Abstract
p21-loss has been implicated in conferring oncogenic activity to known tumor suppressor gene KLF4 and cancer drug tamoxifen. Regulators of p21 therefore play critical roles in tumorigenesis. Here we report that X-linked tumor suppressor FOXP3 is essential for p21 expression in normal epithelia and that lack of FOXP3 associated with p21 down-regulation in breast cancer samples. A specific FOXP3 binding site in the intron 1 is essential for p21 induction by FOXP3. FOXP3 specifically inhibited binding of histone deacetylase (HDAC) 2 and 4 to the site and increased local histone H3 acetylation. ShRNA silencing of either HDAC2 or HDAC4 is sufficient to induce p21 expression. Our data provides a novel mechanism for transcriptional activation by FOXP3 and a genetic mechanism for lack of p21 in a large proportion of breast cancer.
Introduction
As a universal CDK inhibitor, p21 plays an important role in preventing cell cycle progression by acting at G1 checkpoint (1–4). p21 is down-regulated in many type of cancer including the majority of breast cancer (5–7). Absence of p21 has been shown to confer oncogenic properties to KLF4 (8). Moreover, p21-loss is causatively related to tamoxifen-stimulated growth of breast cancer (9). Surprisingly, p21 mutation is rarely observed in cancer (10). Instead, p21 has emerged as a major down-stream targets of tumor suppressor genes, including p53 (1, 11, 12), BRCA1 (13), CHK2 (14), KLF4 (15, 16) and KLF6 (17). Although p53-mediated regulation has been established as a classical example, the lack of correlation between p53 protein levels (usually used as an indication of p53 mutation) and down-regulation of p21 would argue strongly that p53 mutation is perhaps not the major underlying cause for p21 loss in breast cancer (5–7). Likewise, while it has been demonstrated that BRCA1 (13), Chk2 (14)-mediated tumor cell cycle-arrest and senescence require p21 function, mutations of these two genes had not been established as the genetic cause for lack of p21 in the tumors. On the other hand, epigenetic factors have been suggested as possible mechanisms of p21 silencing in the breast cancer cells (18–21).
We reported recently that heterozygous FOXP3 mutation leads to spontaneous development of mammary tumors (22). The significance of FOXP3 mutation in human is demonstrated by both widespread somatic mutation and deletion of the gene in human breast cancer samples (22). Ectopic expression of the FoxP3 gene caused profound growth inhibition for breast cancer cell lines, both in vivo and in vitro. Since FoxP3 is a transcription factor, an important issue is to identify critical targets of FoxP3 that are responsible for FoxP3’s tumor suppressor activity. In this context, we have reported that FoxP3 is a repressor for the HER-2/ErbB2 and Skp2 oncogenes (22, 23). Alternatively, it is possible that FoxP3 may activate additional tumor suppressor genes. To test this hypothesis, we used a gene array analysis to identify genes affected by FOXP3. We uncovered several tumor suppressor genes that were induced more than 2-fold following induction of FOXP3. We focused on p21 as it is the most highly induced tumor suppressor and because of its unique role in breast cancer biology. Here we report that FOXP3 is a potent inducer of p21 in both normal epithelial cells and malignant breast cancer cell lines. Our data provide a novel mechanism for FOXP3-mediated activation of tumor suppressor gene.
Materials and Methods
Mice
Rag2−/−FoxP3+/+ and Rag2−/−FoxP3sf/sf BALB/c mice have been described previously (24). Two months-old virgin mice were used to analyze the impact of FoxP3 mutation on p21 expression and hyperplasia of mammary epithelia. All animal experiments were conducted in accordance with accepted standards of animal care and were approved by the Institutional Animal Care and Use Committee of the University of Michigan.
Cell culture
Breast cancer cell line MCF-7 and immortalized mammary epithelial cell line MCF-10A were purchased from the American Type Culture Collection. The HO15.19 cell line, which is the c-Myc-null derivative of TGR-1 (25, 26), was a kind gift from Dr. John M. Sedivy, Brown University. A previously established Tet-off FOXP3 expression system in the MCF-7 cells was also used (22, 23).
Microarray analysis of FoxP3-regulated genes
The FoxP3-tet-off MCF7 cells (22, 23) were seeded in 6 well plates and cultured with (2.0μg/ml) and without Doxycyclin in the culture media. After 48 hours of incubation, cells were washed with ice-cold PBS twice and RNA extraction was performed with RNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to manufacturer’s protocol. Contaminated genomic DNA was eliminated with DNase I (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. We conducted mRNA microarray analyses using HG-U133 Plus 2.0 (Affymetrix, Santa Clara, CA, USA) according to the manufacturer’s protocols. We used the most current version of ENTREZ Gene-based CDFs at a time of July 2008 that has been maintained at the University of Michigan in order for the accurate analysis (27). dChip software (UCLA Clinical Microarray Core, CA, USA) was used to make a heat map of miroarray profiles according to the instruction of the software. Gene expression profiles of FoxP3-tet-off cells cultured with and without Doxycyclin were compared. Differences of mRNA expression levels between FoxP3+ and FoxP3− cells were calculated by Student’s t-test.
FOXP3, p21 and HDAC silencing
Two FOXP3-short hairpin RNA (shRNA) constructs are FOXP3-993-shRNA and FOXP3-1355-shRNA (GenBank accession number, NM_014009). Oligonucleotides encoding small interfering RNA (siRNA) directed against FOXP3 are 5′-GCTTCATCTGTGGCATCATCC -3′ for FOXP3-993-shRNA (993 to 1013 nucleotides from TSS) and 5′-GAGTCTGCACAAGTGCTTTGT -3′ for FOXP3-1355-shRNA (1355 to 1375 from TSS). The selected shRNA oligonucleotides were cloned into pSIREN-RetroQ vectors (Clontech, Mountain View, CA) to generate siRNA according to manufacturer’s protocol. The human p21shRNA (CGCCTCTGGCATTAGAATTATT), human shHDAC2 (shHDAC2-1, CCGACGGTGATATTGGAAATTA), (shHDAC2-2, CGGGCAGATATTTAAGCCTATT), human shHDAC4 (shHDAC4-1, ACGGCATGACTTTATATTGTAT), (shHDAC4-2, AGACCGGCATGACTTTATATTG) and control lentiviral vectors were purchased from Open Biosystems (Huntsville, AL).
Western blot
The anti-FOXP3 (Abcom, 1:1000), anti-hFOXY (eBioscience, 1: 100), anti-p21 (Cell Signaling, 1:1000), and anti-β-actin (Sigma, 1:3000) were used as the primary antibodies. A 1:3,000–5,000 dilution of the anti-rabbit or mouse IgG HRP-linked secondary antibody (Cell Signaling). To ensure equal loading of proteins, the membraneswere stripped under the same conditions as describedabove. They were then incubated with enhanced chemiluminescence (ECL) reagents (Amersham Biosciences) and exposed to X-ray film for 1–5 min.
