Abstract
During gametogenesis, germ cells must undergo meiosis in order to become viable haploid gametes. Successful completion of this process is dependent upon the expression of genes whose protein products function specifically in meiosis. Failure to express these genes in meiotic cells often results in infertility, whereas aberrant expression in somatic cells may lead to mitotic catastrophe. The mechanisms responsible for regulating the timely expression of meiosis-specific genes have not been fully elucidated. Here we demonstrate that E2F6, a member of the E2F family of transcription factors, is essential for the repression of the newly identified meiosis-specific gene, Slc25a31 (also known as Ant4, Aac4), in somatic cells. This discovery, along with previous studies, prompted us to investigate the role of E2F6 in the regulation of meiosis-specific genes in general. Interestingly, the core E2F6-binding element (TCCCGC) was highly conserved in the proximal promoter regions of 19 out of 24 (79.2%) meiosis-specific genes. This was significantly higher than the frequency found in the promoters of all mouse genes (15.4%). In the absence of E2F6, only a portion of these meiosis-specific genes was derepressed in somatic cells. However, endogenous E2F6 bound to the promoters of these meiosis-specific genes regardless of whether they required E2F6 for their repression in somatic cells. Further, E2F6 overexpression was capable of reducing their transcription. These findings indicate that E2F6 possesses a broad ability to bind to and regulate the meiosis-specific gene population..
Keywords: E2F6, gene regulation, meiosis, repression, Slc25a31
Meiosis-specific genes often harbor an E2F6-binding element in their proximal promoter regions upon which E2F6 binds and reduces expression, indicating that E2F6 may broadly serve as a repressor of meiosis-specific genes in somatic cells.
INTRODUCTION
Successful germ cell development requires that many sophisticated mechanisms coordinately regulate and precisely control the temporal and stage-specific expression patterns of a multitude of genes. Failure to properly regulate the expression of any one of these genes frequently results in a partial or complete loss of fertility (reviewed in [1–2]). Of interest, many of these critical genes are germ-cell-specific and are therefore repressed in all somatic cells in the body. The mechanisms controlling germ-cell-specific gene expression are diverse (reviewed in [3–5]). Studies have found these genes to be regulated extrinsically by hormones secreted from the endocrine system, interactively by factors released from neighboring supportive cells in the gonads, and intrinsically by factors affecting transcription, translation, DNA methylation, and histone modifications. Among such germ-cell-specific genes, it is especially important that those genes that are highly expressed and critical during the meiotic phase of gametogenesis be appropriately regulated. Aberrant expression of these meiotic genes in somatic cells is presumed to be associated with disruptions in the mitotic cell cycle, which may lead to dire consequences such as oncogenic transformation [6–7].
Much of our current understanding regarding the transcriptional regulation of meiotic genes can be attributed to the use of transgenic mouse models. These studies have demonstrated that short proximal promoter regions of meiotic genes are sufficient to specify their silencing in somatic cells and activation in germ cells [8–13]. A more detailed analysis of these minimal promoter regions revealed DNA regulatory elements harboring transcription factor binding sites. Further, incubation of these sequences with nuclear extracts from either somatic or testicular tissue indicated that soma/germ-specific nuclear proteins were indeed binding to these elements. For instance, Mos, a gene found to contain a negative regulatory element in its proximal promoter, is bound by a protein present only in nuclear extracts from somatic cells and not from pachytene spermatocytes [14]. This observation, coupled with the absence of expression of Mos in somatic cells, infers that this protein may be serving as a transcriptional repressor. Further studies revealed that this protein was COUP-TF [15]. Additional transcription factors that bind to various germ cell-specific genes expressed during meiosis are beginning to be uncovered and include SP1, SP3, CTF1, RFX1, RFX2, CTCF, and MYBL2 [16–20]. However, the existence and identity of a master regulatory protein or protein family that binds to the proximal promoters and coordinately regulates the expression of multiple meiotic genes uniformly as a group remains to be elucidated.
The E2F family of transcription factors is traditionally known for regulating the expression of genes whose protein products are essential for cell cycle progression, DNA repair, cellular proliferation, and differentiation (reviewed in [21–22]). The E2F family is composed of eight members, E2F1–E2F8, and two heterodimeric binding partners, DP1 and DP2. E2F1–E2F5 contain transactivation domains and pocket protein binding domains. E2F1, E2F2, and E2F3a serve as transcriptional activators, whereas E2F3b, E2F4, and E2F5 function as transcriptional repressors. E2F6–E2F8 lack transactivation domains and are pocket protein-independent transcriptional repressors [23–26]. Recently, E2F6 was shown to be required for the repression of a limited number of germ cell-specific-genes, including Tuba3a, Tuba3b, Gm1564, Tex12, Stag3, and Smc1b, in somatic cells [27–28]. These six genes all contain the core E2F6-binding element, TCCCGC, within their proximal promoter regions [29]. However, the functions of these genes are diverse, and it appears that the only common feature by which to classify them into a category of E2F6-target genes is their germ cell-specific expression pattern. In the present study, we demonstrate that E2F6 is required for the repression of the newly identified germ cell-specific gene, Slc25a31, in somatic cells. Interestingly, Slc25a31 contains the core E2F6-binding element within its proximal promoter region and, similar to Tex12, Stag3, and Smc1b, its protein product is selectively expressed in meiotic cells [30]. The meiosis-specific expression pattern of these four genes prompted us to investigate whether additional genes in this subcategory of germ cell-specific genes were downstream targets of E2F6.
MATERIALS AND METHODS
Cell Culture and Creation of Stable Cell Lines
The cell lines used in this study were murine wild-type (wt) R1 embryonic stem (ES) cells, HA-E2F6 ES cells, HA-ΔC-E2F6 ES cells, and NIH3T3 cells (American Type Culture Collection, Manassas, VA). Both HA-E2F6 and HA-ΔC-E2F6 ES cell lines were established by stable transfection of linearized HA-E2F6 and HA-ΔC-E2F6 expression vectors [31] into parental R1 ES cells. Stable clones were subsequently confirmed to express E2F6 by Western blotting with anti-HA antibody (Cell Signaling Technology, Danvers, MA). All ES cells were maintained in an undifferentiated state on gelatin-coated dishes in Knock-out Dulbecco modified Eagle medium (DMEM), low glucose (Invitrogen, Carlsbad, CA), containing 10% Knockout Serum Replacement (Invitrogen), 1% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA), and 1000 U/ml recombinant mouse LIF (ESGRO; Millipore, Temecula, CA). NIH3T3 cells were maintained in DMEM (Invitrogen) containing 10% FBS.
Gel Mobility Shift Assays
Gel mobility shift assay was performed as described previously [31]. Briefly, 5 μl of reticulocyte lysates with or without in vitro-translated proteins (see below) were incubated with 0.1 pmol of PAGE-purified, γ32P-end-labeled, annealed oligonucleotides (5′-TCAGCGCCCGCTTTCCCGCCAGGGTAAAGCT-3′) corresponding to the wt E2F6-binding element (underlined) in the murine Slc25a31 promoter. Competition experiments were performed with wt and mutant (5′-TCAGCGCCCGCTTTCTTAACAGGGTAAAGCT-3′) unlabeled, double-stranded oligonucleotides corresponding to the E2F6 site derived from the Slc25a31 promoter, whereby the amount of unlabeled probe relative to the amount of labeled probe was either 20-, 50-, or 100-fold in excess. For preparation of in vitro-translated proteins, 1 μg of HA-E2F6 and Myc-DP2 expression vectors (constructed as described previously [31]) were cotranslated in vitro using a coupled transcription/translation reticulocyte lysate system (Promega, Madison, WI) according to the manufacturer's instructions.
Chromatin Immunoprecipitation Assays
R1 ES cells were plated on 100-mm plates at a density of 1 × 106 cells per plate and then cross-linked with formaldehyde solution and lysed. Nuclei were sonicated for 10 repetitions of 10-sec bursts with 1-min rests on ice using a power setting of 4 on a Fisher Scientific Sonicator Dismembrator Model 100 (Fisher, Waltham, MA). Lysates were centrifuged for 10 min and supernatants were collected and diluted, and a sample was kept as Input DNA. After preclearing with Protein G agarose (Invitrogen), approximately 2 μg of each antibody, control IgG (Sigma-Aldrich, St. Louis, MO) and mouse monoclonal E2F6 [32], were added to the supernatant and incubated overnight at 4°C. The following day, 60 μl of Protein G Agarose-50% slurry was added to the reaction, the agarose complex was collected and washed seven times with LiCl wash buffer (0.25 M LiCl, 0.5% NP-40, 0.5% DOC, 1 mM EDTA, and 10 mM Tris [pH 8.0]), and DNA was eluted off with elution buffer (50 mM Tris [pH 8.0], 1% SDS, and 10 mM EDTA). Cross-linking was reversed and DNA was recovered using phenol/chloroform extraction followed by PCR Purification (Qiagen, Valencia, CA). PCR was performed with either semiquantitative PCR using the Taq DNA polymerase kit (Eppendorf, Westbury, NY) or with real-time quantitative PCR (as described below). The primer sequences used for semiquantitative PCR analysis of chromatin immunoprecipitation (ChIP) are described in Supplemental Table 1 (available online at www.biolreprod.org ).
