Abstract
RAD51, a eukaryotic recombinase, catalyzes homologous-pairing and strand-exchange reactions, which are essential steps in homologous recombination and recombinational repair of double strand breaks. On the other hand, human PSF was originally identified as a component of spliceosomes, and its multiple functions in RNA processing, transcription and DNA recombination were subsequently revealed. In the present study, we found that PSF directly interacted with RAD51. PSF significantly enhanced RAD51-mediated homologous pairing and strand exchange at low RAD51 concentrations; however, in contrast, it inhibited these RAD51-mediated recombination reactions at the optimal RAD51 concentration. Deletion analyses revealed that the N-terminal region of PSF possessed the RAD51- and DNA-binding activities, but the central region containing the RNA-recognition motifs bound neither RAD51 nor DNA. These results suggest that PSF may have dual functions in homologous recombination and RNA processing through its N-terminal and central regions, respectively.
INTRODUCTION
Homologous recombination functions in accurate chromosome segregation during meiotic cell division I (1,2). Defects in meiotic homologous recombination in mice cause infertility, indicating its required function in meiosis (3–5). Homologous recombination also has essential functions in the repair of DNA double strand breaks (DSBs) and the rescue of stalled replication forks during mitosis (6–8). The homologous-recombination defects in mitosis cause instability of the genomic DNA (8,9), leading to chromosomal aberrations and tumorigenesis. Therefore, homologous recombination is essential for both meiotic and mitotic cell divisions.
RAD51, a eukaryotic homologue of bacterial RecA, is a central DNA recombinase, which functions in both meiotic and mitotic homologous recombination (10). During the homologous-recombination process, RAD51 promotes the key recombination reactions, homologous pairing and strand exchange, in an ATP-dependent manner (11–15). To promote homologous pairing, RAD51 first binds to single-stranded DNA (ssDNA), which is produced at the DSB site, and forms helical nucleoprotein filaments. The RAD51–ssDNA filaments then bind to intact double-stranded DNA (dsDNA). The homologous sequences are aligned between ssDNA and dsDNA, and new Watson–Crick base pairs (heteroduplex) are formed between the ssDNA and the complementary strand of the dsDNA within the RAD51–ssDNA–dsDNA filament. This step is called homologous pairing. The heteroduplex region formed by homologous pairing is further expanded by the RAD51-mediated strand exchange reaction. RAD51 requires cofactors for efficient promotion of the homologous-pairing and strand-exchange reactions. In humans, RAD52, RAD54, RAD54B, RAD51B–RAD51C, RAD51AP1 and BRCA2 have been reported as such cofactors for the RAD51-mediated homologous pairing and/or strand exchange reactions in vitro (7–10,16–18).
Polypyrimidine tract-binding protein-associated splicing factor (PSF) was first identified as a component of the spliceosome (19). A tandem repeat of RNA-recognition motifs was found in the central region of PSF, suggesting that PSF functions in RNA processing through its RNA-binding activity. Subsequently, the DNA-binding activity of PSF and its regulatory function for transcription were reported (20–24). PSF was also identified in a complex form with its paralog, p54(nrb). The PSF–p54(nrb) complex reportedly binds to mRNAs, and functions in pre-mRNA processing (25,26). A biochemical study revealed that the PSF–p54(nrb) complex also stimulates the DNA end joining reaction by Ku70/80, DNA ligase IV and XRCC4 (27). These facts suggest that PSF may be a multifunctional protein, which functions in RNA processing, transcription regulation and DNA recombination.
Interestingly, PSF alone reportedly promotes homologous pairing between ssDNA and dsDNA in vitro (28), suggesting that PSF may also be involved in homologous recombination. To evaluate the functions of PSF in homologous recombination, in the present study, we tested whether PSF affects the recombination reactions promoted by RAD51, which is an essential recombinase in both meiotic and mitotic homologous recombination. We found that purified human PSF directly binds to RAD51. Biochemical analyses revealed that PSF synergistically stimulates RAD51-mediated homologous pairing and strand exchange under low RAD51 conditions. In contrast, the RAD51-mediated recombination reactions were significantly inhibited by PSF, when the RAD51 concentrations were optimal for the homologous-pairing and strand-exchange reactions without PSF. Domain mapping analyses showed that the N-terminal region (amino acid residues 1–266) of PSF and the central ATPase domain (amino acid residues 82–339) of RAD51 are involved in the PSF–RAD51 binding. The PSF central region (amino acid residues 267–468), containing a tandem repeat of RNA-recognition motifs, did not bind to DNA, while in contrast, the N-terminal 1–266 fragment of PSF bound to ssDNA and dsDNA. Therefore, the N-terminal region of PSF may be the functional domain for binding RAD51 and regulating the RAD51-mediated recombination reactions.
