Abstract
Many human genes have adapted to the constant threat of exposure to infectious agents; according to the “hygiene hypothesis,” lack of exposure to parasites in modern settings results in immune imbalances, augmenting susceptibility to the development of autoimmune and allergic conditions. Here, by estimating the number of pathogen species/genera in a specific geographic location (pathogen richness) for 52 human populations and analyzing 91 interleukin (IL)/IL receptor genes (IL genes), we show that helminths have been a major selective force on a subset of these genes. A population genetics analysis revealed that five IL genes, including IL7R and IL18RAP, have been a target of balancing selection, a selection process that maintains genetic variability within a population. Previous identification of polymorphisms in some of these loci, and their association with autoimmune conditions, prompted us to investigate the relationship between adaptation and disease. By searching for variants in IL genes identified in genome-wide association studies, we verified that six risk alleles for inflammatory bowel (IBD) or celiac disease are significantly correlated with micropathogen richness. These data support the hygiene hypothesis for IBD and provide a large set of putative targets for susceptibility to helminth infections.
It is commonly believed that infectious diseases have represented one of the major threats to human populations and have therefore acted as a powerful selective force. Even today, despite the advances in treatment and prevention, infectious diseases account for ∼48% of worldwide deaths among people >45 yr of age (1). These figures do not include the heavy burden imposed by helminth infections, which have recently been designated as the “great neglected tropical diseases” (2). With an estimated 2 billion individuals infected worldwide (3), helminths represent the prevalent chronic infectious diseases of humans. Although parasitic worms determine severe clinical symptoms in a minority of heavily infected individuals, even apparently subclinical parasite burdens can result in impaired nutritional status and growth retardation (4, 5). It is therefore conceivable that several human genes have evolved in response to both microbial/viral infectious agents and parasitic worms; indeed, it has been suggested (6, 7) that human populations may have adapted to parasites to such a degree that the lower exposure to infectious agents in modern developed societies results in immune imbalances, with autoimmune and allergic conditions being the outcome.
Genes involved in immunity and inflammation are known to be frequent targets of natural selection; balancing selection, which is thought to be a relatively rare phenomenon in humans, has particularly shaped the evolutionary fate of genes involved in immune responses (8, 9). Balancing selection is the situation whereby genetic variability is maintained in a population via selection. The best known example in the human genome affects the MHC genes, which are characterized by extreme polymorphism levels. Recently, Prugnolle et al. (10) demonstrated that populations from areas with high pathogen diversity display increased MHC genetic variability, indicating the action of pathogen-driven balancing selection.
Quite obviously, the presence of a functional variant is a prerequisite for selection to act, and the identification of nonneutrally evolving genes has been regarded as a strategy complementary to classical clinical and epidemiological studies to provide insight into the mechanisms of host defense (11). Similarly, analysis of the evolutionary history of genes involved in immune defense might provide novel insights into the delicate balance between efficient response to pathogens and autoimmune/allergic manifestations.
In this study, we focused our attention on a large gene family that includes ILs and their receptors (hereafter referred to as IL genes). ILs are small secreted molecules that regulate most aspects of immune and inflammatory responses and exert their effects through binding to specific receptors expressed on target cells. Various IL genes have been associated with differential susceptibility to specific infections (12, 13), and with an augmented likelihood to develop autoimmune or allergic/atopic diseases (for review see reference [14]). Finally, whereas previous reports have demonstrated nonneutral evolution at single IL genes (15–17), no comprehensive analysis has been performed and no attempt to take into account pathogen richness across different human populations has ever been described.
RESULTS
Pathogen-driven selection acts on IL genes
Pathogen-driven selection is a situation whereby the genetic diversity at a specific locus is influenced by pathogens; this is expected to occur because one or more alleles are associated with the modulation of susceptibility to infectious agents. One way to identify loci or variants subjected to pathogen-driven selection is to search for correlations between genetic variability and pathogen richness (10, 18). The latter is a measure of pathogen diversity, and is basically calculated as the number of different pathogen species/genera in a specific geographic location (see Materials and methods for further details). The choice to use pathogen richness rather than more conventional epidemiological parameters such as prevalence/burden stems from several considerations, as follows: (a) comprehensive data on prevalence are impossible to retrieve for many infections; (b) even when prevalence data are available, they may vary considerably within the same country depending on the surveyed regions, the survey period (e.g., before or after eradication campaigns), the population surveyed (e.g., city dwellers rather than farmers/bushmen/nomads or children rather than adults); (c) the prevalence of specific infections might have changed greatly over recent years as a result of eradication campaigns, and historical prevalence data are rarely available; (d) we were not interested in a single species/genus. As a consequence, the prevalence of all (or at least the most common) pathogen species would be required. Still, prevalence data are difficult to combine; e.g., in endemic regions, individuals can be infected with multiple parasite species (2), and these subjects tend to harbor the most intense infections, possibly because of an additive and/or multiplicative impact on nutrition and organ pathology (19).
To verify whether pathogen-driven selection has been acting on IL genes, we exploited the fact that a set of >650,000 single-nucleotide polymorphisms (SNPs) has been genotyped in 52 populations (Human Genome Diversity Project-Centre d'Etude du Polymorphisme Humain [HGDP-CEPH] panel: http://www.cephb.fr/en/hgdp/) distributed worldwide (Fig. S1) (20). Pathogen richness was evaluated by gathering information on the number of different pathogen species/genera present in different geographical areas of the world from the Gideon database. Specifically, pathogens were divided into two major groups: micro- and macropathogens. Micropathogens include viruses, bacteria, fungi, and protozoa. Macropathogens include insects, arthropods, and helminths; in this group parasitic worms were by far the most abundant class (90% of species/genera), so when we refer to macropathogens we basically mean helminths. Analyses were also separately performed for viruses, bacteria, protozoa, and fungi; data are available as supplemental material (Table S1). After data organization in Gideon, we calculated both micro- and macropathogen richness on a country by country basis (i.e., they represent the number of different micro- and macropathogen species/genera per country; Fig. S1). As expected, we observed that micro- and macropathogen richness strongly correlated across geographic locations (Kendall's rank correlation coefficient = 0.67; P < 2−10); this is likely caused by the major impact of climatic factors on the spatial distribution of both pathogen classes (21).
IL genes were retrieved from the HGNC Gene Families/Grouping Nomenclature web site (http://www.genenames.org/genefamily.html). From the resulting 99 genes, IL3RA and IL9R were removed because they are located on the pseudoautosomal regions of sexual chromosomes; IL15RB and IL11RB were not analyzed because their sequence and chromosomal locations are not present in public databases. Finally, four IL9R pseudogenes were discarded. The remaining 91 genes (Table S2) included most known ILs, their receptors, and receptor-accessory proteins.
A total of 1,052 SNPs in IL genes had been typed in the HGDP-CEPH panel, allowing analysis of all genes except for IL2, IL6RL1, IL6STP, IL8RBP, IL17C, IL23A, IL28A, IL28B, IL31, and IL34.
For all 1,052 IL gene SNPs in the dataset, we calculated Kendall's rank correlation coefficient (τ) between allele frequencies in HGDP-CEPH populations and micro- or macropathogen richness; a normal approximation with continuity correction to account for ties was used for p-value calculations. After Bonferroni correction for multiple tests, we observed that 48 and 94 SNPs significantly correlate with micro- and macropathogen richness, respectively, with 32 SNPs correlating with both. These variants map to a total of 44 IL genes (Table I). We next verified whether the correlations between IL SNP frequencies and pathogens could be secondary to associations with other environmental variables (e.g., climatic factors). Hence, for each geographic location corresponding to HGDP-CEPH populations, the following parameters were obtained: average annual mean and maximum temperature, precipitation rate, and short-wave radiation flux. None of the SNPs reported in Table I significantly correlated with any of these variables (Table S3).
Table I.