Chromatin Immunoprecipitation (ChIP)
ChIP was carried out according to published procedure (28). Briefly, the FOXP3-transfected Tet-off cells were sonicated and fixed with 1% paraformaldehyde. The anti-FOXP3, anti-acetyl-H3 (cell signaling), anti-HDAC1, 2, 3, 4, 5, 7 (cell signaling) and anti-IgG (Santa Cruz Biotechnology, Santa Cruz, CA) antibodies were used to pull down chromatin associated with FOXP3. The amounts of the specific DNA fragment were quantitated by real-time PCR and normalized against the genomic DNA preparation from the same cells. The ChIP real-time PCR primers are listed in Table S3.
Quantitative Real-Time PCR
Relative quantities of mRNA expression were analyzed using real-time PCR (Applied Biosystems ABI Prism 7500 Sequence Detection System, Applied Biosystems). The SYBR (Applied Biosystems) green fluorescence dye was used in this study. The primer sequences are listed in Table S3.
Immunohistochemistry
Immunohistochemistry was performed by the avidin-biotin-peroxidase complex (ABC) method. Expression of FOXP3 in human breast cancer or normal tissue samples were determined using immunohistochemistry as described (22, 23). The p21 mouse monoclonal antibody (cell signaling, 1:100) and biotin goat-anti-mouse IgG (Santa Cruz, 1:200) were used as the secondary antibody. FOXP3 and p21 staining were scored double blind.
Statistical Analysis
Data are shown as means ± SD. Statistical analysis was performed with Student’s t-test for means from two groups. ANOVA test was used for analysis of variance between several groups. CHI-square test was used to compare the relationship between the expression of FOXP3 and p21.
Results
1. p21 is upregulated after FOXP3 induction and contributes to its tumor suppressor activity
We used the MCF-7 cell lines engineered to express FOXP3 in the absence Doxycyclin. The cells cultured in the presence or absence of Doxyclin for 48 hours were compared by gene array analysis, with five independent RNA isolates in each group. A summary of the gene array data, depicting genes that are induced by more than 2-folds, is shown in Fig. 1A. The full data set is shown in supplemental Table S1 and Table S2, and the raw data are deposited to MIAExpress (Accession No. E-MTAB-73).
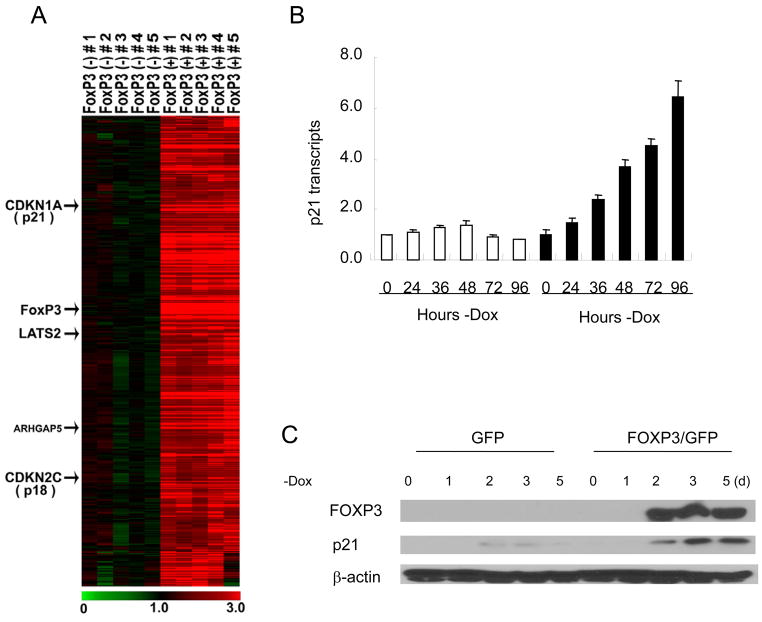
Fig. 1. Identification of p21 as a FOXP3-induced tumor suppressor gene.
A. Gene expression profiles in a panel of 5 flasks of FoxP3+ cells and 5 flasks of FoxP3− cells. Expression values of each row were normalized to the average expression value of each gene among the FoxP3− cells. Color scales of gene expression levels are indicated at the bottom of the figure. Known tumor suppressor genes are indicated at the left side of the heat map. B and C, confirmation of p21 upregulation by FOXP3. After removing doxycycline from medium, the cells were collected at 0, 24, 36, 48, 72, and 96 hours, and measured the FOXP3 expression by realtime-PCR (B) and western blotting (C). (B) The mRNA levels of p21 were measured by real-time PCR for the pBI-GFP vector control cells and pBI-FOXP3/GFP cells. The means of the 0 hour is artificially defined as 1.0. Data shown are means ± SD of 3 independent experiments. (C) The protein levels of FOXP3 and p21 were detected in the GFP control and FOXP3/GFP cells by Western blot without doxycycline from 0 to 5 days.
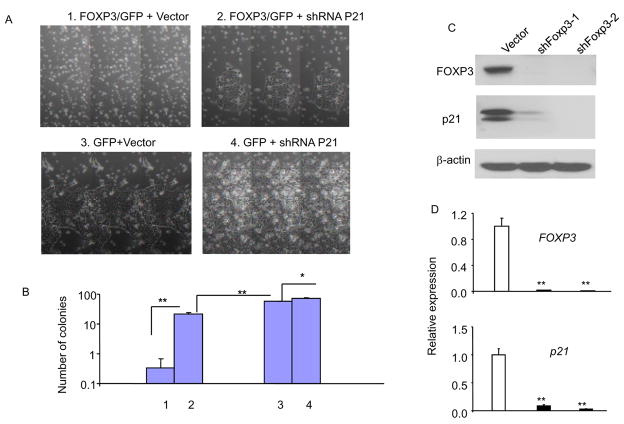
Among the FOXP3-induced genes are several tumor suppressors, including p18, p21, LAT2, and ARHGAPS (Fig. 1A). We have chosen p21 as the prototype to study the mechanism by which FOXP3 activates tumor suppressors as the relevance of defective p21 in breast cancer is well established. In addition, real-time PCR showed induction of p18 is less than 2-fold (data not shown). We first used real-time PCR and Western blot to confirm the induction of p21 following the inducible expression of FOXP3. As shown in Fig. 1B, p21 transcripts was induced by 7-fold in the MCF-7-pBI-FOXP3/GFP cell line after removal of Doxycyclin, but not the MCF-7-pBI-GFP/control cell line under the same culture condition. Western blot analysis confirmed that accumulation of p21 protein followed that of FOXP3 (Fig. 1C). In order to determine whether induction of p21 contributed to tumor suppression, we transfected the MCF-7 cell lines with either control vector or p21 shRNA. The transfectants were cultured in the absence of doxycycline for 10 days and stained by crystal violets. As shown in Fig. 2B, p21 shRNA specifically increased the number of colonies in the cell line that expressed FOXP3, but barely so for those that expressed GFP. Microscopically, the sizes of colonies were usually larger in the shRNA group, even for those that expressed GFP only, consistent with the notion that endogenous p21 in the MCF-7 cells limited its growth potential (Fig. 2A). Even p21-silenced group, FOXP3 transfection still reduces the number of colonies by nearly 60%, which is consistent with the contribution of other FOXP3 targets, including those that we have reported recently (22, 23). Nevertheless, the partial restoration of the colonies indicated that p21 induction contribute to the tumor suppressor activity of the FOXP3 gene.
