotes generally express multiple RecQ enzymes (Bachrati and Hickson, 2003; Bachrati and Hickson, 2008; Hanada and Hickson, 2007; Mandell et al., 2005). For example, four RecQ helicase genes have been found in Caenorhabditis elegans, five in Drosophila melanogaster and Homo sapiens, and seven in the plant species Arabidopsis thaliana and Oryza sativa (Hartung and Puchta, 2006). Biochemical studies have demonstrated that RecQ helicases unwind DNA with a 3′–5′ polarity and, although with some differences, are capable of unwinding a variety of DNA structures other than standard B-form DNA duplexes. Briefly, the substrate specificity of RecQ enzymes includes forked duplexes, displacement loops (D-loops, an intermediate in homologous recombination reactions), triple helices, three- or four-way junctions, and G-quadruplex DNA (Constantinou et al., 2000; Fry and Loeb, 1999; Karow et al., 2000; Mohaghegh et al., 2001; Popuri et al., 2008; Sun et al., 1999; Sun et al., 1998; Wu and Maizels, 2001). For a detailed description of the biochemical properties of RecQ helicases see other reviews (Bachrati and Hickson, 2003; Bachrati and Hickson, 2008; Bennett and Keck, 2004; Opresko et al., 2004). Consistent with an ability to unwind various DNA structures, several cellular functions have been attributed to RecQ proteins, including roles in stabilization and repair of damaged DNA replication forks, telomere maintenance, homologous recombination, and DNA damage checkpoint signaling (Bachrati and Hickson, 2008; Bohr, 2008; Ouyang et al., 2008; Sharma et al., 2006). Moreover, recent studies demonstrated that several RecQ enzymes are also able to promote the annealing of complementary single-stranded DNA molecules (Cheok et al., 2005; Garcia et al., 2004; Machwe et al., 2005; Macris et al., 2005; Sharma et al., 2005). In this regard, it is tempting to speculate that this activity might be required to promote either branch migration of DNA junctions or the regression of stalled replication forks, which are characteristic activities of several RecQ enzymes and which might be facilitated by “active” DNA strand annealing. The exact function of the annealing activity of RecQ helicases in vivo has, however, yet to be demonstrated.
In this review, we discuss conserved and nonconserved motifs and structural elements that characterize the RecQ proteins. We describe putative roles of these domains in DNA substrate and protein recognition, and in the regulation of the DNA unwinding, strand annealing, and branch migration activities of these enzymes. On the basis of the recent findings that the activity of several RecQ enzymes is regulated by protein oligomerization, we also review the putative roles of the different multimeric states described for several RecQ enzymes and suggest a possible conserved mechanism by which RecQ enzymes might switch between these multiple enzymatic activities.
CONSERVED AND UNIQUE SEQUENCE MOTIFS∕DOMAINS IN RECQ HELICASES
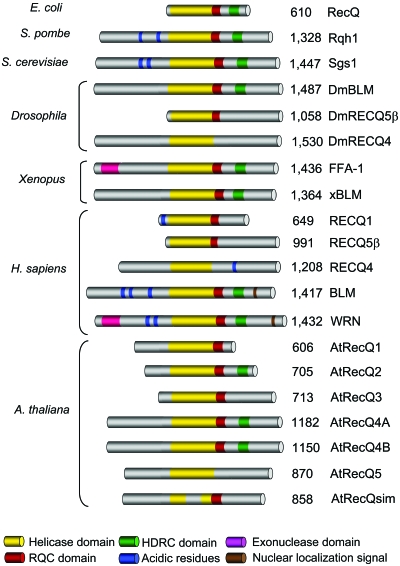
RecQ enzymes have three conserved domains that are commonly found in most helicases of this family: the core helicase domain, the RecQ-C-terminal (RQC) domain, and the helicase-and-RNaseD-like-C-terminal (HRDC) domain (Fig. 1). The helicase domain is present in all RecQ enzymes, while the RQC and∕or HRDC domains are missing in some representatives of the family. In addition, RecQ enzymes vary greatly in the length of their N- and C-terminal domains that flank the presumed “catalytic core” (Fig. 1). These flanking domains are involved in heterologous protein interactions, in the regulation of protein subcellular localization, in directing oligomerization, or in conferring additional enzymatic activities, such as the exonuclease domain in the N-terminal region of WRN.
Figure 1. Schematic of selected members of the RecQ family of DNA helicases.
The respective organism is shown on the left, and the size (in amino acids) and name of each protein are indicated on the right. All proteins are aligned according to the conserved helicase domain, which is shown in yellow. The conserved RQC and HRDC domains are shown in red and green, respectively. The exonuclease domain in the amino-terminal region of WRN and its orthologs is shown in pink. Regions containing patches of acidic residues are shown in blue. The nuclear localization signal sequences identified at the extreme carboxyl terminus of certain family members is shown as a brown bar. The remaining portions of each protein (gray) represent regions that are poorly conserved. The sizes of the individual domains are not to scale. At least three splice variants of the human RECQ5 protein are expressed, of which only the largest (β isoform) is shown.
The core helicase domain
The core helicase domain is characterized by the presence of seven conserved motifs (I, Ia, II, III, IV, V, VI) that are present in all superfamily 1 and 2 (SF1 and SF2) helicases, and has a total size of approximately 300–450 amino acids. These motifs are required for the enzyme to bind NTP and couple the energy derived from NTP hydrolysis to the process of nucleic acid unwinding (Lohman et al., 2008; Matson et al., 1994; Singleton and Wigley, 2002; von Hippel, 2004). The determination of crystal structures of various helicases of the SF1 and SF2 family has shown that these motifs form the core of two RecA-like domains that serve as the ATP-driven “motor” of the helicase. Structural data for this domain are available for two RecQ helicases, the E. coli RecQ and human RECQ1 proteins, which show that the general fold of the core helicase domain of RecQ enzymes is similar to that of the other known SF1 and SF2 helicases (Bernstein et al., 2003; Pike et al., 2008). In SF1 helicases, which probably represent the best-characterized helicase family, binding of the nucleotide in the cleft between the two RecA-like domains induces a structural change that is responsible for the translocation of the enzyme along the single-stranded DNA, via a so-called “inchworm” mechanism (PcrA, Rep, and UvrD) (Korolev et al., 1997; Lee and Yang, 2006; Velankar et al., 1999). A β-hairpin, located after motif VI, and also known as the strand separation pin, abuts the end of the DNA duplex, and is important in DNA strand separation (see also below). The SF2 family, to which RecQ enzymes belong, is a large family that includes functionally diverse enzymes, such as viral helicases (hepatitis C virus NS3), the repair helicases Mus308∕HEL308∕Polθ, the bacterial enzyme RecG, several RNA helicases, and the RecQ helicases (Fry and Loeb, 1999; McGlynn and Lloyd, 2002; Oshige et al., 1999; Seki et al., 2003). The members of this family differ from the SF1 enzymes in the primary sequence of the seven conserved motifs, although they share the same ATP-dependent translocation module comprising two RecA-like domains. Although the nucleotide bound structures of E. coli RecQ and human RECQ1 show that RecQ enzymes bind ATP in a conventional way, we currently have no detailed knowledge of how ATP binding∕hydrolysis cycles are coupled to unidirectional translocation along ssDNA (which is in the 3′–5′ direction in the case of RecQ enzymes).
Besides the seven conserved motifs, the core helicase domain of RecQ helicases is characterized by an additional sequence element, termed motif 0, which is located N-terminally to motif I (Bernstein and Keck, 2003). This motif is well conserved in all RecQ enzymes from different organisms and is composed of four invariant and two conserved amino acids spaced by eight nonconserved residues: Lx3(F∕Y∕W)Gx3F(R∕K)x2Q. The structures of E. coli RecQ and human RECQ1 proteins show that the motif 0 is involved in nucleotide binding, and mutagenesis studies have confirmed that this motif is important for core helicase domain function. For example, substitution of the C-terminal Gln with Ala in RECQ5β significantly reduces ATPase activity, and the replacement of the same residue with Arg inactivates the ATPase and helicase activity of murine BLM (Bahr et al., 1998; Garcia et al., 2004). The same mutation is also listed among the naturally occurring mutations of BLM associated with Bloom’s syndrome (Ellis et al., 1995). Moreover, budding yeast cells carrying this mutation in the SGS1 gene show the same DNA-damaging agent sensitivity as that of an sgs1 deletion strain (Onoda et al., 2000).
