Abstract
DNA methylation plays a crucial role in the regulation of gene expression and chromatin organization within normal eukaryotic cells. In cancer, however, global patterns of DNA methylation are altered with global hypomethylation of repeat-rich intergenic regions and hypermethylation of a subset of CpG-dense gene-associated regions (CpG islands). Extensive research has revealed the cellular machinery that catalyzes DNA methylation, as well as several large protein complexes that mediate the transcriptional repression of hypermethylated genes. However, research is only just beginning to uncover the molecular mechanisms underlying the origins of cancer-specific DNA methylation. Herein, we present several recent advances regarding these mechanisms and discuss the relationship between histone modifications (i.e. H3K4me2/3, H4K16Ac, H3K9me2/3, H3K27me3, H4K20me3), chromatin-modifying enzymes (G9a, EZH2, hMOF, SUV4−20H), and aberrant DNA methylation. Additionally, the role played by inflammation, DNA damage, and miRNAs in the etiology of aberrant DNA methylation is considered. Finally, we discuss the clinical implications of aberrant DNA methylation and the utility of methylated biomarkers in cancer diagnosis and management.
DNA Methylation, CpG Islands, and Gene Silencing
DNA methylation plays an essential role in normal development through its effects on gene imprinting, X-chromosome inactivation, and transcriptional silencing of repetitive elements. In mammalian species, DNA methylation occurs on the number 5 carbon of the pyrimidine ring of cytosines within the context of the CpG dinucleotide (1). In normal human cells, the majority of CpGs are methylated. In general, CpGs are under-represented within the genome as a result of the increased frequency with which methyl-cytosines undergo CpG to TpG transition mutations. CpGs can, however, be found near the expected frequency in clusters referred to as CpG islands (2, 3). CpG islands have presumably retained their CpG content throughout evolution by virtue of their unmethylated, and thus more stable, status within the germ line. Consistent with this hypothesis, organisms whose genomes exhibit little CpG methylation, such as Drosophila and C. elegans, possess CpGs at the expected frequency and show little variation in CpG distribution (4).
Originally identified by Bird et al as regions of CpG-dense DNA that could be cleaved by the methylation-sensitive restriction enzyme HpaII (5), multiple mathematical algorithms have subsequently been proposed for the classification of CpG islands (2, 3, 6). One of the most commonly used set of criteria that minimize the identification of repetitive-elements requires a minimum observed/expected CpG ratio of 0.65 and GC content greater than 55% over a distance of 500bp (3). By this definition, the human genome contains nearly 38,000 CpG islands (or ∼28,000 for the non-repetitive portion of human genome; unpublished data). A large fraction of these islands (37%) localize to the 5’ regulatory regions (promoters) of genes with approximately 70% of known genes having a CpG island within −2kb to +1kb of their transcription start site (unpublished data).
DNA methylation is mediated by a family of highly-related DNA methyltransferase enzymes (DNMT1, DNMT3A, and DNMT3B) which transfer a methyl group from S-adenosyl-L-methionine to cytosines in CpG dinucleotides (1, 7). Typically, the “maintenance” of DNA methylation patterns in somatic cells is attributed to DNMT1, whereas de novo DNA methylation during embryonic development is credited to DNMT3A and DNMT3B (1, 7, 8). This clear delineation of functions is an over-simplification though, as DNMT1 can also contribute to de novo DNA methylation both in vitro and in vivo (1, 7, 9, 10) and the maintenance of methylation in certain regions of the genome requires DNMT3A and DNMT3B (11).
The methyl-cytosines established by the DNMTs serve as binding sites for the methyl-CpG binding domain (MBD) proteins MeCP2, MBD1, MBD2, MBD3, MBD4 (12) and Kaiso, a methyl-cytosine binding protein composed of a POZ-domain and C2H2 zinc finger-domain (13). Through interactions with histone deacetylases, histone methyltransferases, and ATP-dependent chromatin remodeling enzymes, the MBDs translate methylated DNA into a compacted chromatin environment that is repressive for transcription (14).
The Cancer “DNA Methylome”
DNA methylation patterns in human cancer cells are considerably distorted (Figure 1A). Typically, cancer cells exhibit hypomethylation of intergenic regions which normally comprise the majority of a cell's methyl-cytosine content (15). Consequently, transposable elements may become active and contribute to the genomic instability observed in cancer cells (16). Simultaneously, cancer cells exhibit hypermethylation within the promoter regions of many CpG island-associated tumor suppressor genes, such as the retinoblastoma gene (Rb1), glutatione S-transferase pi (GSTP1), and E-cadherin (CDH1). As a result, these regulatory genes are transcriptionally silenced resulting in a loss-of-function. Thus, through the effects of both hypo- and hyper-methylation, DNA methylation significantly affects the genomic landscape of cancer cells, potentially to an even greater extent than coding region mutations which are relatively rare (17).
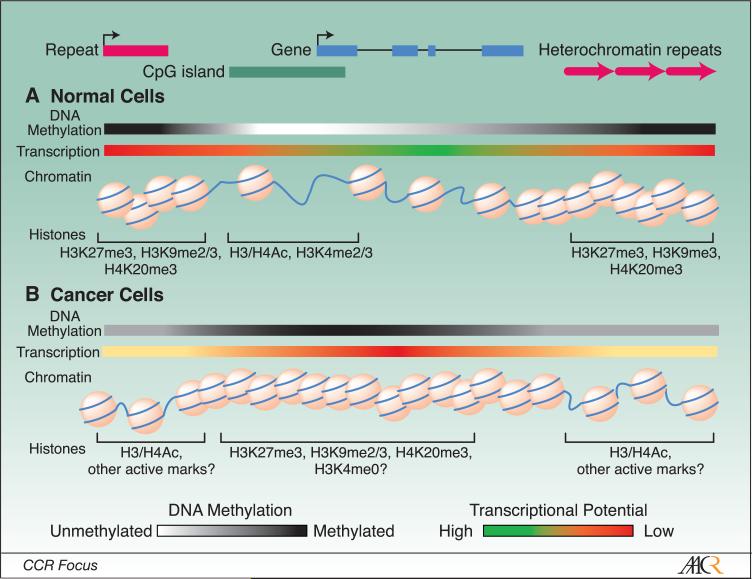
Figure 1. DNA methylation and histone modification patterns are altered in cancers.
A, Approximately 70% of genes possess promoter-associated CpG islands that mostly remain unmethylated in normal cells unlike the remainder of the genome which tends to be heavily methylated. Maintenance of an unmethylated promoter CpG island positively contributes to a high transcriptional potential and is associated with active histone modifications including histone H3 and H4 acetylation and methylation at H3K4. B, Cancer cells, on the other hand, exhibit dense hypermethylation of up to 10% of CpG islands as well as hypomethylation of bulk chromatin including intergenic regions and repetitive elements. A densely methylated CpG island is capable of driving chromatin compaction and repressing gene expression in association with repressive modifications including H3K9me3, H3K27me3, and/or H4K20me3.
The recent development of several genome-scale methylation screening technologies (reviewed in (18)) has considerably expanded our understanding of DNA methylation patterns, both in normal and cancerous cells. In addition to confirming 1) that the repetitive portion of the genome is heavily methylated while most CpG islands remain unmethylated in normal cells, and 2) that cancer cells exhibit widespread loss of intergenic DNA methylation with gain of methylation at many gene-associated CpG islands, these studies have generated significant novel information regarding DNA methylation patterns. For example, within the DNA “methylome” of individual tumors approximately 1−10% of CpG islands are aberrantly hypermethylated (19, 20). Contrary to previous notions, one study found that nearly 5% of gene-associated CpG islands are methylated in normal peripheral blood leukocytes (PBLs) and that a fraction of these normally methylated CpG islands become hypomethylated and transcriptionally active in cancer cells (21). Genome-wide studies also revealed that promoter-associated CpG islands are not the only islands affected by aberrant DNA methylation. Some CpG islands located within the 3’ ends of genes (22) and in intergenic regions (23) exhibit hypermethylation in cancer cells. However, unlike promoter methylation, it is unclear to what extent methylation of these non-promoter CpG islands might affect gene expression. In fact, analysis of several genes with 3’ CpG islands demonstrated increased gene expression upon hypermethylation suggesting a novel function for DNA methylation in this location (22). Thus, cancer-associated DNA methylation patterns are more complicated than previously thought and may have as yet unanticipated effects on gene expression and cellular function.