Fig. 2. p21 induction is an underlying mechanism for the tumor suppressor activity of FOXP3.
(A) MCF-7 cells with inducible expression of either FOXP3 (1, 2) or GFP (3, 4) were supertransfected with either vector control (1, 3) or p21 shRNA (2, 4). After removing untransfected cells by drug selection, the cultures were maintained in Doxycyclin-free conditions for 10 days. Upper and lower panels show photographs of viable (2, 3, 4) or apoptotic MCF-7 cells (1). Magnification, 100×.
(B) The colony number per 60-mm2 plate. Data shown are means of SD of triplicates and are representative of 3 independent experiments. C&D. Silencing of FOXP3 resulted in down-regulation of p21 protein (C) and p21 mRNA (D) in human mammary epithelial cell line MCF10A. MCF10A was transfected with either control vector or FOXP3 shRNAs. The untransfected cells were removed by selection with puromycin. At 2 weeks after transfection, the protein levels were determined by Western blot, using specific anti-FOXP3, anti-p21, and β-actin antibodies as loading control. The mRNA levels of the FOXP3 and p21 transcripts were quantitated by real-time PCR, The RNA inputs were normalized against housekeeping gene GAPDH. The vector control was defined as 1.0. Data shown are means ± SD of triplicates and represent 3 independent experiments.
2. FOXP3 maintains p21 levels in normal mammary epithelial cells
An important issue is whether FOXP3 expression contributes to expression of p21 in normal mammary epithelial cells. As shown in Fig. 2C, the FOXP3 protein can be identified by Western blot in immortalized human mammary epithelial cell line MCF-10A. To determine the role for FOXP3 in p21 expression, we used FOXP3 shRNA to silence FOXP3 expression and measured the levels of p21 transcripts. As shown in Fig. 2D, FOXP3 silencing caused 5–10-fold reduction of the p21 transcripts, which revealed a critical role for FOXP3 in maintaining p21 expression in mammary epithelial cells. Similar effect was observed when FOXP3 was silenced in early passage of primary human mammary epithelial culture (Supplemental Fig. S1).
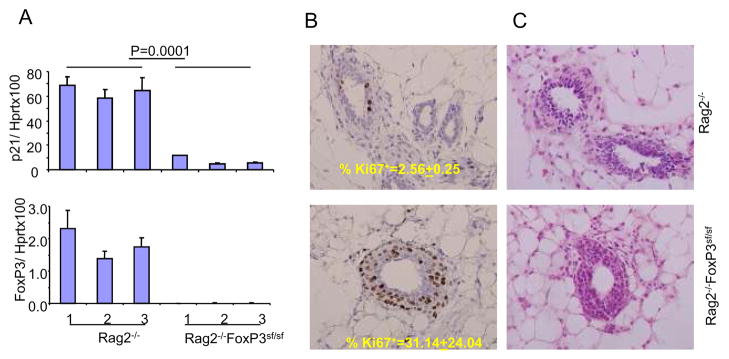
To test the role of FoxP3 in p21 expression in vivo, we micro-dissected mammary epithelium from 2-month old Rag2−/−FoxP3sf/sf and Rag2−/−FoxP3+/+ mice. The amounts of p21 transcripts were determined by real-time PCR. As shown in Fig. 3A, the FoxP3 mutation caused approximately 6-fold reduction in p21 transcripts. Perhaps due to non-mediated decay caused by frameshift mutation, mammary epithelia from FoxP3 mutant mice lacked FoxP3 transcripts. Correspondingly, dramatically increased numbers of breast epithelial cells in the Rag2−/−FoxP3sf/sf mice have entered the cell cycle as judged by Ki67 staining (Fig. 3B). H&E staining of the mammary tissue indicated extensive ductal hyperplasia in the Rag2−/−FoxP3sf/sf mice (Fig. 3C). These data demonstrated that the FoxP3 mutation leads to reduced p21 expression and increased proliferation of normal epithelium in vivo. Since the young mice had yet to develop mammary tumor at this age, down-regulation of p21 is not due to secondary effect of malignant transformation.
Fig. 3. FoxP3 mutation in benign mammary epithelial increased p21 transcripts and cause increased proliferation of epithelial cells.
A. The FoxP3 mutation increased p21 transcripts. Mammary epithelia from virgin Rag2−/−FoxP3+/+ and Rag2−/−FoxP3sf/sf BALB/c mice were isolated by microdissection. The p21 and FoxP3 transcripts were measured by real-time PCR. The data shown were means and SEM of % of house keeping gene Hprt. Three independent mice were used in each group. B. Increased proliferation of the mammary epithelial cells as revealed by Ki67 staining. The data shown are representative fields from each group. The means and SD from groups of 3 mice are shown in the insert (p<0.05). C. H&E staining of 2-month old virgin Rag2−/−FoxP3+/+ and Rag2−/−FoxP3sf/sf BALB/c mice. Data shown are representative of three mice per group.
3. Correlation between expressions of FOXP3 and p21 in human breast cancer
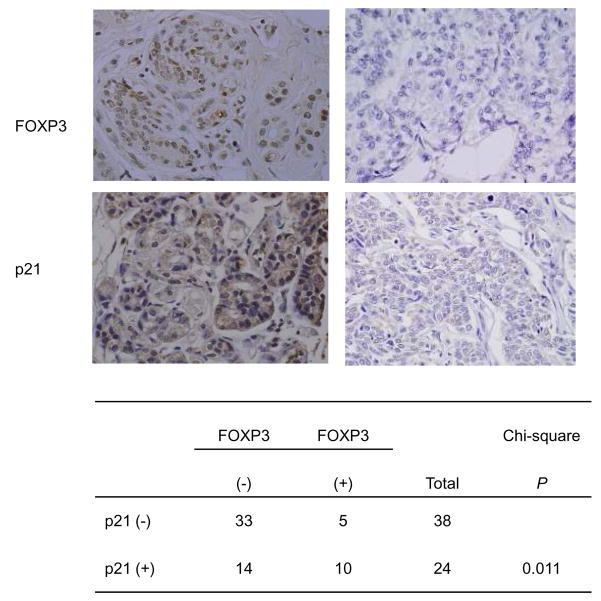
The majority of breast cancer samples lack p21 and FOXP3 expression (5–7) (22, 23). An important issue is therefore whether the expression of the two genes is inter-related among human breast cancer samples. To address this issue, we analyzed 62 cases of breast cancer samples in TMA for expression of FOXP3. As shown in Fig. 4, among the FOXP3+ samples, 66% are also p21+. In contrast, only 30% of the FOXP3− samples expressed p21. The strong correlation between FOXP3 and p21 expression suggests that FOXP3 down-regulation may be an important factor for the lack of p21 among breast cancer tissue.