MATERIALS AND METHODS
Protein purification
The human PSF gene was isolated from a human cDNA pool by polymerase chain reaction, and was cloned into the pET-15b vector (Novagen, Darmstadt, Germany). In this construct, the His6 tag sequence was fused at the N-terminal end of the gene. The DNA fragments encoding PSF(1–266) and PSF(267–468) were cloned into the pET-15b vector. The His6-tagged PSF protein, the His6-tagged PSF(1–266) mutant and the His6-tagged PSF(267–468) mutant were each expressed in the Escherichia coli BL21(DE3) strain, which also carried an expression vector for the minor tRNAs (Codon(+)RP, Stratagene, La Jolla, CA, USA). The cells producing the proteins were resuspended in buffer A [50 mM Tris-HCl (pH 8.0), 500 mM NaCl, 2 mM 2-mercaptoethanol and 10% glycerol], and were disrupted by sonication. The cell debris was removed by centrifugation for 20 min at 27 700×g, and the supernatant was mixed gently with 3 ml of Ni–NTA agarose beads (Qiagen, Hilden, Germany) at 4°C for 1 h. The protein-bound beads were then packed into an Econo-column (Bio-Rad Laboratories, Hercules, CA, USA), and were washed with 120 ml of buffer A, containing 20 mM imidazole. The proteins were eluted by a 60 ml linear gradient of imidazole from 20 to 300 mM. The peak fractions were collected, and 2 units of thrombin protease (GE Healthcare Biosciences, Uppsala, Sweden) per mg of protein were added to remove the His6 tag. The samples were immediately dialyzed against buffer containing 20 mM Tris-HCl (pH 8.0), 200 mM NaCl, 0.25 mM EDTA, 2 mM 2-mercaptoethanol and 10% glycerol. After removal of the His6 tag, PSF and PSF(267–468) were loaded onto a 3 ml hydroxyapatite column (Bio-Rad Laboratories, Hercules, CA, USA), which was immediately eluted with 40 ml of buffer containing 20 mM sodium phosphate (pH 8.0), 200 mM NaCl, 0.25 mM EDTA, 2 mM 2-mercaptoethanol and 10% glycerol. On the other hand, PSF(1–266) was loaded onto a 3 ml hydroxyapatite column, and the flow through fractions were collected. PSF was further purified by chromatography on a 1.5 ml SP-Sepharose column (GE Healthcare Biosciences, Uppsala, Sweden). The column was washed with 15 ml of buffer B [20 mM Tris-HCl (pH 8.5), 0.25 mM EDTA, 2 mM 2-mercaptoethanol and 10% glycerol] containing 250 mM KCl, and PSF was eluted with a 30 ml linear gradient of KCl from 250 to 800 mM. PSF(1–266) was dialyzed against buffer B containing 100 mM KCl, and was further purified by chromatography on a MonoS column (GE Healthcare Biosciences, Uppsala, Sweden). The column was washed with 10 ml of buffer B containing 100 mM KCl and PSF(1–266) was eluted with 10 ml of buffer B containing 800 mM KCl. PSF(267–468) was dialyzed against buffer B, and was further purified by chromatography on a MonoQ column (GE Healthcare Biosciences, Uppsala, Sweden). The column was washed with 10 ml of buffer B, and PSF(267–468) was eluted with a 10 ml linear gradient of KCl from 0 to 800 mM. PSF, PSF(1–266) and PSF(267–468) were dialyzed against buffer containing 20 mM Tris-HCl (pH 8.0), 200 mM NaCl, 0.1 mM EDTA, 1 mM DTT and 10% glycerol, and were stored at −80°C.
Human RAD51 and human RPA were expressed in E. coli cells (29,30), and were purified as described previously (30–32). The RAD51 used in the present study was the K313-type isoform, which is highly conserved among eukaryotes (33).
The DNA fragments encoding the N-terminal domain (1–114 amino acid residues) and the ATPase domain (82–339 amino acid residues) of RAD51 were cloned into the pET-15b vector (Novagen, Darmstadt, Germany). The His6-tagged N-terminal and ATPase domains of RAD51 were each expressed in the E. coli JM109(DE3) strain, which also carried an expression vector for the minor tRNAs (Codon(+)RIL, Stratagene, La Jolla, CA, USA). The cells producing the proteins were resuspended in buffer A, containing 5 mM imidazole, and were disrupted by sonication. The cell debris was removed by centrifugation for 20 min at 27 700×g, and the supernatant was mixed gently with 1.5 ml of Ni–NTA agarose beads (Qiagen, Hilden, Germany) at 4°C for 1 h. The protein-bound beads were then packed into an Econo-column (Bio-Rad Laboratories, Hercules, CA, USA). The RAD51 N-terminal domain-bound beads were washed with 120 ml of buffer A containing 5 mM imidazole, and the RAD51 ATPase domain-bound beads were washed with 120 ml of buffer A containing 30 mM imidazole. The RAD51 N-terminal domain was eluted by an 80 ml linear gradient of imidazole from 5 to 500 mM. The RAD51 ATPase domain was eluted by an 80 ml linear gradient of imidazole from 30 to 500 mM. The RAD51 N-terminal and ATPase domains were stored at 4°C, and were dialyzed before use against 20 mM sodium phosphate buffer (pH 8.0), containing 300 mM NaCl, 0.1 mM EDTA, 2 mM 2-mercaptoethanol and 10% glycerol.