Mechanisms of Aberrant CpG Island Methylation
Genome-wide studies are also revealing the relationships between the DNA methylomes of different tumor types. Tumors derived either from different tissues or from the same tissue, but with different histology, exhibit unique methylation profiles (19, 24-26). Despite the considerable variation among tumors, a subset of CpG islands are frequently methylated in multiple tumor types (19, 26). One recent study found that while approximately 40% of hypermethylation events occurred in only one tumor-type, more than 10% were methylated in at least 50% of tumor types (26). Thus, it appears that one mechanism driving the hypermethylation of some CpG islands is an inherently elevated susceptibility to de novo methylation. However, it remains unclear what contributes to this susceptibility. Several mechanisms, centering around two basic themes, have been proposed: selective advantage and selective targeting. The selective advantage hypothesis posits that aberrant DNA hypermethylation begins with random seeding of DNA methylation throughout the genome, perhaps resulting from deregulation of the DNA methylation machinery. Those methylation events occurring within the promoters of genes that function to limit cell survival and proliferation (i.e. tumor suppressor genes) are then selected for during tumor progression. Support for this hypothesis was recently provided by mouse models of cancer in which MYC over-expression was coupled with inactivation of Pten, Trp53, or E2f2 (27). Nearly 4 dozen CpG islands were found to exhibit late-stage differential methylation that occurred in a genotype-specific manner. Since the different genotypes generate unique selective pressures, it can be argued that these genotype-specific methylation events resulted from the outgrowth of cells that harbored advantageous hypermethylation events. However, since not all hypermethylated genes confer a growth or survival advantage, and many are not expressed in normal tissues, selective advantage can not be the only mechanism.
The second hypothesis suggests that hypermethylation results from the aberrant targeting of DNMTs to certain regions and/or that these regions possess intrinsic, cis-acting features that make them better substrates for de novo DNA methylation. An example of a specific trans-acting factor that might target methylation is the oncogenic fusion protein PML-RAR which is capable of directing de novo DNA methylation to its target genes (28). On the other hand, the hypothesis that cis-acting mechanisms play a role in the targeting of DNA methylation is supported by the findings that hypermethylated genes tend to cluster in the genome (29, 30) and that they exhibit common sequence signatures (30-33). Several approaches have been utilized to identify cis-acting DNA sequences that might contribute to methylation susceptibility. First, the DNA methylation machinery has been found to have a target site preference that extends at least 4bp 5’ and 3’ of the CpG site (34). In vitro validation experiments demonstrated a 500-fold difference in the methylation rates of preferred substrates (e.g. CTTACGCAAG) compared to non-preferred substrates (e.g. TGTTCGGTGG) (34). A related approach utilizing massively parallel sequencing of bisulfite modified DNA from leukemia and lymphoma samples identified a 30bp motif that was capable of predicting methylation susceptibility for CpG dinucleotides with up to 75% accuracy in cross-validation studies (35). Second, several groups, including ours, have utilized pattern recognition and/or motif elicitation to discover DNA sequence signatures of CpG dinucleotides or CpG islands with increased susceptibility to DNA methylation, both in normal or cancer cells (30-33). Despite the identification of several sequences that correlate with methylation susceptibility, the specific functions of these patterns remain unknown. Additionally, little consensus exists between the patterns identified by different groups, which likely results from the use of different datasets and varied computational approaches. Similar analyses of CpG islands resistant to hypermethylation have identified motifs that correlate with zinc-finger transcription factor binding (36) and, perhaps unexpectedly, with Alu repetitive elements (33). Identification of these sequence signatures has permitted the development of classification algorithms which predict the methylation status of CpG islands either in normal cells (31, 37), a cell culture model of de novo methylation (32, 33), or cancer cells (38, 39). Through further pattern recognition and supervised learning approaches, we may discover additional features of CpG islands that regulate methylation susceptibility.
The Histone Code and DNA Methylation Connection
DNA methylation is only part of a broader epigenetic code that dictates transcriptional potential of genomic domains. DNA is wrapped around an octamer of histone proteins to form the nucleosome, the smallest unit of chromatin. The amino terminal tails of the histones protrude from the nucleosome body and are subject to considerable post-translational modifications including acetylation, methylation, phosphorylation, ubiquitination, and sumoylation (40). The constellation of specific modifications, referred to as the “histone code”, influences interactions with the DNA backbone, neighboring nucleosomes, and non-histone chromatin proteins (e.g. modification-specific binding factors) to mediate the assembly of a chromatin environment that is either permissive or repressive for transcription (41). In general, permissive regions exhibit an open chromatin structure marked by hyperacetylation of histones H3 and H4 and di- and tri-methylation of histone H3 at lysine 4 (H3K4me2/3) (42) (Figure 1). In contrast, repressed regions exhibit a compact chromatin structure that lacks H3/H4 acetylation and H3K4 methylation, and instead is enriched in the “repressive” modifications, di-and tri-methylation of H3K9 (H3K9me2/3), tri-methylation of H3K27 (H3K27me3), and trimethylation of H4K20 (H4K20me3) (Figure 1) (42, 43). While the code is not yet fully deciphered, it is apparent that DNA methylation can both influence, and be influenced by, histone modifications.
Like DNA methylation, the histone portion of the epigenome undergoes both widespread and gene-specific changes in cancer. Overall, cancer cells exhibit a global decrease in the levels of H4K20me2/3, H3K9me2, and H4 acetylation (Ac), particularly at H4K16 (44, 45). Like cancer-associated DNA hypomethylation, the loss of H4K16Ac and H4K20me2/3 derives primarily from the repetitive fraction of the genome, occurs in premalignant lesions, and increases in magnitude during tumor progression (44). The loss of DNA methylation, H3K9me2, and H4K20me3 speaks to a global dysregulation of transcriptional repression in cancer cells, which may promote tumorigenesis through the de-repression of endogenous transposons (e.g. Alu) or miRNAs (see below), an impaired DNA damage response (46), loss of checkpoint controls (47), and increased chromosomal instability (48).
The mechanisms by which CpG islands remain unmethylated in normal cells and acquire DNA methylation in cancer cells is likely intimately linked to the underlying histone code. The unmethylated CpG islands of active genes are enriched in acetylated H3 and H4 and H3K4me2(42) (Figure 1). Nucleosomes are strongly positioned across CpG islands in their active and unmethylated state, except at the transcription start site where there is often a one nucleosome gap (49, 50). In contrast, the CpG islands of genes that are aberrantly methylated in cancer cells are remodeled such that nucleosomes are more randomly positioned and there is a shift from H3/H4 acetylation and H3K4 methylation, to H3K9me2/3 and/or H3K27me3 (Figure 1). This is achieved through the recruitment of MBDs, histone deacetylases (HDACs), histone methyltransferases, and H3K9me2/3 binding proteins (eg. HP1) which lock the domain into a heterochromatin-like state that is mitotically heritable and essentially irreversible.