The RecQ-C-terminal domain
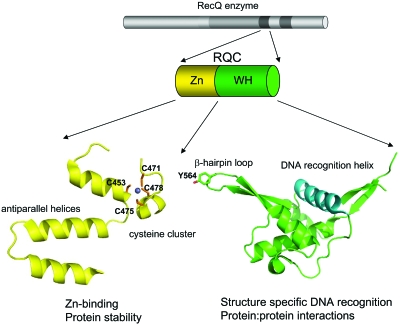
The second most highly conserved domain in RecQ helicases is the RQC domain (Fig. 2). It is present in all RecQ enzymes with the exception of human RECQ4 and its orthologs. This domain is unique to RecQ enzymes and can be divided into two subdomains; a Zn2+-binding domain and a so-called winged helix (WH) domain. The Zn2+-binding domain is characterized by a pair of antiparallel α-helices and four Cys residues that are needed to coordinate a single Zn2+ atom. The structures of the Zn2+-binding domains of human RECQ1 and E. coli RecQ are highly similar, although the two α-helices are longer in bacterial RecQ than in the human protein (15 and 19 amino acids, versus 12 and 15, respectively) (Bernstein et al., 2003; Pike et al., 2008). Single amino acid substitutions in the Zn2+-binding domain of the human BLM and bacterial RecQ proteins generate variants that are either insoluble or very prone to degradation, suggesting that this domain plays an important role in maintaining the structural integrity of the protein and hence protein stability (Guo et al., 2005; Janscak et al., 2003; Liu et al., 2004). Missense mutations affecting the Cys residues of the Zn2+-binding of BLM are found in Bloom’s syndrome cases, and a similar mutation in the budding yeast SGS1 gene confers enhanced DNA-damage sensitivity and a hyperrecombination phenotype (Ellis et al., 1995; Onoda et al., 2000). In addition to playing a role in protein stability, the Zn2+-binding domain might be also involved in DNA and∕or protein interactions, as already suggested for other proteins that contain the same domain (Berg and Shi, 1996); however, further studies will be required to test this hypothesis.
Figure 2. Schematic of the structural and functional properties of the RQC domain.
The RQC domain is divided into two subdomains, the Zn2+-binding domain (yellow) and the WH domain (green). The ribbon representations of a Zn2+ and WH domain of human RECQ1 are shown. The four Cys residues of the Zn2+ domain that coordinate the Zn ion (light blue sphere) are indicated. The Tyr residue (Y564) at the tip of the hairpin loop of the WH domain of RECQ1 is highlighted, and the DNA recognition helix is shown in blue. Putative functional properties of the Zn2+ and WH domains are indicated below.
The WH helix domain is characterized by a poor degree of primary sequence similarity between the different RecQ helicases. However, the crystal structures of the WH domains of E. coli RecQ and human RECQ1, and the NMR derived structure of the equivalent domain in human WRN, show a very well conserved domain organization (Bernstein et al., 2003; Hu et al., 2005; Pike et al., 2008). This structural unit is characterized by a helix-turn-helix fold that is also present in variety of DNA binding proteins, such as the transcription factors CAP and hRFX1, and the human DNA repair protein AGT (Daniels et al., 2004; Gajiwala and Burley, 2000; Gajiwala et al., 2000; Schultz et al., 1991). Interestingly, the WH domain of RECQ1 is characterized by the presence of a prominent β-hairpin loop, with an aromatic residue at the tip, which is significantly shorter in the equivalent structures of E. coli RecQ and WRN. Mutagenesis studies on RECQ1 showed that this loop is essential for DNA unwinding, as the substitution of only the Tyr residue at the tip of the loop is sufficient to abolish the unwinding activity of RECQ1 (Pike et al., 2008). These results suggest that the Tyr residue may act as a pin that abuts the end of the DNA duplex and hence promotes strand separation, as previously observed for other helicases of the SF2 family (Buttner et al., 2007). The fact that this loop is significantly shorter in E. coli RecQ and WRN suggests that these helicases might utilize a different mechanism to unwind DNA from that of RECQ1. Indeed, mutations in the hairpin loop of E. coli RecQ do not affect enzymatic activity (Pike et al., 2008).
Several studies have shown that the WH domain is also important for dsDNA recognition. More specifically, gel mobility shifts experiments showed that the RQC domain of E. coli RecQ and BLM is required for G-quadruplex DNA binding, and similar studies with a WRN fragmentencompassing the RQC domain demonstrated that this domain is required for the interaction of WRN with Holliday junctions and forked-duplex substrates (Huber et al., 2006; von Kobbe et al., 2003). The importance of the WH domain in DNA binding is also underscored by the fact that the homologous region of a variety of other proteins has been identified as a critical DNA-binding domain (Daniels et al., 2004; Gajiwala and Burley, 2000; Gajiwala et al., 2000; Schultz et al., 1991). Perhaps surprisingly, given its small size and critical role in mediating interactions with DNA, studies with WRN have shown that many of its interactions with other binding partners are mediated by the WH domain, suggesting that this helix-turn-helix motif might also be involved in protein-protein interactions (Lee et al., 2005).
The helicase-and-RNaseD-like-C-terminal domain
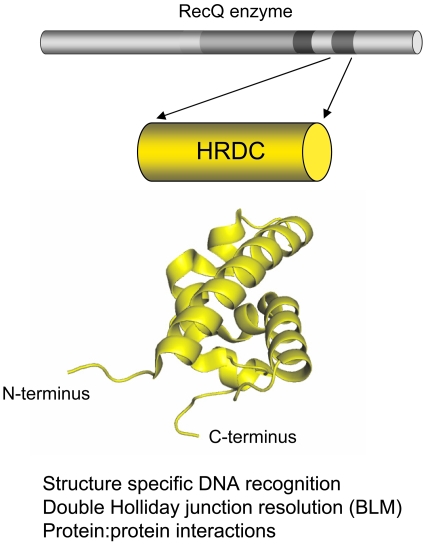
The third conserved region of RecQ helicases derives its name from its similarity with the C-terminal region of the RNaseD protein. For this reason, it is called the helicase-and-RNaseD-like-C-terminal domain (Morozov et al., 1997). The HRDC region is much less well conserved than the helicase and RQC domains among RecQ helicases. Moreover, it is also absent from several RecQ enzymes. For example, among the five human RecQ helicases, only BLM and WRN possess a recognizable HRDC domain, which is located toward the C-terminus. Interestingly, the RecQ enzyme from Rhodobacter sphaeroides contains two HRDC domains, while other RecQ helicases from Deinococcus radiodurans, Neisseria meningitidis, and Neisseria gonorrhea are characterized by possessing three HRDC repeats. These multiple HRDC domains regulate the enzymatic activity of Deinococcus radiodurans RecQ and differentially affect the ability of the enzyme to bind and unwind DNA (Killoran and Keck, 2006) (see Fig. 3).
Figure 3. Schematic of the structural and functional properties of the HRDC domain.
The structure of the HRDC domain of WRN is shown. Putative functional properties of HRDC domain are indicated below.
Structural and biochemical studies confirmed that the HRDC domain is associated with structure-specific recognition of DNA substrates and plays a crucial role in differentiating the activity and functions of the various RecQ homologs. The structures of the isolated HRDC domains of E. coli RecQ, S. cerevisiae Sgs1, WRN, and Deinococcus radiodurans RecQ reveal a similar overall fold (Bernstein and Keck, 2005; Killoran and Keck, 2008; Kitano et al., 2007; Liu et al., 1999). However, different regions of these domains seem to be required for DNA binding. For example, the HRDC domain of S. cerevisiae Sgs1 is characterized by a lysine- and arginine-rich patch that forms an electropositive surface important for the interaction with ssDNA (Liu et al., 1999). This surface is missing in the structure of the HRDC domain of WRN, which is instead distinguished by the presence of a cluster of acidic and hydrophobic residues (Kitano et al., 2007). The HRDC domain of E. coli RecQ also uses an electropositive surface to interact with DNA, but the residues that form this surface are located on a different face of the domain from that in the Sgs1 protein (Bernstein and Keck, 2005). Moreover, the HRDC of E. coli RecQ is characterized by a 310 helix with a Tyr (residue 555) on its surface, which is essential for binding to ssDNA and partial duplex DNA. These structural differences are likely associated with the distinct DNA binding specificity observed for individual HRDC domains. For example, the isolated HRDC domain of E. coli RecQ binds preferentially to ssDNA over other DNA structures, and the S. cerevisiae Sgs1 HRDC domain can bind both ssDNA and partially double-stranded DNA. In contrast, the HRDC domain of WRN does not appear to interact with DNA in vitro (Bernstein et al., 2003; Kitano et al., 2007; Liu et al., 1999; von Kobbe et al., 2003). However, a WRN fragment containing the HRDC domain and additional residues at the C-terminus (fragment 1072-1432) binds forked-duplex DNA and Holliday junctions with high affinity (von Kobbe et al., 2003).