Fig. 4. A positive correlation between FOXP3 and p21 expression in human breast cancer.
Tissue micro-array samples were stained with either anti-FOXP3 antibody or anti-p21 antibody and were scored by a double-blind fashion. Samples with nuclear staining by the anti-FOXP3 antibody were scored as positive. Samples with greater than 10% p21 staining in nuclear and/or cytosolic were scored as positive. Magnification, 600×. Summary data from 62 independent cases is presented in the lower panel. The P value of the χ2 test is listed.
4. Specific binding of FOXP3 to the p21 locus is essential for activation of p21
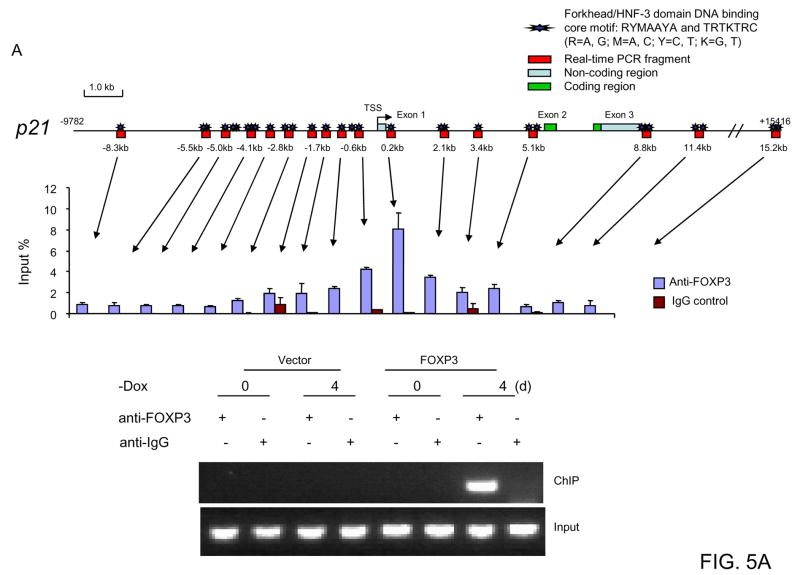
Two p21 mRNA isoforms (1: NM_078467 and 2: NM_000389) have been reported with different exon1 exon 2 junctions. In order to properly align the genomic structure of the locus, we sequenced the p21 RNA from the MCF-7 cells after the induction of FOXP3. As shown in supplemental Fig. S2, only isoform 2 was produced in FOXP3-transfected MCF-7 cells. This allowed us to assign the position of intron 1 for the p21 locus. As illustrated in Fig. 5A upper panels, large number of forkhead binding motifs RYMAAYA (29, 30) and TRTKTRC (31, 32) (R=A, G; M=A, C; Y=C, T; K=G, T) can be identified throughout the p21 gene. In order to identify the sites that bind to FOXP3, we induced FOXP3 by culturing the cell line in the absence of Doxycyclin and then used ChIP to determine whether FOXP3 interact with the p21 locus. In order to normalize the efficiency of PCR primers, the products were compared to input DNA amplified by the same primers. As shown in Fig. 5A middle panel, quantitative analysis demonstrate that peak binding activity localized at the forkhead/HNF-3 binding motif at 0.2 kb 3′ of the transcription starting site (TSS). Low, but detectable levels of DNA are observed over an 8 kb fragment, which could be due to either low resolution of ChIP or existence of multiple weaker binding sites. To confirm the specific requirement for FOXP3 for the signal at the 0.2 kb, we also compared the signal to uninduced pBI-FOXP3/GFP cell lines and the pBI-GFP control cell lines cultured in the presence or absence of doxycycline. As shown in Fig. 5A lower panel, the p21 region is precipitated, if and only if, FOXP3 was induced.
Fig. 5. FOXP3 as a transcriptional activator for p21.
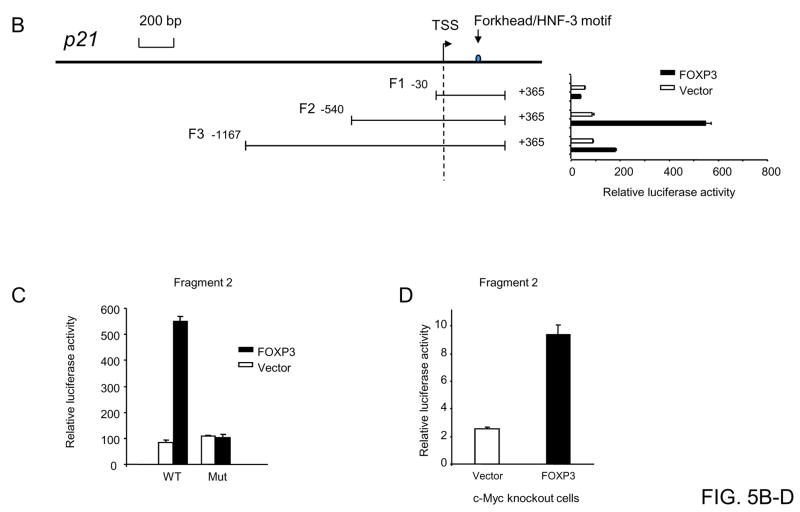
(A). Chromatin immunoprecipitation. A diagram of the p21 gene, including the promoter and exon 1–3 (NM_000389) is depicted on the top. The forkhead binding motifs are illustrated with black asterisks, while the regions surveyed by real-time PCR are marked in red bars. The middle panel shows the amount of DNA precipitated by either control IgG or anti-FOXP3 mAb expressed as percentage of the total input genomic DNA. Data shown are means and standard deviation (SD) of triplicates. This experiment has been repeated twice with similar results. The lower panel shows specificity of the ChIP assay, as demonstrated by the requirements for both FOXP3 induction and anti-FOXP3 antibody. (B). Identification of the promoter region most responsive to FOXP3-mediated induction. The HEK293 cells were transfected with either vector control or FOXP3 (1 μg/well) in conjunction with the luciferase reporter driven by different 5′ promoter regions of the p21 gene (0.5 μg/well). pRL-TK was used as internal control. The luciferase activity from the cells transfected with the pGL2-basic vector was arbitrarily defined as 1.0. Data shown are means and SD of triplicates and have been repeated at least three times. (C) Site-directed mutagenesis of one candidate forkhead/HNF-3 binding motif in the P21 promoter abrogated the induction of the p21 transcription activity by FOXP3. The wild-type forkhead-binding motif TGTGTGC were mutated into CCCAAAA. The promoter activity was measured and normalized as detailed in (B). Data shown are means and SD of triplicates. This experiment has been repeated twice with similar results. (D) p21 transcription is directly induced by FOXP3 by a c-Myc-independent mechanism. Transfection of FOXP3 into c-Myc−/− cells increased p21 activity, as by a luciferase assay. Data shown are means and SD of triplicates. This experiment has been repeated three times.