Real-Time Quantitative PCR
The primers and probes used for real-time PCR were designed with Primer Express software and synthesized at Applied Biosystems (Foster City, CA). Each real-time PCR reaction was performed using TaqMan Universal PCR Master Mix (Applied Biosystems) and TaqMan Gene Expression Assay Mix (Applied Biosystems). The reaction was carried out using the Applied Biosystems 7900HT Real-Time PCR System. The TaqMan primers and probes for monitoring transcription levels were assay-on-demand gene expression products labeled with FAM reporter dye (Applied Biosystems). A mixture of mouse Actb primers with VIC-labeled Actb probe was used as endogenous control (catalog number 4352341E; Applied Biosystems). The primer sequences and probes for quantifying E2F6 enrichment after performing ChIP analysis were custom ordered: Actb: sense primer 5′-CGGAGGCTATTCCTGTACATCTG-3′, antisense primer 5′-CGAGATTGAGGAAGAGGATGAAGAG-3′, FAM-labeled probe 5′-CCAGCACCCATCGCC-3′; Slc25a31: sense primer 5′-GCTGTTCTCCCAGCATCCT-3′, antisense primer 5′-GAGAACTGGAAAACCGCTTCAG-3′, FAM-labeled probe 5′-CTTTCCCGCCAGGGTAA-3′; Dmc1: sense primer 5′-GGCCCCGCCCATCAA-3′, antisense primer 5′-CGCCCTGTCGTCGAACA-3′, and FAM-labeled probe 5′-CCGCGGCCCTCATT-3′. All samples were tested in triplicate and expressed as means ± SD. Analysis of results was performed using SDS v2.3 software (Applied Biosystems) according to the manufacturer's instructions. The comparative CT method (ΔΔCT) was used for quantification of gene expression.
Plasmid Construction and Site-Directed Mutagenesis
The Slc25a31 promoter region (from −263 bp to +25 bp) was PCR-amplified from mouse R1 ES cells' genomic DNA with high fidelity LA-Taq (Takara, Otsu, Japan) using the following Slc25a31 primers: sense primer 5′-AATCACCGGGTTGGTGTAG-3′, antisense primer 5′-GCCACACCAACACTCAAGC-3′. The 288-bp fragment was excised with Xho1 and HindIII (New England Biolabs, Ipswich, MA) using a QIAquick gel extraction kit (Qiagen). The Takara DNA ligation kit was then used to ligate the fragment of the Slc25a31 promoter into a PGL2-basic vector (Promega) containing a luciferase reporter gene. Mutations in the Slc25a3-luciferase reporter vector at the location of the E2F6-binding element were generated using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) in combination with the following PAGE-purified primers: Slc25a31: sense, 5′-CCCAGCATCCTCAGCGCCCGCGCCAGGGTAAAGCTGAAGCGG-3′, antisense, 5′-CCGCTTCAGCTTTACCCTGGCGCGGGCGCTGAGGATGCTGGG-3′ (site of mutation is underlined).
Transient Transfection and Reporter Assays
NIH3T3 cells were plated at a density of 1 × 105 cells per well in six-well plates. After 24 h, FuGENE 6 (Roche, Basel, Switzerland) was used to transfect the following according to the manufacturer's instructions: 0.5 μg Slc25a31-luciferase reporter vector (wt or mutant), 0.1 μg pRL-TK-Renilla internal control vector (Promega), and 1.0 μg of one of the following vectors: HA-E2F6, HA-ΔC-E2F6, HA-E68-E2F6, HA-ΔN-E2F6 (constructed as we previously described [31]), or pCMVTag2 empty control vector (Stratagene). Twenty-four hours after transfection, the cells were harvested. Firefly and Renilla luciferase activities were measured using a dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions. The firefly luciferase data for each sample was normalized based on transfection efficiency as measured by Renilla luciferase activity. Data from at least three independent experiments were analyzed and expressed as means ± SD. Statistical analysis was performed by Student t-test, and P values of less than 0.05 were considered significant.
Immunoblotting
NIH3T3 cells transiently transfected, using Fugene 6, with one of four E2F6 expression vectors—HA-E2F6, HA-ΔC-E2F6, HA-E68-E2F6, or HA-ΔN-E2F6—were lysed in RIPA buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% NP-40, 0.5% Na-Deoxycholate, and 0.1% SDS) plus protease inhibitors, and then harvested. Total protein was normalized by Lowry assay (Bio-Rad, Hercules, CA). Next, 25 μg of total protein was separated on a 12% SDS-PAGE gel and transferred to a nitrocellulose membrane. The membrane was probed with primary antibody directed against HA (Cell Signaling Technology).
Reverse Transcription PCR
Total RNA was extracted using an RNAqueous kit (Ambion, Austin, TX). The cDNA was synthesized using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer's instructions. PCR was performed using Taq DNA polymerase (Eppendorf) with the primer sequences listed in Supplemental Table 2 (available online at www.biolreprod.org ). The sense primers were always designed in different exons than the antisense primers in order to ensure that the PCR product represented the specific mRNA species and not genomic DNA background.
Computational Analysis
All genomic DNA sequences of meiosis-specific genes were downloaded from the University of California at Santa Cruz (UCSC) Genome Bioinformatics Web site ( http://genome.ucsc.edu ) [33]. An interspecies comparison of promoter sequences was also carried out using UCSC Genome Bioinformatics tools. A genome-scale DNA pattern-matching algorithm was used to identify all core E2F6-binding elements within the proximal promoter regions of all genes in the entire mouse genome (based on 31 113 genes in the database) using Regulatory Sequence Analysis Tools (RSAT) software ( http://rsat.scmbb.ulb.ac.be/rsat/ ) [34].
Preparation of Stage-Specific Spermatogenic Cells
Stage-specific spermatogenic cells were prepared as we described previously [30].
RESULTS
E2F6 Binds to the Proximal Promoter Region of the Slc25a31 Gene
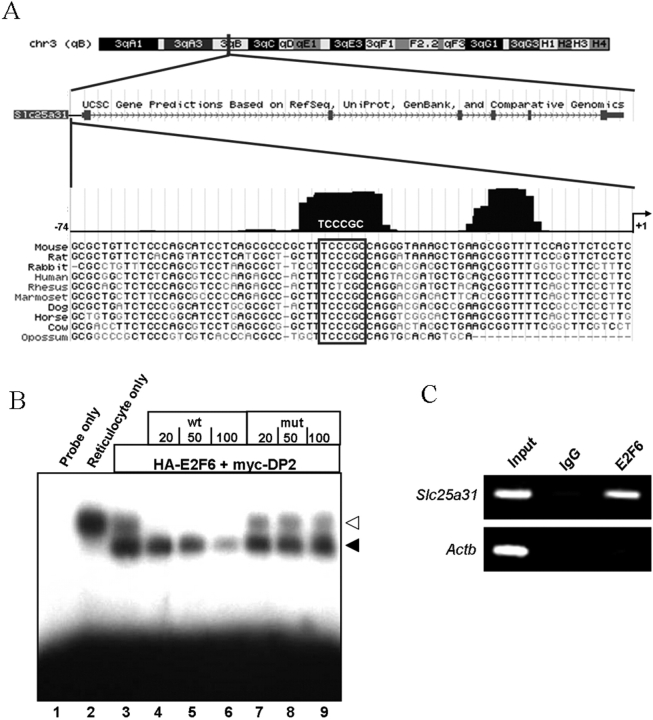
Slc25a31 (also known as Ant4, Aac4) is a newly identified germ cell-specific gene and is a member of the adenine nucleotide translocase family [35–37]. Slc25a31 is selectively expressed in meiotic male germ cells and is essential for male meiosis [30]. Although our previous studies determined that the repression of Slc25a31 in somatic cells is dependent upon CpG methylation of its proximal promoter region [36], its transcriptional regulation has remained largely undetermined. By comparing Slc25a31 genomic sequences from ten mammalian species using the UCSC Genome Browser [34], here we identified two conserved regions within 74 bp upstream of the transcription initiation site (Fig. 1A). Further, we located one E2F family transcription factor binding site (TFBS) with the core E2F6-binding sequence, TCCCGC, within one of these conserved regions (Fig. 1A). This potential E2F6 TFBS was located 36 bp upstream of the transcription initiation site and was conserved in 8 out of 10 mammalian species (Fig. 1A).