Regarding the impact of the HRDC domain on the activity of full-length RecQ proteins, experiments with an E. coli RecQΔC mutant lacking the HRDC domain indicated that the HRDC domain is dispensable for the ATPase and unwinding activity of the enzyme but is required for stable binding to partial duplex DNA (Bernstein and Keck, 2003). These results led the authors to suggest that, although it is dispensable for the catalytic activity of bacterial RecQ, the HRDC domain might facilitate the unwinding of long DNA duplexes by stably binding DNA (Bernstein and Keck, 2003). A HRDC truncation mutant of Sgs1 protein is also active as a helicase and ATPase in vitro (Bennett et al., 1998; Lu et al., 1996). Conversely, studies with a BLM mutant lacking the HRDC region demonstrated that, although this domain has a minor effect on forked-duplex unwinding and is not required for ATP hydrolysis, it does play an important role in Holliday junction disruption (Janscak et al., 2003; Wu et al., 2005). Furthermore, the lysine-1270 residue of BLM, which resides in the HRDC domain, plays an important role in the double Holliday junction dissolution activity of BLM, which is an activity that is highly specific for BLM among human RecQ helicases (Wu et al., 2005; Wu and Hickson, 2003).
Other studies have also shown that the C-terminal fragment of BLM, encompassing the RQC and HRDC domains, is necessary for the interaction with the telomere-associated protein, TRF2, which stimulates BLM-mediated unwinding of two telomere substrates in vitro; a 3′-overhang and a telomere D-loop structure (Lillard-Wetherell et al., 2004). Collectively, these studies indicate that the HRDC domain plays an important role both in conferring some specific enzymatic activities to the individual RecQ enzymes and in DNA structure-specific recognition. In addition, it may mediate some protein-protein interactions. The question of how the HRDC domain interacts functionally with the other two conserved regions of RecQ enzymes (the core helicase and the RQC domain) and, as a result, modulates the specific enzymatic properties of the individual helicases, is still open.
Exonuclease domain of WRN
Human WRN and its orthologs in other organisms, such asX. laevis FFA-1, are unique among RecQ family proteins in possessing a clearly defined exonuclease domain located in the N-terminal region of the proteins (Fig. 1) (Huang et al., 1998; Mushegian et al., 1997; Shen et al., 1998). Recombinant WRN possesses 3′–5′ exonuclease activity that can act on a wide variety of different substrates, much the same as its helicase function (Choudhary et al., 2004). This exonuclease domain can function independently of the rest of WRN polypeptide in that a recombinant fragment encompassing residues 1–333 displays exonuclease activity (Huang et al., 1998). Nevertheless, it seems plausible that the helicase and exonuclease activities of WRN will be coordinated in vivo, as has been suggested from some in vitro studies (Opresko et al., 2001).
The crystal structure of the WRN exonuclease domain (residues 38–236) has been determined (Perry et al., 2006). The catalytic mechanism of phosphodiester bond hydrolysis involves two metal ions. Interestingly, the structure places the WRN exonuclease into the DnaQ family of nucleases, which includes the 3′–5′ proofreading domain of E. coli DNA polymerase I. Indeed, Perry et al. proposed that the WRN exonuclease might act in a proofreading capacity during nonhomologous end-joining reactions mediated via the interaction of WRN with Ku70∕80 and DNA-dependent protein kinase (Perry et al., 2006). This interaction stimulates the activity of the isolated WRN exonuclease domain to a similar extent to that of the full-length WRN protein, suggesting a direct interaction between this domain and the Ku70∕80 heterodimer.
Nonconserved N- and C-terminal domains of RecQ helicases
There is little or no indication from making an initial inspection of the primary sequences of the N- and C-terminal domains of different RecQ helicases exactly what role these domains might play. Exceptions to this broad statement include the exonuclease domain of WRN and the known nuclear localization signal sequences (Fig. 1). Interestingly, the N-terminus of RECQ4 bears homology to the N-termini of the yeast proteins Sld2 (S. cerevisiae) and DRC1 (S. pombe), which play a central role for the establishment of DNA replication forks in yeast and do not have any apparent homolog in vertebrates. Consistently, a recent study provided evidence for a previously unrecognized role of the X. laevis homolog of human RECQ4 in the initiation of DNA replication (Sangrithi et al., 2005). Of less obvious significance, but nevertheless a conserved feature, is the presence of blocks of acidic residues. While these are superficially reminiscent of similar domains in transcriptional activators, there is no suggestion in RecQ enzymes that they perform a similar role. One possibility, however, is that they mediate interactions with a common partner protein, one possible candidate being replication protein A (Brosh et al., 2000; Brosh et al., 1999).
Other than the above sequence motifs, the N- and C-terminal domains of RecQ helicases are quite featureless. Nevertheless, the domains are highly conserved among the different mammalian orthologs of each human RecQ family member, suggesting that they are functionally important. Studies on recombinant proteins suggest that they play little role in catalytic activity, although as noted below they likely influence DNA substrate specificity by influencing the oligomeric state of certain RecQs. One clear role for the N- and C-terminal domains is to direct interactions with heterologous proteins. For example, BLM binds directly to topoisomerase IIIα via residues in the first 212 (likely 1–133) amino acids of the N-terminal domain, and to the RAD51 recombinase via both the N-terminal 1–212 region and the final C-terminal 150 residues (Hu et al., 2001; Johnson et al., 2000; Wu et al., 2001; Wu et al., 2000). Interestingly, both of these interactions and their approximate location in the RecQ helicase polypeptide are conserved between BLM and its yeast ortholog, Sgs1. Numerous other protein interaction sites have been mapped within the N- and∕or C-terminal domains of BLM, WRN, and other human RecQ helicases, the details of which can be found in recent review articles (Bachrati and Hickson, 2008; Bohr, 2008; Sharma et al., 2006).
The other main role of the N- and C-terminal flanking domains is to perform regulatory duties. This involves serving as a target site for post-translational modifications, including phosphorylation and sumoylation. The best-characterized RecQs in this regard are BLM and WRN. BLM is phosphorylated on several serine∕theonine residues in the N-terminal domain (as well as in the catalytic core). The ATM kinase targets BLM in Thr-99, and ATR targets both Thr-99 and Thr-122, following γ-irradiation and replication blockade, respectively. The modification of Thr-99 is critical for BLM function, since a T99A mutant BLM is unable to complement the hydroxyurea sensitivity of BLM cells (Davies et al., 2007). The N-terminal domain of BLM is also targeted by the Mps1 kinase on Ser-144, which appears important for regulating BLM function in mitosis (Leng et al., 2006).
WRN is also a phosphoprotein in vivo. The major serine-theonine kinase that modifies WRN appears to be DNA-dependent protein kinase (DNA-PK) (Karmakar et al., 2002; Yannone et al., 2001). Consistent with this, WRN phosphorylation in vivo is sensitive to wortmannin, which preferentially targets DNA-PK, and cells lacking DNA-PK show little or no WRN phosphorylation (Karmakar et al., 2002). However, other studies suggested that WRN is also phosphorylated in an ATR∕ATM dependent manner, and that WRN is targeted by the Abl kinase on tyrosine residues (Cheng et al., 2003; Pichierri et al., 2003). WRN binds C-Abl in vivo via the N-terminal domain (residues 1–120) and in the catalytic core comprising the helicase and RQC domains. Tyrosine phosphorylation appears to inhibit the helicase and exonuclease activities of WRN (Cheng et al., 2003).
BLM is sumoylated in vivo, probably on multiple lysine residues, including K317 and K331. Sumoylation appears primarily to influence the subcellular localization of BLM. A mutant BLM carrying K317R and K331R substitutions fails to be sumoylated and does not localize to PML nuclear bodies, unlike wild-type BLM. Instead, the double mutant induces a cellular response reminiscent of that seen following DNA damage, including focal accumulation of γ-H2AX (Eladad et al., 2005). Despite the above discussion, Hu et al., 2001 found that the region of BLM required for PML body localization was in the N-terminal domain between residues 133 and 237 (Hu et al., 2001).