Correlations with micro- and macropathogen richness
| SNP | Gene | Micropathogens | Macropathogens | ||||
| τ | P-value (corrected)a | Rankb | τ | P-value (corrected)a | Rankb | ||
| rs17561 | IL1A | 0.38 | 0.151 | 0.854 | 0.47 | 0.006 | 0.937 |
| rs1143634 | IL1B | 0.55 | <0.0001 | 0.996 | 0.58 | <0.0001 | 0.996 |
| rs6761276 | IL1F10 | −0.31 | 2.011 | 0.740 | −0.51 | 0.001 | 0.980 |
| rs10496447 | IL1F8 | 0.54 | <0.0001 | 0.995 | 0.57 | <0.0001 | 0.994 |
| rs3917304 | IL1R1 | −0.53 | <0.0001 | 0.994 | −0.51 | <0.0001 | 0.979 |
| rs6444444 | IL1RAP | −0.55 | <0.0001 | 0.998 | −0.50 | 0.003 | 0.969 |
| rs6800609 | IL1RAP | 0.39 | 0.092 | 0.865 | 0.47 | 0.005 | 0.930 |
| rs2885373 | IL1RAP | 0.55 | <0.0001 | 0.998 | 0.50 | 0.003 | 0.969 |
| rs17196143 | IL1RAP | −0.49 | 0.003 | 0.974 | −0.51 | 0.002 | 0.966 |
| rs6444435 | IL1RAP | −0.47 | 0.003 | 0.972 | −0.54 | <0.0001 | 0.989 |
| rs6630730 | IL1RAPL1 | −0.41 | 0.079 | 0.883 | −0.53 | <0.0001 | 0.980 |
| rs7052954 | IL1RAPL1 | −0.48 | 0.009 | 0.970 | −0.52 | 0.002 | 0.976 |
| rs6526833 | IL1RAPL1 | 0.43 | 0.020 | 0.923 | 0.52 | <0.0001 | 0.982 |
| rs7890572 | IL1RAPL1 | 0.44 | 0.054 | 0.917 | 0.50 | 0.007 | 0.951 |
| rs10521948 | IL1RAPL1 | 0.49 | 0.003 | 0.974 | 0.54 | <0.0001 | 0.988 |
| rs7056388 | IL1RAPL1 | 0.41 | 0.131 | 0.875 | 0.51 | 0.003 | 0.964 |
| rs196990 | IL1RAPL1 | 0.41 | 0.057 | 0.899 | 0.56 | <0.0001 | 0.994 |
| rs7881819 | IL1RAPL1 | 0.38 | 0.406 | 0.830 | 0.51 | 0.003 | 0.966 |
| rs10521946 | IL1RAPL1 | −0.49 | 0.003 | 0.974 | −0.62 | <0.0001 | 0.999 |
| rs1318832 | IL1RAPL1 | −0.51 | <0.0001 | 0.987 | −0.55 | <0.0001 | 0.995 |
| rs5943559 | IL1RAPL1 | −0.45 | 0.007 | 0.955 | −0.56 | <0.0001 | 0.995 |
| rs12387961 | IL1RAPL1 | 0.37 | 0.261 | 0.832 | 0.46 | 0.009 | 0.928 |
| rs721953 | IL1RAPL2 | 0.46 | 0.004 | 0.967 | 0.44 | 0.017 | 0.918 |
| rs1384360 | IL1RAPL2 | −0.45 | 0.009 | 0.952 | −0.44 | 0.018 | 0.908 |
| rs6621992 | IL1RAPL2 | −0.45 | 0.010 | 0.955 | −0.46 | 0.008 | 0.940 |
| rs7583215 | IL1RL2 | −0.55 | <0.0001 | 0.996 | −0.54 | <0.0001 | 0.991 |
| rs2287041 | IL1RL2 | 0.51 | <0.0001 | 0.985 | 0.47 | 0.004 | 0.945 |
| rs3771188 | IL1RL2 | −0.54 | <0.0001 | 0.993 | −0.52 | <0.0001 | 0.976 |
| rs315931 | IL1RN | −0.41 | 0.040 | 0.916 | −0.46 | 0.007 | 0.941 |
| rs2637988 | IL1RN | −0.44 | 0.011 | 0.953 | −0.47 | 0.004 | 0.954 |
| rs2029582 | IL1RN | −0.30 | 2.565 | 0.728 | −0.46 | 0.008 | 0.936 |
| rs3087266 | IL1RN | −0.34 | 0.728 | 0.780 | −0.46 | 0.008 | 0.928 |
| rs2386841 | IL2RA | 0.48 | 0.002 | 0.970 | 0.47 | 0.004 | 0.942 |
| rs11256497 | IL2RA | −0.49 | 0.002 | 0.973 | −0.49 | 0.002 | 0.953 |
| rs2284034 | IL2RB | 0.37 | 0.244 | 0.834 | 0.48 | 0.003 | 0.950 |
| rs1003694 | IL2RB | 0.32 | 1.570 | 0.734 | 0.46 | 0.006 | 0.936 |
| rs228966 | IL2RB | −0.32 | 1.705 | 0.757 | −0.48 | 0.003 | 0.963 |
| rs2284033 | IL2RB | 0.37 | 0.245 | 0.853 | 0.56 | <0.0001 | 0.996 |
| rs228973 | IL2RB | 0.44 | 0.012 | 0.943 | 0.52 | <0.0001 | 0.980 |
| rs228975 | IL2RB | −0.49 | 0.001 | 0.980 | −0.59 | <0.0001 | 0.999 |
| rs2235330 | IL2RB | 0.41 | 0.059 | 0.889 | 0.54 | <0.0001 | 0.987 |
| rs2243268 | IL4 | −0.51 | <0.0001 | 0.987 | −0.58 | <0.0001 | 0.997 |
| rs2243288 | IL4 | −0.43 | 0.021 | 0.932 | −0.47 | 0.004 | 0.948 |
| rs2243290 | IL4 | 0.49 | 0.001 | 0.978 | 0.54 | <0.0001 | 0.991 |
| rs2070874 | IL4 | −0.50 | 0.001 | 0.984 | −0.55 | <0.0001 | 0.993 |
| rs3024672 | IL4R | −0.40 | 0.189 | 0.866 | −0.53 | 0.001 | 0.975 |
| rs3024607 | IL4R | −0.48 | 0.006 | 0.974 | −0.53 | 0.001 | 0.987 |
| rs17026370 | IL5RA | 0.47 | 0.011 | 0.951 | 0.55 | <0.0001 | 0.990 |
| rs2066992 | IL6 | −0.41 | 0.049 | 0.899 | −0.46 | 0.006 | 0.933 |
| rs2069835 | IL6 | −0.46 | 0.021 | 0.949 | −0.53 | 0.002 | 0.982 |
| rs17505589 | IL7 | 0.50 | 0.006 | 0.986 | 0.43 | 0.115 | 0.851 |
| rs11567697 | IL7R | 0.43 | 0.086 | 0.904 | 0.51 | 0.005 | 0.972 |
| rs1554286 | IL10 | −0.45 | 0.006 | 0.956 | −0.56 | <0.0001 | 0.994 |
| rs3024490 | IL10 | −0.39 | 0.103 | 0.880 | −0.52 | <0.0001 | 0.988 |
| rs2512144 | IL10RA | −0.38 | 0.343 | 0.832 | −0.47 | 0.009 | 0.938 |
| rs999261 | IL10RB | 0.39 | 0.140 | 0.854 | 0.49 | 0.002 | 0.958 |
| rs2243115 | IL12A | −0.46 | 0.008 | 0.952 | −0.43 | 0.045 | 0.843 |
| rs17129789 | IL12RB2 | −0.43 | 0.026 | 0.910 | −0.51 | 0.001 | 0.971 |
| rs10521698 | IL13RA2 | −0.40 | 0.164 | 0.860 | −0.50 | 0.003 | 0.956 |
| rs1589241 | IL15 | 0.48 | 0.005 | 0.970 | 0.48 | 0.006 | 0.950 |
| rs2322262 | IL15 | 0.47 | 0.008 | 0.962 | 0.49 | 0.005 | 0.953 |
| rs13106911 | IL15 | −0.36 | 0.361 | 0.811 | −0.46 | 0.007 | 0.925 |
| rs8177636 | IL15RA | 0.39 | 0.179 | 0.858 | 0.47 | 0.006 | 0.942 |
| rs8177685 | IL15RA | −0.41 | 0.057 | 0.881 | −0.47 | 0.004 | 0.931 |
| rs3136614 | IL15RA | −0.47 | 0.005 | 0.957 | −0.40 | 0.125 | 0.798 |
| rs2296139 | IL15RA | 0.44 | 0.011 | 0.933 | 0.53 | <0.0001 | 0.985 |
| rs12437819 | IL16 | −0.50 | 0.001 | 0.981 | −0.41 | 0.071 | 0.846 |
| rs12438640 | IL16 | 0.50 | 0.001 | 0.982 | 0.41 | 0.060 | 0.854 |
| rs10484879 | IL17A | −0.49 | 0.008 | 0.970 | −0.39 | 0.425 | 0.770 |
| rs6518661 | IL17RA | −0.45 | 0.009 | 0.944 | −0.43 | 0.026 | 0.896 |
| rs879576 | IL17RA | 0.45 | 0.025 | 0.931 | 0.50 | 0.004 | 0.953 |
| rs999514 | IL17RB | 0.47 | 0.004 | 0.964 | 0.45 | 0.015 | 0.916 |
| rs708567 | IL17RC | −0.40 | 0.066 | 0.890 | −0.51 | 0.001 | 0.976 |
| rs6445854 | IL17RD | −0.43 | 0.030 | 0.912 | −0.52 | <0.0001 | 0.982 |
| rs4535195 | IL17RD | −0.44 | 0.013 | 0.942 | −0.48 | 0.003 | 0.957 |
| rs12487790 | IL17RD | 0.43 | 0.018 | 0.935 | 0.52 | <0.0001 | 0.983 |
| rs17216900 | IL17RD | 0.45 | 0.009 | 0.946 | 0.36 | 0.368 | 0.779 |
| rs12496746 | IL17RD | −0.46 | 0.006 | 0.951 | −0.42 | 0.048 | 0.866 |
| rs455863 | IL17RE | −0.46 | 0.005 | 0.957 | −0.57 | <0.0001 | 0.996 |
| rs279581 | IL17RE | 0.39 | 0.116 | 0.878 | 0.50 | 0.001 | 0.974 |
| rs279572 | IL17RE | 0.37 | 0.208 | 0.854 | 0.48 | 0.002 | 0.960 |
| rs172155 | IL17RE | −0.37 | 0.244 | 0.847 | −0.48 | 0.003 | 0.953 |
| rs2272128 | IL18RAP | −0.47 | 0.004 | 0.965 | -0.39 | 0.125 | 0.829 |
| rs2243193 | IL19 | 0.42 | 0.031 | 0.919 | 0.50 | 0.001 | 0.973 |
| rs12044804 | IL19 | −0.48 | 0.002 | 0.973 | −0.62 | <0.0001 | 1.000 |
| rs4845143 | IL19 | 0.41 | 0.042 | 0.910 | 0.49 | 0.001 | 0.969 |
| rs12409415 | IL19 | −0.45 | 0.012 | 0.928 | −0.49 | 0.002 | 0.945 |
| rs12046559 | IL19 | 0.43 | 0.021 | 0.923 | 0.54 | <0.0001 | 0.989 |
| rs12145973 | IL19 | 0.59 | <0.0001 | 1.000 | 0.42 | 0.106 | 0.820 |
| rs2138992 | IL19 | 0.46 | 0.006 | 0.959 | 0.52 | <0.0001 | 0.984 |
| rs2232360 | IL20 | −0.38 | 0.166 | 0.858 | −0.46 | 0.007 | 0.934 |
| rs1322393 | IL20RA | 0.39 | 0.098 | 0.876 | 0.49 | 0.002 | 0.963 |
| rs1322394 | IL20RA | −0.38 | 0.206 | 0.844 | −0.48 | 0.003 | 0.948 |
| rs75977 | IL20RB | -0.43 | 0.019 | 0.934 | −0.47 | 0.005 | 0.940 |
| rs835634 | IL20RB | 0.43 | 0.020 | 0.932 | 0.46 | 0.008 | 0.929 |
| rs747842 | IL20RB | 0.43 | 0.023 | 0.929 | 0.46 | 0.008 | 0.930 |
| rs12934152 | IL21R | −0.57 | <0.0001 | 0.999 | −0.50 | 0.005 | 0.952 |
| rs10903022 | IL22RA1 | −0.42 | 0.037 | 0.920 | −0.49 | 0.002 | 0.971 |
| rs10751768 | IL22RA1 | 0.42 | 0.030 | 0.926 | 0.50 | 0.001 | 0.974 |
| rs3795302 | IL22RA1 | 0.39 | 0.087 | 0.898 | 0.49 | 0.001 | 0.971 |
| rs4486393 | IL22RA1 | 0.39 | 0.146 | 0.863 | 0.50 | 0.002 | 0.968 |
| rs4292900 | IL22RA1 | 0.40 | 0.109 | 0.882 | 0.47 | 0.005 | 0.947 |
| rs16829209 | IL22RA1 | 0.39 | 0.156 | 0.856 | 0.50 | 0.002 | 0.959 |
| rs11570915 | IL26 | 0.40 | 0.132 | 0.852 | 0.53 | 0.001 | 0.980 |
| rs3814240 | IL26 | 0.42 | 0.026 | 0.929 | 0.49 | 0.002 | 0.964 |
| rs3814241 | IL26 | 0.50 | 0.001 | 0.982 | 0.47 | 0.004 | 0.946 |
| rs10878789 | IL26 | 0.47 | 0.004 | 0.962 | 0.41 | 0.052 | 0.868 |
| rs9632389 | IL31RA | −0.48 | 0.005 | 0.960 | −0.43 | 0.058 | 0.837 |
| rs10055201 | IL31RA | 0.39 | 0.120 | 0.869 | 0.45 | 0.009 | 0.927 |
| rs1554999 | IL32 | −0.48 | 0.002 | 0.965 | −0.55 | <0.0001 | 0.988 |
Bonferroni-corrected p-value.