DNA and RNA substrates
In the D-loop formation assay, superhelical dsDNA (pB5Sarray DNA) was prepared by a method avoiding alkaline treatment of the cells harboring the plasmid DNA, to prevent the dsDNA substrates from undergoing irreversible denaturation. Instead, the cells were gently lysed using sarkosyl, as described previously (31). The pB5Sarray DNA contained 11 repeats of a sea urchin 5S rRNA gene (207-bp fragment) within the pBlueScript II SK(+) vector. For the ssDNA substrate used in the D-loop assay, the following high-performance liquid chromatography (HPLC)-purified oligonucleotide was purchased from Nihon Gene Research Laboratory: 50-mer, 5′-GGA ATT CGG TAT TCC CAG GCG GTC TCC CAT CCA AGT ACT AAC CGA GCC CT-3′. The 5′ ends of the oligonucleotide were labeled with T4 polynucleotide kinase (New England Biolabs, Ipswich, MA, USA) in the presence of [γ-32P]ATP at 37°C for 30 min. Single-stranded ϕX174 viral (+) strand DNA and double-stranded ϕX174 replicative form I DNA were purchased from New England Biolabs. The linear dsDNA was prepared from the ϕX174 replicative form I DNA by PstI digestion. For the strand exchange assay with olignucleotides, the following HPLC-purified DNA and RNA oligonucleotides were purchased from Nihon Gene Research Laboratory and Gene Design: 63-mer 5′-TCC TTT TGA TAA GAG GTC ATT TTT GCGGAT GGC TTA GAG CTT AAT TGC TGA ATC TGG TGCTGT-3′,
32-mer top strand, 5′-CCA TCC GCA AAA ATG ACC TCT TAT CAA AAG GA-3′,
32-mer bottom strand, 5′-TCC TTT TGA TAA GAG GTC ATT TTT GCG GAT GG-3′,
32-mer RNA, 5′-UCC UUU UGA UAA GAG GUC AUU UUU GCG GAU GG-3′. The 5′ ends of the oligonucleotide (32-mer top strand) were labeled with T4 polynucleotide kinase in the presence of [γ-32P]ATP.
For the pull-down assay with the ssDNA beads, the following HPLC-purified 5′ biotinylated DNA oligonucleotide was purchased from Nihon Gene Research Laboratory: 80-mer 5′-CTG CTT TAT CAA GAT AAT TTT TCG ACT CAT CAG AAA TAT CCG AAA GTG TTA ACT TCT GCG TCA TGG AAG CGA TAA AAC TC-3′. DNA concentrations are expressed in moles of nucleotides.
The pull-down assay with PSF-conjugated beads
PSF (40 µg) was covalently conjugated to Affi-Gel 10 beads (75 µl, Bio-Rad Laboratories, Hercules, CA, USA), according to the manufacturer's instructions, and the Affi-Gel 10-protein matrices were adjusted to 50% slurries. The His6-tagged RAD51, the His6-tagged RAD51 N-terminal domain, or the His6-tagged RAD51 ATPase domain (4 µg) was incubated with PSF-conjugated Affi-Gel 10 beads (20 µl, 50% slurry) in 100 µl of binding buffer, containing 20 mM sodium phosphate (pH 8.0), 60 mM NaCl, 0.1 mM EDTA, 2 mM 2-mercaptoethanol, 0.05% Triton X-100 and 10% glycerol. After an incubation at room temperature for 2.5 h, the PSF beads were washed two times with 100 µl of wash buffer, containing 20 mM sodium phosphate (pH 8.0), 60 mM NaCl, 0.1 mM EDTA, 2 mM 2-mercaptoethanol and 10% glycerol. The proteins bound to the PSF beads were fractionated by 15% SDS-polyacrylamide gel electrophoresis (PAGE), and the bands were visualized by Coomassie Brilliant Blue staining.