In order to directly demonstrate the function and specificity of the FOXP3-mediated induction of p21, we first produced three constructs consisting of overlapping fragments of the 5′ of the p21 locus (Fig. 5B). Using duo-luciferase assay, we found that FOXP3-mediated induction of p21 requires sequences that are both 5′ and 3′ to the TSS, with the maximal activity requiring −540 bp and +365 bp at the 5′ and 3′ respectively. Further extension in the 3′ significantly reduced the p21 induction (Fig. 5B). We therefore used the optimal reporter to confirm the function of the forkhead binding site at the 0.2 kb 3′ of TSS. As shown in Fig. 5C, while WT reporter is induced by FOXP3 expression, mutation of the forkhead binding site abrogated the induction. These data demonstrated the specific cis-element is essential for FOXP3-mediated activation of the p21 locus.
It has been demonstrated that c-Myc can target the p21 promoter and inhibits its expression (33–35). To determine whether the FOXP3 gene regulate p21 directly, we measured the effects of FOXP3 on the p21 promoter activity in the c-Myc knockout cell line. As shown in Fig. 5D, the promoter activity of p21 was significantly induced by FOXP3 in c-Myc knockout cells. The relative low induction, in comparison to HEK 293 cells, is likely due to drastically reduced transfection efficiency of the Myc-deficient cell line (our unpublished observation).
4. Localized chromatin modification as a mechanism for FOXP3-induced expression of p21
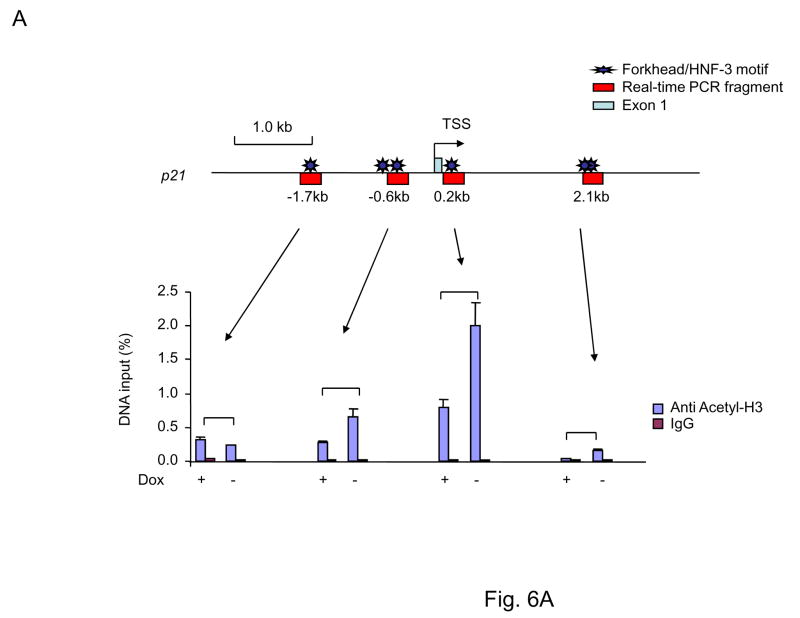
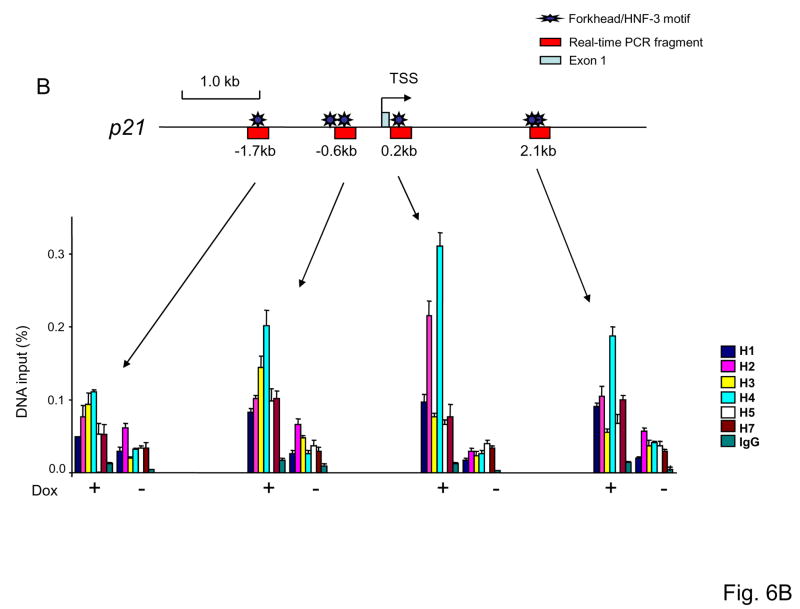
Recent studies demonstrated that FOXP3-mediated induction of gene expression is associated with histone acetylation (36). We therefore used anti-acetyl-H3 antibodies to monitor local chromatin changes associated with FOXP3 binding. As shown in Fig. 6A and B, in cells expressing FOXP3, H3 acetylation in the +0.2 kb site of p21 was increased by more than 2-fold. The increase in the neighboring areas mirrored what was observed with FOXP3 binding. These data demonstrated that FOXP3 enhance H3 acetylation of p21, especially at the 0.2 kb region. We carried out ChIP analysis using antibodies specific for HDAC1-7. The MCF-7 cells with or without FOXP3 induction were compared. As shown in Fig. 6B, a generalized reduction of HDAC association to the p21 locus was observed following FOXP3 induction. However, by far, the strongest effect was observed at the 0.2 kb site where FOXP3 associate to the p21 locus. Moreover, although a reduction of HDAC1-7 was observed following FOXP3 binding, the most significant reduction was observed on HDAC 2 and 4 as these two HDAC showed the strongest association prior to FOXP3 induction.