Whereas considerable effort has gone into defining the characteristics of these beginning and end stages, the exact sequence of events and underlying molecular mechanisms are not yet resolved. Of considerable recent interest is the inter-dependent relationship between DNA methylation and histone methylation. Methylation of H3K9 and DNA methylation are tightly associated in heterochromatin and transcriptionally-repressed euchromatic regions. H3K9 methylation is absolutely required for DNA methylation in fungi and directs CpNpG methylation in plants (51, 52). The SUV39H1/2 histone methyltransferases catalyze the trimethylation of H3K9 at pericentric heterochromatin, and are necessary for the maintenance of DNA methylation in these regions (53). Dimethylation of H3K9 plays an equally important role in gene silencing in euchromatin and is catalyzed by distinct H3K9 methyltransferases, G9a and the related GLP/Eu-HMTase1 (54). G9a plays an important role in the silencing and subsequent de novo DNA methylation of embryonic and germline genes during normal development (55), and is necessary for the maintenance of DNA methylation at endogenous retrotransposons, imprinted loci, and other genes in differentiated cells (56). Interestingly, G9a-mediated DNA methylation does not require its catalytic activity (55), suggesting that it may have additional functions in directing DNA methylation, such as the recruitment of DNMTs or the recognition of methyl-lysine residues (57). A model has recently emerged for the coordinated regulation of DNA methylation and H3K9me2 involving their co-deposition during DNA replication through direct or indirect interactions between DNMT1, G9a, the H3K9me2/3 binding factor HP1, and UHRF1, a recently described DNMT1 co-factor that binds preferentially to hemi-methylated DNA (Figure 2A) (58-61).
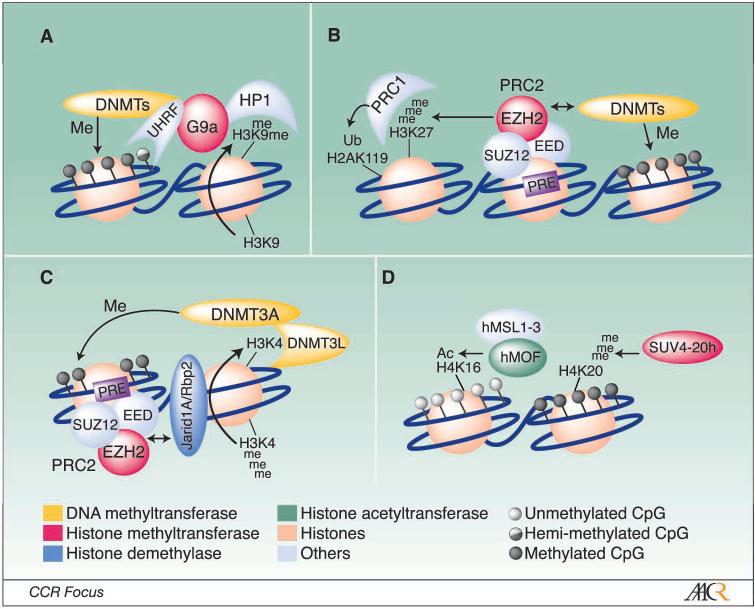
Figure 2. Chromatin-mediated repression mechanisms associated with DNA methylation.
A, G9a links H3K9me2 and DNA Methylation. The H3K9 methyltransferase G9a may stimulate de novo DNA methylation through either the direct recruitment of DNMTs, or indirectly, through interactions between the DNMTs and the methylated H3K9 binding factor, HP1. Recent evidence linking G9a to the DNMT1 cofactor UHRF1 suggests that H3K9me2 and DNA methylation may be coordinately regulated during DNA replication. B, PRC2 mediates H3K27me3 and recruits the DNMTs. In addition to mediating the repressive H3K27me3 modification, PRC2 may recruit the DNA methyltransferases to its target genes thereby stimulating de novo DNA methylation. Via this mechanism, PRC2 drives the irreversible silencing of many genes normally involved in stem cell maintenance, development, and differentiation. C, PRC2 stimulates demethylation of H3K4. PRC2 may also have an indirect effect on DNA methylation through the recruitment of the Rbp2 (Jarid1a) H3K4me2/3 demethylase to its target genes. Rbp2 catalyzes the demethylation of H3K4me2/3 to H3K4me0 which further recruits DNMT3L and the DNMT3A de novo methyltransferase. D, Relationship between H4K16Ac, H4K20me3, and DNA Methylation. The histone H4 modifications H4K20me3 and H4K16Ac may play opposing roles in gene repression. Recent work indicates that some genes that undergo aberrant DNA methylation in cancer lose H4K16Ac and gain H4K20me3, a mark typically associated with repressed genes. Unfilled circles, unmethylated CpGs. Filled circles, methylated CpGs.
G9a-mediated H3K9 methylation may be one of the key factors in the maintenance of transcriptionally-silent gene promoters in cancer. Reactivation of silenced tumor suppressor genes in response to 5-aza-2’-deoxycytidine-induced DNA demethylation is accompanied by a concomitant decrease in H3K9me2, but not other silencing marks such as H3K9me3 or H3K27me3 (62, 63). Furthermore, G9a is enriched at the promoters of aberrantly methylated genes in cancer cells, and co-recruitment of G9a, DNMT1, and HP1 to the promoter of the survivin gene stimulates H3K9me2 and DNA hypermethylation (61). Indeed, recent evidence suggests that inhibition of G9a alone is sufficient to induce the reactivation of silenced metastasis suppressor genes in cancer cells, an effect that is potentiated by concurrent inhibition of DNMT1 (64), thus paving the way for novel therapeutic approaches aimed at the combined inhibition of H3K9 methylation and DNA methylation (see below).
Polycomb-mediated repression is another chromatin-based silencing mechanism with ties to aberrant DNA methylation in cancer. The EZH2 histone methyltransferase is a component of the Polycomb Repressive Complex 2 (PRC2) and represses developmental regulatory genes through establishment of the H3K27me3 mark (Figure 2B). A second complex, Polycomb Repressive Complex 1 (PRC1), which consists of HPH, HPC, RING1, and BMI1, binds H3K27me3 and stimulates transcriptional silencing through nucleosome compaction mediated by exclusion of chromatin remodeling enzymes and ubiquitylation of H2AK119 (65, 66). Several links have now been established between this important developmental transcriptional regulator and DNA methylation: 1) both the DNA methylation and polycomb machinery are required for early embryonic differentiation and development (67, 68), 2) PRC components interact with the DNMTs either directly, as in the case of EZH2, or indirectly through the DNMT1-associated protein 1 (Dmap1), as in the case of BMI1 (69, 70), and 3) EZH2, Bmi1, and Dmap1 are necessary for the maintenance of some CpG island methylation patterns in both normal and cancer cells (69, 70).
PRC2 is an attractive candidate for the targeting of aberrant hypermethylation. Several of its components, including EZH2, are over-expressed in cancer, and a ‘polycomb repression’ signature is observed in metastatic prostate cancer (71, 72). PRC2 is necessary for de novo methylation of p16 during immortalization of mammary epithelial cells (73) and is required, along with DNMT1, to maintain epigenetic silencing of the Fas gene in K-ras-transformed cells (74). Furthermore, several recent studies have demonstrated that genes marked by PRC2 in embryonic stem cells and/or normal cell types are predisposed to future hypermethylation in cancers (39, 43, 75, 76). Interestingly, a sequence signature which predicts methylation-prone CpG islands also identifies PRC2 binding sites and incorporation of PRC2 binding information into the prediction algorithm improved prediction accuracy (39). However, the complexity of this relationship is highlighted by recent studies that have reported genes that acquire H3K27me3 and EZH2 binding de novo in cancer cells without DNA methylation (77), genes that lose the H3K27me3 mark after acquiring de novo DNA methylation and H3K9 methylation (77, 78), and genes whose aberrant DNA methylation is maintained in the absence of H3K27me3 or EZH2 binding (79). Thus, the simple notion of one repressive mark equaling future DNA hypermethylation appears over-simplified.