FIG. 1.
The proximal promoter region of the Slc25a31 gene has a conserved E2F6 binding site. A) UCSC Genome Browser prediction of the genomic location and structure of the mouse Slc25a31 gene (labeled Slc25a31) followed by a 10-way mammalian species comparison of the Slc25a31 proximal promoter region spanning −74 bp to +1 bp. Conserved regions are indicated as black peaks above the sequences. An E2F TFBS with the preferred E2F6 binding sequence (TCCCGC) was found in one of the two conserved regions (a box is drawn around the E2F6 TFBS sequence). B) Gel mobility shift assay showing that E2F6 binds to the E2F6 TFBS in the murine Slc25a31 promoter. A labeled oligonucleotide corresponding to the E2F6 TFBS in the mouse Slc25a31 promoter (lane 1) was incubated with control rabbit reticulocyte lysate alone (lane 2) or in vitro cotranslated HA-E2F6 and myc-DP2 proteins (lanes 3–9). Unlabeled Slc25a31 wt (lanes 4–6) or mutated (mut) (lanes 7–9) oligonucleotides were added in excess amounts over the amount of labeled probe as indicated. Filled arrow, position of the specific E2F6/DP2 complex; open arrow, endogenous binding activity in the reticulocyte lysate. C) The proximal promoter region of Slc25a31 is bound by endogenous E2F6 in undifferentiated wt R1 ES cells. ChIP was performed using mouse-anti E2F6 antibody. Primers amplifying the Slc25a31 proximal promoter region containing the E2F6 TFBS and Actb control primers lacking the E2F6 TFBS were used for semiquantitative PCR. Samples from each primer set were precipitated with IgG to control for nonspecific enrichment. Input samples represent 1% of the starting amount of chromatin and were analyzed to confirm that the different chromatin preparations contain equal amounts of DNA.
Next, we examined the functional significance of the E2F6 TFBS in the murine Slc25a31 promoter by analyzing the ability of the E2F6 protein to interact with this site by gel retardation assay. As shown in Figure 1B, addition of the recombinant E2F6/DP2 protein complex, which was produced using reticulocyte lysates, to radiolabeled Slc25a31 probe encompassing the E2F6 TFBS in the Slc25a31 promoter (−54 bp to −24 bp) generated an additional retarded band. The specificity of this interaction was verified by competition with wt and mutant cold competitors in 20-, 50-, or 100-fold excess amounts over radiolabeled Slc25a31 probe, whereby only the wt competitor was able to efficiently compete for binding to the E2F6 TFBS (Fig. 1B, filled arrow). It should be noted that the reticulocyte lysate used for in vitro translation (IVT) contains an endogenous protein that interacts with the Slc25a31 probe. This interaction was evidenced by the presence of a retarded band when the probe was incubated with control rabbit reticulocyte lysate in the absence of IVT (Fig. 1B, open arrow). However, addition of specific antibodies against HA and Myc into the protein/probe mixture selectively reduced the intensity of the bands indicated by the filled arrow as previously demonstrated [31] (data not shown).
Subsequently, we analyzed the occupancy of this E2F6 TFBS in vivo using ChIP. Here we found that endogenous E2F6 bound to the proximal promoter of Slc25a31 but not to Actb in undifferentiated R1 ES cells (Fig. 1C). Collectively, these observations show that Slc25a31 contains a conserved E2F6 TFBS that binds E2F6 both in vitro and in vivo.
E2F6 Is Required for the Repression of Slc25a31
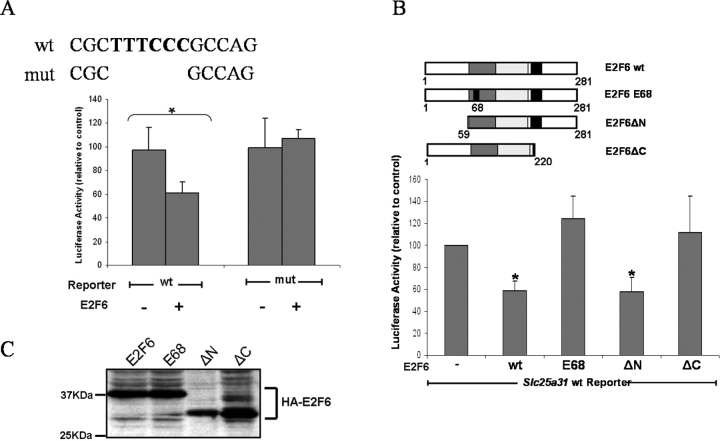
To clarify the role that E2F6 plays in the regulation of Slc25a31 transcription, we then performed luciferase reporter assays using an Slc25a31-promoter-luciferase reporter vector in combination with various E2F6 expression vectors. The Slc25a31-luciferase reporter contains a 288-bp region of the Slc25a31 promoter (−263 bp to +25 bp) that includes the E2F6 TFBS. For transient transfection, we used NIH3T3 cells due to their high transfection efficiency and their apparent absence of endogenous E2F6 expression (data not shown). The activity of the wt Slc25a31-luciferase reporter significantly decreased upon cotransfection with an HA-E2F6 expression vector (Fig. 2A, left bars). However, when the E2F6 TFBS in the Slc25a31-luciferase reporter was mutated using site-directed mutagenesis, cotransfection with HA-E2F6 failed to repress Slc25a31 transcription (Fig. 2A, right bars). Similarly, when the wt Slc25a31 reporter vector was cotransfected with HA-E2F6 expression vectors in which either the DNA binding domain was mutated (HA-E68-E2F6) or the repression domain was mutated (HA-ΔC-E2F6), reporter activity was not repressed (Fig. 2B). E2F6 proteins containing a mutation in the N-terminal domain (HA-ΔN-E2F6), which have been shown to retain their repressor activity, had similar repressive effects to that of wt HA-E2F6 [31]. Western blotting confirmed that the E2F6 proteins were expressed at comparable levels when equivalent amounts of DNA were transfected (Fig. 2C). These reporter assays indicate that the E2F6 protein can repress Slc25a31, provided its DNA binding and repressor domains are intact, and that this repression depends on sequence-specific DNA binding.
FIG. 2.
Slc25a31 transcription is repressed by transient E2F6 overexpression in NIH-3T3s. A) Slc25a31 promoter (−263 bp to +25 bp) luciferase reporter plasmids containing wt or mutated (mut) E2F6 TFBS were transiently cotransfected with an empty vector (-) or an E2F6 expression vector (+) into NIH-3T3 cells. Luciferase activity was measured relative to the Renilla internal control vector as described in Materials and Methods. Sequences for both wt and mut E2F6 TFBS reporters are shown above the bar graph. B) Luciferase reporter assay in NIH-3T3 cells transiently cotransfected with either empty vector (-), wt E2F6, or mutant (E68, ΔN, or ΔC) HA-E2F6 expression vectors and the Slc25a31 wt reporter vector. A schematic representation of the HA-tagged E2F6 vectors is shown above the graph. E68, DNA binding domain point mutation; ΔN, N-terminus deletion mutant; ΔC, C-terminus deletion mutant; white boxes, N- and C-terminal domains; dark gray boxes, DNA binding domains; light gray boxes, dimerization domains; black boxes, marked boxes; black box inside dark gray box, site of DNA binding domain mutation. C) Western blot analysis using anti-HA antibody, showing that all the E2F6 expression vectors transiently transfected for luciferase reporter assays in (B) expressed their respective E2F6 proteins at comparable levels. KDa, Kilodalton. For (A) and (B), *P < 0.05 and data from at least three independent experiments were analyzed and expressed as mean ± SD.