The pull-down assay with Ni–NTA beads
Purified His6-tagged PSF, His6-tagged PSF(1–266), or His6-tagged PSF(267–468) (3.8 µg) was mixed with RAD51 (7.4 µg) in 100 µl of binding buffer, containing 15 mM sodium phosphate (pH 7.5), 95 mM NaCl, 5 mM imidazole, 0.21 mM EDTA, 2 mM 2-mercaptoethanol, 0.025% Triton X-100 and 9% glycerol, and Ni–NTA agarose beads (3 µl, 50% slurry) were then added. Purified His6-tagged PSF was also tested with RPA (10 µg) in 100 µl of binding buffer, containing 15 mM sodium phosphate (pH 7.5), 80 mM NaCl, 5 mM KCl, 5 mM imidazole, 0.2 mM EDTA, 2 mM 2-mercaptoethanol, 0.025% Triton X-100 and 9% glycerol, and then Ni–NTA agarose beads (3 µl, 50% slurry) were added. After an incubation at room temperature for 1 h, the beads were washed with 1 ml of wash buffer, containing 20 mM sodium phosphate (pH 7.5), 100 mM NaCl, 0.25 mM EDTA, 2 mM 2-mercaptoethanol and 10% glycerol. The proteins bound to the beads and those in the supernatant were fractionated by 12% SDS–PAGE, and the bands were visualized by Coomassie Brilliant Blue staining.
The pull-down assay with the ssDNA beads
PSF (1 µM) and RAD51 (5 µM) were incubated with magnetic streptavidin beads conjugated with the biotinylated ssDNA 80-mer (16 µM) at 37°C for 20 min, in 10 µl of reaction buffer, containing 28 mM Tris-HCl (pH 8.0), 4 mM HEPES-NaOH (pH 7.5), 110 mM NaCl, 0.06 mM EDTA, 0.4 mM 2-mercaptoethanol, 6% glycerol, 1 mM MgCl2, 1.4 mM DTT and 1 mM ATP. After an incubation, the beads were washed with 10 µl of reaction buffer. The proteins bound to the beads were fractionated by 12% SDS–PAGE, and the bands were visualized by Coomassie Brilliant Blue staining.
The D-loop formation assay
PSF and RAD51 were incubated with the 32P-labeled 50-mer oligonucleotide (1 µM) at 37°C for 10 min, in 7 µl of reaction buffer, containing 24 mM Tris-HCl (pH 8.0), 2.0 mM HEPES-NaOH (pH 7.5), 55 mM NaCl, 0.03 mM EDTA, 0.2 mM 2-mercaptoethanol, 3% glycerol, 1 mM MgCl2, 1.2 mM DTT, 2 mM AMPPNP and 0.1 mg/ml BSA. The reactions were then initiated by the addition of the pB5Sarray superhelical dsDNA (30 µM) along with 9 mM MgCl2, and were continued at 37°C for 30 min. The reactions were stopped by the addition of 0.2% SDS and 1.5 mg/ml proteinase K (Roche Applied Science, Basel, Switzerland), and were further incubated at 37°C for 15 min. After adding six-fold loading dye, the deproteinized reaction products were separated by 1% agarose gel electrophoresis in 1×TAE buffer at 3.3 V/cm for 2.5 hr. The gels were dried, exposed to an imaging plate, and visualized using an FLA-7000 imaging analyzer (Fujifilm, Tokyo, Japan).
The strand-exchange assay
The ϕX174 circular ssDNA (20 µM), RAD51 (0.5–6.6 µM) and RPA (1.3 µM) were incubated with or without PSF (1.0 µM) at 37°C for 10 min, in the reaction buffer, containing 30 mM Tris-HCl (pH 8.0), 95 mM NaCl, 5 mM KCl, 0.05 mM EDTA, 0.2 mM 2-mercaptoethanol, 6% glycerol, 1 mM MgCl2, 1 mM CaCl2, 1.5 mM DTT, 1 mM ATP, 0.1 mg/ml BSA, 20 mM creatine phosphate and 75 µg/ml creatine kinase. The reactions were then initiated by the addition of ϕX174 linear dsDNA (20 µM), and were continued at 37°C. At the indicated times, a 10 µl aliquot of the reaction mixture was mixed with 0.2% SDS and 1.5 mg/ml proteinase K to stop the reaction, and was further incubated at 37°C for 15 min. After adding six-fold loading dye, the deproteinized reaction products were separated by 1% agarose gel electrophoresis in 1×TAE buffer at 3.3 V/cm for 4 h. The products were visualized by SYBR Gold (Invitrogen, Carlsbad, CA, USA) staining.