Interestingly, the connection between PRCs and DNA methylation may not be limited to an effect on H3K27me3. Recent work has shown that PRC2 recruits to its target genes the H3K4me2/3 demethylase Rbp2 (Jarid1a) which may promote DNA methylation through demethylation of H3K4 (Figure 2C) (80). Methylated CpG islands universally exhibit loss of H3K4 methylation both in the context of normal differentiation and cancer-associated silencing (81, 82). Unmethylated H3K4 (H3K4me0) is recognized by the catalytically-inactive DNMT regulatory factor DNMT3L, which may stimulate de novo DNA methylation via its binding partners, DNMT3A or DNMT3B (83) (Figure 2C). While this mechanism was reported to function during gene imprinting, it may also contribute to cancer-associated hypermethylation.
Alterations in histone H4 modifications may also contribute to the aberrant silencing of certain genes in cancer. H4K20me3 is a repressive mark found in constitutive heterochromatin and at imprinted genes where it is selectively enriched on the DNA methylated allele (47, 84, 85). Recent work from our lab indicates that H4K20me3 localizes to the promoter of the TMS1/ASC gene in human breast cancer cells in which it is methylated and transcriptionally silent, suggesting that H4K20me3 also plays a role in the repression of selected genes in cancer (50). Currently, little is known about the targeting of H4K20me3 to individual genes, but it may involve an interaction between SUV4−20H, the histone methyltransferase responsible for H4K20me2/3, and the retinoblastoma tumor suppressor which is necessary for its localization to heterochromatin (86). H4K20me3 may repress transcription in part by antagonizing H4K16Ac (Figure 2D). H4K16Ac is associated with active genes, but also plays an important role in mediating euchromatin/heterochromatin boundaries in yeast (87). Similarly, we find that H4K16Ac selectively marks the nucleosomes flanking the unmethylated CpG island and maintains nucleosome positioning and gene activity at the TMS1/ASC locus. Thus, loss of H4K16Ac may be a pre-requisite to epigenetic silencing in cancer cells. Down-regulation of hMOF, the histone acetyltransferase responsible for H4K16Ac, has been observed in human breast cancers and medulloblastomas (88), and its loss of function leads to defects in the cell cycle and genome instability (89). Together, these data point to the dysregulation of an epigenetic switch involving H4K16Ac and H4K20me3 that may be involved in the aberrant silencing of at least some tumor suppressor genes in cancer.
While studies regarding the role of repressive histone modifications in cancer have focused primarily on lysine methylation, recent work also suggests a potential role for histone arginine methylation. Arginine methylation is mediated by a family of protein arginine methyltransferase (PRMTs) which are classified into two general types based on whether they catalyze dimethylation asymmetrically (me2a) or symmetrically (me2s) (90). PRMT6 and CARM1 are type I PRMTs that are responsible for H3R2me1 and H3R2me2a (90). PRMT5, on the other hand, is a type II PRMT that mono- methylates and symmetrically dimethylates H4R3 and H3R8 (90). H3R2me2a represses transcription by inhibiting both H3K4 methylation (91) and binding of the basal transcription machinery (92). Since loss of the H3K4me3 and CpG island hypermethylation are closely correlated, it is possible that methylation of H3R2 also has an impact on de novo DNA methylation. A more direct effect on DNA methylation may be mediated by H4R3me2s as DNMT3A was recently shown to bind histone tails bearing this modification (93). Furthermore, modulation of PRMT5 levels positively correlated with DNA methylation at the γ-globin promoter (93). A role for H4R3me2 in cancer is supported by immunohistochemical studies demonstrating that the levels of H4R3me2 (along with 4 other histone modifications), was capable of predicting the risk of local recurrence following prostatectomy in low-grade prostate cancers (94).
The above discussion underscores the complex relationship that exists between the histone code and susceptibility to DNA methylation and suggests that this relationship may be crucial to the development and targeting of DNA hypermethylation in cancer cells. From a clinical/translational point of view, a multi-faceted strategy targeting multiple components of the epigenetic machinery may be more effective for the re-awakening of silenced tumor suppressor genes. Thus far, clinical applications of “epigenetic therapy” have primarily focused on nucleoside analog inhibitors of the DNMTs, alone (95) and in combination with HDAC inhibitors (95-98). Histone methyltransferase inhibitors have not yet been widely explored in cancer therapy, but small molecule inhibitors of these enzymes are beginning to reach preclinical testing. In particular, a small molecule inhibitor of G9a and GLP, BIX01294, has demonstrated efficacy in reducing global and gene-specific H3K9me2 levels, resulting in the reactivation of several known G9a targets (99). The structure of BIX01294 in complex with GLP and the cofactor S-adenosyl-methionine was recently solved (100), and should facilitate lead optimization to generate new, more effective compounds. Similarly, an inhibitor of polycomb complexes, DzNEP, was identified in a screen of the NCI small molecule library for agents that induce E2F-mediated apoptosis (101). A known inhibitor of adenosyl-homocysteine hydrolase, DzNEP induces cell death at doses in the μM range in a number of cancer cell lines, but not in normal cells. DzNEP induces hypomethylation of H3K27me3 and to a lesser extent H4K20me3, but has no impact on H3K9me3. Interestingly, DzNEP treatment was more effective than 5-aza-2’-deoxycytidine alone, or in combination with an HDAC inhibitor, at inducing the reactivation of PRC2 target genes (101). However, only a fraction of the genes induced by DzNEP treatment were also up-regulated in response to knockdown of PRC2 components suggesting that DzNEP likely affects other histone methyltransferases in addition to EZH2. As the area of cancer epigenetics continues to grow, it is very likely that additional histone methyltransferase inhibitors and agents that target other components of silencing complexes (e.g. histone demethylases and DNA/histone methylation binding factors) will continue to surface.
Drivers of Hypermethylation: Carcinogens, DNA Damage, and microRNAs
While the precise molecular mechanisms underlying the establishment of aberrant hypermethylation remain elusive, recent studies have identified some of the contributing etiologic factors. For example, chronic exposure of human bronchial epithelial cells to tobacco-derived carcinogens drives hypermethylation of several tumor suppressor genes including E-cadherin and RASSF2A (102). Stable knockdown of DNMT1 prior to carcinogen exposure prevented methylation of several of these genes indicating a necessary role for this enzyme in the molecular mechanism underlying hypermethylation (102). The reactive oxygen species (ROS) associated with chronic inflammation is another source of DNA damage with the potential to affect DNA methylation as.halogenated pyrimidines, one form of ROS-induced damage, mimic 5-methylcytosine and stimulate DNMT1-mediated CpG methylation in vitro and in vivo (103, 104). Indeed, study of the glutatione peroxidase 1 and 2 double knockout model of inflammatory bowel disease found that 60% of genes that are hypermethylated in colon cancers also exhibit aberrant methylation in the inflamed non-cancerous precursor tissues (105). Although the mechanisms by which DNA damage mediates DNA methylation are not fully understood, O'Hagan et al (106) have examined the process with an engineered cell culture model in which a unique restriction site was incorporated into the CpG island of the E-cadherin promoter. Following induction of a double-strand DNA break in this model, the SIRT1 histone deacetylase was recruited to the site along with components of PRC2, DNMT1, and DNMT3B (106). The region was subsequently deacetylated at H4K16, methylated at H3K27, transcriptionally silenced, and in some cases DNA hypermethylated. Taken together, these studies suggest that one source of de novo DNA methylation during carcinogenesis may be the DNA damage generated during persistent cancer-associated inflammation.