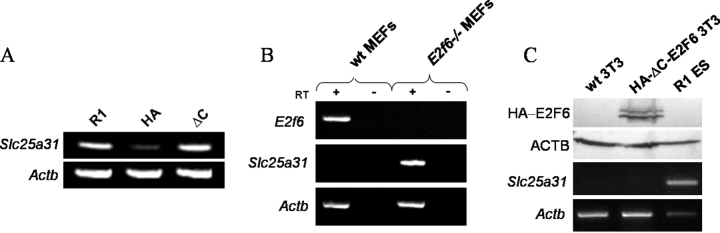
To further elucidate the potential role of E2F6 in the repression of the Slc25a31 promoter, we next created stable ES cell lines overexpressing either HA-E2F6 or HA-ΔC-E2F6 expression vectors. In contrast to somatic cells, many germ cell-specific genes are considered to be transcriptionally permissive in ES cells and therefore have detectable mRNA transcripts [38]. For instance, the CpG island of the Slc25a31 gene promoter is hypomethylated in ES cells, and a low level of Slc25a31 transcription is detectable [36]. Using semiquantitative RT-PCR, we found that overexpression of HA-E2F6 but not mutant HA-ΔC-E2F6 was able to reduce Slc25a31 transcription relative to parental R1 ES cells (Fig. 3A). This indicates that increasing the amount of E2F6 protein present in a cell is sufficient to trigger E2F6-mediated repression. Based on this result, it is reasonable to believe that the opposite scenario, in which E2F6 protein has been depleted from a cell, could potentially result in the derepression of Slc25a31 transcription.
FIG. 3.
Slc25a31 transcription is repressed by stable E2F6 overexpression in ES cells and is derepressed in E2f6−/− MEFs. A) Semiquantitative RT-PCR analysis of Slc25a31 and Actb expression in parental R1, HA-E2F6 (HA), and HA-ΔC-E2F6 (ΔC) ES cell lines. B) Semiquantitative RT-PCR analysis of E2f6, Slc25a31, and Actb expression in wt MEFs and E2f6−/− MEFs. Reverse transcriptase minus samples were also amplified to eliminate the possibility of genomic DNA contamination. C) Further semiquantitative RT-PCR analysis of Slc25a31 and Actb expression in parental NIH 3T3 (wt 3T3), HA-ΔC-E2F6-tranfected NIH 3T3, and R1 ES cell lines. Western blot is shown in the top two rows to confirm HA-ΔC-E2F6 expression in NIH 3T3 cells. R1 ES cells were included as a positive control for Slc25a31 expression. Transfection of HA-ΔC-E2F6 construct did not subsequently restore Slc25a31 expression in somatic cells.
To this end, we examined the expression of Slc25a31 in mouse embryonic fibroblast (MEF) cell lines derived from both wt and E2f6-null mice. Using semiquantitative PCR, we found that Slc25a31 was in fact aberrantly expressed in E2f6−/− MEFs (Fig. 3B). Taken together, these findings indicate that E2F6 is an essential repressor for the Slc25a31 gene in somatic cells.
In addition, since HA-ΔC-E2F6 was suggested to work as a dominant negative mutant [31], we also examined whether HA-ΔC-E2F6 can derepress Slc25a31 transcription in somatic cells using a mouse fibroblastic cell line (NIH3T3). As shown in Figure 3C, HA-ΔC-E2F6 was not able to derepress the Slc25a31 gene in the cells, which may imply that E2F6 inactivation may not be sufficient for derepressing Slc25a31 once the gene repression has already been established.
The E2F6-Binding Element Is Conserved Within the Proximal Promoter Regions of Meiosis-Specific Genes
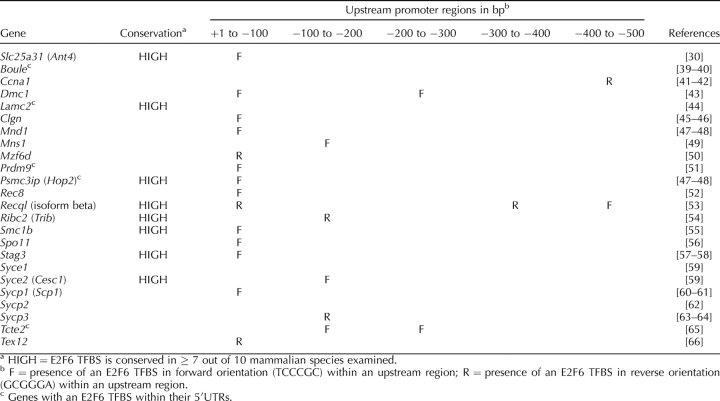
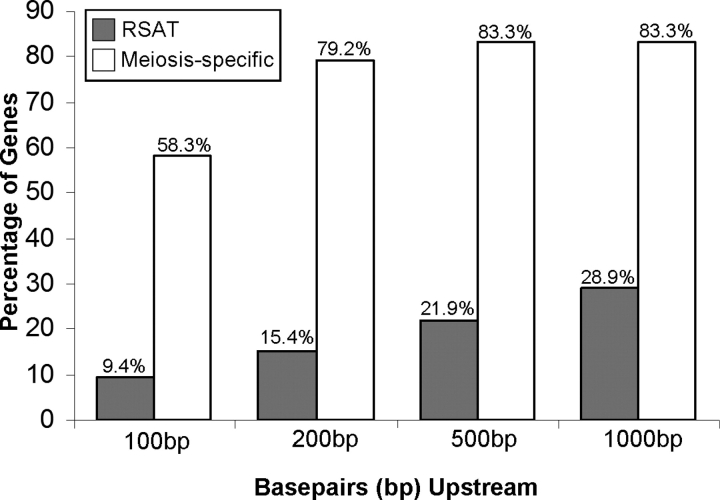
Our discovery that Slc25a31 is regulated by E2F6, along with previous studies suggesting a role for E2F6 in germ cell gene repression [27–28], prompted us to investigate whether additional genes involved in germ cell development require E2F6 for their repression. In particular, we focused on genes that are similar to Slc25a31 in that their expression is restricted to germ cells undergoing meiosis. We compiled a list of 24 genes that were previously reported in the literature to have meiosis-specific expression (Table 1). It should be noted here that we excluded genes from the list that continue to have predominant levels of expression postmeiotically, such as Pgk2. For each of these meiosis-specific genes, we screened a genomic region spanning 1 kb upstream of their transcription initiation sites for the presence of E2F6-binding elements (TCCCGC or GCGGGA, depending on the direction of binding) using the UCSC Genome Browser [33]. As shown in Table 1, potential E2F6-binding elements are accumulated within the proximal promoter region of meiosis-specific genes. In total, 19 out of the 24 meiosis-specific genes (79.2%) contain at least one E2F6-binding element within 200 bp upstream of their transcription initiation site (Fig. 4, white bars). The relevance of this finding was examined through the application of a genome-scale DNA pattern-matching software tool, RSAT [34]. The RSAT software was used to identify the presence of E2F6-binding elements within promoter regions (−1000 bp to +1 bp) of all genes in the entire mouse genome (based on 31 113 genes in the Ensembl database; Fig. 4, gray bars). A comparison between the meiosis-specific genes and all genes in the mouse genome indicates that the proximal promoter regions of these meiosis-specific genes are indeed enriched with E2F6-binding elements. Further, the selectivity of the E2F6-binding element to the proximal promoter, a genomic region that is known to harbor binding sites of critical transcriptional regulators, suggests that this E2F6-binding element possesses a high degree of functional significance. It should be noted here that 5 of the 24 meiosis-specific genes also had an E2F6-binding element within their 5′UTR (Table 1). Given that many genes harbor several putative transcription initiation sites, it is possible that those E2F6 binding sites found in the 5′UTR could also play an important role in modulating transcription initiation.
TABLE 1.
Conservation and locations of E2F6 TFBS within upstream promoter regions of meiosis-specific genes relative to their transcription initiation sites.
FIG. 4.
Frequency of appearance of E2F6 TFBS within upstream regions of genes relative to their transcription initiation sites. Bar graph indicates the occurrence rate of E2F6 TFBS for the following categories: RSAT, the actual frequency that the E2F6 TFBS occurs at least once within the proximal promoter regions of genes (31 113 genes) in the mouse genome; meiosis-specific, the actual frequency that the E2F6 TFBS occurs at least once within a population of meiosis-specific genes (those listed in Table 1). The x-axis indicates the location of the E2F6 TFBS to be within 100 bp, 200 bp, 500 bp, or 1000 bp upstream of the transcription initiation site; the y-axis denotes the percentage of genes from each category (RSAT or Meiosis-specific) that have at least one E2F6 TFBS within a given upstream region.
As an additional measure of significance, for each meiosis-specific gene we analyzed the mammalian conservation of the E2F6-binding element (Table 1). Interestingly, one-third of the meiosis-specific genes had an E2F6-binding element that was conserved in more than 7 out of 10 species examined (species are listed in Fig. 1A).