The strand-exchange assay with oligonucleotides
RAD51 (3 µM) and the indicated amount of PSF were incubated with a 63-mer ssDNA (15 µM) at 37°C for 10 min, in 10 µl of reaction buffer, containing 28 mM Tris-HCl (pH 8.0), 4 mM HEPES-NaOH (pH 7.5), 110 mM NaCl, 0.06 mM EDTA, 0.4 mM 2-mercaptoethanol, 6% glycerol, 1 mM MgCl2, 1 mM CaCl2, 1.4 mM DTT, 1 mM ATP, 0.1 mg/ml BSA, 20 mM creatine phosphate and 75 µg/ml creatine kinase. The reactions were initiated by the addition of the 32-mer dsDNA or RNA/DNA hybrid (1.5 µM), which shared sequence homology with the 63-mer ssDNA. The 32-mer dsDNA and the RNA/DNA hybrid used in this assay were purified by polyacrylamide gel electrophoresis. After a 30 min incubation at 37°C, the reaction was stopped by the addition of 0.2% SDS and 1.5 mg/ml proteinase K, and was further incubated at 37°C for 15 min. After six-fold loading dye was added, the reaction mixtures were subjected to 15% polyacrylamide gel electrophoresis in 0.5×TBE buffer (45 mM Tris, 45 mM boric acid and 1 mM EDTA) at 10 V/cm for 4 h. The gels were dried, exposed to an imaging plate, and visualized using an FLA-7000 imaging analyzer.
The DNA-binding assay
The ϕX174 circular ssDNA (20 µM) or the linearized ϕX174 dsDNA (20 µM) was mixed with the indicated amount of PSF, PSF(1–266) or PSF(267–468) in 10 µl of a standard reaction solution, containing 30 mM Tris–HCl (pH 8.0), 1.5 mM DTT, 100 mM NaCl, 1 mM MgCl2, 5% glycerol and 0.1 mg/ml BSA. The reaction mixtures were incubated at 37°C for 10 min, and were then separated by 0.8% agarose gel electrophoresis in 1×TAE buffer (40 mM Tris-acetate and 1 mM EDTA) at 3.3 V/cm for 2.5 h. The bands were visualized by ethidium bromide staining.
MALDI-TOF-MS analysis
A 2 µl aliquot of 5% trifluoroacetic acid (TFA) was added to 18 µl of a 0.4 mg/ml PSF solution, or 1 µl of 5% TFA was added to 9 µl of a 1.0 mg/ml PSF(1–266) or 1.3 mg/ml PSF(267–468) sample, and the mixture was bound to a ZipTip (C4) pipette tip (Millipore, Billerica, MA, USA). After the protein was bound, the tip was first rinsed with 0.1% TFA and 5% methanol, and then rinsed with 0.1% TFA. PSF, PSF(1–266), or PSF(267–468) was eluted with 0.1% TFA and 80% acetonitrile, and was deposited onto the sample plate, which was precoated with a dried layer of sinapic acid (Sigma-Aldrich, St Louis, MO, USA). A MALDI-TOF mass spectrometry analysis was performed with an AXIMA-CFR mass spectrometer (Kratos Analytical, Manchester, UK). Bovine carbonic anhydrase II and BSA were used as standard proteins for external calibration. Each single spectrum was obtained as an accumulation of 50 laser shots.
RESULTS
Human PSF binds to RAD51
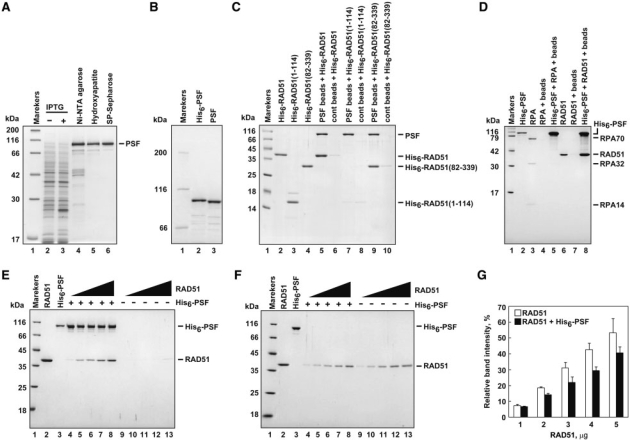
To test whether PSF binds to RAD51, we performed pull-down assays with the PSF-conjugated beads. To prepare the PSF beads, human PSF was expressed in E. coli cells as a His6-tagged protein, and was purified by chromatography on a Ni-NTA agarose column (Figure 1A, lane 4). The His6 tag was removed by thrombin protease treatment (Figure 1B), and the protein was further purified by chromatography on a hydroxyapatite and SP-Sepharose columns (Figure 1A, lanes 5 and 6). The purified PSF was conjugated to Affi-Gel 10 beads, and RAD51 binding to the PSF beads was tested by the pull-down assay. As shown in Figure 1C (lane 5), substantial amounts of RAD51 were associated with the PSF beads, indicating that PSF directly binds to RAD51.
Figure 1.