Another area of recent interest is the contribution of microRNA (miRNA) species to DNA methylation. These single-stranded non-coding RNA molecules of ∼21 nucleotides regulate gene expression through partial complementary hybridization with the 3’ untranslated regions of mRNAs with subsequent mRNA cleavage or translational inhibition. As many as 1000 miRNAs may be encoded within the human genome (107) and, with each miRNA possibly controlling the expression of multiple targets, it is estimated that more than 25% of human genes may be regulated by miRNAs. MicroRNAs are processed and matured by the DICER RNase III family nuclease and studies of cells deficient in DICER have implicated miRNAs in de novo DNA methylation (108-110). DICER−/− ES cells exhibit considerable loss of DNA methylation at H4K20me3- and H3K9me3-enriched heterochromatin, suggesting that DICER is necessary for the maintenance of DNA methylation in these regions (108). The miR-290 family miRNAs normally target the retinoblastoma-like 2 (Rbl2) gene whose protein product represses E2F-mediated transcription of all three active DNMTs (Figure 3). These miRNAs fail to mature in DICER−/− cells, thus leading to down-regulation of the DNMTs (108). Functional miRNA processing was also required for the hypermethylation of at least 8 CpG islands, including that of SFRP4, in a human colon cancer cell line lacking DICER (110). Finally, Fabbri et al (111) identified another family of miRNAs (miR-29) that repress the DNMT3A and DNMT3B genes directly (Figure 3). Interestingly, this family of miRNAs is itself hypermethylated in lung cancers leading to overexpression of the de novo DNA methyltransferases (111). Restoration of the miR-29 family in lung cancer cell lines induced DNA demethylation and reduced cell proliferation and tumorigenicity (111). Recent studies utilizing miRNA profiling approaches have demonstrated that a significant fraction of miRNAs are regulated by epigenetic mechanisms (112, 113). Pharmacologic ‘unmasking’ of miRNAs hypermethylated in cell lines derived from lymph node metastases identified three miRNAs that normally function to suppress metastasis by targeting the c-MYC, CDK6, E2F3, and TGIF2 transcripts (113). Thus, it is clear that a complex inter-dependent relationship exists between miRNAs and DNA methylation with important implications for both gene silencing and tumor progression (i.e. metastasis).
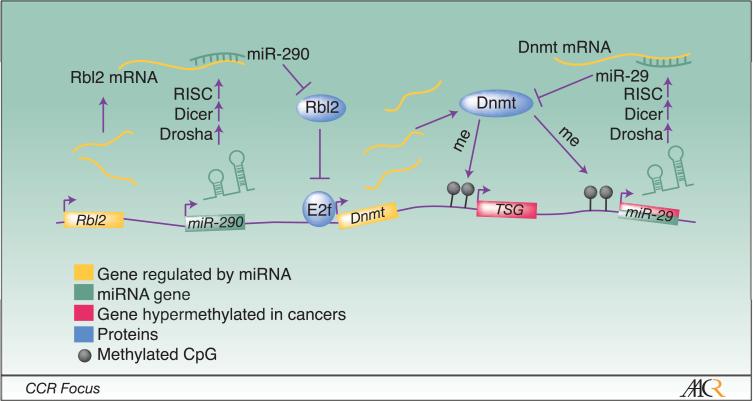
Figure 3. MicroRNAs (miRNAs) play a complex role in the regulation of genome-wide DNA methylation patterns.
Micro RNAs are ∼21 nucleotide single-stranded non-coding RNA molecules that are transcribed as a primary microRNA (pri-miRNA) transcript before undergoing two processing events by Drosha and Dicer. The mature miRNA interacts with the RNA-induced silencing complex (RISC) which can mediate both translational repression and mRNA transcript cleavage depending on the extent of homology between the miRNA and its target. At least two families of miRNAs affect the expression of DNMTs. The miR-290 family stimulates DNMT expression by targeting Rbl2, a retinoblastoma family protein that represses E2F-mediated activation of the DNMT genes. In contrast, the miR-29 family directly represses DNMT3A and DNMT3B transcripts. However, in cancer cells, the miR-29 locus is hypermethylated leading to transcriptional silencing of miR-29 and elevated expression of DNMT3A and DNMT3B.
Clinical Applications of DNA Methylation
Recent advances in our understanding of cancer-associated DNA methylation underlie many promising clinical applications including the development of molecular markers for early detection of cancer, prediction of prognosis, and prediction of treatment outcomes. The ability of methylation markers to detect cancers has been evaluated in multiple body fluids including sputum, plasma, stool, urine and nipple aspirates (Table 1). While the results have been encouraging, limitations have thus far prevented wide-spread clinical application. First, the methylation frequency of many candidate genes is not high enough to achieve the sensitivity required for a clinical test. Second, a methylation assay that exhibits suitable sensitivity in primary tumors may not perform as well when applied to bodily fluids. Technical advances in the near future are expected to dramatically reduce these problems. Current genome-wide methylation profiling technology which permits the rapid and simultaneous analysis of thousands of loci will likely help identify novel, superior methylation markers with higher sensitivity and specificity.
Table 1.
Current testing and clinical application of DNA methylation for cancer.
| Diagnosis and early detection | |||||
|---|---|---|---|---|---|
| Study |
Cancer type |
Gene |
Tissue |
Sensitivity |
Specificity |
| Belinsky et al (129) | Lung | p16, PAX5-b, MGMT, DAPK, GATA5, RASSF1A | sputum | 64% | 64% |
| Gonzalgo et al (130) | Prostate | GSTP1 | urine | 58% | 67% |
| Hoque et al (131) | Prostate | GSTP1/p16/ARF/MGMT | urine | 87% | near 100% |
| Chen et al (132) | Colon | Vimentin exon1 | stool | 43% | 90% |
| Lenhard et al (133) | Colon | HIC1 | stool | 42% | near 100% |
| Krassenstein et al (134) | Breast | DAPK, RAR-b, p16,p14, RASSF1, GSTP1 | nipple aspirate | 82% | 100% |
| Prognosis | |||
|---|---|---|---|
| Study |
Cancer type |
Gene |
Outcome |
| Lu et al (114) | Lung | DAPK | HR for death (M vs. U) 1.69 |
| Brock et al (117) | Lung | p16, H-cadherin, APC, RASSF1A | HR for death (M vs. U) up to 15.5 |
| Harbeck et al (115) | Breast | PITX2 | HR for distant recurrence (M vs.U)2.35 |
| Alumkal et al (119) | Prostate | ASC, CDH-13 | HR for PSA recurrence (M vs.U) 5.64 |
| Prediction of response | ||||
|---|---|---|---|---|
| Study |
Cancer type |
Gene |
Therapy |
Outcome |
| Esteller et al (121) | Glioma | MGMT | Carmustine | HR for death U vs M: 9.5 |
| Hegi et al. (122) | Glioma | MGMT | Temozolomide | HR for death U vs M: 2.2 |
| Taniguchi et al (126) | Ovarian | FANCF | Cisplatin | in vitro assays IC50 ,<1.0 uM(S),>1.0uM(RS) |
| Satoh et al (125) | Gastric | CHFR | Taxane | increased sensitivity in in vitro assays |
| Agrelo et al. (128) | Colon | Werner-1 | Irinotecan | OS 39.4(M) vs 20.7 (U) months p<0.05 |
Abnormal promoter methylation can also provide prognostic information (Table 1). In resected early stage lung cancer, methylation of the pro-apoptotic gene DAPK has been associated with significantly shorter disease-free and overall survival (OR for death 1.69) (114). In hormone receptor (+) Her-2 (−) breast cancer, PITX2 methylation was both an independent risk factor for recurrence in node-negative patients treated with tamoxifen (115) and node-positive patients treated with anthracycline-based adjuvant chemotherapy (116). Combinations of multiple methylation markers may provide even more prognostic potential. A 4-marker panel including p16, H-cadherin, APC, and RASSF1A was associated with a higher risk of recurrence in resected early stage lung cancer, particularly when simultaneous methylation of two markers (p16 and H-cadherin) was present in both primary tumor and histologically negative mediastinal lymph nodes (OR for recurrence 15.5) (117). Similar relationships exist between the number of methylated genes and risk of relapse in breast and prostate cancer (118-120).