Only a Portion of Meiosis-Specific Genes Are Derepressed in E2f6−/− MEFs
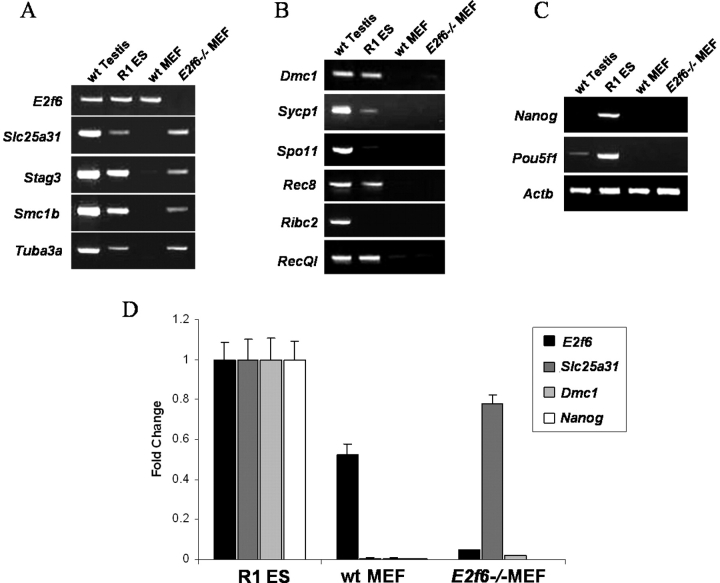
Given that 19 of the 24 meiosis-specific genes examined here contain an E2F6 binding site within 200 bp upstream of their transcription initiation sites, we predicted that similar to Slc25a31, these genes would become derepressed in E2f6−/− MEFs. Surprisingly, with the exception of Slc25a31 and the genes previously found to be derepressed in the E2f6−/− MEFs (Fig. 5A), all other meiosis-specific genes containing the E2F6-binding element remained repressed in the E2f6−/− MEFs (Fig. 5B). As a control, we confirmed that ES cell-pluripotency genes (Nanog, Pou5f1) remained repressed in E2f6−/− MEFs (Fig. 5C). A representative sampling of this data was further quantified using real-time PCR (Fig. 5D). It should be noted here that the expression levels of those genes that were derepressed in E2f6−/− MEFs were lower when compared to physiological expression of these genes in the testis (Fig. 5).
FIG. 5.
Limited meiosis-specific genes are derepressed in E2f6−/− MEFs. Semiquantitative RT-PCR analysis of meiosis-specific gene expression in wt testis from 6-wk-old mice, R1 ES cells, wt MEFs, and E2f6−/− MEFs. A) E2f6 and genes showing derepression in E2f6−/− MEFs. B) Other meiosis-specific genes. C) Nonmeiotic ES cell pluripotency genes and Actb genes. D) Quantitative real-time PCR analysis of a representative group of genes from A, B, and C. All samples were tested in triplicate and expressed as mean ± SD.
Meiosis-Specific Gene Promoters Are Bound by E2F6
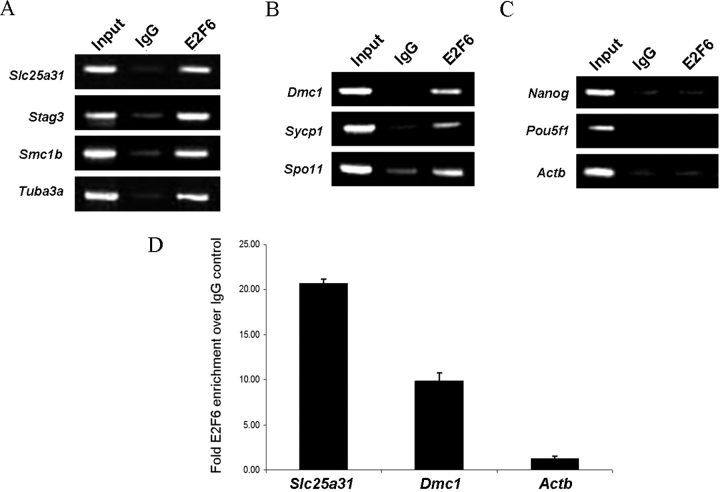
To test the functional significance of the conserved E2F6-binding elements, we performed ChIP analysis using undifferentiated ES cells and observed that endogenous E2F6 binds to meiosis-specific genes (Fig. 6). Figure 6A shows that E2F6 indeed binds to the promoters of meiotic genes that have been shown to be derepressed in E2f6−/− MEFs. To clarify, Tuba3a is one of these derepressed genes that is highly expressed in meiotic cells, but its expression is not restricted to meiotic cells and is therefore not included in Table 1. Interestingly, Figure 6B demonstrates that E2F6 also binds to those meiosis-specific genes that remain repressed in E2f6−/− MEFs. Additionally, the inability of E2F6 to bind to the promoters of both housekeeping and ES cell-pluripotency genes validates that E2F6 binding is targeted to meiosis-specific gene promoters (Fig. 6C). Lastly, we selected a representative set of genes and verified using quantitative real-time PCR that E2F6 was indeed binding to meiosis-specific genes regardless of their expression status in E2f6−/− MEFs (Fig. 6D).
FIG. 6.
E2F6 binds to meiosis-specific genes. ChIP of endogenous E2F6 binding activity in wt R1 ES cells. ChIP was performed using mouse-anti-E2F6 antibody and analyzed by semiquantitative PCR. Input and IgG are shown as controls. E2F6 binding activity on: (A) the promoters of meiotic genes that are known to be derepressed in E2f6−/− MEFs, (B) the promoters of other meiosis-specific genes, and (C) the promoters of nonmeiotic ES cell pluripotency and Actb genes. D) Real-time PCR quantification of binding as determined by the ratio of specific ChIP/IgG ChIP relative to input. All samples were tested in triplicate and expressed as mean ± SD.
E2F6 Overexpression Represses Meiosis-Specific Genes
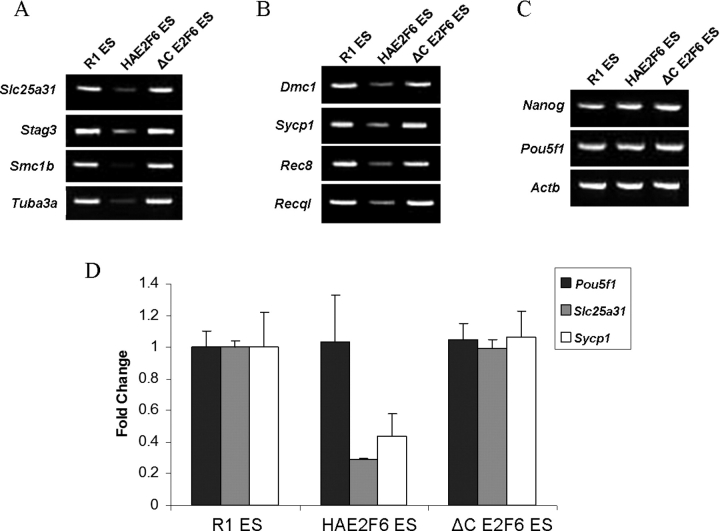
Next, we looked to see whether E2F6 would repress the meiosis-specific genes to which it was bound. In both semiquantitative (Fig. 7, A–C) and quantitative real-time PCR analyses (Fig. 7D), expression of meiosis-specific transcripts was reduced by overexpression of wt E2F6 but not mutant E2F6. In agreement with the status of E2F6 binding, E2F6 overexpression reduced meiosis-specific gene transcription regardless of the genes' expression status in E2f6−/− MEFs. Additionally, E2F6 overexpression did not reduce the expression of housekeeping and ES cell-pluripotency genes.
FIG. 7.
Meiosis-specific genes are repressed by E2F6. Semiquantitative RT-PCR analysis of meiosis-specific gene expression in R1, HA-E2F6, and HA-ΔC-E2F6 ES cells. A) Genes derepressed in E2f6−/− MEFs. B) Other meiosis-specific genes. C) Nonmeiotic ES cell pluripotency and Actb genes. D) Quantitative real-time PCR analysis of a representative group of genes from A, B, and C. All samples were tested in triplicate and expressed as mean ± SD.