Human PSF binds to RAD51. (A) Purification of human PSF. Proteins from each purification step were analyzed by 12% SDS–PAGE with Coomassie Brilliant Blue staining. Lane 1 indicates the molecular mass markers. Lanes 2 and 3 are the whole cell lysates before and after induction with isopropyl-1-thio-β-d-galactopyranoside, respectively. Lanes 4–6 indicate the samples from the peak Ni–NTA agarose fraction, the peak hydroxyapatite fraction, and the peak SP-Sepharose fraction. (B) A 6% SDS–PAGE image of the Ni–NTA agarose fraction before (lane 2) and after (lane 3) the removal of the hexahistidine tag. (C) Pull-down assay with the PSF-conjugated beads. Proteins bound to the PSF-conjugated beads were analyzed by 15% SDS–PAGE with Coomassie Brilliant Blue staining. Lane 1 indicates the molecular mass markers. Lanes 2–4 represent His6-tagged RAD51, His6-tagged RAD51(1–114), and His6-tagged RAD51(82–339), respectively. Lanes 5 and 6 indicate experiments with RAD51 with the PSF-conjugated beads and the Affi-Gel 10 beads, respectively. Lanes 7 and 8 indicate experiments with His6-tagged RAD51(1–114) with the PSF-conjugated beads and the Affi-Gel 10 beads, respectively. Lanes 9 and 10 indicate experiments with His6-tagged RAD51(82–339) with the PSF-conjugated beads and the Affi-Gel 10 beads, respectively. (D) Pull-down assay with the Ni–NTA agarose beads. Proteins bound to His6-tagged PSF were captured by the Ni–NTA agarose beads, and were analyzed by 15% SDS–PAGE with Coomassie Brilliant Blue staining. In the Ni–NTA pull-down assay, RPA and RAD51 did not contain His6-tags. Lane 1 indicates the molecular mass markers. Lanes 2, 3 and 6 represent His6-tagged PSF, RPA and RAD51, respectively. Lanes 4 and 5 indicate experiments with RPA in the absence and presence of His6-tagged PSF, respectively. Lanes 7 and 8 indicate experiments with RAD51 in the absence and presence of His6-tagged PSF, respectively. (E) The RAD51-titration experiments. Proteins bound to His6-tagged PSF were captured by the Ni–NTA agarose beads, and were analyzed by 15% SDS–PAGE with Coomassie Brilliant Blue staining. Lane 1 indicates the molecular mass markers. Lanes 2 and 3 represent RAD51 and His6-tagged PSF, respectively. Lanes 4–8 and 9–13 indicate experiments in the presence and absence of His6-tagged PSF, respectively. RAD51 were 1 µg (lanes 4 and 9), 2 µg (lanes 5 and 10), 3 µg (lanes 6 and 11), 4 µg (lanes 7 and 12), and 5 µg (lanes 8 and 13). (F) Supernatant fractions of the experiments shown in panel E were analyzed by 15% SDS–PAGE with Coomassie Brilliant Blue staining. Lane numbers correspond to those in panel E. (G) Graphic representation of the experiments shown in panel F. Average relative intensities of the RAD51 bands from three independent experiments are represented with the SD values.
Structural studies revealed that RAD51 is composed of two distinct domains, the N-terminal and ATPase domains (34–38). To identify the PSF-binding domain of RAD51, we prepared two RAD51 fragments, amino acid residues 1–114 and 82–339, which contained the N-terminal and ATPase domains, respectively (Figure 1C, lanes 3 and 4). The pull-down assay revealed that the ATPase domain of RAD51 bound to PSF with the almost same efficiency as the full-length RAD51 (Figure 1C, lane 9). In contrast, the RAD51 N-terminal fragment containing amino acid residues 1–114 did not show significant binding to PSF (Figure 1C, lane 7). These results indicate that PSF directly binds to the ATPase domain of RAD51.
The PSF–RAD51 binding was also detected by the Ni-bead pull-down assay with His6-tagged PSF (Figure 1D, lane 8). In contrast, PSF did not bind to RPA (Figure 1D, lane 5), suggesting that the PSF–RAD51 interaction is specific. The Ni-bead pull-down assay, in which all of the proteins could be visualized on the SDS–PAGE gel, allowed us to estimate the binding stoichiometry between PSF and RAD51. As shown in Figure 1E, His6-tagged PSF copelleted RAD51 in a RAD51 concentration-dependent manner. Consistently, the amount of free RAD51 in the supernatant was also reduced in the presence of His6-tagged PSF (Figure 1F and G). The PSF–RAD51 binding stoichiometry was about 1:1, in the presence of an excess of RAD51 (Figure 1E, lane 8).