Since genes involved in the repair of DNA damage are frequently targeted by hypermethylation in cancers, the study of these loci may be useful in predicting response to chemotherapy (Table 1). The best correlation reported to date is between methylation of the MGMT gene and response to alkylating agents in gliomas (121, 122). Loss of MGMT renders cells unable to repair alkylating agent chemotherapy-induced DNA damage and induces cell death. These findings have been validated in prospective randomized trials in patients with gliomas (122). Analogously, correlations have been observed between CHFR silencing/methylation and sensitivity to taxanes (123-125), FANCF methylation and sensitivity to cisplatin in ovarian cancer cell lines (126), p73 methylation and sensitivity to cisplatin in the NCI60 cell line panel (127), and methylation of the premature aging syndrome Werner-1 gene and sensitivity to the topoisomerase II inhibitor irinotecan in colon cancer (128). With increasing application of genome-wide methylation profiling, it is anticipated that additional methylation markers with improved prognostic potential will soon be available. When combined with other profiling techniques, such as gene expression profiling, a patient's DNA methylome may play a crucial role in the development of personalized medicine.
REFERENCES
- 1.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 2.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–82. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 3.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002;99:3740–5. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elango N, Yi SV. DNA methylation and structural and functional bimodality of vertebrate promoters. Mol Biol Evol. 2008;25:1602–8. doi: 10.1093/molbev/msn110. [DOI] [PubMed] [Google Scholar]
- 5.Bird A, Taggart M, Frommer M, Miller OJ, Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985;40:91–9. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- 6.Bock C, Walter J, Paulsen M, Lengauer T. CpG island mapping by epigenome prediction. PLoS Comput Biol. 2007;3:e110. doi: 10.1371/journal.pcbi.0030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hermann A, Gowher H, Jeltsch A. Biochemistry and biology of mammalian DNA methyltransferases. Cell Mol Life Sci. 2004;61:2571–87. doi: 10.1007/s00018-004-4201-1. [DOI] [PubMed] [Google Scholar]
- 8.Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–20. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 9.Graff JR, Herman JG, Myohanen S, Baylin SB, Vertino PM. Mapping patterns of CpG island methylation in normal and neoplastic cells implicates both upstream and downstream regions in de novo methylation. J Biol Chem. 1997;272:22322–9. doi: 10.1074/jbc.272.35.22322. [DOI] [PubMed] [Google Scholar]
- 10.Vertino PM, Yen RW, Gao J, Baylin SB. De novo methylation of CpG island sequences in human fibroblasts overexpressing DNA (cytosine-5-)-methyltransferase. Mol Cell Biol. 1996;16:4555–65. doi: 10.1128/mcb.16.8.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang G, Chan MF, Tomigahara Y, et al. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol Cell Biol. 2002;22:480–91. doi: 10.1128/MCB.22.2.480-491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhasarathy A, Wade PA. The MBD protein family-Reading an epigenetic mark? Mutat Res. 2008;647:39–43. doi: 10.1016/j.mrfmmm.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prokhortchouk A, Hendrich B, Jorgensen H, et al. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 2001;15:1613–8. doi: 10.1101/gad.198501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bird AP, Wolffe AP. Methylation-induced repression--belts, braces, and chromatin. Cell. 1999;99:451–4. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 15.Feinberg AP, Gehrke CW, Kuo KC, Ehrlich M. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer Res. 1988;48:1159–61. [PubMed] [Google Scholar]
- 16.Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2008;27:404–8. doi: 10.1038/sj.onc.1210631. [DOI] [PubMed] [Google Scholar]
- 17.Schuebel KE, Chen W, Cope L, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–23. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zilberman D, Henikoff S. Genome-wide analysis of DNA methylation patterns. Development. 2007;134:3959–65. doi: 10.1242/dev.001131. [DOI] [PubMed] [Google Scholar]
- 19.Costello JF, Fruhwald MC, Smiraglia DJ, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–8. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 20.Rauch TA, Zhong X, Wu X, et al. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci U S A. 2008;105:252–7. doi: 10.1073/pnas.0710735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen L, Kondo Y, Guo Y, et al. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007;3:2023–36. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith JF, Mahmood S, Song F, et al. Identification of DNA methylation in 3' genomic regions that are associated with upregulation of gene expression in colorectal cancer. Epigenetics. 2007;2:161–72. doi: 10.4161/epi.2.3.4805. [DOI] [PubMed] [Google Scholar]
- 23.Weber M, Davies JJ, Wittig D, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–62. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 24.Chung W, Kwabi-Addo B, Ittmann M, et al. Identification of novel tumor markers in prostate, colon and breast cancer by unbiased methylation profiling. PLoS ONE. 2008;3:e2079. doi: 10.1371/journal.pone.0002079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai Z, Lakshmanan RR, Zhu WG, et al. Global methylation profiling of lung cancer identifies novel methylated genes. Neoplasia. 2001;3:314–23. doi: 10.1038/sj.neo.7900162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ehrich M, Turner J, Gibbs P, et al. Cytosine methylation profiling of cancer cell lines. Proc Natl Acad Sci U S A. 2008;105:4844–9. doi: 10.1073/pnas.0712251105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Opavsky R, Wang SH, Trikha P, et al. CpG island methylation in a mouse model of lymphoma is driven by the genetic configuration of tumor cells. PLoS Genet. 2007;3:1757–69. doi: 10.1371/journal.pgen.0030167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Croce L, Raker VA, Corsaro M, et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science. 2002;295:1079–82. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- 29.Frigola J, Song J, Stirzaker C, Hinshelwood RA, Peinado MA, Clark SJ. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat Genet. 2006;38:540–9. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- 30.Keshet I, Schlesinger Y, Farkash S, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38:149–53. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 31.Fang F, Fan S, Zhang X, Zhang MQ. Predicting methylation status of CpG islands in the human brain. Bioinformatics. 2006;22:2204–9. doi: 10.1093/bioinformatics/btl377. [DOI] [PubMed] [Google Scholar]
- 32.Feltus FA, Lee EK, Costello JF, Plass C, Vertino PM. Predicting aberrant CpG island methylation. Proc Natl Acad Sci U S A. 2003;100:12253–8. doi: 10.1073/pnas.2037852100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feltus FA, Lee EK, Costello JF, Plass C, Vertino PM. DNA motifs associated with aberrant CpG island methylation. Genomics. 2006;87:572–9. doi: 10.1016/j.ygeno.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 34.Handa V, Jeltsch A. Profound flanking sequence preference of Dnmt3a and Dnmt3b mammalian DNA methyltransferases shape the human epigenome. J Mol Biol. 2005;348:1103–12. doi: 10.1016/j.jmb.2005.02.044. [DOI] [PubMed] [Google Scholar]
- 35.Kim S, Li M, Paik H, et al. Predicting DNA methylation susceptibility using CpG flanking sequences. Pac Symp Biocomput. 2008:315–26. doi: 10.1142/9789812776136_0031. [DOI] [PubMed] [Google Scholar]
- 36.Fan S, Fang F, Zhang X, Zhang MQ. Putative zinc finger protein binding sites are over-represented in the boundaries of methylation-resistant CpG islands in the human genome. PLoS ONE. 2007;2:e1184. doi: 10.1371/journal.pone.0001184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bock C, Paulsen M, Tierling S, Mikeska T, Lengauer T, Walter J. CpG island methylation in human lymphocytes is highly correlated with DNA sequence, repeats, and predicted DNA structure. PLoS Genet. 2006;2:e26. doi: 10.1371/journal.pgen.0020026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goh L, Murphy SK, Muhkerjee S, Furey TS. Genomic sweeping for hypermethylated genes. Bioinformatics. 2007;23:281–8. doi: 10.1093/bioinformatics/btl620. [DOI] [PubMed] [Google Scholar]