E2f6 Expression Did Not Decrease in Meiotic Spermatocytes
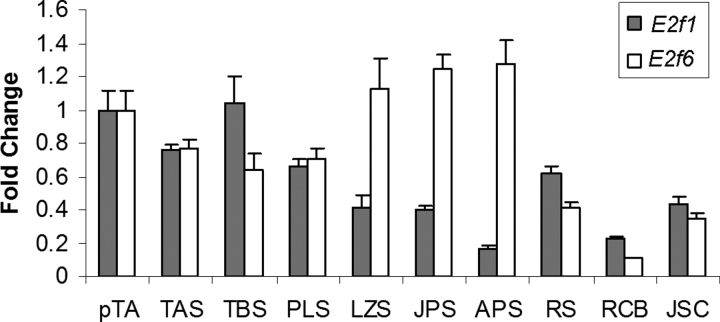
To gain further insight regarding the role of E2F6 in the control of meiosis-specific genes, we also examined the RNA expression status of the E2f6 gene in stage-specific spermatogenic cells (Fig. 8). E2f1, a well-known activating member of the E2F family, was also shown for the purpose of comparison. Of interest, in contrast to our prediction based on the proposed role of E2F6 in the repression of meiosis-specific genes, E2f6 expression did not decrease during meiosis but rather increased from preleptotene to pachytene spermatocyte stages.
FIG. 8.
E2f1 and E2f6 mRNA transcript levels in stage-specific spermatogenic cells. Real-time PCR quantification of E2f1 (gray bars) and E2f6 (white bars) transcript levels in the following purified spermatogenic cell types: pTA, primitive Type A spermatogonia; TAS, Type A spermatogonia; TBS, Type B spermatogonia; PLS, preleptotene spermatocytes; LZS, leptotene/zygotene spermatocytes; JPS, juvenile pachytene spermatocytes; APS, adult pachytene spermatocytes; RS, round spermatids; RCB, residual cytoplasmic bodies; JSC, juvenile (D6) Sertoli cells. The relative transcript levels are shown with the transcript level of pTA for E2f1 and E2f6 set as 1. All samples were tested in triplicate and expressed as mean ± SD.
DISCUSSION
There are several notable examples of coordinated gene regulation whereby genes whose protein products function in the same physiological processes are concomitantly expressed. Typically, a common transcription factor binding site can be found in the promoter regions of such genes [67–69]. For instance, previous studies have screened DNA sequences from the EMBL and GenBank data banks and found that liver-specific gene promoters harbor binding sites for the HNF1 transcription factor 2.5 times more frequently than do other genes [67]. Likewise, NFAT/AP-1 binding sites are present at rates 10 times higher in the promoters of immune response genes than in random sequences pulled from EPD and GenBank databases [68]. Further, the promoter regions of genes whose protein products are known to play a role in cell cycle progression were found to have a high frequency of E2F-binding site (the traditional TCGCGC site common to E2F1–E2F5 rather than the E2F6-preferred TCCCGC site) occurrence in comparison to the promoters of functionally different genes [69]. Here we have taken a defined population of critical germ cell-specific genes, the meiosis-specific genes, and identified a regulatory element that is commonly shared among members of this population. This E2F6-binding element occurs in the proximal promoter regions of meiosis-specific genes at a significantly higher frequency (79.2%) than that found in the promoters of all mouse genes (15.4%). Further, we have shown that E2F6 binds to this element and is capable of reducing meiosis-specific gene expression. The identification of a transcription factor that may coordinately regulate the timely expression of the meiosis-specific gene population is notable given that failure to express any one of these genes often results in infertility [1–2] and aberrant expression in somatic cells can lead to mitotic catastrophe [6–7].
In agreement with previous studies, E2F6 deficiency alone derepressed only a portion of these meiosis-specific genes in somatic cells, including the newly identified Slc25a31 as well as Stag3, Smc1b, and Tex12. Prior reports have described cDNA microarray experiments with mRNA from wt and E2f6−/− MEFs and shown that very few germ cell-specific genes were upregulated in E2f6−/− MEFs [27–28]. By the same token, these studies failed to identify more than three meiosis-specific genes as E2F6-target genes because the parameters used during the experiments only allowed for identification of genes that were derepressed in E2f6−/− MEFs. Under such criteria, the discovery of additional E2F6-target genes may have been overlooked as a result of a functional redundancy, whereby in the absence of E2F6, another factor can compensate to maintain repression. It is also possible that not all meiosis-specific genes that are regulated by E2F6 require E2F6 for their repression in somatic cells. Our decision to more closely examine the promoter regions of meiosis-specific genes regardless of their expression status in E2f6−/− MEFs revealed 16 additional E2F6-target genes, all of which are essential to successful germ cell development.
Future studies should be directed at distinguishing the differences between those meiosis-specific genes that are and those that are not derepressed in E2f6−/− MEFs. It is conceivable that functional redundancies may only exist for those genes that remain repressed. Interestingly, E2F family members have been shown to compensate for each other when other E2F family members are rendered nonfunctional. In particular, E2F4 can compensate for the loss of E2F6 by binding to the promoters of G1/S-regulated genes (that under normal circumstances are bound by E2F6) in the absence of E2F6 [70]. In agreement with this concept, ChIP analysis of the Stag3 promoter in E2f6−/− MEFs indicates that E2F4 does not bind [28]. It is possible that this lack of redundancy with E2F4 could explain why Stag3 is aberrantly expressed in E2f6−/− MEFs. It would be naïve, however, to infer that the only transcription factors capable of compensating for the loss of E2F6 are other E2F family members. One example of a redundancy that most likely does not involve other E2F family members may be Ribc2. Ribc2 is one of the meiosis-specific genes that remains repressed in E2f6−/− MEFs (Fig. 5). Ribc2 and Smc1b genes overlap on chromosome 15 and are transcribed in opposite orientations, with their promoter regions embedded within each other [54]. These genes are not only transcribed at similar times during meiosis, but they share an E2F6-binding element in their overlapping promoter region that we have shown binds E2F6 in vivo (shown as Smc1b, Fig. 6A). DNA elements existing within introns and 3′ regions specific to each gene may be accountable for the differential regulation. Although the mechanisms responsible for this distinction in gene regulation remain unclear, a redundancy of an E2F family member at the location of this shared E2F6 binding site would most likely not account for such a phenomenon.
Lastly, the unexpected increase in E2F6 expression during male germ cell meiosis may imply an additional role for E2F6 in meiosis-specific gene regulation. E2F6 may in fact have dual roles as both an activator and a repressor. These roles may be determined by cell type and by differences in the complexes by which E2F6 is bound. For instance, if E2F6 represses transcription by serving as a platform on which factors such as polycomb protein complexes can bind, then the actual repression activity is coming from the proteins that are recruited by E2F6 rather than from E2F6 itself. In meiotic cells, E2F6 may instead be recruiting an activation complex to meiosis-specific genes. Such an activation complex could consist of components that have already been shown to play a role in meiotic gene activation, such as MYB, SP1, PRDM9, and tTAFs. Indeed, E2F6 knockout mice demonstrated a moderately impaired spermatogenesis, supporting this idea [32]. Relationships between these proteins and E2F6 need to be further elucidated.
In summary, the present study has revealed a common E2F6-binding element in the proximal promoter regions of meiosis-specific genes. These findings suggest that E2F6 may play an even broader role in the regulation of critical germ cell-specific genes than was originally thought. Further, this study has provided new insights regarding coordinated gene control and has laid the groundwork for future studies on elucidating the exact function of E2F6 in the context of meiosis-specific gene regulation.
Supplementary Material
Acknowledgments
We thank J. Kehoe for assistance with statistical analysis of E2F6-binding elements. We also thank J. Bungert and J.L. Resnick for critical reading of the manuscript.
Footnotes
1Supported in part by NIH grants DK59699 and RR17001 to N.T. and a DFG (GA575/3-2) grant to S. Gaubatz.