Percentile rank relative to the distribution of SNP control sets matched for allele frequency.
Allele frequency spectra in human populations are affected by selective and nonselective events; whereas selection acts on specific genes, nonselective forces (e.g., demography or distance from Africa [22]) are expected to affect all loci equally. We thus compared the strength of IL gene correlations to sets of control SNPs in the dataset. In particular, for each IL SNP in Table I, we extracted from the full HGDP-CEPH dataset all SNPs having an overall minor allele frequency (averaged over all populations) differing by <0.01 from its frequency; for all SNPs in the frequency-matched groups, we calculated Kendall's rank correlation coefficient (τ) between micro- and macropathogen richness and allele frequencies. We next calculated the percentile rank of IL gene SNPs in the distribution of Kendall's τ obtained for the control sets. In most cases (46 for micro- and 66 for macropathogens), percentile ranks higher than the 95th were obtained for IL SNPs. These data indicate that SNPs in IL genes are clearly more strongly influenced by pathogen richness compared with control SNPs, suggesting that selective forces (i.e., pathogen-driven selection), and not nonselective forces, are responsible for the observed associations. All data are gathered in Table I, which shows the compilation of all SNPs that significantly correlate with either micro- or macropathogen richness; the value of τ for both correlations, as well as Bonferroni-corrected P values and percentile ranks, are reported. It is worth noting that data in Table I have been organized to keep together all SNPs in the same gene and to group ligands and receptors; therefore, the order does not reflect greater association with micro- or macropathogens, which can instead be inferred from correlation values.
Another issue that deserves attention relates to the genomic organization of IL genes, because many of them are located in clusters. This raises the possibility that many of the observed allele associations are spurious and derive from linkage to a single selected allele. Yet analysis of linkage disequilibrium (LD; Fig. S2) indicates that linkage is not extensive across gene clusters, with the exception of IL17RE and IL17RC. Therefore, with the exception of these two genes, the remaining loci are independent targets of pathogen-driven selection.
Balancing selection acts on IL1F5, IL1F7, IL1F10, IL7R, and IL18RAP
Pathogen-driven variations in allele frequencies can occur under different selection scenarios, such as directional or balancing selection. Given the aforementioned results and the role of IL genes in regulating immune responses, we next verified whether selection signatures could be identified at IL genes by using classical population genetic analyses. To this aim, we exploited the observation that 68 out of 91 IL loci have been included in genetic variation projects (i.e., the SeattleSNPs program and the Innate Immunity in Heart, Lung and Blood Disease Programs for Genomic Applications) so that resequencing data (although with some gaps) are available in at least two populations: one with European ancestry (EU) and one with African ancestry (either Yorubans [YRI] or African Americans [AA]). Common population genetics tests include Tajima's D (DT) (23) and Fu and Li's D* and F* (24). DT tests the departure from neutrality by comparing two nucleotide diversity indexes: θW (25), which is an estimate of the expected per site heterozygosity, and π (26), which is the average number of pairwise sequence nucleotide differences. Positive values of DT indicate an excess of intermediate frequency variants and are a hallmark of balancing selection; negative DT values indicate either purifying selection or a high representation of rare variants as a result of a selective sweep. Fu and Li's F* and D* (24) are also based on SNP frequency spectra and differ from DT in that they also take into account whether mutations occur in external or internal branches of a genealogy. Population history, in addition to selective processes, affects frequency spectra and all related statistics; for this reason, statistical significance was evaluated by performing coalescent simulations using a population genetics model that incorporates demographic scenarios (27). Simulations were performed using the cosi package (27) and were used to derive a p-value that indicates whether or not the value obtained for a given IL locus is expected under a given demographic scenario; a significant P value indicates that the obtained value is unlikely under the specified conditions and, therefore, that neutrality can be rejected.
Another method of figuring out the effects of selection and population history again lies in the assumption that selection acts on a single locus, whereas demography affects the whole genome; therefore, we calculated test statistics for 238 genes resequenced by the National Institute of Environmental Health Science (NIEHS) program (Table S4). These empirical distributions were used to calculate the percentile rank of DT, D*, and F* values for analyzed IL genes; this procedure provides a direct comparison of the locus under analysis with a sample of human genes and allows an estimation of how unusual the value obtained is (e.g., a percentile rank of 0.99 suggests that neutral evolution is unlikely).
5 of the 68 IL genes available for population genetic analysis gave significant results in at least one population; three of them (IL1F10, IL7R, and IL18RAP) also display at least one SNP correlating with pathogen richness (Table I). Data concerning θW and π, as well as DT, D*, and F*, are reported in Table II. For IL1F5, IL1F7, and IL1F10, θW and π were close to or higher than the 95th percentile in the distribution of NIEHS genes in both populations, indicating an excess of polymorphisms at these loci; conversely, no exceptional θW and π were observed for IL18RAP in AA and for IL7R in either population. Calculation of DT, D*, and F* for the five genes indicated that one or more statistics rejected neutrality in both Africans and Europeans for IL1F5, IL1F10, and IL18RAP; conversely, unusually high values were obtained only for YRI and EU in the case of IL1F7 and IL7R, respectively. Overall, these data suggest the action of balancing selection. It should be noted that IL7R is encompassed by a copy number variant (CNV; http://projects.tcag.ca/variation/); yet the CNV only occurs in 1 out of 110 chromosomes, and thus should not affect our results.
Table II.
Summary statistics for five IL genes
| Gene | La | Pb | Nc | Sd | θe | πf | DT | D* | F* | ||||||
| Value | P-valueg | Rankh | Value | P-valueg | Rankh | Value | P-valueg | Rankh | |||||||
| IL1F5 | 6.4 | YRI | 48 | 50 | 17.61 | 21.73 | 0.81 | 0.033 | 0.97 | 0.43 | 0.11 | 0.85 | 0.68 | 0.045 | 0.93 |
| EU | 46 | 39 | 13.87 | 25.33 | 2.84 | 0.0004 | >0.99 | 1.68 | 0.0018 | >0.99 | 2.49 | 0.0002 | >0.99 | ||
| IL1F7 | 6.1 | YRI | 48 | 45 | 16.57 | 22.07 | 1.14 | 0.016 | >0.99 | 0.82 | 0.037 | 0.95 | 1.11 | 0.012 | 0.98 |
| EU | 46 | 35 | 13.02 | 8.69 | −1.13 | 0.10 | 0.15 | 1.18 | 0.046 | 0.96 | 0.43 | 0.28 | 0.69 | ||
| IL1F10 | 4 | YRI | 48 | 34 | 19.36 | 18.98 | −0.066 | 0.21 | 0.76 | 1.15 | 0.012 | 0.98 | 0.85 | 0.027 | 0.95 |
| EU | 46 | 16 | 9.20 | 12.68 | 1.19 | 0.070 | 0.89 | 1.59 | 0.0041 | >0.99 | 1.72 | 0.0090 | 0.99 | ||
| IL18RAP | 17.8 | AA | 48 | 97 | 12.30 | 14.24 | 0.56 | 0.039 | 0.95 | 0.69 | 0.013 | 0.94 | 0.77 | 0.011 | 0.96 |
| EU | 46 | 88 | 11.27 | 15.56 | 1.36 | 0.057 | 0.91 | 1.26 | 0.0099 | 0.97 | 1.54 | 0.013 | 0.98 | ||
| IL7R | 21 | AA | 48 | 119 | 12.80 | 9.94 | −0.80 | 0.42 | 0.39 | 0.26 | 0.10 | 0.82 | −0.16 | 0.21 | 0.71 |
| EU | 46 | 70 | 7.60 | 10.33 | 1.28 | 0.084 | 0.90 | 1.53 | 0.0029 | >0.99 | 1.71 | 0.011 | 0.99 | ||
Length of analyzed resequenced region (in kilobasepairs).