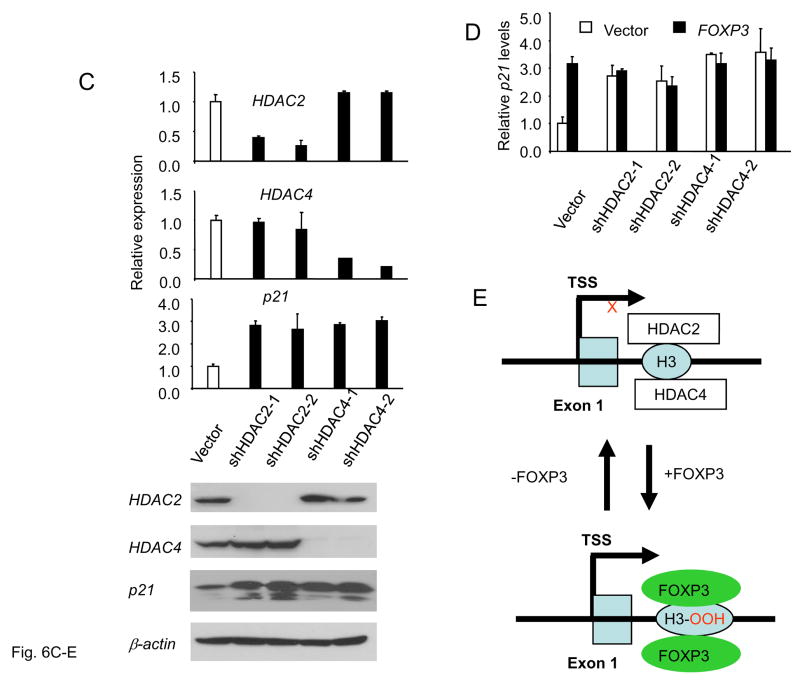
Fig. 6. FOXP3 specifically increased acetylation of the histone H3 associated with the FOXP3 binding site by inhibiting association of HDAC 2 and 4.
(A). FOXP3 increases acetylation of histone H3 associated with the FOXP3 binding site. (B). The increased acetylation of H3 is due to FOXP3-mediated inhibition association of HDAC2 and 4 to the site. The MCF-7 cells with inducible FOXP3 expression were cultured with (−FOXP3) or without (+FOXP3) doxycycline for 4 days and subjected to ChIP analysis using acetyl-H3 (A) and HDAC1,2,3,4,5 and 7 (B) antibody or control IgG. Precipitated genomic DNA was probed for the promoter/enhancer regions of the p21 locus by realtime-PCR. The amounts of DNA precipitated were expressed as percentage of the total input genomic DNA. Data shown are means of SD of triplicates. Results depicted are representative of three separate experiments. (C). ShRNA silencing of either HDAC2 or 4 is sufficient to induce p21 expression. Data shown in bar graphs are from real-time PCR quantitation of shRNA efficacy and the effect of shRNA silencing, using the means of the vector group as 1.0. Those in the bottom are from Western blot. Note that some p21 proteins in HDAC4 or HDAC2 shRNA groups are degraded products with a molecular weight of 15 kD, while those in the control group consisted of mostly intact p21. These experiments have been repeated 4 times with similar results. D. Silencing HDAC4 abrogates induction of p21 by FOXP3. Control or HDAC2/4-silenced MCF-7 cells were transfected with FOXP3 cDNA. After removal of non-transfected cells by blasticidin, the levels of p21 transcripts were determined by real-time PCR. Data shown have been repeated 2 times. E. Diagram of a proposed mechanism of FOXP3-mediated gene activation. FOXP3 removes association of HDAC2, 4 and thereby increase H3 acetylation and gene activation.
To determine whether the HDAC2 and 4 are involved in p21 up-regulation, we used shRNAs to modulate their expression. As shown in Fig. 6C, two independent shRNAs specifically silencing the expression of either HDAC2 or 4. Correspondingly, the levels of p21 transcripts were increased by 2–3-fold following the silence of either gene. If is of note that some of the p21 protein induced by HDAC shRNAs had a molecular weight of 15kD rather than 21 kD. This is likely due to cleavage of p21 associated with a general increase in histone acetylation, as reported by others (37). To test is HDAC2/4 are necessary mediators of p21 induction by FOXP3, we transfected FOXP3 into MCF7 cells in which HDAC2/4 are silenced and compared the levels of p21 in either FOXP3- or vector control-transfected cells. As shown in Fig. 6C, FOXP3-mediated induction of p21 is abrogated in the HDAC4-silenced cell lines. These data further support the notion that FOXP3-mediated induction of p21 is mediated by disruption of HDAC2/4-mediated repression.
Discussion
Mechanism of p21 regulation in normal and cancerous epithelial cells may hold keys to molecular mechanism of carcinogenesis. Here we present several lines of evidence demonstrating a critical role for p21 as a down-stream target of FOXP3 and its expression contributes to FOXP3-mediated growth inhibition of a breast cancer cell line.
First, in confirming the cDNA microarray data, we showed that inducible expression of FOXP3 induced the p21 transcripts and protein in breast cancer cell line MCF-7. The induction is mediated by transcriptional regulation as it is reflected in luciferase assay. ChIP analysis revealed that a specific site at 0.2 kb down-stream of TSS is necessary for FOXP3-mediated induction by FOXP3. Moreover, the induction is not an artifact of FOXP3 over-expression as shRNA silencing of the FOXP3 gene leads to a dramatic reduction of p21 in primary mammary epithelial cells.
Second, in order to determine whether induction of p21 contribute to growth inhibition of the tumor cell line, we tested whether blunting p21 induction by shRNA abrogate growth inhibition by FOXP3. Our data demonstrated that significant, albeit incomplete, rescue of FOXP3-mediated growth inhibition. The significant rescue demonstrates an important role for p21 induction in FOXP3-mediated growth inhibition of MCF-7 cell line. Other recent studies indicate that FOXP3 also inhibition growth by repressing expression of HER-2 and SKP2 (22, 23). Thus, depending on tumor cell lines used, FOXP3-mediated inhibition of oncogenes and induction of tumor suppressor may work either independently or in concert to cause growth inhibition of breast cancer cell line.
Thirdly, our analysis of 62 cases of breast cancer samples demonstrated a significant correlation between expression of FOXP3 and p21. Nevertheless, not unlike other tumor suppressor targets, there was no 1:1 correlation between expression of FOXP3 and p21. For instance, approximately 30% of cases that stained positive for FOXP3 still lack detectable p21. This can in part due to the fact that nearly 1/3 of breast cancer samples show somatic missense mutation of FOXP3 (22, 23). Conversely, nearly 1/3 of the FOXP3 negative tumor cells still express p21. This can be due to either to false-negative staining of FOXP3, perhaps relating to the quality of tumor tissues and or levels of FOXP3 expression in the first place. In addition, since p53 can induce p21 expression, it is possible that the p21 expression in FOXP3-negative tumor samples was due to functional p53. The limited sample set used in this study cannot distinguish these possibilities. Regardless of how the discrepancies are explained, the positive association between p21 and FOXP3 in clinical samples, when viewed in the context of the data in mice with FoxP3 mutation and the in vitro analysis of normal and malignant tumor cells, made a compelling case that FOXP3 is a major regulator for p21 expression in breast cancer. Recent studies revealed an interesting role of p21 loss and tamoxifen-stimulated growth of breast cancer (9). It is of great interest to determine whether genetic lesion to FOXP3 may account for the p21 loss and therefore the unusual response to a widely used drug.