REFERENCES
- Matzuk MM, Lamb DJ.Genetic dissection of mammalian fertility pathways. Nat Cell Biol 2002; 4: S41–S49.. [DOI] [PubMed] [Google Scholar]
- Roy A, Matzuk MM.Deconstructing mammalian reproduction: using knockouts to define fertility pathways. Reproduction 2006; 131: 207–219.. [DOI] [PubMed] [Google Scholar]
- Dejong J.Basic mechanisms for the control of germ cell gene expression. Gene 2006; 366: 39–50.. [DOI] [PubMed] [Google Scholar]
- Eddy EM.Male germ cell gene expression. Recent Prog Horm Res 2002; 57: 103–128.. [DOI] [PubMed] [Google Scholar]
- MacLean JA, II, Wilkinson MF.Gene regulation in spermatogenesis. Curr Top Dev Biol 2005; 71: 131–197.. [DOI] [PubMed] [Google Scholar]
- Sagata N.What does Mos do in oocytes and somatic cells? Bioessays 1997; 19: 13–21.. [DOI] [PubMed] [Google Scholar]
- Simpson AJG, Caballero OL, Jungbluth A, Chen Y, Old LJ.Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer 2005; 5: 615–625.. [DOI] [PubMed] [Google Scholar]
- Bartell JG, Davis T, Kremer EJ, Dewey MJ, Kistler WS.Expression of the rat testis-specific histone H1t gene in transgenic mice. J Biol Chem 1996; 271: 4046–4054.. [DOI] [PubMed] [Google Scholar]
- Han SY, Xie W, Kim SH, Yue L, DeJong J.A short core promoter drives expression of the ALF transcription factor in reproductive tissues of male and female mice. Biol Reprod 2004; 71: 933–941.. [DOI] [PubMed] [Google Scholar]
- Iannello RC, Young J, Sumarsono S, Tymms MJ, Dahl HM, Gould J, Hedger M, Kola I.Regulation of Pdha-2 expression is mediated by proximal promoter sequences and CpG methylation. Mol Cell Biol 1997; 17: 612–619.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lele KM, Wolgemuth DJ.Distinct regions of the mouse Cyclin A1 gene, Ccna1, confer male germ-cell-specific expression and enhancer function. Biol Reprod 2004; 71: 1340–1347.. [DOI] [PubMed] [Google Scholar]
- Robinson MO, McCarrey JR, Simon MI.Transcriptional regulatory regions of testis-specific PGK-2 defined in transgenic mice. Proc Natl Acad Sci U S A 1989; 86: 8437–8441.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage J, Martin L, Meuwissen R, Heyting C, Cuzin F, Rassoulzadegan M.Temporal and spatial control of the Sycp1 gene transcription in the mouse meiosis: regulatory elements active in the male are not sufficient for expression in the female gonad. Mech Dev 1999; 80: 29–39.. [DOI] [PubMed] [Google Scholar]
- Xu W, Cooper GM.Identification of a candidate c-mos repressor that restricts transcription of germ cell-specific genes. Mol Cell Biol 1995; 1715: 5369–5375.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Jurk M, Gulick T, Cooper GM.Identification of COUP-TF as a transcriptional repressor of the c-mos proto-oncogene. J Biol Chem 1999; 274: 36796–36800.. [DOI] [PubMed] [Google Scholar]
- Bartusel T, Schubert S, Klempnauer K.Regulation of the cyclin D1 and cyclin A1 promoters by B-Myb is mediated by Sp1 binding sites. Gene 2005; 351: 171–180.. [DOI] [PubMed] [Google Scholar]
- Gebara MM, McCarrey JR.Protein-DNA interactions associated with the onset of testis-specific expression of the mammalian Pgk-2 gene. Mol Cell Biol 1992; 12: 1422–1431.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath GC, Kistler WS, Kistler MK.RFX2 is a potential transcriptional regulatory factor for histone H1t and other genes expressed during the meiotic phase of spermatogenesis. Biol Reprod 2004; 71: 1551–1559.. [DOI] [PubMed] [Google Scholar]
- Kim M, Li D, Cui Y, Mueller K, Chears WC, DeJong J.Regulatory factor interactions and somatic silencing of the germ cell-specific ALF gene. J Biol Chem 2006; 281: 34288–34298.. [DOI] [PubMed] [Google Scholar]
- Wilkerson DC, Wolfe SA, Grimes SR.H1t/GC-box and H1t/TE1 element are essential for promoter activity of the testis-specific histone H1t gene. Biol Reprod 2002; 67: 1157–1164.. [DOI] [PubMed] [Google Scholar]
- Dimova DK, Dyson NJ.The E2F transcriptional network: old acquaintances with new faces. Oncogene 2005; 24: 2810–2826.. [DOI] [PubMed] [Google Scholar]
- Trimarchi JM, Lees JA.Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol 2002; 3: 11–20.. [DOI] [PubMed] [Google Scholar]
- Bruin A, Maiti B, Jakoi L, Timmers C, Buerki R, Leone G.Identification and characterization of E2F7, a novel mammalian E2F family member capable of blocking cellular proliferation. J Biol Chem 2003; 278: 42041–42049.. [DOI] [PubMed] [Google Scholar]
- Maiti B, Li J, Bruin A, Gordon F, Timmers C, Opavsky R, Patil K, Tuttle J, Cleghorn W, Leone G.Cloning and characterization of mouse E2F8, a novel mammalian E2F family member capable of blocking cellular proliferation. J Biol Chem 2005; 280: 18211–18220.. [DOI] [PubMed] [Google Scholar]
- Morkel M, Wenkel J, Bannister AJ, Kouzarides T, Hagemeier C.An E2F-like repressor of transcription. Nature 1997390: 567–568.. [DOI] [PubMed] [Google Scholar]
- Trimarchi JM, Fairchild B, Verona R, Moberg K, Andon N, Lees JA.E2F-6, a member of the E2F family that can behave as a transcriptional repressor. Proc Natl Acad Sci U S A 1998; 95: 2850–2855.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlers M, Truss M, Frede U, Scholz A, Strehle M, Kuban RJ, Hoffmann B, Morkel M, Birchmeier C, Hagemeier C.A role for E2F6 in the restriction of male-germ-cell-specific gene expression. Curr Biol 2005; 15: 1051–1057.. [DOI] [PubMed] [Google Scholar]
- Storre J, Schäfer A, Reichert N, Barbero JL, Hauser S, Eilers M, Gaubatz S.Silencing of meiotic genes SMC1β and STAG3 in somatic cells by E2F6. J Biol Chem 2005; 280: 41380–41386.. [DOI] [PubMed] [Google Scholar]
- Cartwright P, Müller H, Wagener C, Holm K, Helin K.E2F-6: a novel member of the E2F family is an inhibitor of E2F-dependent transcription. Oncogene 1998; 17: 611–623.. [DOI] [PubMed] [Google Scholar]
- Brower JV, Rodic N, Seki T, Jorgensen M, Fliess N, Yachnis AT, McCarrey JR, Oh SP, Terada N.Evolutionarily conserved mammalian adenine nucleotide translocase 4 is essential for spermatogeneis. J Biol Chem 2007; 282: 29658–29666.. [DOI] [PubMed] [Google Scholar]
- Gaubatz S, Wood JG, Livingston DM.Unusual proliferation arrest and transcriptional control properties of a newly discovered E2F family member, E2F6. Proc Natl Acad Sci U S A 1998; 95: 9190–9195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storre J, Elsässer H, Fuchs M, Ullmann D, Livingston DM, Gaubatz S.Homeotic transformations of the axial skeleton that accompany a targeted deletion of E2f6. EMBO Rep 2002; 3: 695–700.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, Weber RJ, Haussler D, et al. The UCSC Genome Browser Database. Nucleic Acids Res 2003; 31: 51–54.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van-Heldin J.Regulatory sequence analysis tools. Nucleic Acids Res 2003; 31: 3593–3596.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolce V, Scarcia P, Iacopetta D, Palmieri F.A fourth ADP/ATP carrier isoform in man: identification, bacterial expression, functional characterization, and tissue distribution. FEBS Lett 2005; 579: 633–637.. [DOI] [PubMed] [Google Scholar]
- Rodic N, Oka M, Hamazaki T, Murawski MR, Jorgensen M, Maatouk DM, Resnick JL, Li E, Terada N.DNA methylation is required for silencing of ant4, an adenine nucleotide translocase selectively expressed in mouse embryonic stem cells and germ cells. Stem Cells 2005; 23: 1314–1323.. [DOI] [PubMed] [Google Scholar]
- Kim YH, Haidl G, Schaefer M, Egner U, Mandal A, Herr JC.Compartmentalization of a unique ADP/ATP carrier protein SFEC (Sperm Flagellar Energy Carrier, AAC4) with glycolytic enzymes in the fibrous sheath of the human sperm flagellar principal piece. Dev Biol 2006; 302: 463–476.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geijsen N, Horoschak M, Kim K, Gribnau J, Eggan K, Daley GQ.Derivation of embryonic germ cells and male gametes from embryonic stem cells. Nature 2004; 427: 148–154.. [DOI] [PubMed] [Google Scholar]