Population.
Sample size (chromosomes).
Number of segregating sites.
θw estimation per site (×10−4).
π estimation per site (×10−4).
P-values obtained by applying a calibrated population genetics model, as described in the text.
Percentile rank relative to the distribution of values obtained for 238 NIEHS genes (i.e., comparison with an empirical distribution).
Another commonly used test to verify departure from selective neutrality is the Hudson, Kreitman, and Aguade (HKA) test (28); it is based on the assumption that under neutral evolution, the amount of within-species diversity correlates with levels of between-species divergence because both depend on the neutral mutation rate. An excess of intraspecific diversity compared with divergence (k > 1) is considered a signature of balancing selection. Here, we performed a maximum likelihood HKA test (MLHKA) (29) by comparing each IL gene in Table II to 16 neutrally evolving genes resequenced in the same individuals (see Materials and methods for details). The MLHKA test rejected the neutral evolution model for IL1F5 and IL7R (in both populations), as well as IL1F10 (in YRI), but not for IL1F7 and IL18RAP. Indeed, in the latter case, interspecific divergence higher than the genome average paralleled the high levels of intraspecific diversity (unpublished data).
Population genetic diversity, measured as FST, can also provide information on selective processes. Under selective neutrality, FST is determined by genetic drift, which is mainly accounted for by demographic history and similarly affects all genomic loci. Conversely, natural selection being a locus-specific force, it can affect FST values for specific genes. Balancing selection may lead to a decrease in FST compared with neutrally evolving loci (8); specifically, low FST values among continental populations strongly suggests the action of balancing selection worldwide (i.e., irrespective of local environmental pressures), whereas reduced population differentiation within continents is consistent with a locally exerted selective pressure resulting in balancing selection. We calculated FST for the 5 IL genes and compared it to the distribution of this parameter among 238 NIEHS genes. Unusual FST values were only observed for IL18RAP; in this case, a negative FST was obtained, indicating that the real value is close to 0, therefore corresponding to a percentile rank lower than the 2.5th.
One other effect of balancing selection is the maintenance of divergent haplotype clades separated by deep coalescence times (30). We therefore studied haplotype genealogies by constructing median-joining networks. In particular, regions displaying low recombination rates were selected (see Materials and methods and Fig. S3); in the case of IL18RAP and IL1F10, network analysis was not performed because of the low LD throughout the gene region.
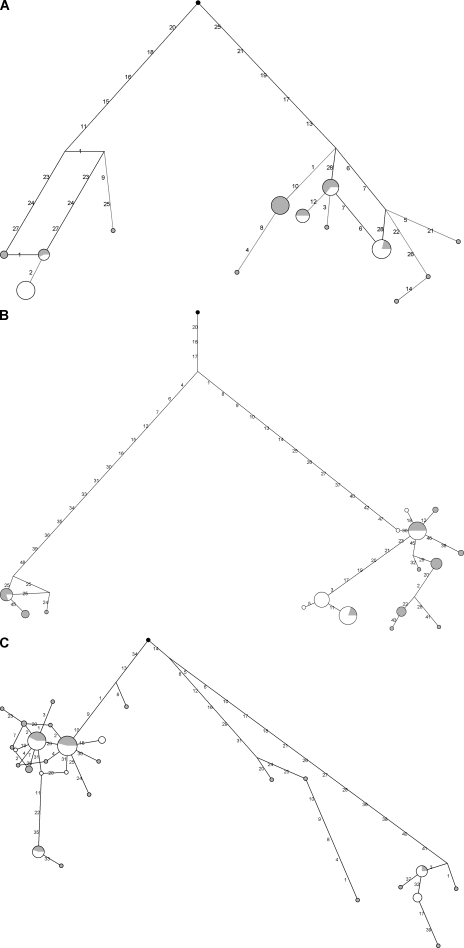
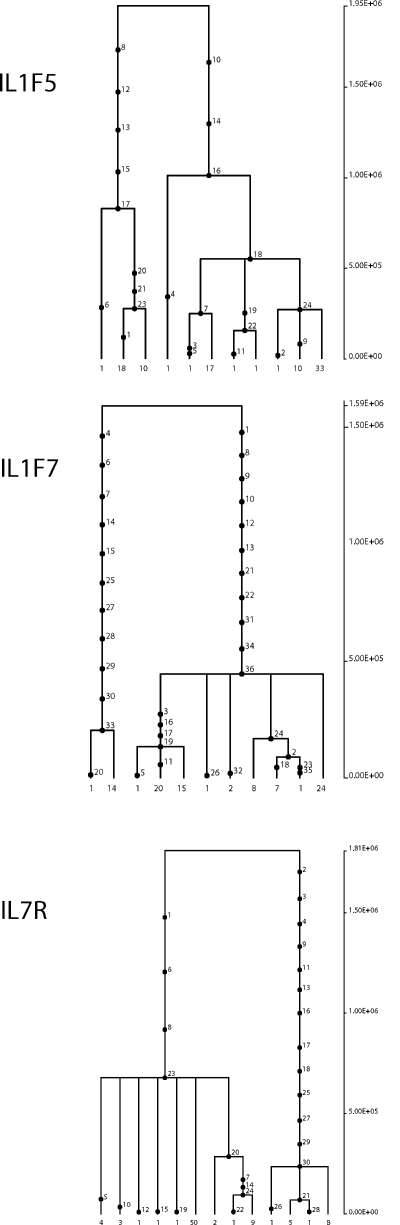
Haplotype genealogies for IL1F5, IL1F7, and IL7R revealed two major clades separated by long branches (Fig. 1), each containing common haplotypes. To estimate the time to the most recent common ancestor (TMRCA) of the haplotype clades, we applied a phylogeny-based method (31) based on the measure ρ, which is the average pairwise difference between haplotypes and a root. TMRCA estimates of 2.76 million years (MY; SD = 660 KY), 2.10 MY (SD = 404 KY), and 1.68 MY (SD = 408 KY) were obtained for IL1F5, IL1F7, and IL7R, respectively. In all cases, we verified these results using GENETREE, which is based on a maximum-likelihood coalescent analysis (32). Consistently, the resulting gene trees, rooted using the chimpanzee sequences, are partitioned into two deep branches (Fig. 2). Using this method, a second estimate of the TMRCAs for the three genes was obtained (Fig. 2 and Table S5). Estimates of coalescence times for neutrally evolving autosomal human loci range between 0.8 and 1.5 MY (33); the TMRCAs we estimated for IL1F5, IL1F7, and IL7R, although not exceptional, are deeper than for most neutrally evolving loci. Again, this finding suggests the action of balancing selection (30).
Figure 1.
Haplotype genealogy for IL1F5, IL1F7, and IL7R gene regions. The analyzed regions correspond to the largest LD block for each gene (Fig. S2). Each node represents a different haplotype, with the size of the circle proportional to the haplotype frequency. Nucleotide differences between haplotypes are indicated on the branches of the network. Circles are color-coded according to population (gray, AA or YRI; white, EU). The chimpanzee sequence is also shown (black). Fig. S2 is available at http://www.jem.org/cgi/content/full/jem.20082779/DC1.
Figure 2.
Estimated trees for IL1F5, IL1F7, and IL7R gene regions. The analyzed regions correspond to the largest LD block for each gene (Fig. S2). Mutations are represented as black dots and named for their physical position along the regions. The absolute frequency of each haplotype is also reported. Fig. S2 is available at http://www.jem.org/cgi/content/full/jem.20082779/DC1.
Heterozygote advantage (also known as overdominance) is one of the possible causes of balancing selection. To verify whether this is the case for the five selected IL genes, for each population we calculated the ratio of observed heterozygosity to expected gene diversity. This same ratio calculation has recently been applied to human HapMap SNPs, and threshold values for inference of overdominance have been set to 1.160 and 1.165 for YRI and EU, respectively (34). To obtain an additional estimate of this parameter distribution in the human genome, ratios were also calculated for NIEHS genes. Whereas all other genes showed nonexceptional values, the observed heterozygosity to expected gene diversity ratio for IL1F5 amounted to 1.07 and 1.20 for YRI and EU, respectively. This value is higher than the previously set threshold for EU and falls above the 99th percentile value obtained from NIEHS genes in this same population.
Overall, these data strongly suggest the action of balancing selection on these 5 IL genes reported in Tables II and III; it is worth mentioning that the MLHKA test failed to reject neutrality for IL1F7 and IL18RAP, suggesting that relaxed constraint rather than balancing selection might be responsible for the observed high DT, D*, and F* values. Still, we noticed that the 6 polymorphisms within IL1F7 exons in YRI are all accounted for by nonsynonymous substitutions; application of the MK tests (35) using orangutan for divergence yielded a p-value of 0.058. Although not fully significant, this result indicates an unusual frequency of replacement polymorphic variants and, together with the deeper-than-average TMRCA, is in line with balancing selection rather than with functional relaxation.
Table III.
MLHKA test and FST for five IL genes
| Gene | Pa | MLHKA | FST (rankb) | |
| kc | P-value | |||
| IL1F5 | YRI | 2.92 | 0.0043 | 0.13 (0.53) |
| EU | 2.85 | 0.0032 | ||
| IL1F7 | YRI | 1.82 | 0.29 | 0.28 (0.89) |
| EU | 1.91 | 0.29 | ||
| IL1F10 | YRI | 2.38 | 0.017 | 0.16 (0.62) |
| EU | 1.46 | 0.56 | ||
| IL18RAP | AA | 1.18 | 0.22 | 0 (<0.025) |
| EU | 1.62 | 0.38 | ||
| IL7R | AA | 2.53 | 0.0042 | 0.019 (0.084) |
| EU | 2.14 | 0.028 | ||
Population.