DIFFERENT OLIGOMERIC STATES OF RECQ HELICASES
Assembly states ranging from monomers to hexamers have been described previously for various RecQ enzymes. The role of these different oligomeric forms is, however, still uncertain in most cases. For E. coli RecQ protein, there is compelling evidence to indicate that the active species is a monomer. The crystal structure of a RecQ1–516 variant, lacking the HRDC domain, indicates that the catalytic core ofE. coli RecQ is monomeric in its nucleotide bound and unbound forms (Bernstein et al., 2003). Although a previous study suggested that the full-length E. coli RecQ helicase might exist as a multimer comprising at least three subunits in solution (Harmon and Kowalczykowski, 2001), more recent biochemical and biophysical studies provided evidence that this enzyme remains in a monomeric form also when bound to single-stranded DNA and when unwinding DNA, in agreement with the crystallographic results (Xu et al., 2003; Zhang et al., 2006).
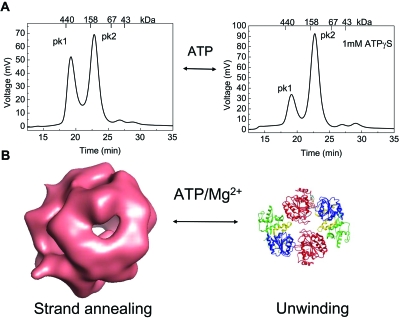
Quaternary structures ranging from monomers to higher-order oligomers have been reported for the human RECQ1, WRN, and BLM helicases. The best characterized in this regard is the human RECQ1 protein, for which a crystal structure of a truncated form lacking the first 48 and the last 33 amino acids is available (PDB id: 2v1x) (Pike et al., 2008). A combination of electron microscopy and size exclusion chromatography suggested that RECQ1 exists in two different oligomeric states: higher-order oligomers consistent with a pentamer or hexamer, and smaller oligomers consistent with a monomer or dimer (Muzzolini et al., 2007). These forms are apparently associated with the strand annealing and unwinding activities of RECQ1, respectively (Fig. 4). The equilibrium between these different assembly states appears to be controlled by ssDNA and ATP binding, with ATP favoring the smaller oligomeric form. Further studies identified the N-terminal region of the protein (residues 1–48) as essential for higher-order oligomer formation, and confirmed that smaller oligomers are involved in DNA unwinding (Popuri et al., 2008). Interestingly, the N-terminal region is also necessary for the ability of RECQ1 to disrupt model Holliday junction substrates, suggesting that this domain, or the higher-assembly state promoted by it, is essential for the ability of the protein to interact with and∕or branch migrate Holliday junctions (Popuri et al., 2008). Consistent with this, the RECQ148–616 mutant used for the crystallographic studies is able to unwind forked-duplex substrates with a similar efficiency to that of the full-length protein, but it is not active on Holliday junctions and has no detectable strand annealing activity (Pike et al., 2008).
Figure 4. The human RECQ1 helicase exists in different oligomeric forms.
(A) Chromatographic profiles of the human RECQ1 in the absence (left) and presence (right) of ATPγS eluting from a Superdex200 HR 10∕30 gel filtration column. The protein species were detected by protein fluorescence (λexcitation=290 nm and λemission=340 nm). Approximately 40 μg of recombinant RECQ1 were loaded at a final concentration of 1 μM. The protein elutes in two main peaks. The first peak (PK1) corresponds to a calculated molecular mass of approximately 400 kDa, whereas the second peak (PK2) corresponds to a calculated molecular mass of approximately 155 kDa. (Note that ATPγS shifts the equilibrium toward PK2). (B) Schematic of the two quaternary forms of RECQ1: higher-order oligomers (probably tetramers) exhibit DNA strand annealing as well as four-way Holliday junction resolution activity, while the lower-oligomers, probably dimers, are required for DNA unwinding. The surface rendering of 3D EM reconstructions of the higher RECQ1 assembly state were obtained as previously described (Muzzolini et al., 2007). The ribbon representation of RECQ1 was generously provided by Opher Gileadi from the Structural Genomics Consortium (SGC), Oxford, UK. The structure of the Mg2+-ADP bound RECQ1 molecule (PDB id: 2V1X) shows the two RecA-like domains (colored in red and blue), the Zn2+-binding domain (yellow), and the WH domain (green).
The structure of RECQ148–616 showed that the protein exists as a dimer in the crystals. Although this could reflect a crystallographic artifact, the existence of RECQ1 dimers in solution was confirmed by size exclusion chromatography, equilibrium ultracentrifugation, and chemical cross-linking experiments (O Gileadi, personal communication). Interestingly, preliminary fitting experiments using the coordinates of the crystal structure and the density maps of RECQ1 derived by electron microscopy suggest that the higher-order oligomers might represent tetramers, rather than pentamers or hexamers as was inferred previously from gel filtration studies. Collectively, these data suggest that RECQ1 exists in different oligomeric forms: higher-order oligomers, which are probably tetramers, and are required strand annealing and Holliday junction resolution, and smaller oligomers (likely dimers) that are involved in DNA unwinding.
Interestingly, a similar mixture of oligomeric forms was also reported for the WRN helicase. A recent electron microscopy study revealed that WRN exists predominantly as a dimer in solution and as a tetramer when bound to Holliday junction DNA or forked-duplexes (Compton et al., 2008). Moreover, mutagenesis studies suggested that the region of WRN between the RQC and HRDC domains (residues 1072–1150) is important for higher-order oligomer formation, and that the deletion of this region eliminates strand annealing activity (Muftuoglu et al., 2008). Previous size-exclusion experiments suggested, however, that full-length recombinant WRN, as well as a WRN1–333 fragment, elutes at a size consistent with a trimeric structure (Huang et al., 2000). Similarly, atomic force microscopy analysis of a 171-amino acid fragment of WRN comprising the exonuclease domain of the enzyme revealed a trimer-hexamer equilibrium in the absence of DNA, with the equilibrium being significantly shifted toward the hexamer form in the presence of DNA (Xue et al., 2002). Thus, there remains an open question about the function and exact stoichiometry of the different WRN assembly states, even though recent kinetic experiments support a model where a monomeric unit is involved in DNA unwinding (Choudhary et al., 2004).
Electron microscopy experiments have shown that the full-length BLM protein can form hexameric ring-like structures in the absence of ATP and DNA (Karow et al., 1999). Interestingly, the same study also described the presence of fourfold symmetric structures that might represent an oligomeric form distinct from the hexameric ring (such as a tetramer or an octamer). Gel filtration studies showed that BLM has a native molecular mass of approximately 700 kDa, consistent with an oligomeric state comprising tetramers or hexamers. However, studies of a BLM deletion mutant, BLM642–1290, revealed that it retains helicase activity and elutes as a monomer from a gel filtration column, both in free solution and in its ssDNA-bound form (Janscak et al., 2003). Gel retardation assays showed that a BLM mutant lacking 60 residues in its C-terminal tail (residues 1290–1350) fails to form higher-order protein-DNA complexes. In addition, this truncated mutant lacks strand annealing activity, suggesting that oligomerization might be required for the strand annealing function of BLM, as was observed for RECQ1 and WRN (Cheok et al., 2005). Nevertheless, Beresten et al. (1999) showed that an N-terminal fragment of BLM (residues 1–431) can form oligomers in solution (Beresten et al., 1999).
Collectively, the data available thus far indicate that human RecQ helicases are dynamic enzymes able to adopt different structures when bound to different cofactors such as DNA and ATP, with the exception of RECQ5 that remains monomeric both in its free and DNA∕ATP-bound forms (Garcia et al., 2004). The domains required for higher-order oligomer formation seem to be located in different regions of the different family members, i.e., in the first 48 residues at the N-terminus of RECQ1, but in the C-terminal domain of BLM and WRN. The function of the different assembly states of RecQ helicases is still the subject of debate. However, on the basis of the results obtained so far, it is tempting to speculate that they might share a common mechanism whereby smaller oligomers, which might be monomers in the case of BLM and WRN, and dimers in the case of RECQ1, are required for DNA unwinding, while higher-order oligomers are required for more specialized activities, such as Holliday junction branch migration∕disruption and DNA strand annealing. Further studies are however required to accurately measure the size of different quaternary structures of the various RecQ enzymes and achieve a better understanding of the mechanism(s) that regulates the switch between these different oligomeric forms.