- Eberhart CG, Maines JZ, Wasserman SA.Meiotic cell cycle requirement for a fly homologue of human Deleted in Azoospermia. Nature 1996; 381: 783–785.. [DOI] [PubMed] [Google Scholar]
- Xu EY, Moore FL, Reijo-Pera RA.A gene family required for human germ cell development evolved from an ancient meiotic gene conserved in metazoans. Proc Natl Acad Sci U S A 2001; 98: 7414–7419.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravnik SE, Wolgemuth DJ.Regulation of meiosis during mammalian spermatogenesis: the A-type Cyclins and their associated Cyclin-dependent kinases are differentially expressed in the germ-cell lineage. Dev Biol 1999; 207: 408–418.. [DOI] [PubMed] [Google Scholar]
- Sweeney C, Murphy M, Kubelka M, Ravnik SE, Hawkins CF, Wolgemuth DJ, Carrington M.A distinct cyclin A is expressed in germ cells in the mouse. Development 1996; 122: 53–64.. [DOI] [PubMed] [Google Scholar]
- Habu T, Takuyu T, West A, Nishimune Y, Morita T.The mouse and human homologs of DMC1, the yeast meiosis-specific homologous recombination gene, have a common unique form of exon-skipped transcript in meiosis. Nucleic Acids Res 1996; 24: 470–477.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsheimer M, Benavente R.Change of karyoskeleton during mammalian spermatogenesis: expression pattern of nuclear Lamin C2 and its regulation. Exp Cell Res 1996; 228: 181–188.. [DOI] [PubMed] [Google Scholar]
- Don J, Wolgemuth DJ.Identification and characterization of the regulated pattern of expression of a novel mouse gene, meg1, during the meiotic cell cycle. Cell Growth Differ 1992; 3: 495–505.. [PubMed] [Google Scholar]
- Watanabe D, Yamada K, Nishina Y, Tajima Y, Koshimizu U, Nagata A, Nishimune Y.Molecular cloning of a novel Ca2+-binding protein (Calmegin) specifically expressed during male meiotic germ cell development. J Biol Chem 1994; 269: 7744–7749.. [PubMed] [Google Scholar]
- Petukhova GV, Pezza RJ, Vanevski F, Ploquin M, Masson J, Camerini-Otero RD.The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat Struct Mol Biol 2005; 12: 449–453.. [DOI] [PubMed] [Google Scholar]
- Pezza RJ, Petukhova GV, Ghirlando R, Camerini-Otero RD.Molecular activities of meiosis-specific proteins Hop2, Mnd1, and the Hop2-Mnd1 complex. J Biol Chem 2006; 281: 18426–18434.. [DOI] [PubMed] [Google Scholar]
- Furukawa K, Inagaki H, Naruge T, Tabata S, Tomida T, Yamaguchi A, Yoshikuni M, Nagahama Y, Hotta Y.cDNA cloning and functional characterization of a meiosis-specific protein (MNS1) with apparent nuclear association. Chromosome Res 1994; 2: 99–113.. [DOI] [PubMed] [Google Scholar]
- Looman C, Mark C, Abrink M, Hellman L.MZF6D, a novel KRAB zinc-finger gene expressed exclusively in meiotic male germ cells. DNA Cell Biol 2003; 22: 489–496.. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Yoshida K, Matsui Y.A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 2005; 438: 374–378.. [DOI] [PubMed] [Google Scholar]
- Lee J, Iwai T, Yokota T, Yamashita M.Temporally and spatially selective loss of Rec8 protein from meiotic chromosomes during mammalian meiosis. J Cell Sci 2003; 116: 2781–2790.. [DOI] [PubMed] [Google Scholar]
- Wang WS, Seki M, Yamaoka T, Seki T, Tada S, Katada T, Fujimoto H, Enomoto T.Cloning of two isoforms of mouse DNA helicase Q1/RecQL cDNA; alpha form is expressed ubiquitously and beta form specifically in the testis. Biochem Biophys Res Commun 1998; 1443: 198–202.. [DOI] [PubMed] [Google Scholar]
- Arango NA, Pearson EJ, Donahoe PK, Teixeira J.Genomic structure and expression analysis of the mouse testis-specific ribbon protein (Trib) gene. Gene 2004; 343: 221–227.. [DOI] [PubMed] [Google Scholar]
- Revenkova E, Eupe M, Heyting C, Gross B, Jessberger R.Novel meiosis-specific isoform of mammalian SMC1. Mol Cell Biol 2001; 1721: 6984–6998.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon M, Richardson L, Christian A, Handel MA, Thelen MP.Differential gene expression of mammalian SPO11/TOP6A homologs during meiosis. FEBS Lett 1999; 462: 329–334.. [DOI] [PubMed] [Google Scholar]
- Pezzi N, Prieto I, Kremer L, Perez Jurado LA, Valero C, Del Mazo J, Martinez AC, Barbero JL.STAG3, a novel gene encoding a protein involved in meiotic chromosome pairing and location of STAG3-related genes flanking the Williams-Beuren syndrome deletion. FASEB J 2000; 14: 581–592.. [DOI] [PubMed] [Google Scholar]
- Prieto I, Suja JA, Pezzi N, Kremer L, Martinez AC, Rufas JS, Barbero JL.Mammalian STAG3 is a cohesion specific to sister chromatid arms in meiosis 1. Nat Cell Biol 2001; 3: 761–766.. [DOI] [PubMed] [Google Scholar]
- Costa Y, Speed R, Ollinger R, Alsheimer M, Semple CA, Gautier P, Maratou K, Novak I, Hoog C, Benavente R, Cooke HJ.Two novel proteins recruited by synaptonemal complex protein 1 (SYCP1) are at the centre of meiosis. J Cell Sci 2005; 118: 2755–2762.. [DOI] [PubMed] [Google Scholar]
- Meuwissen RLJ, Offenberg HH, Dietrich AJJ, Riesewijk A, Van Lersel M, Heyting C.A coiled-coil related protein specific for synapsed regions of meiotic prophase chromosomes. EMBO J 1992; 11: 5091–5100.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Brundell E, Hoog C.Expression of the meiosis-specific Synaptonemal Complex Protein 1 in a heterologous system results in the formation of large protein structures. Exp Cell Res 1996; 229: 272–275.. [DOI] [PubMed] [Google Scholar]
- Offenberg HH, Schalk JA, Meuwissen RL, van Aalderen M, Kester HA, Dietrich AJ, Heyting C.SCP2: a major protein component of the axial elements of synaptonemal complexes of the rat. Nucleic Acids Res 1998; 26: 2572–2579.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Carlo AD, Travia G, Felici MD.The meiotic specific synaptonemal complex protein SCP3 is expressed by female and male primordial germ cells of the mouse embryo. Int J Dev Biol 2000; 44: 241–244.. [PubMed] [Google Scholar]
- Lammers JHM, Offenberg HH, Van Aalderen M, Vink ACG, Dietrich AJJ, Heyting C.The gene encoding a major component of the lateral elements of synaptonemal complexes of the rat is related to X-linked lymphocyte-regulated genes. Mol Cell Biol 1994; 1714: 1137–1146.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braidotti G, Barlow DP.Identification of a male meiosis-specific gene, Tcte2, which is differentially spliced in species that form sterile hybrids with laboratory mice and deleted in t chromosomes showing meiotic drive. Dev Biol 1997; 186: 85–99.. [DOI] [PubMed] [Google Scholar]
- Hamer G, Gell K, Kouznetsova A, Novak I, Benavente R, Höög C.Characterization of a novel meiosis-specific protein within the central element of the synaptonemal complex. J Cell Sci 2006; 119: 4025–4032.. [DOI] [PubMed] [Google Scholar]
- Tronche F, Ringeisen F, Blumenfeld M, Yaniv M, Pontoglio M.Analysis of the distribution of binding sites for a tissue-specific transcription factor in the vertebrate genome. J Mol Biol 1997; 266: 231–245.. [DOI] [PubMed] [Google Scholar]
- Kel AE, Kel-Margoulis OV, Farnham PJ, Bartley SM, Wingender E, Zhang MQ.Computer-assisted identification of cell cycle-related genes: new targets for E2F transcription factors. J Mol Biol 2001; 309: 99–120.. [DOI] [PubMed] [Google Scholar]
- Wasserman WW, Fickett JW.Identification of regulatory regions which confer muscle-specific gene expression. J Mol Biol 1998; 278: 167–181.. [DOI] [PubMed] [Google Scholar]
- Giangrande PH, Zhu W, Schlisio S, Sun X, Mori S, Gaubatz S, Nevins JR.A role for E2F6 in distinguishing G1/S and G2/M-specific transcription. Genes Dev 2004; 18: 2941–2951.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.