Finally, while a number of studies have addressed the mechanism of FOXP3-mediated gene repression, the mechanism by which FOXP3 directly induce gene expression remained largely obscure. A recent report showed association between FOXP3-induced gene activation and histone acetylation (36), although the mechanism and significance of such acetylation has not been addressed. Our data demonstrated that FOXP3 binding to a specific site in intron 1 of p21 increased histone H3 acetylation by reducing binding of HDAC4 and HDAC2 to the same site (Fig. 6E). Gene silencing with shRNA confirmed the significance of these two HDACs in p21 expression, although FOXP3 did not repress expression of either HDAC2 or 4 (Supplemental Fig. S4). Therefore, our data provide a novel mechanism for FOXP3-mediated transcriptional activation. Since FOXP3 has been shown to recruit histone acetyl transferases (HATs) (38), it is of interest to investigate whether this interaction contributes, either directly or indirectly, to increased H3-acetylation in the p21 locus.
Taken together, our data demonstrated that p21 as a down-stream target for FOXP3, the first X-linked tumor suppressor in breast cancer. Since p21 serves as an important target for all major tumor suppressor genes of breast cancer and since irreversible genetic lesion to p21 is relatively rare, it might be possible to reactivate p21 in cancer by inducing FOXP3. While p21 induction can be achieved by a general silencing of HDAC2 and 4, the induced p21 induced are rapidly degraded, presumably due to simultaneous induction of other proteins involved in p21 cleavage (37). On the other hand, our data showed that p21 induced by FOXP3 remained intact and mediates tumor suppression. Therefore reactivating FOXP3 may prove to be a more relevant approach.
Supplementary Material
Acknowledgments
This study is supported by the National Institute of Health (CA120910 to YL) and Department of Defense (W81XWH08-1-0537 to YL) and American Cancer Society (RSG-06-072-01-TBE to PZ). The raw data for microarray analyses have been deposited to MIAExpress (Accession No. E-MTAB-73).
Footnotes
The authors have no conflict of financial interest.
References
- 1.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75(4):817–25. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 2.Gu Y, Turck CW, Morgan DO. Inhibition of CDK2 activity in vivo by an associated 20K regulatory subunit. Nature. 1993;366(6456):707–10. doi: 10.1038/366707a0. [DOI] [PubMed] [Google Scholar]
- 3.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75(4):805–16. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 4.Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366(6456):701–4. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 5.Pellikainen MJ, Pekola TT, Ropponen KM, Kataja VV, Kellokoski JK, Eskelinen MJ, Kosma VM. p21WAF1 expression in invasive breast cancer and its association with p53, AP-2, cell proliferation, and prognosis. J Clin Pathol. 2003;56(3):214–20. doi: 10.1136/jcp.56.3.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinto AE, Andre S, Laranjeira C, Soares J. Correlations of cell cycle regulators (p53, p21, pRb and mdm2) and c-erbB-2 with biological markers of proliferation and overall survival in breast cancer. Pathology (Phila) 2005;37(1):45–50. doi: 10.1080/00313020400011250. [DOI] [PubMed] [Google Scholar]
- 7.Tiezzi DG, Andrade JM, Ribeiro-Silva A, Zola FE, Marana HR, Tiezzi MG. HER-2, p53, p21 and hormonal receptors proteins expression as predictive factors of response and prognosis in locally advanced breast cancer treated with neoadjuvant docetaxel plus epirubicin combination. BMC cancer. 2007;7:36. doi: 10.1186/1471-2407-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol. 2005;7(11):1074–82. doi: 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]
- 9.Abukhdeir AM, Vitolo MI, Argani P, De Marzo AM, Karakas B, Konishi H, Gustin JP, Lauring J, Garay JP, Pendleton C, Konishi Y, Blair BG, Brenner K, Garrett-Mayer E, Carraway H, Bachman KE, Park BH. Tamoxifen-stimulated growth of breast cancer due to p21 loss. Proc Natl Acad Sci U S A. 2008;105(1):288–93. doi: 10.1073/pnas.0710887105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shiohara M, el-Deiry WS, Wada M, Nakamaki T, Takeuchi S, Yang R, Chen DL, Vogelstein B, Koeffler HP. Absence of WAF1 mutations in a variety of human malignancies. Blood. 1994;84(11):3781–4. [PubMed] [Google Scholar]
- 11.Dulic V, Kaufmann WK, Wilson SJ, Tlsty TD, Lees E, Harper JW, Elledge SJ, Reed SI. p53-dependent inhibition of cyclin-dependent kinase activities in human fibroblasts during radiation-induced G1 arrest. Cell. 1994;76(6):1013–23. doi: 10.1016/0092-8674(94)90379-4. [DOI] [PubMed] [Google Scholar]
- 12.Slebos RJ, Lee MH, Plunkett BS, Kessis TD, Williams BO, Jacks T, Hedrick L, Kastan MB, Cho KR. p53-dependent G1 arrest involves pRB-related proteins and is disrupted by the human papillomavirus 16 E7 oncoprotein. Proc Natl Acad Sci U S A. 1994;91(12):5320–4. doi: 10.1073/pnas.91.12.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Somasundaram K, Zhang H, Zeng YX, Houvras Y, Peng Y, Wu GS, Licht JD, Weber BL, El-Deiry WS. Arrest of the cell cycle by the tumour-suppressor BRCA1 requires the CDK-inhibitor p21WAF1/CiP1. Nature. 1997;389(6647):187–90. doi: 10.1038/38291. [DOI] [PubMed] [Google Scholar]
- 14.Aliouat-Denis CM, Dendouga N, Van den Wyngaert I, Goehlmann H, Steller U, van de Weyer I, Van Slycken N, Andries L, Kass S, Luyten W, Janicot M, Vialard JE. p53-independent regulation of p21Waf1/Cip1 expression and senescence by Chk2. Mol Cancer Res. 2005;3(11):627–34. doi: 10.1158/1541-7786.MCR-05-0121. [DOI] [PubMed] [Google Scholar]