- 39.McCabe MT, Lee EK, Vertino PM. A multifactorial signature of DNA sequence and polycomb binding predicts aberrant CpG island methylation. Cancer Res. 2009;69:282–91. doi: 10.1158/0008-5472.CAN-08-3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garcia BA, Shabanowitz J, Hunt DF. Characterization of histones and their post-translational modifications by mass spectrometry. Curr Opin Chem Biol. 2007;11:66–73. doi: 10.1016/j.cbpa.2006.11.022. [DOI] [PubMed] [Google Scholar]
- 41.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 42.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 43.Schlesinger Y, Straussman R, Keshet I, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–6. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 44.Fraga MF, Ballestar E, Villar-Garea A, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 45.Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41:246–50. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schotta G, Sengupta R, Kubicek S, et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 2008;22:2048–61. doi: 10.1101/gad.476008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benetti R, Gonzalo S, Jaco I, et al. Suv4−20h deficiency results in telomere elongation and derepression of telomere recombination. J Cell Biol. 2007;178:925–36. doi: 10.1083/jcb.200703081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 49.Lin JC, Jeong S, Liang G, et al. Role of nucleosomal occupancy in the epigenetic silencing of the MLH1 CpG island. Cancer Cell. 2007;12:432–44. doi: 10.1016/j.ccr.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kapoor-Vazirani P, Kagey JD, Powell DR, Vertino PM. Role of hMOF-dependent histone H4 lysine 16 acetylation in the maintenance of TMS1/ASC gene activity. Cancer Res. 2008;68:6810–21. doi: 10.1158/0008-5472.CAN-08-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamaru H, Zhang X, McMillen D, et al. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat Genet. 2003;34:75–9. doi: 10.1038/ng1143. [DOI] [PubMed] [Google Scholar]
- 52.Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–60. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 53.Lehnertz B, Ueda Y, Derijck AA, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 54.Tachibana M, Ueda J, Fukuda M, et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19:815–26. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dong KB, Maksakova IA, Mohn F, et al. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. Embo J. 2008;27:2691–701. doi: 10.1038/emboj.2008.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ikegami K, Iwatani M, Suzuki M, et al. Genome-wide and locus-specific DNA hypomethylation in G9a deficient mouse embryonic stem cells. Genes Cells. 2007;12:1–11. doi: 10.1111/j.1365-2443.2006.01029.x. [DOI] [PubMed] [Google Scholar]
- 57.Collins RE, Northrop JP, Horton JR, et al. The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules. Nat Struct Mol Biol. 2008;15:245–50. doi: 10.1038/nsmb.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Esteve PO, Chin HG, Smallwood A, et al. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20:3089–103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim JK, Esteve PO, Jacobsen SE, Pradhan S. UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res. 2009;37:493–505. doi: 10.1093/nar/gkn961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharif J, Muto M, Takebayashi S, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–12. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 61.Smallwood A, Esteve PO, Pradhan S, Carey M. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev. 2007;21:1169–78. doi: 10.1101/gad.1536807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McGarvey KM, Fahrner JA, Greene E, Martens J, Jenuwein T, Baylin SB. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 2006;66:3541–9. doi: 10.1158/0008-5472.CAN-05-2481. [DOI] [PubMed] [Google Scholar]
- 63.Nguyen CT, Weisenberger DJ, Velicescu M, et al. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2'-deoxycytidine. Cancer Res. 2002;62:6456–61. [PubMed] [Google Scholar]
- 64.Wozniak RJ, Klimecki WT, Lau SS, Feinstein Y, Futscher BW. 5-Aza-2'-deoxycytidine-mediated reductions in G9A histone methyltransferase and histone H3 K9 di-methylation levels are linked to tumor suppressor gene reactivation. Oncogene. 2007;26:77–90. doi: 10.1038/sj.onc.1209763. [DOI] [PubMed] [Google Scholar]
- 65.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–96. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 66.Wang H, Wang L, Erdjument-Bromage H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–8. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 67.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–26. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 68.O'Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21:4330–6. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vire E, Brenner C, Deplus R, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–4. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 70.Negishi M, Saraya A, Miyagi S, et al. Bmi1 cooperates with Dnmt1-associated protein 1 in gene silencing. Biochem Biophys Res Commun. 2007;353:992–8. doi: 10.1016/j.bbrc.2006.12.166. [DOI] [PubMed] [Google Scholar]
- 71.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. Embo J. 2003;22:5323–35. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu J, Yu J, Rhodes DR, et al. A polycomb repression signature in metastatic prostate cancer predicts cancer outcome. Cancer Res. 2007;67:10657–63. doi: 10.1158/0008-5472.CAN-07-2498. [DOI] [PubMed] [Google Scholar]
- 73.Reynolds PA, Sigaroudinia M, Zardo G, et al. Tumor suppressor p16INK4A regulates polycomb-mediated DNA hypermethylation in human mammary epithelial cells. J Biol Chem. 2006;281:24790–802. doi: 10.1074/jbc.M604175200. [DOI] [PubMed] [Google Scholar]
- 74.Gazin C, Wajapeyee N, Gobeil S, Virbasius CM, Green MR. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007;449:1073–7. doi: 10.1038/nature06251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Widschwendter M, Fiegl H, Egle D, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–8. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 76.Ohm JE, McGarvey KM, Yu X, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–42. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gal-Yam EN, Egger G, Iniguez L, et al. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci U S A. 2008;105:12979–84. doi: 10.1073/pnas.0806437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kondo Y, Shen L, Cheng AS, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40:741–50. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 79.McGarvey KM, Greene E, Fahrner JA, Jenuwein T, Baylin SB. DNA methylation and complete transcriptional silencing of cancer genes persist after depletion of EZH2. Cancer Res. 2007;67:5097–102. doi: 10.1158/0008-5472.CAN-06-2029. [DOI] [PubMed] [Google Scholar]
- 80.Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev. 2008;22:1345–55. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–71. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mohn F, Weber M, Rebhan M, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–66. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 83.Ooi SK, Qiu C, Bernstein E, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–7. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pannetier M, Julien E, Schotta G, et al. PR-SET7 and SUV4−20H regulate H4 lysine-20 methylation at imprinting control regions in the mouse. EMBO Rep. 2008;9:998–1005. doi: 10.1038/embor.2008.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schotta G, Lachner M, Sarma K, et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–62. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gonzalo S, Garcia-Cao M, Fraga MF, et al. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol. 2005;7:420–8. doi: 10.1038/ncb1235. [DOI] [PubMed] [Google Scholar]
- 87.Suka N, Luo K, Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat Genet. 2002;32:378–83. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- 88.Pfister S, Rea S, Taipale M, et al. The histone acetyltransferase hMOF is frequently downregulated in primary breast carcinoma and medulloblastoma and constitutes a biomarker for clinical outcome in medulloblastoma. Int J Cancer. 2008;122:1207–13. doi: 10.1002/ijc.23283. [DOI] [PubMed] [Google Scholar]
- 89.Taipale M, Rea S, Richter K, et al. hMOF histone acetyltransferase is required for histone H4 lysine 16 acetylation in mammalian cells. Mol Cell Biol. 2005;25:6798–810. doi: 10.1128/MCB.25.15.6798-6810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wysocka J, Allis CD, Coonrod S. Histone arginine methylation and its dynamic regulation. Front Biosci. 2006;11:344–55. doi: 10.2741/1802. [DOI] [PubMed] [Google Scholar]