Percentile rank relative to the distribution of FST values calculated for 238 NIEHS genes.
Selection parameter.
With respect to IL18RAP, the extremely low FST value we observed (Table III) is not consistent with relaxed functional constraints, but instead supports the idea that a balanced polymorphism is maintained in AA and EU, suggesting response to a widespread selective pressure (i.e., not local adaptation). Finally, it should be noted that IL1F5 and IL1F10 are located nearby, raising the possibility that the signatures we observe at both gene regions are caused by hitchhiking with a single functional variant; yet, LD is low across the region because of the presence of a recombination hotspot within the IL1F10 gene region. Moreover, although hitchhiking has the potential to affect large genomic regions, the signatures of balancing selection are predicted to extend over relatively short distances (36, 37). We therefore consider that the two genes might be independent selection targets.
Risk alleles for inflammatory bowel and celiac disease correlate with pathogen-richness
Previous analyses showed that a polymorphism (rs917997) located ∼1,500 bp downstream of IL18RAP is significantly associated with both celiac disease (CeD) and inflammatory bowel disease (IBD; with the SNP also influencing the level of gene expression (38, 39). This variant was not included in our analysis of pathogen correlations because of its location outside the gene region (as defined in Materials and methods). Still, we observed that rs917997 is in strong LD (D' = 1 and 0.87 in EU and YRI, respectively) with rs2272128, which we found to correlate with macro- and micropathogen richness. We therefore checked the presence of rs917997 and verified that the risk allele for CeD and IBD also correlates significantly with pathogen richness (Table IV). Stimulated by this finding, we next verified whether the frequency distribution of other susceptibility alleles in IL genes has been influenced by pathogen richness. To analyze only variants that have been identified in an unbiased manner (i.e., without a priori hypothesis on the genes involved), we searched among published genome-wide association studies for SNPs in IL genes that have been associated with any trait. For available variants in the CEPH-HGDP panel, we next calculated correlations with micro- and macropathogen richness. Results reported in Table IV indicate that six out of nine risk variants for CeD or IDB/Crohn's disease (CD) were associated with micro- and macropathogen richness; in particular, two of them located within IL23R are in tight LD, and thus do not represent independent observations. Noticeably, in all six cases, the risk allele correlates with pathogen richness and correlations with micropathogens were stronger compared with macropathogens (with the exception of rs11465804), a situation different from the general observation that macropathogens have represented a more powerful selective pressure on IL genes.
Table IV.
Correlations with micro- and macropathogen richness for SNPs associated with different traits
| SNP | Gene/location | Allele | Micropathogen | Macropathogen | Trait/disease | |||
| Risk | Anc.a | τb | P-valuec | τb | P-valuec | |||
| rs6897932 | IL7R | C | C | 0.22 | n.s.d | 0.21 | n.s.d | Multiple sclerosis/Type 1 diabetes |
| rs917997 | IL18RAP (downstream) | A | G | 0.42 | <0.001 | 0.35 | 0.008 | CeD/IBD |
| rs10045431 | IL12B | C | C | 0.43 | <0.001 | 0.34 | 0.015 | CD |
| rs7517847 | IL23R | C | A | 0.23 | n.s.d | 0.26 | n.s.d | IBD |
| rs11209026 | IL23R | G | G | 0.47 | <0.001 | 0.44 | 0.001 | IBD |
| rs11465804 | IL23R | T | T | 0.39 | 0.004 | 0.44 | <0.001 | CD |
| rs6822844 | IL2/IL21 (intergenic) | G | G | 0.40 | 0.004 | 0.39 | 0.006 | CeD |
| rs3024505 | IL10 | T | C | −0.28 | n.s.d | −0.28 | n.s.d | Ulcerative colitis |
| rs17810546 | IL12A | G | A | 0.03 | n.s.d | −0.11 | n.s.d | CeD |
| rs13015714 | IL18R1 | C | A | 0.47 | <0.001 | 0.39 | 0.002 | CeD |
| rs2250417 | IL18 | A | A | 0.23 | n.s.d | 0.26 | n.s.d | Protein quantitative trait loci |
| rs7626795 | IL1RAP | G | A | 0.20 | n.s.d | 0.04 | n.s.d | Lung cancer |
| rs4129267 | IL6R | C | C | −0.17 | n.s.d | −0.24 | n.s.d | Protein quantitative trait loci/pulmonary function |
| rs6761276 | IL1F10e | n.r.f | T | −0.31 | 0.030 | −0.51 | <0.001 | Protein quantitative trait loci |
| rs12251307 | IL2RA | T | T | −0.16 | n.s.d | −0.13 | n.s.d | Type 1 diabetes |
Ancestral state based on chimpanzee sequence.
The correlation coefficient is calculated between pathogen richness and the risk allele frequency;.
Bonferroni-corrected p-value (15 tests).
Nonsignificant (P > 0.05).
This SNP is located within IL1F10, but affects IL1RN protein levels.
Not reported.
DISCUSSION
We analyzed the recent evolutionary history of IL genes in humans by integrating information on environmental variables with classical population genetics and association studies. Results herein suggest that microbes and parasitic worms played a relevant role as selective agents, but the pressure imposed by helminths on IL genes has been stronger than the one caused by viral and microbial agents. Helminths were present among our ancestors before the emergence of humans as a species (for review see reference [40]). These parasites evolve at lower rates than viruses and bacteria and, in contrast to most viral/microbial agents, are able to maintain themselves in small human communities (41). Notably, by establishing chronic infections, parasitic worms affect the susceptibility of their host to viruses, bacteria, and protozoa (for review see reference [2]). Therefore, helminths might have represented a stable threat to human populations and their distribution, which is not associated with sudden epidemics and, as in the case of micropathogens, might have left stronger genetic signatures.
A limitation of our study is that we implicitly assumed that the number of different pathogen species/genera per country has been maintained proportionally unchanged along human evolutionary history. Although clearly an oversimplification, this might reflect reality to some degree, given that climatic variables (e.g., precipitation rates and temperature) have a primary importance in driving the spatial distribution of human micro- and macropathogens (21). Therefore, while the fitness cost imposed by specific species/genera might have evolved rapidly, the relative number of pathogen species per country might have changed proportionally less.
Another possible caveat of our results concerns the definition of “pathogen,” in that we included any organism that can cause a disease irrespective of its virulence or pathogenicity. The reason for this choice is that the fitness of a pathogen is a direct measure of the ability of such pathogen to replicate within a given environment. Fitness is dependent on both the features of the pathogen and of the host. The features of the pathogen include its abilities to (a) replicate within the host, (b) select a proper cell target (tropism), (c) avoid the immune response, and (d) escape the obstacles posed by drugs. The features of the host include the immune response, the genetic background, and the availability of target cells. The reciprocal balance between these factors determines the virulence of a pathogen, a feature that, therefore, cannot be considered as a constant, but rather evolves dynamically over time.
Despite these limitations, we were able to identify several variants that are likely candidates for pathogen-driven selection, and the suitability of this approach is confirmed by the previous demonstration that variability at HLA genes correlates with a similar measure of pathogen richness (10). Our data point to the SNPs in Table I as good candidates for experimental analyses aimed at inferring their role in IL gene function, given that a signature of natural selection necessarily implies the presence of a functional variant (either the correlated variant itself or a linked one). Also, genes subjected to a selective pressure from infectious diseases should be regarded (42) as obvious candidates for genetic epidemiology studies (e.g., case-control studies).
It is worth noting that most members of the IL-1 signaling pathway were observed to correlate with pathogen richness. IL-1A and IL-1B are pleiotropic cytokines with central roles in immune and inflammatory responses (43) required for the development of Th2-mediated immunity and protection against chronic infection in mice (44). The observation that SNPs strongly correlated with micro- and macro-pathogen richness map to IL1RAPL1 is more puzzling, as this gene is not known to be associated with infection and immune response, but is involved in brain development and function (45); nonetheless, this gene is expressed in tissues that are different from brain (46), suggesting that it plays other roles apart from neurodevelopment.
Strong correlations with macropathogen richness were obtained for IL4, IL4R, and IL10. These molecules are pivotal to the elicitation of Th2 response, which is the immune response central to helminth resistance (40, 47). The strongest correlation with macropathogens was observed for rs12044804 in IL19 (τ = −0.62). This gene is located within an IL cluster, which also comprises IL10 and IL20, and is encompassed by a low-frequency CNV. The low levels of LD across the IL cluster (Fig. S2) suggest that IL19, IL20, and IL10 SNPs are independently associated with pathogen richness. IL19, IL20, IL22, IL24, and IL26 all belong to the IL-20 subfamily, are all produced by different leukocyte populations, and all bind to partially shared receptors that are mainly expressed by epithelial cells and known to promote keratinocyte growth and to induce skin inflammatory responses (48, 49). The correlation of SNPs in most of these genes with micropathogen and helminth richness suggests that modulation of skin immunological properties might represent an adaptive response to parasite species that infect humans through the skin.
SNPs in IL2RB, IL15, and IL15RA also correlated with macropathogen richness; these genes converge on the same pathway as IL-2RB and IL-15RA are part of a trimeric complex that binds IL-15 (50). IL-15 has a central role in intestinal inflammatory processes and in the pathogenesis of CD and CeD (51), possibly participating in the immune protection of gut tissues. An interesting observation is that IL-2RB, via IL-15 binding, regulates the intestinal epithelial barrier by transducing signals that result in tight junction formation (52). Variation of mucosal permeability after nematode infection is a host defense response and is important for efficient parasite expulsion (53); the correlation we observed between SNPs in these genes and macropathogen richness might indicate selection for improved intestinal clearance of nematodes. Again, mouse models will prove central in addressing the role of these loci in parasite expulsion; e.g., in the case of IL4R, which also correlates with helminth richness (Table I), treatment with antibodies or use of il4r-deficient mice prevented expulsion of Heligmosomoides polygyrus, Trichuris muris, and Nippostrongylus brasiliensis (54).