In addition to the biochemical and structural approaches already used for several RecQ enzymes, a productive avenue for future research might be to utilize biophysical techniques, such as dynamic light scattering or analytical ultracentrifugation, to study the different assembly states of RecQ enzymes present in solution. Dynamic light scattering permits a precise determination of the hydrodynamic radius of a macromolecule in solution and, in combination with results derived from size exclusion chromatography experiments, might offer valuable information on the oligomeric state of a RecQ enzyme in the presence and absence of different cofactors (Cui et al., 2004; Matsui et al., 2006). In analytical ultracentrifugation, the macromolecules are visualized in real time during sedimentation allowing an accurate determination of thermodynamic parameters associated with the quaternary state of a protein (Dong et al., 1995; Galletto et al., 2004; Lee et al., 2003). The transmission electron microscopy (TEM) studies on negatively stained particles of the RECQ1 and BLM helicases described above provided the first low-resolution images on the higher-order assembly of these two RecQ enzymes, suggesting that they can form species comprising four to six RecQ protomers (Karow et al., 1999; Muzzolini et al., 2007). Cryoelectron microscopy experiments might also be used to obtain higher resolution images compared to the negative staining approach, and provide more accurate data on the size of the different oligomeric forms. This technique was applied successfully to elucidate the mechanism of double-hexamer formation of the SV40 large T antigen at viral replication origins and showed that the N-terminal region of the large T antigen mediates the interaction between hexamers (Valle et al., 2006). Finally, mass spectrometry is emerging as a very valuable new tool to define the stoichiometry of protein-DNA complexes and determine the active oligomeric state of an enzyme. The key aspect in such mass spectrometry analysis is to maintain interactions during the transition from the solution to the gas phase by using the appropriate ionization energy (Hanson and Robinson, 2004). As an example, the development of gentle ionization techniques led to the demonstration, using electrospray ionization (ESI) mass spectrometry, that the E. coli endoribonuclease E forms tetrameric structures able to bind up to four RNA molecules (Callaghan et al., 2003). None of the above approaches has been applied to a study of a RecQ helicase to date, but we believe that they could provide invaluable information on the size and stoichiometry of different RecQ-DNA complexes.
How the equilibrium between the different multimeric states and enzymatic activities of RecQ enzymes might be controlled in cells is still an open question. The results obtained for the human RECQ1 helicase indicate that ATP binding triggers a switch from the strand annealing or branch migration mode to a DNA unwinding mode (Muzzolini et al., 2007). However, additional cofactors that interact with a specific oligomeric form might stabilize a particular protein structure associated with a specific enzymatic activity of the RecQ enzyme in vivo. For example, human replication protein A (hRPA) is known to stimulate the helicase activity of RECQ1, WRN, and BLM, and might conceivably do so by binding to smaller oligomers with higher affinity than to higher-order oligomers. Similarly, other proteins, yet to be identified, may interact with the large oligomeric complexes with higher affinity and in this way promote the other specialized enzymatic activities of these helicases. A challenging avenue for future studies will be to apply the biochemical and biophysical approaches described above to test if newly discovered RecQ helicase binding proteins regulate the different enzymatic activities of these enzymes by specifically interacting with one or other oligomeric forms (Aygun et al., 2008; Selak et al., 2008; Singh et al., 2008; Xu et al., 2008). One candidate in the case of BLM is the RMI1∕RMI2 complex that, like RPA, is a multi-OB-fold containing a complex that is capable of stimulating BLM to disrupt Holliday junctions (Raynard et al., 2006; Wu et al., 2006).
It is a common observation that gene families tend to increase in complexity the higher up the evolutionary path one travels. The RecQs represent a classical example of this, as discussed above. However, the added complexity in, for example, human RecQ helicases compared to yeast does not appear simply to be a reflection of gene duplication followed by some minor divergence. Some human RecQs have acquired new catalytic activities; most notably, the exonuclease activity of WRN, while others seem to have dropped functions—or at least functional domains (RECQ4 only possesses the central helicase domain). This strong divergence of function seems to have been accompanied, not surprisingly, by the specialization of certain RecQs. It seems likely that human BLM has retained many if not most of the core activities∕roles of yeast Sgs1p or Rqh1p, while the other four human RecQs have diverged, acquired new partner proteins, and established roles in DNA replication, and transcription. Biochemical analysis of isolated RecQs will never be sufficient to tease out these new roles. A combination of genetic analysis in rodent models and cell biological studies will be needed to fully elucidate how this protein family impacts so dramatically on human disease. Such studies are complicated in the case of BLM or RECQ4, for example, by an apparent requirement for the protein during embryonic development. A major challenge for the future will be to create mouse models containing hypomorphic alleles of these genes that should provide definitive data on at least some of the roles of the encoded RecQ enzyme.
ACKNOWLEDGMENTS
We thank members of the Vindigni and Hickson laboratories for helpful comments, and O. Gileadi (Oxford, UK) for communicating results prior to publication. A. Vindigni is supported by grants from the Associazione Italiana per la Ricerca sul Cancro (AIRC) and HYPO Alpe-Adria bank, and I. D. Hickson is supported by Cancer Research UK and the Bloom’s Syndrome Foundation.
References
- Aygun, O, Svejstrup, J, and Liu, Y (2008). “A RECQ5-RNA polymerase, I. I. association identified by targeted proteomic analysis of human chromatin.” Proc. Natl. Acad. Sci. U.S.A. 105, 8580–8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrati, C Z, and Hickson, I D (2003). “RecQ helicases: suppressors of tumorigenesis and premature aging.” Biochem. J. 374, 577–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrati, C Z, and Hickson, I D (2008). “RecQ helicases: guardian angels of the DNA replication fork.” Chromosoma 117, 219–233. [DOI] [PubMed] [Google Scholar]
- Bahr, A, De Graeve, F, Kedinger, C, and Chatton, B (1998). “Point mutations causing Bloom’s syndrome abolish ATPase and DNA helicase activities of the BLM protein.” Oncogene 17, 2565–2571. [DOI] [PubMed] [Google Scholar]
- Bennett, R J, and Keck, J L (2004). “Structure and function of RecQ DNA helicases.” Crit. Rev. Biochem. Mol. Biol. 39, 79–97. [DOI] [PubMed] [Google Scholar]
- Bennett, R J, Sharp, J A, and Wang, J C (1998). “Purification and characterization of the Sgs1 DNA helicase activity of Saccharomyces cerevisiae.” J. Biol. Chem. 273, 9644–9650. [DOI] [PubMed] [Google Scholar]
- Beresten, S F, Stan, R, van Brabant, A J, Ye, T, Naureckiene, S, and Ellis, N A (1999). “Purification of overexpressed hexahistidine-tagged BLM N431 as oligomeric complexes.” Protein Expression Purif. 17, 239–248. [DOI] [PubMed] [Google Scholar]
- Berg, J M, and Shi, Y (1996). “The galvanization of biology: a growing appreciation for the roles of zinc.” Science 271, 1081–1085. [DOI] [PubMed] [Google Scholar]