PSF modulates homologous pairing by RAD51
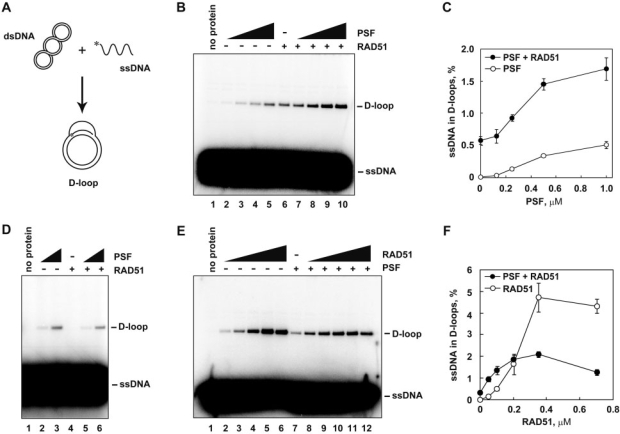
We next tested the effect of PSF in the RAD51-mediated homologous pairing. PSF reportedly promotes the formation of D-loops (28), which are the products of the homologous pairing reaction between ssDNA fragments and superhelical dsDNA. Therefore, we performed the D-loop formation assay (Figure 2A). The superhelical dsDNA used in this assay was prepared by a method without alkali treatment, to avoid denaturation of the double helix of the dsDNA (31). Consistent with the previous observations, PSF or RAD51 alone promoted homologous pairing (Figure 2B, lanes 2–6, and 2C) (28). When PSF and a low amount of RAD51 (0.1 µM) were present together, the amount of D-loops increased, in a PSF-concentration-dependent manner (Figure 2B, lanes 7–10, and Figure 2C). This stimulation of D-loop formation was not observed when the reactions were conducted in the absence of an ATP analog, AMPPNP, which was required for the RAD51-mediated D-loop formation, but not for the PSF-mediated D-loop formation (Figure 2D). Therefore, PSF may function as an activator for RAD51 in homologous pairing under low RAD51 conditions.
Figure 2.
Human PSF stimulates the RAD51-mediated homologous pairing. (A) A schematic diagram of the D-loop formation assay. Asterisks indicate the 32P-labeled end of the 50-mer ssDNA. (B) The D-loop formation assay with AMPPNP. The reactions were conducted without RAD51 (lanes 1–5) or with 0.1 µM RAD51 (lanes 6–10) in the presence of increasing amounts of PSF. The PSF concentrations were 0 µM (lanes 1 and 6), 0.125 µM (lanes 2 and 7), 0.25 µM (lanes 3 and 8), 0.5 µM (lanes 4 and 9), and 1.0 µM (lanes 5 and 10). (C) Graphic representation of the experiments shown in panel B. The average values of three independent experiments are shown with the SD values. Closed and open circles represent the experiments with and without RAD51, respectively. (D) The D-loop formation assay without AMPPNP. The reactions were conducted without RAD51 (lanes 1–3) or with 0.1 µM RAD51 (lanes 4–6) in the presence of increasing amounts of PSF. The PSF concentrations were 0 µM (lanes 1 and 4), 0.25 µM (lanes 2 and 5) and 0.5 µM (lanes 3 and 6). (E) The RAD51-titration experiments of the D-loop formation assay with AMPPNP. Lane 1 indicates a negative control experiment without RAD51 and PSF, and lane 7 indicates a control experiment without RAD51 in the presence of PSF. Lanes 2–6 and 7–12 indicate experiments in the absence and presence of PSF, respectively. The reactions were conducted with 0.5 µM PSF (lanes 7–12). The RAD51 concentrations were 0.05 µM (lanes 2 and 8), 0.1 µM (lanes 3 and 9), 0.2 µM (lanes 4 and 10), 0.35 µM (lanes 5 and 11) and 0.7 µM (lanes 6 and 12). (F) Graphic representation of the experiments shown in panel E. The average values of three independent experiments are shown with the SD values. Closed and open circles represent the experiments with and without PSF, respectively.
In contrast to the PSF-mediated homologous-pairing activation in the presence of 0.1 µM RAD51, PSF significantly inhibited the RAD51-mediated homologous pairing, when the reaction was conducted in the presence of RAD51 concentrations above 0.35 µM, which are optimal for the reaction without PSF. As shown in Figure 2E and F, PSF stimulated the RAD51-mediated homologous pairing in the presence of 0.05 and 0.1 µM RAD51 (compare lane 2 with lane 8, and lane 3 with lane 9). In contrast, PSF inhibited the homologous pairing in the presence of 0.35 and 0.7 µM RAD51 (Figure 2E, compare lane 5 with lane 11, and lane 6 with lane 12, and Figure 2F). These results indicated that PSF stimulates homologous pairing in the presence of a suboptimal concentration of RAD51, but inhibits the reaction in the presence of an optimal concentration of RAD51.