A major locus for susceptibility to Schistosoma mansoni infection (SM1) has previously been mapped to 5q31-q33 (55). This region covers IL4, IL5, IL3, IL13, IL9, and IL12B, as well as other candidate loci (IRF1 and CSF2). Although our data do not allow inference on which gene is responsible for increased susceptibility to schistosomiasis, it is worth noting that four SNPs in IL4 display very strong correlation with helminth richness. IL4 might therefore be regarded as a candidate locus for susceptibility to S. mansoni infection, with dedicated association studies being required to verify this prediction. Conversely, the SM2 region (6q22-q23), where a locus controlling hepatic fibrosis in S. mansoni infection has been mapped (56), harbors no IL genes.
Pathogen-driven variations in allele frequencies can occur under different selection scenarios, such as directional or balancing selection; the latter itself is the result of an initial stage of positive selection that favors the spread in a population of a new allele until selection opposes its fixation and a balanced situation is established. Common causes of balancing selection include heterozygote advantage, changing environmental conditions, and frequency-dependent selection, all of these possibly applying to host–pathogen interactions. Also, given the pleiotropic roles of many IL genes, selective pressures different from pathogen richness might affect the evolutionary fate of these loci. Classical population genetics analyses indicated that five IL genes are likely targets of balancing selection. IL1F5, IL1F7, and IL1F10 are recently discovered IL-1 family members (57) that are located within the IL1 gene cluster; based on comparative analyses, IL-1F5 and IL-1F10 are predicted to act as antagonists (58). IL1F5 is a regulator of skin and brain inflammation (59, 60) and it is expressed in many different human tissues (57). Interestingly, an excess of heterozygotes was observed for IL1F5, suggesting that overdominance might underly the maintenance of a balanced polymorphism in the gene. Overdominance is rare in humans (36, 61) and is hypothesized to enhance immune response flexibility by modulating allele-specific gene expression in different cell types and in response to diverse stimuli/cytokines (62). Whether this is the case for IL1F5 remains to be verified.
IL1F10 is a still relatively unknown protein mainly expressed in skin, proliferating B cells, and tonsils (63). One of the intermediate frequency SNPs in the gene is accounted for by a missense substitution that replaces an aspartic acid residue with an alanine (Asp51Ala); the presence of a negatively charged residue at this position is conserved among mammals (Fig. S4), possibly suggesting functional significance and awaiting experimental testing.
In analogy to IL1F5, the other IL1 family member we identified as rejecting neutral evolution, i.e., IL1F7, also acts as an antiinflammatory molecule (64).
IL1F5, IL1F7, and IL1F10 are not known to be involved in human diseases; in contrast, IL18RAP and IL7R play a role in triggering immune responses. The IL-7/IL-7R ligand-receptor pair is central to the proliferation and survival of B and T leukocytes. We identified one SNP in the gene as highly correlated with macropathogen richness (Table I). This is not surprising given the role of Th2 responses in helminth infection and the involvement of IL-7R in the TSLP signaling pathway (65), which in turn regulates Th2-mediated inflammatory responses (66).
Similar to IL7R, IL18RAP plays a known role in human pathology. The gene encodes a component of the protein complex involved in transducing IL-18 signal, resulting in the activation of NF-κB (67). The IL-18 receptor complex is expressed in the intestine (38), and one SNP immediately downstream IL18RAP (rs917997) has been associated with both CeD and IBD (38, 39). We found the predisposing allele of rs917997 and a linked variant (rs2272128) to correlate with pathogen richness. The location of rs2272128 in the 5′ gene region and its strong correlation with pathogens might suggest that it (rather than rs917997) represents or is in close LD with the functional allele. Moreover, the correlation of a risk allele for autoimmune diseases with pathogen-richness suggests an interesting link between adaptation and disease. Indeed, we observed that five more risk alleles for either IBD or CeD significantly correlate with micro- and macropathogen richness. Albeit preliminary, these data suggest that infectious agents have shaped the genetic variation at IL loci involved in intestinal inflammatory processes and, as a consequence, the genetic predisposition to both CeD and CD/IBD.
A north–south gradient for IBD prevalence has been described in both the US and Europe (68). This observation, together with the increase of IBD prevalence in the last 40 yr (68) and the hypothesis that helminths elicit Th2-mediated responses, led to the proposal that lower exposure to parasitic worms in the setting of industrialized countries results in unbalanced immune response, and eventually predisposes to IBD (68, 69). The so-called hygiene hypothesis, which clearly implies evolutionary considerations concerning human–pathogen interactions, has been supported by recent studies in both humans and mice (40, 68–70). Data herein seem to indicate that a portion of CeD- and IBD-predisposing alleles have been selected by micropathogen richness, pointing to an adaptive role for these variants. Although not directly supportive of the “IBD hygiene hypothesis,” these results indicate a higher disease predisposition in subjects carrying IL SNP variants that confer stronger protection against viruses/bacteria and therefore likely elicit more vigorous Th1 responses. Living conditions in industrialized countries have resulted in a reduction of both helminth and bacterial/viral infection. The effect of this environmental change on the homeostasis of immune responses might be difficult to reconcile with simple theories (71). In this complex scenario, we consider that evolutionary studies and population genetics approaches, such as the one proposed here, provide some insight into the genetic basis of predisposition to infectious and autoimmune diseases.
MATERIALS AND METHODS
Data retrieval and haplotype construction.
Data concerning the HGDP-CEPH panel derive from a previous work (20). A SNP was ascribed to a specific gene if it was located within the transcribed region or no further than 500 bp upstream the transcription start site.
Genotype data for resequenced IL genes were retrieved from the SeattleSNPs (http://pga.mbt.washington.edu) and Innate Immunity PGA (http://innateimmunity.net/) web sites. A total of 68 genes were available for analysis. For each gene, genotypes deriving from 24 subjects of African ancestry and 23 of Caucasian ancestry were retrieved.
Genotype data for 238 resequenced human genes were derived from the NIEHS SNPs Program web site (http://egp.gs.washington.edu). We specifically selected genes that had been resequenced in populations of defined ethnicity (NIEHS panel 2).
Haplotypes were inferred using PHASE version 2.1 (72), a program for reconstructing haplotypes from unrelated genotype data through a Bayesian statistical method.
Recombination rates were derived from the University of California at Santa Cruz genome browser web site (http://genome.ucsc.edu). Information concerning CNVs was derived from the database of genomic variants (http://projects.tcag.ca/variation/).
Variants and risk alleles identified in genome-wide association studies were retrieved from the National Human Genome Research Institute web site (http://www.genome.gov/) updated on December 1, 2008.
Statistical analysis.
DT (23), Fu and Li's D* and F* (24) statistics, and diversity parameters θW (25) and π (26) were calculated using libsequence (73). Coalescent simulations were performed using the cosi package (27) and its best-fit parameters for YRI, AA, and EU populations with 104 iterations. cosi is a simulation package based on a population genetics model calibrated on empirical data; it therefore allows incorporation of demographic scenarios in simulations.
The FST statistic (74) estimates genetic differentiation among populations and was calculated as previously proposed (75).
The maximum likelihood ratio HKA test was performed using the MLHKA software (29) as previously described (18). In brief, we used multilocus data of 16 selected genes and Pan troglodytes (NCBI panTro2) as an outgroup (except for IL1F7, where Pongo pygmaeus abelii, NCBI ponAbe2, was used as the outgroup). The 16 reference genes were randomly selected among NIEHS loci <20 kb that have been resequenced across panel 2; the only criterion was that no reference gene rejected the neutral model (i.e., that no gene yielded significant DT). The reference loci used were as follows: VNN3, PLA2G2D, MB, MAD2L2, HRAS, CYP17A1, ATOX1, BNIP3, CDC20, NGB, TUBA1, MT3, NUDT1, PRDX5, RETN, and JUND.
LD analyses were performed using Haploview (76), and haplotype blocks were identified through an implemented method.
Median-joining networks to infer haplotype genealogy were constructed using NETWORK 4.5 (31). Estimate of the TMRCA was obtained using a phylogeny-based approach implemented in NETWORK using a mutation rate based on the number of fixed differences between human and chimpanzee or orangutan and assuming a separation time from humans of 6 MY and 13 MY ago, respectively. A second TMRCA estimate was derived from application of a maximum-likelihood coalescent method implemented in GENETREE (32). Again, the mutation rate μ was obtained on the basis of the divergence between human and a primate, assuming a generation time of 25 yr. Using this μ and the maximum likelihood θ (θML), we estimated the effective population size parameter (Ne). With these assumptions, the coalescence time, scaled in 2Ne units, was converted into years. For the coalescence process, 106 simulations were performed. All calculations were performed in the R environment (www.r-project.org).
Environmental variables.
Pathogen absence/presence matrices for the 21 countries where HGDP-CEPH populations are located were derived from the Gideon database (http://www.gideononline.com) following previous methods (10, 18). Information in Gideon is updated weekly and derives from WHO reports, National Health Ministries, PubMed searches, and epidemiology meetings. The Gideon Epidemiology module follows the status of known infectious diseases globally, as well as in individual countries, with specific notes indicating the disease's history, incidence, and distribution per country. We manually curated pathogen absence/presence matrices by extracting information from single Gideon entries. These may refer to either species or genera (in case data are not available for different species of a same genus). Following previous suggestions (10, 18), we recorded only species/genera that are transmitted in the 21 countries, meaning that cases of transmission caused by tourism and immigration were not taken into account; also, species that have recently been eradicated as a result, for example, of vaccination campaigns, were recorded as present in the matrix. A total of 283 pathogen species were retrieved (Table S6). Other environmental variables such as average annual mean and maximum temperature, precipitation rate, and short-wave radiation flux were derived for the geographic coordinates corresponding to HGDP-CEPH populations from the NCEP/NCAR database (http://www.cdc.noaa.gov/PublicData/).