- Bernstein, D A, and Keck, J L (2003). “Domain mapping of Escherichia coli RecQ defines the roles of conserved N- and C-terminal regions in the RecQ family.” Nucleic Acids Res. 31, 2778–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, D A, and Keck, J L (2005). “Conferring substrate specificity to DNA helicases: role of the RecQ HRDC domain.” Structure (London) 13, 1173–1182. [DOI] [PubMed] [Google Scholar]
- Bernstein, D A, Zittel, M C, and Keck, J L (2003). “High-resolution structure of the E. coli RecQ helicase catalytic core.” EMBO J. 22, 4910–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohr, V A (2008). “Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance.” Trends Biochem. Sci. 33, 609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh, R M, Jr., Li, J L, Kenny, M K, Karow, J K, Cooper, M P, Kureekattil, R P, Hickson, I D, and Bohr, V A (2000). “Replication protein A physically interacts with the Bloom’s syndrome protein and stimulates its helicase activity.” J. Biol. Chem. 275, 23500–23508. [DOI] [PubMed] [Google Scholar]
- Brosh, R M, Jr., Orren, D K, Nehlin, J O, Ravn, P H, Kenny, M K, Machwe, A, and Bohr, V A (1999). “Functional and physical interaction between WRN helicase and human replication protein A.” J. Biol. Chem. 274, 18341–18350. [DOI] [PubMed] [Google Scholar]
- Buttner, K, Nehring, S, and Hopfner, K P (2007). “Structural basis for DNA duplex separation by a superfamily-2 helicase.” Nat. Struct. Mol. Biol. 10.1038/nsmb1246 14, 647–652. [DOI] [PubMed] [Google Scholar]
- Callaghan, A J, Grossmann, J G, Redko, Y U, Ilag, L L, Moncrieffe, M C, Symmons, M F, Robinson, C V, McDowall, K J, and Luisi, B F (2003). “Quaternary structure and catalytic activity of the Escherichia coli ribonuclease E amino-terminal catalytic domain.” Biochemistry 42, 13848–13855. [DOI] [PubMed] [Google Scholar]
- Cheng, W H, von Kobbe, C, Opresko, P L, Fields, K M, Ren, J, Kufe, D, and Bohr, V A (2003). “Werner syndrome protein phosphorylation by abl tyrosine kinase regulates its activity and distribution.” Mol. Cell. Biol. 23, 6385–6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheok, C F, Wu, L, Garcia, P L, Janscak, P, and Hickson, I D (2005). “The Bloom’s syndrome helicase promotes the annealing of complementary single-stranded DNA.” Nucleic Acids Res. 33, 3932–3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary, S, Sommers, J A, and Brosh, R M, Jr., (2004). “Biochemical and kinetic characterization of the DNA helicase and exonuclease activities of Werner syndrome protein.” J. Biol. Chem. 279, 34603–34613. [DOI] [PubMed] [Google Scholar]
- Compton, S A, Tolun, G, Kamath-Loeb, A S, Loeb, L A, and Griffith, J D (2008). “The Werner syndrome protein binds replication fork and Holliday junction DNAs as an oligomer.” J. Biol. Chem. 283, 24478–24483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinou, A, Tarsounas, M, Karow, J K, Brosh, R M, Bohr, V A, Hickson, I D, and West, S C (2000). “Werner’s syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest.” EMBO Rep. 1, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, S, Arosio, D, Doherty, K M, Brosh, R M, Jr., Falaschi, A, and Vindigni, A (2004). “Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A.” Nucleic Acids Res. 32, 2158–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels, D S, Woo, T T, Luu, K X, Noll, D M, Clarke, N D, Pegg, A E, and Tainer, J A (2004). “DNA binding and nucleotide flipping by the human DNA repair protein AGT.” Nat. Struct. Mol. Biol. 11, 714–720. [DOI] [PubMed] [Google Scholar]
- Davies, S L, North, P S, and Hickson, I D (2007). “Role for BLM in replication-fork restart and suppression of origin firing after replicative stress.” Nat. Struct. Mol. Biol. 14, 677–679. [DOI] [PubMed] [Google Scholar]
- Dong, F, Gogol, E P, and von Hippel, P H (1995). “The phage T4-coded DNA replication helicase (gp41) forms a hexamer upon activation by nucleoside triphosphate.” J. Biol. Chem. 270, 7462–7473. [DOI] [PubMed] [Google Scholar]
- Eladad, S, Ye, T Z, Hu, P, Leversha, M, Beresten, S, Matunis, M J, and Ellis, N A (2005). “Intranuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification.” Hum. Mol. Genet. 14, 1351–1365. [DOI] [PubMed] [Google Scholar]
- Ellis, N A, Groden, J, Ye, T Z, Straughen, J, Lennon, D J, Ciocci, S, Proytcheva, M, and German, J (1995). “The Bloom’s syndrome gene product is homologous to RecQ helicases.” Cell 83, 655–666. [DOI] [PubMed] [Google Scholar]
- Fry, M, and Loeb, L A (1999). “Human Werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n.” J. Biol. Chem. 274, 12797–12802. [DOI] [PubMed] [Google Scholar]
- Gajiwala, K S, and Burley, S K (2000). “Winged helix proteins.” Curr. Opin. Struct. Biol. 10, 110–116. [DOI] [PubMed] [Google Scholar]
- Gajiwala, K S, Chen, H, Cornille, F, Roques, B P, Reith, W, Mach, B, and Burley, S K (2000). “Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding.” Nature (London) 403, 916–921. [DOI] [PubMed] [Google Scholar]
- Galletto, R, Maillard, R, Jezewska, M J, and Bujalowski, W (2004). “Global conformation of the Escherichia coli replication factor DnaC protein in absence and presence of nucleotide cofactors.” Biochemistry 43, 10988–11001. [DOI] [PubMed] [Google Scholar]
- Garcia, P L, Liu, Y, Jiricny, J, West, S C, and Janscak, P (2004). “Human RECQ5 beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide.” EMBO J. 23, 2882–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, R B, Rigolet, P, Zargarian, L, Fermandjian, S, and Xi, X G (2005). “Structural and functional characterizations reveal the importance of a zinc binding domain in Bloom’s syndrome helicase.” Nucleic Acids Res. 33, 3109–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada, K, and Hickson, I D (2007). “Molecular genetics of RecQ helicase disorders.” Cell. Mol. Life Sci. 64, 2306–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson, C L, and Robinson, C V (2004). “Protein-nucleic acid interactions and the expanding role of mass spectrometry.” J. Biol. Chem. 279, 24907–24910. [DOI] [PubMed] [Google Scholar]
- Harmon, F G, and Kowalczykowski, S C (2001). “Biochemical characterization of the DNA helicase activity of the Escherichia coli RecQ helicase.” J. Biol. Chem. 276, 232–243. [DOI] [PubMed] [Google Scholar]
- Hartung, F, and Puchta, H (2006). “The RecQ gene family in plants.” Plant Physiol. 163, 287–296. [DOI] [PubMed] [Google Scholar]
- Hickson, I D (2003). “RecQ helicases: caretakers of the genome.” Nat. Rev. Cancer 3, 169–178. [DOI] [PubMed] [Google Scholar]
- Hu, J S, Feng, H, Zeng, W, Lin, G X, and Xi, X G (2005). “Solution structure of a multifunctional DNA- and protein-binding motif of human Werner syndrome protein.” Proc. Natl. Acad. Sci. U.S.A. 102, 18379–18384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, P, Beresten, S F, van Brabant, A J, Ye, T Z, Pandolfi, P P, Johnson, F B, Guarente, L, and Ellis, N A (2001). “Evidence for BLM and topoisomerase IIIα interaction in genomic stability.” Hum. Mol. Genet. 10, 1287–1298. [DOI] [PubMed] [Google Scholar]
- Huang, S, Beresten, S, Li, B, Oshima, J, Ellis, N A, and Campisi, J (2000). “Characterization of the human and mouse WRN 3′→5′ exonuclease.” Nucleic Acids Res. 28, 2396–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S, Li, B, Gray, M D, Oshima, J, Mian, I S, and Campisi, J (1998). “The premature ageing syndrome protein WRN is a 3′→5′ exonuclease.” Nat. Genet. 20, 114–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber, M D, Duquette, M L, Shiels, J C, and Maizels, N (2006). “A conserved G4 DNA binding domain in RecQ family helicases.” J. Mol. Biol. 358, 1071–1080. [DOI] [PubMed] [Google Scholar]
- Janscak, P, Garcia, P L, Hamburger, F, Makuta, Y, Shiraishi, K, Imai, Y, Ikeda, H, and Bickle, T A (2003). “Characterization and mutational analysis of the RecQ core of the bloom syndrome protein.” J. Mol. Biol. 330, 29–42. [DOI] [PubMed] [Google Scholar]
- Johnson, F B, et al. (2000). “Association of the Bloom syndrome protein with topoisomerase IIIα in somatic and meiotic cells.” Cancer Res. 60, 1162–1167. [PubMed] [Google Scholar]