- 91.Kirmizis A, Santos-Rosa H, Penkett CJ, et al. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449:928–32. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vermeulen M, Mulder KW, Denissov S, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 93.Zhao Q, Rank G, Tan YT, et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol. 2009;16:304–11. doi: 10.1038/nsmb.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Seligson DB, Horvath S, Shi T, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–6. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 95.Issa JJ, Kantarjian HM. Targeting DNA methylation. Clin Cancer Res. 2009;15 doi: 10.1158/1078-0432.CCR-08-2783. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bots M, Johnstone RW. Rational combinations using HDAC inhibitors. Clin Cancer Res. 2009;15 doi: 10.1158/1078-0432.CCR-08-2786. In press. [DOI] [PubMed] [Google Scholar]
- 97.Prince HM, Bishton MJ, Harrison SJ. Clinical studies of histone deacetylase inhibitors. Clin Cancer Res. 2009;15 doi: 10.1158/1078-0432.CCR-08-2785. In press. [DOI] [PubMed] [Google Scholar]
- 98.Schrump DS, et al. Cytotoxicity mediated by histone deacetylsae inhibitors in cancer cells: mechanisms and potential clinical implications. Clin Cancer Res. 2009;15 doi: 10.1158/1078-0432.CCR-08-2787. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kubicek S, O'Sullivan RJ, August EM, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25:473–81. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 100.Chang Y, Zhang X, Horton JR, et al. Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tan J, Yang X, Zhuang L, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–63. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Damiani LA, Yingling CM, Leng S, Romo PE, Nakamura J, Belinsky SA. Carcinogen-induced gene promoter hypermethylation is mediated by DNMT1 and causal for transformation of immortalized bronchial epithelial cells. Cancer Res. 2008;68:9005–14. doi: 10.1158/0008-5472.CAN-08-1276. [DOI] [PubMed] [Google Scholar]
- 103.Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–50. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- 104.Lao VV, Herring JL, Kim CH, Darwanto A, Soto U, Sowers LC. Incorporation of 5-chlorocytosine into mammalian DNA results in heritable gene silencing and altered cytosine methylation patterns. Carcinogenesis. 2009 doi: 10.1093/carcin/bgp060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hahn MA, Hahn T, Lee DH, et al. Methylation of polycomb target genes in intestinal cancer is mediated by inflammation. Cancer Res. 2008;68:10280–9. doi: 10.1158/0008-5472.CAN-08-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.O'Hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet. 2008;4:e1000155. doi: 10.1371/journal.pgen.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–4. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 108.Benetti R, Gonzalo S, Jaco I, et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15:268–79. doi: 10.1038/nsmb.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sinkkonen L, Hugenschmidt T, Berninger P, et al. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008;15:259–67. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- 110.Ting AH, Suzuki H, Cope L, et al. A requirement for DICER to maintain full promoter CpG island hypermethylation in human cancer cells. Cancer Res. 2008;68:2570–5. doi: 10.1158/0008-5472.CAN-07-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fabbri M, Garzon R, Cimmino A, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–10. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Han L, Witmer PD, Casey E, Valle D, Sukumar S. DNA methylation regulates MicroRNA expression. Cancer Biol Ther. 2007;6:1284–8. doi: 10.4161/cbt.6.8.4486. [DOI] [PubMed] [Google Scholar]
- 113.Lujambio A, Calin GA, Villanueva A, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A. 2008;105:13556–61. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lu C, Soria JC, Tang X, et al. Prognostic factors in resected stage I non-small-cell lung cancer: a multivariate analysis of six molecular markers. J Clin Oncol. 2004;22:4575–83. doi: 10.1200/JCO.2004.01.091. [DOI] [PubMed] [Google Scholar]
- 115.Harbeck N, Nimmrich I, Hartmann A, et al. Multicenter study using paraffin-embedded tumor tissue testing PITX2 DNA methylation as a marker for outcome prediction in tamoxifen-treated, node-negative breast cancer patients. J Clin Oncol. 2008;26:5036–42. doi: 10.1200/JCO.2007.14.1697. [DOI] [PubMed] [Google Scholar]
- 116.Hartmann O, Spyratos F, Harbeck N, et al. DNA methylation markers predict outcome in node-positive, estrogen receptor-positive breast cancer with adjuvant anthracycline-based chemotherapy. Clin Cancer Res. 2009;15:315–23. doi: 10.1158/1078-0432.CCR-08-0166. [DOI] [PubMed] [Google Scholar]
- 117.Brock MV, Hooker CM, Ota-Machida E, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med. 2008;358:1118–28. doi: 10.1056/NEJMoa0706550. [DOI] [PubMed] [Google Scholar]
- 118.Carraway HE, Wang S, Blackford A, et al. Promoter hypermethylation in sentinel lymph nodes as a marker for breast cancer recurrence. Breast Cancer Res Treat. 2008 doi: 10.1007/s10549-008-0004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Alumkal JJ, Zhang Z, Humphreys EB, et al. Effect of DNA methylation on identification of aggressive prostate cancer. Urology. 2008;72:1234–9. doi: 10.1016/j.urology.2007.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ellinger J, Bastian PJ, Jurgan T, et al. CpG island hypermethylation at multiple gene sites in diagnosis and prognosis of prostate cancer. Urology. 2008;71:161–7. doi: 10.1016/j.urology.2007.09.056. [DOI] [PubMed] [Google Scholar]
- 121.Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–4. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 122.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 123.Banno K, Yanokura M, Kawaguchi M, et al. Epigenetic inactivation of the CHFR gene in cervical cancer contributes to sensitivity to taxanes. Int J Oncol. 2007;31:713–20. [PubMed] [Google Scholar]
- 124.Koga Y, Kitajima Y, Miyoshi A, Sato K, Sato S, Miyazaki K. The significance of aberrant CHFR methylation for clinical response to microtubule inhibitors in gastric cancer. J Gastroenterol. 2006;41:133–9. doi: 10.1007/s00535-005-1732-7. [DOI] [PubMed] [Google Scholar]
- 125.Satoh A, Toyota M, Itoh F, et al. Epigenetic inactivation of CHFR and sensitivity to microtubule inhibitors in gastric cancer. Cancer Res. 2003;63:8606–13. [PubMed] [Google Scholar]
- 126.Taniguchi T, Tischkowitz M, Ameziane N, et al. Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat Med. 2003;9:568–74. doi: 10.1038/nm852. [DOI] [PubMed] [Google Scholar]
- 127.Shen L, Kondo Y, Ahmed S, et al. Drug sensitivity prediction by CpG island methylation profile in the NCI-60 cancer cell line panel. Cancer Res. 2007;67:11335–43. doi: 10.1158/0008-5472.CAN-07-1502. [DOI] [PubMed] [Google Scholar]
- 128.Agrelo R, Cheng WH, Setien F, et al. Epigenetic inactivation of the premature aging Werner syndrome gene in human cancer. Proc Natl Acad Sci U S A. 2006;103:8822–7. doi: 10.1073/pnas.0600645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Belinsky SA, Liechty KC, Gentry FD, et al. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Res. 2006;66:3338–44. doi: 10.1158/0008-5472.CAN-05-3408. [DOI] [PubMed] [Google Scholar]
- 130.Gonzalgo ML, Pavlovich CP, Lee SM, Nelson WG. Prostate cancer detection by GSTP1 methylation analysis of postbiopsy urine specimens. Clin Cancer Res. 2003;9:2673–7. [PubMed] [Google Scholar]
- 131.Hoque MO, Topaloglu O, Begum S, et al. Quantitative methylation-specific polymerase chain reaction gene patterns in urine sediment distinguish prostate cancer patients from control subjects. J Clin Oncol. 2005;23:6569–75. doi: 10.1200/JCO.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 132.Chen WD, Han ZJ, Skoletsky J, et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst. 2005;97:1124–32. doi: 10.1093/jnci/dji204. [DOI] [PubMed] [Google Scholar]
- 133.Lenhard K, Bommer GT, Asutay S, et al. Analysis of promoter methylation in stool: a novel method for the detection of colorectal cancer. Clin Gastroenterol Hepatol. 2005;3:142–9. doi: 10.1016/s1542-3565(04)00624-x. [DOI] [PubMed] [Google Scholar]
- 134.Krassenstein R, Sauter E, Dulaimi E, et al. Detection of breast cancer in nipple aspirate fluid by CpG island hypermethylation. Clin Cancer Res. 2004;10:28–32. doi: 10.1158/1078-0432.ccr-0410-3. [DOI] [PubMed] [Google Scholar]