Online supplemental material.
Table S1 shows correlations between the richness of viruses, bacteria, protozoa, and fungi. Table S2 is a list of IL genes analyzed in the study. Table S3 shows correlations with climatic variables. Table S4 provides diversity indexes and summary statistics for 238 human genes resequenced by the NIEHS program. Table S5 shows GENETREE estimates for IL1F5, IL1F7, and IL7R. Table S6 is a list of pathogen species/genera identified in at least one population. Fig. S1 shows the geographic location and pathogen richness estimates for the 52 HGDP-CEPH populations. Fig. S2 shows LD structure for IL gene clusters. Fig. S3 reports LD blocks for IL1F5, IL1F7, and IL7R. Fig. S4 shows multiple protein alignment for IL-1F10. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20082779/DC1.
Acknowledgments
We are grateful to Dr. Roberto Giorda for helpful discussion about the manuscript. We also wish to thank Dr. Daniele Sampietro for technical assistance in retrieving data on climatic variables.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used: AA, African American; CD, Crohn's disease; CeD, celiac disease; CNV, copy number variant; DT, Tajima's D; EU, European; HGDP-CEPH, Human Genome Diversity Project-Centre d'Etude du Polymorphisme Humain; HKA, Hudson, Kreitman, and Aguade; IBD, inflammatory bowel disease; LD, linkage disequilibrium; MLHKA, maximum likelihood HKA; MY, million years; NIEHS, National Institute of Environmental Health Science; SNP, single-nucleotide polymorphism; TMRCA, time to the most recent common ancestor; YRI, Yoruba.
References
- Kapp C. 1999. WHO warns of microbial threat.Lancet. 353:2222. [DOI] [PubMed] [Google Scholar]
- Hotez P.J., Brindley P.J., Bethony J.M., King C.H., Pearce E.J., Jacobson J. 2008. Helminth infections: the great neglected tropical diseases.J. Clin. Invest. 118:1311–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colley D.G., LoVerde P.T., Savioli L. 2001. Infectious disease. Medical helminthology in the 21st century.Science. 293:1437–1438 [DOI] [PubMed] [Google Scholar]
- Callender J.E., Grantham-McGregor S., Walker S., Cooper E.S. 1992. Trichuris infection and mental development in children.Lancet. 339:181. [DOI] [PubMed] [Google Scholar]
- Nokes C., Grantham-McGregor S.M., Sawyer A.W., Cooper E.S., Bundy D.A. 1992. Parasitic helminth infection and cognitive function in school children.Proc. Biol. Sci. 247:77–81 [DOI] [PubMed] [Google Scholar]
- Strachan D.P. 1989. Hay fever, hygiene, and household size.BMJ. 299:1259–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccone P., Burton O.T., Cooke A. 2008. Interplay of parasite-driven immune responses and autoimmunity.Trends Parasitol. 24:35–42 [DOI] [PubMed] [Google Scholar]
- Charlesworth D. 2006. Balancing selection and its effects on sequences in nearby genome regions.PLoS Genet. 2:e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst L.D. 2009. Fundamental concepts in genetics: genetics and the understanding of selection.Nat. Rev. Genet. 10:83–93 [DOI] [PubMed] [Google Scholar]
- Prugnolle F., Manica A., Charpentier M., Guegan J.F., Guernier V., Balloux F. 2005. Pathogen-driven selection and worldwide HLA class I diversity.Curr. Biol. 15:1022–1027 [DOI] [PubMed] [Google Scholar]
- Quintana-Murci L., Alcais A., Abel L., Casanova J.L. 2007. Immunology in natura: clinical, epidemiological and evolutionary genetics of infectious diseases.Nat. Immunol. 8:1165–1171 [DOI] [PubMed] [Google Scholar]
- Picard C., Casanova J.L., Abel L. 2006. Mendelian traits that confer predisposition or resistance to specific infections in humans.Curr. Opin. Immunol. 18:383–390 [DOI] [PubMed] [Google Scholar]
- Frodsham A.J., Hill A.V. 2004. Genetics of infectious diseases.Hum. Mol. Genet. 13 Spec No 2:R187–R194 [DOI] [PubMed] [Google Scholar]
- Lettre G., Rioux J.D. 2008. Autoimmune diseases: insights from genome-wide association studies.Hum. Mol. Genet. 17:R116–R121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Di Rienzo A., Ober C. 2001. A population genetics study of single nucleotide polymorphisms in the interleukin 4 receptor alpha (IL4RA) gene.Genes Immun. 2:128–134 [DOI] [PubMed] [Google Scholar]
- Sakagami T., Witherspoon D.J., Nakajima T., Jinnai N., Wooding S., Jorde L.B., Hasegawa T., Suzuki E., Gejyo F., Inoue I. 2004. Local adaptation and population differentiation at the interleukin 13 and interleukin 4 loci.Genes Immun. 5:389–397 [DOI] [PubMed] [Google Scholar]
- Akey J.M., Eberle M.A., Rieder M.J., Carlson C.S., Shriver M.D., Nickerson D.A., Kruglyak L. 2004. Population history and natural selection shape patterns of genetic variation in 132 genes.PLoS Biol. 2:e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M., Cagliani R., Pozzoli U., Riva S., Comi G.P., Menozzi G., Bresolin N., Sironi M. 2009. Widespread balancing selection and pathogen-driven selection at blood group antigen genes.Genome Res. 19:199–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullan R., Brooker S. 2008. The health impact of polyparasitism in humans: are we under-estimating the burden of parasitic diseases? Parasitology. 135:783–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.Z., Absher D.M., Tang H., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L., Myers R.M. 2008. Worldwide human relationships inferred from genome-wide patterns of variation.Science. 319:1100–1104 [DOI] [PubMed] [Google Scholar]
- Guernier V., Hochberg M.E., Guegan J.F. 2004. Ecology drives the worldwide distribution of human diseases.PLoS Biol. 2:e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley L.J., Manica A., Goudet J., Balloux F. 2007. Going the distance: human population genetics in a clinal world.Trends Genet. 23:432–439 [DOI] [PubMed] [Google Scholar]
- Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism.Genetics. 123:585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.X., Li W.H. 1993. Statistical tests of neutrality of mutations.Genetics. 133:693–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G.A. 1975. On the number of segregating sites in genetical models without recombination.Theor. Popul. Biol. 7:256–276 [DOI] [PubMed] [Google Scholar]
- Nei M., Li W.H. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases.Proc. Natl. Acad. Sci. USA. 76:5269–5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner S.F., Foo C., Gabriel S., Reich D., Daly M.J., Altshuler D. 2005. Calibrating a coalescent simulation of human genome sequence variation.Genome Res. 15:1576–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R.R., Kreitman M., Aguade M. 1987. A test of neutral molecular evolution based on nucleotide data.Genetics. 116:153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S.I., Charlesworth B. 2004. The HKA test revisited: a maximum-likelihood-ratio test of the standard neutral model.Genetics. 168:1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N. 1990. A simple genealogical structure of strongly balanced allelic lines and trans-species evolution of polymorphism.Proc. Natl. Acad. Sci. USA. 87:2419–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt H.J., Forster P., Rohl A. 1999. Median-joining networks for inferring intraspecific phylogenies.Mol. Biol. Evol. 16:37–48 [DOI] [PubMed] [Google Scholar]
- Griffiths R.C., Tavare S. 1995. Unrooted genealogical tree probabilities in the infinitely-many-sites model.Math. Biosci. 127:77–98 [DOI] [PubMed] [Google Scholar]
- Tishkoff S.A., Verrelli B.C. 2003. Patterns of human genetic diversity: implications for human evolutionary history and disease.Annu. Rev. Genomics Hum. Genet. 4:293–340 [DOI] [PubMed] [Google Scholar]
- Alonso S., Lopez S., Izagirre N., de la Rua C. 2008. Overdominance in the human genome and olfactory receptor activity.Mol. Biol. Evol. 25:997–1001 [DOI] [PubMed] [Google Scholar]
- McDonald J.H., Kreitman M. 1991. Adaptive protein evolution at the Adh locus in Drosophila.Nature. 351:652–654 [DOI] [PubMed] [Google Scholar]
- Bubb K.L., Bovee D., Buckley D., Haugen E., Kibukawa M., Paddock M., Palmieri A., Subramanian S., Zhou Y., Kaul R., et al. 2006. Scan of human genome reveals no new Loci under ancient balancing selection.Genetics. 173:2165–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiuf C., Zhao K., Innan H., Nordborg M. 2004. The probability and chromosomal extent of trans-specific polymorphism.Genetics. 168:2363–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt K.A., Zhernakova A., Turner G., Heap G.A., Franke L., Bruinenberg M., Romanos J., Dinesen L.C., Ryan A.W., Panesar D., et al. 2008. Newly identified genetic risk variants for celiac disease related to the immune response.Nat. Genet. 40:395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova A., Festen E.M., Franke L., Trynka G., van Diemen C.C., Monsuur A.J., Bevova M., Nijmeijer R.M., van 't Slot R., Heijmans R., et al. 2008. Genetic analysis of innate immunity in Crohn's disease and ulcerative colitis identifies two susceptibility loci harboring CARD9 and IL18RAP.Am. J. Hum. Genet. 82:1202–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunne D.W., Cooke A. 2005. A worm's eye view of the immune system: consequences for evolution of human autoimmune disease.Nat. Rev. Immunol. 5:420–426 [DOI] [PubMed] [Google Scholar]