- Karmakar, P, Piotrowski, J, Brosh, R M, Jr., Sommers, J A, Miller, S P, Cheng, W H, Snowden, C M, Ramsden, D A, and Bohr, V A (2002). “Werner protein is a target of DNA-dependent protein kinase in vivo and in vitro, and its catalytic activities are regulated by phosphorylation.” J. Biol. Chem. 277, 18291–18302. [DOI] [PubMed] [Google Scholar]
- Karow, J K, Constantinou, A, Li, J L, West, S C, and Hickson, I D (2000). “The Bloom’s syndrome gene product promotes branch migration of Holliday junctions.” Proc. Natl. Acad. Sci. U.S.A. 97, 6504–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow, J K, Newman, R H, Freemont, P S, and Hickson, I D (1999). “Oligomeric ring structure of the Bloom’s syndrome helicase.” Curr. Biol. 9, 597–600. [DOI] [PubMed] [Google Scholar]
- Killoran, M P, and Keck, J L (2006). “Three HRDC domains differentially modulate Deinococcus radiodurans RecQ DNA helicase biochemical activity.” J. Biol. Chem. 281, 12849–12857. [DOI] [PubMed] [Google Scholar]
- Killoran, M P, and Keck, J L (2008). “Structure and function of the regulatory C-terminal HRDC domain from Deinococcus radiodurans RecQ.” Nucleic Acids Res. 36, 3139–3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano, K, Yoshihara, N, and Hakoshima, T (2007). “Crystal structure of the HRDC domain of human Werner syndrome protein, WRN.” J. Biol. Chem. 282, 2717–2728. [DOI] [PubMed] [Google Scholar]
- Kitao, S, Lindor, N M, Shiratori, M, Furuichi, Y, and Shimamoto, A (1999). “Rothmund–Thomson syndrome responsible gene, RECQL4: genomic structure and products.” Genomics 61, 268–276. [DOI] [PubMed] [Google Scholar]
- Korolev, S, Hsieh, J, Gauss, G H, Lohman, T M, and Waksman, G (1997). “Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP.” Cell 10.1016/S0092-8674(00)80525-5 90, 635–647. [DOI] [PubMed] [Google Scholar]
- Lee, J W, Harrigan, J, Opresko, P L, and Bohr, V A (2005). “Pathways and functions of the Werner syndrome protein.” Mech. Ageing Dev. 126, 79–86. [DOI] [PubMed] [Google Scholar]
- Lee, J Y, and Yang, W (2006). “UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke.” Cell 127, 1349–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y C, Flora, R, McCafferty, J A, Gor, J, Tsaneva, I R, and Perkins, S J (2003). “A tetramer-octamer equilibrium in Mycobacterium leprae and Escherichia coli RuvA by analytical ultracentrifugation.” J. Mol. Biol. 333, 677–682. [DOI] [PubMed] [Google Scholar]
- Leng, M, Chan, D W, Luo, H, Zhu, C, Qin, J, and Wang, Y (2006). “MPS1-dependent mitotic BLM phosphorylation is important for chromosome stability.” Proc. Natl. Acad. Sci. U.S.A. 103, 11485–11490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillard-Wetherell, K, Machwe, A, Langland, G T, Combs, K A, Behbeheni, G K, Schonberg, S A, German, J, Turchi, J J, Orren, D K, and Groden, J (2004). “Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2.” Hum. Mol. Genet. 13, 1919–1932. [DOI] [PubMed] [Google Scholar]
- Liu, J L, Rigolet, P, Dou, S X, Wang, P Y, and Xi, X G (2004). “The zinc finger motif of Escherichia coli RecQ is implicated in both DNA binding and protein folding.” J. Biol. Chem. 279, 42794–42802. [DOI] [PubMed] [Google Scholar]
- Liu, Z, Macias, M J, Bottomley, M J, Stier, G, Linge, J P, Nilges, M, Bork, P, and Sattler, M (1999). “The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins.” Structure (London) 7, 1557–1566. [DOI] [PubMed] [Google Scholar]
- Lohman, T M, Tomko, E J, and Wu, C G (2008). “Non-hexameric DNA helicases and translocases: mechanisms and regulation.” Nat. Rev. Mol. Cell Biol. 9, 391–401. [DOI] [PubMed] [Google Scholar]
- Lu, J, Mullen, J R, Brill, S J, Kleff, S, Romeo, A M, and Sternglanz, R (1996). “Human homologues of yeast helicase.” Nature (London) 383, 678–679. [DOI] [PubMed] [Google Scholar]
- Machwe, A, Xiao, L, Groden, J, Matson, S W, and Orren, D K (2005). “RecQ family members combine strand pairing and unwinding activities to catalyze strand exchange.” J. Biol. Chem. 280, 23397–23407. [DOI] [PubMed] [Google Scholar]
- Macris, M A, Krejci, L, Bussen, W, Shimamoto, A, and Sung, P (2005). “Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome.” DNA Repair 5, 172–180. [DOI] [PubMed] [Google Scholar]
- Mandell, J G, Goodrich, K J, Bahler, J, and Cech, T R (2005). “Expression of a RecQ helicase homolog affects progression through crisis in fission yeast lacking telomerase.” J. Biol. Chem. 280, 5249–5257. [DOI] [PubMed] [Google Scholar]
- Matson, S W, Bean, D W, and George, J W (1994). “DNA helicases: enzymes with essential roles in all aspects of DNA metabolism.” BioEssays 16, 13–22. [DOI] [PubMed] [Google Scholar]
- Matsui, T, Hogetsu, K, Usukura, J, Sato, T, Kumasaka, T, Akao, Y, and Tanaka, N (2006). “Structural insight of human DEAD-box protein rck∕p54 into its substrate recognition with conformational changes.” Genes Cells 11, 439–452. [DOI] [PubMed] [Google Scholar]
- McGlynn, P, and Lloyd, R G (2002). “Genome stability and the processing of damaged replication forks by RecG.” Trends Genet. 18, 413–419. [DOI] [PubMed] [Google Scholar]
- Mohaghegh, P, Karow, J K, Brosh, Jr, R M, Bohr, V A, and Hickson, I D (2001). “The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases.” Nucleic Acids Res. 29, 2843–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov, V, Mushegian, A R, Koonin, E V, and Bork, P (1997). “A putative nucleic acid-binding domain in Bloom’s and Werner’s syndrome helicases.” Trends Biochem. Sci. 22, 417–418. [DOI] [PubMed] [Google Scholar]
- Muftuoglu, M, Kulikowicz, T, Beck, G, Lee, J W, Piotrowski, J, and Bohr, V A (2008). “Intrinsic ssDNA annealing activity in the C-terminal region of WRN.” Biochemistry 47, 10247–10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushegian, A R, Bassett, D E, Jr., Boguski, M S, Bork, P, and Koonin, E V (1997). “Positionally cloned human disease genes: patterns of evolutionary conservation and functional motifs.” Proc. Natl. Acad. Sci. U.S.A. 94, 5831–5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzzolini, L, Beuron, F, Patwardhan, A, Popuri, V, Cui, S, Niccolini, B, Rappas, M, Freemont, P S, and Vindigni, A (2007). “Different quaternary structures of human RECQ1 are associated with its dual enzymatic activity.” PLoS Biol. 5, e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama, H, Nakayama, K, Nakayama, R, Irino, N, Nakayama, Y, and Hanawalt, P C (1984). “Isolation and genetic characterization of a thymineless death-resistant mutant of Escherichia coli K12: identification of a new mutation (recQ1) that blocks the RecF recombination pathway.” Mol. Gen. Genet. 195, 474–480. [DOI] [PubMed] [Google Scholar]
- Onoda, F, Seki, M, Miyajima, A, and Enomoto, T (2000). “Elevation of sister chromatid exchange in Saccharomyces cerevisiae sgs1 disruptants and the relevance of the disruptants as a system to evaluate mutations in Bloom’s syndrome gene.” Mutat Res. 459, 203–209. [DOI] [PubMed] [Google Scholar]
- Opresko, P L, Cheng, W H, and Bohr, V A (2004). “Junction of RecQ helicase biochemistry and human disease.” J. Biol. Chem. 279, 18099–18102. [DOI] [PubMed] [Google Scholar]
- Opresko, P L, Laine, J P, Brosh, R M, Jr., Seidman, M M, and Bohr, V A (2001). “Coordinate action of the helicase and 3′ to 5′ exonuclease of Werner syndrome protein.” J. Biol. Chem. 276, 44677–44687. [DOI] [PubMed] [Google Scholar]
- Oshige, M, Aoyagi, N, Harris, P V, Burtis, K C, and Sakaguchi, K (1999). “A new DNA polymerase species from Drosophila melanogaster: a probable mus308 gene product.” Mutat Res. 433, 183–192. [DOI] [PubMed] [Google Scholar]
- Ouyang, K J, Woo, L L, and Ellis, N A (2008). “Homologous recombination and maintenance of genome integrity: cancer and aging through the prism of human RecQ helicases.” Mech. Ageing Dev. 129, 425–440. [DOI] [PubMed] [Google Scholar]