- 15.Chen X, Johns DC, Geiman DE, Marban E, Dang DT, Hamlin G, Sun R, Yang VW. Kruppel-like factor 4 (gut-enriched Kruppel-like factor) inhibits cell proliferation by blocking G1/S progression of the cell cycle. J Biol Chem. 2001;276(32):30423–8. doi: 10.1074/jbc.M101194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W, Geiman DE, Shields JM, Dang DT, Mahatan CS, Kaestner KH, Biggs JR, Kraft AS, Yang VW. The gut-enriched Kruppel-like factor (Kruppel-like factor 4) mediates the transactivating effect of p53 on the p21WAF1/Cip1 promoter. J Biol Chem. 2000;275(24):18391–8. doi: 10.1074/jbc.C000062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narla G, Heath KE, Reeves HL, Li D, Giono LE, Kimmelman AC, Glucksman MJ, Narla J, Eng FJ, Chan AM, Ferrari AC, Martignetti JA, Friedman SL. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science. 2001;294(5551):2563–6. doi: 10.1126/science.1066326. [DOI] [PubMed] [Google Scholar]
- 18.Duan Z, Zarebski A, Montoya-Durango D, Grimes HL, Horwitz M. Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol Cell Biol. 2005;25(23):10338–51. doi: 10.1128/MCB.25.23.10338-10351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagger G, Doetzlhofer A, Schuettengruber B, Haidweger E, Simboeck E, Tischler J, Chiocca S, Suske G, Rotheneder H, Wintersberger E, Seiser C. The tumor suppressor p53 and histone deacetylase 1 are antagonistic regulators of the cyclin-dependent kinase inhibitor p21/WAF1/CIP1 gene. Mol Cell Biol. 2003;23(8):2669–79. doi: 10.1128/MCB.23.8.2669-2679.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richon VM, Sandhoff TW, Rifkind RA, Marks PA. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc Natl Acad Sci U S A. 2000;97(18):10014–9. doi: 10.1073/pnas.180316197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia W, Nagase S, Montia AG, Kalachikov SM, Keniry M, Su T, Memeo L, Hibshoosh H, Parsons R. BAF180 is a critical regulator of p21 induction and a tumor suppressor mutated in breast cancer. Cancer Res. 2008;68(6):1667–74. doi: 10.1158/0008-5472.CAN-07-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuo T, Wang L, Morrison C, Chang X, Zhang H, Li W, Liu Y, Wang Y, Liu X, Chan MW, Liu JQ, Love R, Liu CG, Godfrey V, Shen R, Huang TH, Yang T, Park BK, Wang CY, Zheng P. FOXP3 is an X-linked breast cancer suppressor gene and an important repressor of the HER-2/ErbB2 oncogene. Cell. 2007;129(7):1275–86. doi: 10.1016/j.cell.2007.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuo T, Liu R, Zhang H, Chang X, Liu Y, Wang L, Zheng P. FOXP3 is a novel transcriptional repressor for the breast cancer oncogene SKP2. J Clin Invest. 2007;117(12):3765–73. doi: 10.1172/JCI32538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang X, Gao JX, Jiang Q, Wen J, Seifers N, Su L, Godfrey VL, Zuo T, Zheng P, Liu Y. The Scurfy mutation of FoxP3 in the thymus stroma leads to defective thymopoiesis. J Exp Med. 2005;202(8):1141–51. doi: 10.1084/jem.20050157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prouty SM, Hanson KD, Boyle AL, Brown JR, Shichiri M, Follansbee MR, Kang W, Sedivy JM. A cell culture model system for genetic analyses of the cell cycle by targeted homologous recombination. Oncogene. 1993;8(4):899–907. [PubMed] [Google Scholar]
- 26.Mateyak MK, Obaya AJ, Adachi S, Sedivy JM. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8(10):1039–48. [PubMed] [Google Scholar]
- 27.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, Watson SJ, Meng F. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33(20):e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Im H, Grass JA, Johnson KD, Boyer ME, Wu J, Bresnick EH. Measurement of protein-DNA interactions in vivo by chromatin immunoprecipitation. Methods Mol Biol. 2004;284:129–46. doi: 10.1385/1-59259-816-1:129. [DOI] [PubMed] [Google Scholar]
- 29.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, Takusagawa KT, Lander ES, Gifford DK, Fraenkel E, Young RA. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431(7004):99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pic A, Lim FL, Ross SJ, Veal EA, Johnson AL, Sultan MR, West AG, Johnston LH, Sharrocks AD, Morgan BA. The forkhead protein Fkh2 is a component of the yeast cell cycle transcription factor SFF. Embo J. 2000;19(14):3750–61. doi: 10.1093/emboj/19.14.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cardinaux JR, Chapel S, Wahli W. Complex organization of CTF/NF-I, C/EBP, and HNF3 binding sites within the promoter of the liver-specific vitellogenin gene. J Biol Chem. 1994;269(52):32947–56. [PubMed] [Google Scholar]
- 32.Kaufmann E, Knochel W. Five years on the wings of fork head. Mech Dev. 1996;57(1):3–20. doi: 10.1016/0925-4773(96)00539-4. [DOI] [PubMed] [Google Scholar]
- 33.Seoane J, Le HV, Massague J. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 2002;419(6908):729–34. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell KO, El-Deiry WS. Overexpression of c-Myc inhibits p21WAF1/CIP1 expression and induces S-phase entry in 12-O-tetradecanoylphorbol-13-acetate (TPA)-sensitive human cancer cells. Cell Growth Differ. 1999;10(4):223–30. [PubMed] [Google Scholar]
- 35.Claassen GF, Hann SR. A role for transcriptional repression of p21CIP1 by c-Myc in overcoming transforming growth factor beta -induced cell-cycle arrest. Proc Natl Acad Sci U S A. 2000;97(17):9498–503. doi: 10.1073/pnas.150006697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen C, Rowell EA, Thomas RM, Hancock WW, Wells AD. Transcriptional regulation by Foxp3 is associated with direct promoter occupancy and modulation of histone acetylation. J Biol Chem. 2006;281(48):36828–34. doi: 10.1074/jbc.M608848200. [DOI] [PubMed] [Google Scholar]
- 37.Chai F, Evdokiou A, Young GP, Zalewski PD. Involvement of p21(Waf1/Cip1) and its cleavage by DEVD-caspase during apoptosis of colorectal cancer cells induced by butyrate. Carcinogenesis. 2000;21(1):7–14. doi: 10.1093/carcin/21.1.7. [DOI] [PubMed] [Google Scholar]
- 38.Samanta A, Li B, Song X, Bembas K, Zhang G, Katsumata M, Saouaf SJ, Wang Q, Hancock WW, Shen Y, Greene MI. TGF-beta and IL-6 signals modulate chromatin binding and promoter occupancy by acetylated FOXP3. Proc Natl Acad Sci U S A. 2008;105(37):14023–7. doi: 10.1073/pnas.0806726105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.