- Dobson A. 1992. People and disease. In The Cambridge Encyclopedia of Human Evolution Jones S., Martin R., Pilbeam D., Cambridge University Press, Cambridge: 411–420 [Google Scholar]
- Burgner D., Jamieson S.E., Blackwell J.M. 2006. Genetic susceptibility to infectious diseases: big is beautiful, but will bigger be even better? Lancet Infect. Dis. 6:653–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarello C.A. 1994. The biological properties of interleukin-1.Eur. Cytokine Netw. 5:517–531 [PubMed] [Google Scholar]
- Helmby H., Grencis R.K. 2004. Interleukin 1 plays a major role in the development of Th2-mediated immunity.Eur. J. Immunol. 34:3674–3681 [DOI] [PubMed] [Google Scholar]
- Carrie A., Jun L., Bienvenu T., Vinet M.C., McDonell N., Couvert P., Zemni R., Cardona A., Van Buggenhout G., Frints S., et al. 1999. A new member of the IL-1 receptor family highly expressed in hippocampus and involved in X-linked mental retardation.Nat. Genet. 23:25–31 [DOI] [PubMed] [Google Scholar]
- Born T.L., Smith D.E., Garka K.E., Renshaw B.R., Bertles J.S., Sims J.E. 2000. Identification and characterization of two members of a novel class of the interleukin-1 receptor (IL-1R) family. Delineation of a new class of IL-1R-related proteins based on signaling.J. Biol. Chem. 275:29946–29954 [DOI] [PubMed] [Google Scholar]
- Maizels R.M., Yazdanbakhsh M. 2003. Immune regulation by helminth parasites: cellular and molecular mechanisms.Nat. Rev. Immunol. 3:733–744 [DOI] [PubMed] [Google Scholar]
- Parrish-Novak J., Xu W., Brender T., Yao L., Jones C., West J., Brandt C., Jelinek L., Madden K., McKernan P.A., et al. 2002. Interleukins 19, 20, and 24 signal through two distinct receptor complexes. Differences in receptor-ligand interactions mediate unique biological functions.J. Biol. Chem. 277:47517–47523 [DOI] [PubMed] [Google Scholar]
- Kotenko S.V., Izotova L.S., Mirochnitchenko O.V., Esterova E., Dickensheets H., Donnelly R.P., Pestka S. 2001. Identification of the functional interleukin-22 (IL-22) receptor complex: the IL-10R2 chain (IL-10Rbeta) is a common chain of both the IL-10 and IL-22 (IL-10-related T cell-derived inducible factor, IL-TIF) receptor complexes.J. Biol. Chem. 276:2725–2732 [DOI] [PubMed] [Google Scholar]
- Burton J.D., Bamford R.N., Peters C., Grant A.J., Kurys G., Goldman C.K., Brennan J., Roessler E., Waldmann T.A. 1994. A lymphokine, provisionally designated interleukin T and produced by a human adult T-cell leukemia line, stimulates T-cell proliferation and the induction of lymphokine-activated killer cells.Proc. Natl. Acad. Sci. USA. 91:4935–4939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel D.A., Franke L., Hunt K.A., Gwilliam R., Zhernakova A., Inouye M., Wapenaar M.C., Barnardo M.C., Bethel G., Holmes G.K., et al. 2007. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21.Nat. Genet. 39:827–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama R., Sakaguchi T., Kinugasa T., Gu X., MacDermott R.P., Podolsky D.K., Reinecker H.C. 2001. Interleukin-2 receptor beta subunit-dependent and -independent regulation of intestinal epithelial tight junctions.J. Biol. Chem. 276:35571–35580 [DOI] [PubMed] [Google Scholar]
- Murray M., Jarrett W.F., Jennings F.W. 1971. Mast cells and macromolecular leak in intestinal immunological reactions. The influence of sex of rats infected with Nippostrongylus brasiliensis.Immunology. 21:17–31 [PMC free article] [PubMed] [Google Scholar]
- Urban J.F., Jr., Noben-Trauth N., Donaldson D.D., Madden K.B., Morris S.C., Collins M., Finkelman F.D. 1998. IL-13, IL-4Ralpha, and Stat6 are required for the expulsion of the gastrointestinal nematode parasite Nippostrongylus brasiliensis.Immunity. 8:255–264 [DOI] [PubMed] [Google Scholar]
- Marquet S., Abel L., Hillaire D., Dessein H., Kalil J., Feingold J., Weissenbach J., Dessein A.J. 1996. Genetic localization of a locus controlling the intensity of infection by Schistosoma mansoni on chromosome 5q31-q33.Nat. Genet. 14:181–184 [DOI] [PubMed] [Google Scholar]
- Dessein A.J., Hillaire D., Elwali N.E., Marquet S., Mohamed-Ali Q., Mirghani A., Henri S., Abdelhameed A.A., Saeed O.K., Magzoub M.M., Abel L. 1999. Severe hepatic fibrosis in Schistosoma mansoni infection is controlled by a major locus that is closely linked to the interferon-gamma receptor gene.Am. J. Hum. Genet. 65:709–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.E., Renshaw B.R., Ketchem R.R., Kubin M., Garka K.E., Sims J.E. 2000. Four new members expand the interleukin-1 superfamily.J. Biol. Chem. 275:1169–1175 [DOI] [PubMed] [Google Scholar]
- Nicklin M.J., Barton J.L., Nguyen M., FitzGerald M.G., Duff G.W., Kornman K. 2002. A sequence-based map of the nine genes of the human interleukin-1 cluster.Genomics. 79:718–725 [DOI] [PubMed] [Google Scholar]
- Blumberg H., Dinh H., Trueblood E.S., Pretorius J., Kugler D., Weng N., Kanaly S.T., Towne J.E., Willis C.R., Kuechle M.K., et al. 2007. Opposing activities of two novel members of the IL-1 ligand family regulate skin inflammation.J. Exp. Med. 204:2603–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costelloe C., Watson M., Murphy A., McQuillan K., Loscher C., Armstrong M.E., Garlanda C., Mantovani A., O'Neill L.A., Mills K.H., Lynch M.A. 2008. IL-1F5 mediates anti-inflammatory activity in the brain through induction of IL-4 following interaction with SIGIRR/TIR8.J. Neurochem. 105:1960–1969 [DOI] [PubMed] [Google Scholar]
- Asthana S., Schmidt S., Sunyaev S. 2005. A limited role for balancing selection.Trends Genet. 21:30–32 [DOI] [PubMed] [Google Scholar]
- Beaty J.S., West K.A., Nepom G.T. 1995. Functional effects of a natural polymorphism in the transcriptional regulatory sequence of HLA-DQB1.Mol. Cell. Biol. 15:4771–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Ho A.S., Haley-Vicente D., Zhang J., Bernal-Fussell J., Pace A.M., Hansen D., Schweighofer K., Mize N.K., Ford J.E. 2001. Cloning and characterization of IL-1HY2, a novel interleukin-1 family member.J. Biol. Chem. 276:20597–20602 [DOI] [PubMed] [Google Scholar]
- Sharma S., Kulk N., Nold M.F., Graf R., Kim S.H., Reinhardt D., Dinarello C.A., Bufler P. 2008. The IL-1 family member 7b translocates to the nucleus and down-regulates proinflammatory cytokines.J. Immunol. 180:5477–5482 [DOI] [PubMed] [Google Scholar]
- Al-Shami A., Spolski R., Kelly J., Fry T., Schwartzberg P.L., Pandey A., Mackall C.L., Leonard W.J. 2004. A role for thymic stromal lymphopoietin in CD4(+) T cell development.J. Exp. Med. 200:159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huston D.P., Liu Y.J. 2006. Thymic stromal lymphopoietin: a potential therapeutic target for allergy and asthma.Curr. Allergy Asthma Rep. 6:372–376 [DOI] [PubMed] [Google Scholar]
- Wu C., Sakorafas P., Miller R., McCarthy D., Scesney S., Dixon R., Ghayur T. 2003. IL-18 receptor beta-induced changes in the presentation of IL-18 binding sites affect ligand binding and signal transduction.J. Immunol. 170:5571–5577 [DOI] [PubMed] [Google Scholar]
- Elliott D.E., Jr., Urban J.F., Argo C.K., Weinstock J.V. 2000. Does the failure to acquire helminthic parasites predispose to Crohn's disease? FASEB J. 14:1848–1855 [DOI] [PubMed] [Google Scholar]
- Oliva-Hemker M., Fiocchi C. 2002. Etiopathogenesis of inflammatory bowel disease: the importance of the pediatric perspective.Inflamm. Bowel Dis. 8:112–128 [DOI] [PubMed] [Google Scholar]
- Weinstock J.V., Elliott D.E. 2008. Helminths and the IBD hygiene hypothesis.Inflamm. Bowel Dis. 15:128–133 [DOI] [PubMed] [Google Scholar]
- Radford-Smith G.L. 2005. Will worms really cure Crohn's disease? Gut. 54:6–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M., Scheet P. 2005. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation.Am. J. Hum. Genet. 76:449–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton K. 2003. Libsequence: a C++ class library for evolutionary genetic analysis.Bioinformatics. 19:2325–2327 [DOI] [PubMed] [Google Scholar]
- Wright S. 1950. Genetical structure of populations.Nature. 166:247–249 [DOI] [PubMed] [Google Scholar]
- Hudson R.R., Slatkin M., Madison W.P. 1992. Estimation of levels of gene flow from DNA sequence data.Genetics. 132:583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J.C., Fry B., Maller J., Daly M.J. 2005. Haploview: analysis and visualization of LD and haplotype maps.Bioinformatics. 21:263–265 [DOI] [PubMed] [Google Scholar]