- Perry, J J, Yannone, S M, Holden, L G, Hitomi, C, Asaithamby, A, Han, S, Cooper, P K, Chen, D J, and Tainer, J A (2006). “WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.” Nat. Struct. Mol. Biol. 13, 414–422. [DOI] [PubMed] [Google Scholar]
- Pichierri, P, Rosselli, F, and Franchitto, A (2003). “Werner’s syndrome protein is phosphorylated in an ATR∕ATM-dependent manner following replication arrest and DNA damage induced during the S phase of the cell cycle.” Oncogene 22, 1491–1500. [DOI] [PubMed] [Google Scholar]
- Pike, A, Shrestha, B, Popuri, V, Burgess-Brown, N, Muzzolini, L, Costantini, S, Vindigni, A, and Gileadi, O (2008). “Structure of the human RECQ1 helicase: identification of a putative strand-separation pin.” Proc. Natl. Acad. Sci. U.S.A. 106, 1039–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popuri, V, Bachrati, C Z, Muzzolini, L, Mosedale, G, Costantini, S, Giacomini, E, Hickson, I D, and Vindigni, A (2008). “The human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities.” J. Biol. Chem. 283, 17766–17776. [DOI] [PubMed] [Google Scholar]
- Raynard, S, Bussen, W, and Sung, P (2006). “A double Holliday junction dissolvasome comprising BLM topoisomerase IIIα and BLAP75.” J. Biol. Chem. 281, 13861–13864. [DOI] [PubMed] [Google Scholar]
- Sangrithi, M N, Bernal, J A, Madine, M, Philpott, A, Lee, J, Dunphy, W G, and Venkitaraman, A R (2005). “Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome.” Cell 121, 887–898. [DOI] [PubMed] [Google Scholar]
- Schultz, S C, Shields, G C, and Steitz, T A (1991). “Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees.” Science 253, 1001–1007. [DOI] [PubMed] [Google Scholar]
- Seki, M, Marini, F, and Wood, R D (2003). “POLQ (Pol θ): a DNA polymerase and DNA-dependent ATPase in human cells.” Nucleic Acids Res. 31, 6117–6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selak, N, Bachrati, C Z, Shevelev, I, Dietschy, T, van Loon, B, Jacob, A, Hübscher, U, Hoheisel, J D, Hickson, I D, and Stagljar, I (2008). “The Bloom’s syndrome helicase (BLM) interacts physically and functionally with p12, the smallest subunit of human DNA polymerase delta.” Nucleic Acids Res. 36, 5166–5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, S, Doherty, K M, and Brosh, R M, Jr. (2006). “Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability.” Biochem. J. 398, 319–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, S, Sommers, J A, Choudhary, S, Faulkner, J K, Cui, S, Andreoli, L, Muzzolini, L, Vindigni, A, and Brosh, R M, Jr. (2005). “Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1.” J. Biol. Chem. 280, 28072–28084. [DOI] [PubMed] [Google Scholar]
- Shen, J C, Gray, M D, Oshima, J, Kamath-Loeb, A S, Fry, M, and Loeb, L A (1998). “Werner syndrome protein. I: DNA helicase and DNA exonuclease reside on the same polypeptide.” J. Biol. Chem. 273, 34139–34144. [DOI] [PubMed] [Google Scholar]
- Siitonen, H A, Kopra, O, Kaariainen, H, Haravuori, H, Winter, R M, Saamanen, A M, Peltonen, L, and Kestila, M (2003). “Molecular defect of RAPADILINO syndrome expands the phenotype spectrum of RECQL diseases.” Hum. Mol. Genet. 12, 2837–2844. [DOI] [PubMed] [Google Scholar]
- Singh, T R, Ali, A M, Busygina, V, Raynard, S, Fan, Q, Du, C H, Andreassen, P R, Sung, P, and Meetei, A R (2008). “BLAP18∕RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome.” Genes Dev. 22, 2856–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton, M R, Dillingham, M S, and Wigley, D B (2007). “Structure and mechanism of helicases and nucleic acid translocases.” Annu. Rev. Biochem. 76, 23–50. [DOI] [PubMed] [Google Scholar]
- Singleton, M R, and Wigley, D B (2002). “Modularity and specialization in superfamily 1 and 2 helicases.” J. Bacteriol. 10.1128/JB.184.7.1819-1826.2002 184, 1819–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H, Bennett, R J, and Maizels, N (1999). “The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G-G paired DNAs.” Nucleic Acids Res. 27, 1978–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H, Karow, J K, Hickson, I D, and Maizels, N (1998). “The Bloom’s syndrome helicase unwinds G4 DNA.” J. Biol. Chem. 273, 27587–27592. [DOI] [PubMed] [Google Scholar]
- Valle, M, Chen, X S, Donate, L E, Fanning, E, and Carazo, J M (2006). “Structural basis for the cooperative assembly of large T antigen on the origin of replication.” J. Mol. Biol. 357, 1295–1305. [DOI] [PubMed] [Google Scholar]
- Van Maldergem, L, et al. (2006). “Revisiting the craniosynostosis-radial ray hypoplasia association: Baller-Gerold syndrome caused by mutations in the RECQL4 gene.” J. Med. Genet. 43, 148–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velankar, S S, Soultanas, P, Dillingham, M S, Subramanya, H S, and Wigley, D B (1999). “Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism.” Cell 10.1016/S0092-8674(00)80716-3 97, 75–84. [DOI] [PubMed] [Google Scholar]
- von Hippel, P H (2004). “Helicases become mechanistically simpler and functionally more complex.” Nat. Struct. Mol. Biol. 11, 494–496. [DOI] [PubMed] [Google Scholar]
- von Kobbe, C, Thoma, N H, Czyzewski, B K, Pavletich, N P, and Bohr, V A (2003). “Werner syndrome protein contains three structure-specific DNA binding domains.” J. Biol. Chem. 278, 52997–53006. [DOI] [PubMed] [Google Scholar]
- Wu, L, Bachrati, C Z, Ou, J, Xu, C, Yin, J, Chang, M, Wang, W, Li, L, Brown, G W, and Hickson, I D (2006). “BLAP75∕RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates.” Proc. Natl. Acad. Sci. U.S.A. 103, 4068–4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, L, Chan, K L, Ralf, C, Bernstein, D A, Garcia, P L, Bohr, V A, Vindigni, A, Janscak, P, Keck, J L, and Hickson, I D (2005). “The HRDC domain of BLM is required for the dissolution of double Holliday junctions.” EMBO J. 24, 2679–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, L, Davies, S L, Levitt, N C, and Hickson, I D (2001). “Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51.” J. Biol. Chem. 276, 19375–19381. [DOI] [PubMed] [Google Scholar]
- Wu, L, Davies, S L, North, P S, Goulaouic, H, Riou, J F, Turley, H, Gatter, K C, and Hickson, I D (2000). “The Bloom’s syndrome gene product interacts with topoisomerase III.” J. Biol. Chem. 275, 9636–9644. [DOI] [PubMed] [Google Scholar]
- Wu, L, and Hickson, I D (2003). “The Bloom’s syndrome helicase suppresses crossing over during homologous recombination.” Nature (London) 10.1038/nature02253 426, 870–874. [DOI] [PubMed] [Google Scholar]
- Wu, X, and Maizels, N (2001). “Substrate-specific inhibition of RecQ helicase.” Nucleic Acids Res. 29, 1765–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, D, Guo, R, Sobeck, A, Bachrati, C Z, Yang, J, Enomota, T, Brown, G W, Hoatlin, M E, Hickson, I E, and Wang, W (2008). “RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability.” Genes Dev. 22, 2843–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H Q, Deprez, E, Zhang, A H, Tauc, P, Ladjimi, M M, Brochon, J C, Auclair, C, and Xi, X G (2003). “The Escherichia coli RecQ helicase functions as a monomer.” J. Biol. Chem. 278, 34925–34933. [DOI] [PubMed] [Google Scholar]
- Xue, Y, Ratcliff, G C, Wang, H, Davis-Searles, P R, Gray, M D, Erie, D A, and Redinbo, M R (2002). “A minimal exonuclease domain of WRN forms a hexamer on DNA and possesses both 3′–5′ exonuclease and 5′-protruding strand endonuclease activities.” Biochemistry 41, 2901–2912. [DOI] [PubMed] [Google Scholar]
- Yannone, S M, Roy, S, Chan, D W, Murphy, M B, Huang, S, Campisi, J, and Chen, D J (2001). “Werner syndrome protein is regulated and phosphorylated by DNA-dependent protein kinase.” J. Biol. Chem. 276, 38242–38248. [DOI] [PubMed] [Google Scholar]
- Yu, C E, et al. (1996). “Positional cloning of the Werner’s syndrome gene.” Science 272, 258–262. [DOI] [PubMed] [Google Scholar]
- Zhang, X D, Dou, S X, Xie, P, Hu, J S, Wang, P Y, and Xi, X G (2006). “Escherichia coli RecQ is a rapid, efficient, and monomeric helicase.” J. Biol. Chem. 281, 12655–12663. [DOI] [PubMed] [Google Scholar]