PSF modulates RAD51-mediated strand exchange
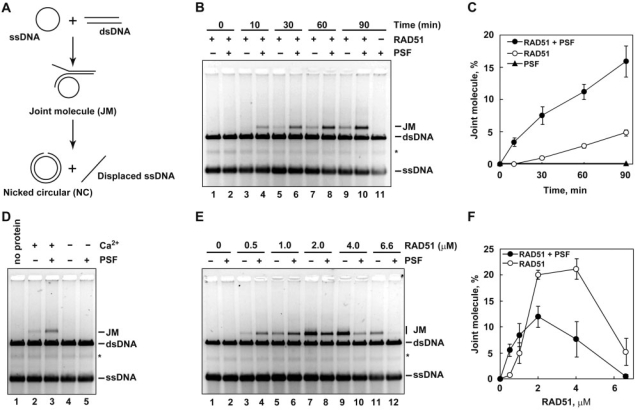
We next tested whether PSF affects strand exchange by RAD51 (Figure 3A). To evaluate the activator function of PSF in the RAD51-mediated strand exchange, we first performed the strand-exchange assay with a suboptimal concentration of RAD51 (0.5 µM). Under these reaction conditions, a small amount of joint molecule (JM) product was detected in the absence of PSF in the 30, 60 and 90 min reactions (Figure 3B, lanes 5, 7 and 9, and Figure 3C). In contrast to the D-loop formation, PSF itself did not promote strand exchange (Figure 3B, lane 11, and Figure 3C). When PSF was added to the reaction mixture, the amount of the JM products formed by the RAD51-mediated strand exchange significantly increased (Figure 3B, lanes 4, 6, 8 and 10, and Figure 3C). These results indicated that PSF stimulates the RAD51 strand-exchange activity, as well as the homologous-pairing stimulating activity, in the presence of a suboptimal concentration of RAD51. This RAD51 activation by PSF was not observed when the strand-exchange reactions were performed without Ca2+, which stabilizes the RAD51–ssDNA filament by inhibiting ATP hydrolysis (39) (Figure 3D).
Figure 3.
Human PSF stimulates the RAD51-mediated strand exchange. (A) A schematic representation of the strand-exchange reaction. The joint molecule product is denoted as JM. (B) The ϕX174 circular ssDNA (20 µM), RAD51 (0.5 µM) and RPA (1.3 µM) were incubated with or without PSF (1.0 µM) at 37°C for 10 min. After this incubation, the reactions were then initiated by the addition of ϕX174 linear dsDNA (20 µM), and were continued at 37°C for the indicated times. The deproteinized products were separated by 1% agarose gel electrophoresis, and were visualized by SYBR Gold staining. The asterisk indicates the self-annealing products of the ssDNA. Lanes 1, 3, 5, 7 and 9 indicate control experiments without PSF. Lanes 2, 4, 6, 8 and 10 indicate experiments with PSF. Lane 11 indicates a negative control experiment with PSF in the absence of RAD51. (C) Graphic representation of the strand-exchange experiments shown in panel B. The band intensities of the JM products were quantified, and the average values of three independent experiments are shown with the SD values. Closed and open circles represent the experiments with and without PSF, respectively. Closed triangles represent the experiments with PSF in the absence of RAD51. (D) Ca2+ requirement. The strand-exchange reactions were conducted with or without Ca2+, and were performed according to the same procedure as shown in panel B. The reaction time was 60 min. Lane 1 indicates a negative control experiment without proteins. Lanes 2 and 3 indicate experiments without and with PSF, respectively, in the presence of CaCl2. Lanes 4 and 5 indicate experiments without and with PSF, respectively, in the absence of CaCl2. (E) The RAD51-titration experiments. The strand-exchange reactions were conducted with Ca2+, and were performed according to the same procedure as shown in panel B. The reaction time was 60 min. Lanes 2, 4, 6, 8, 10 and 12 indicate experiments with 1 µM PSF, and lanes 1, 3, 5, 7, 9 and 11 indicate experiments without PSF. The RAD51 concentrations were 0 µM (lanes 1 and 2), 0.5 µM (lanes 3 and 4), 1 µM (lanes 5 and 6), 2 µM (lanes 7 and 8), 4 µM (lanes 9 and 10) and 6.6 µM (lanes 11 and 12). (F) Graphic representation of the strand-exchange experiments shown in panel E. The band intensities of the JM products were quantified, and the average values of three independent experiments are shown with the SD values. Closed and open circles represent the experiments with and without PSF, respectively.
Under the reaction conditions with Ca2+, a 2–4 µM RAD51 concentration was optimal for strand exchange without PSF (Figure 3E, lanes 7 and 9 and Figure 3F). Consistent with the results from the D-loop formation assay, PSF inhibited the RAD51-mediated strand exchange in the presence of an optimal RAD51 concentration, although it stimulated the reactions under low RAD51 conditions (0.5 and 1.0 µM RAD51) (Figure 3E and F). Therefore, PSF functions as activator or inhibitor of the homologous-pairing and strand-exchange activities of RAD51, in a RAD51 concentration-dependent manner.
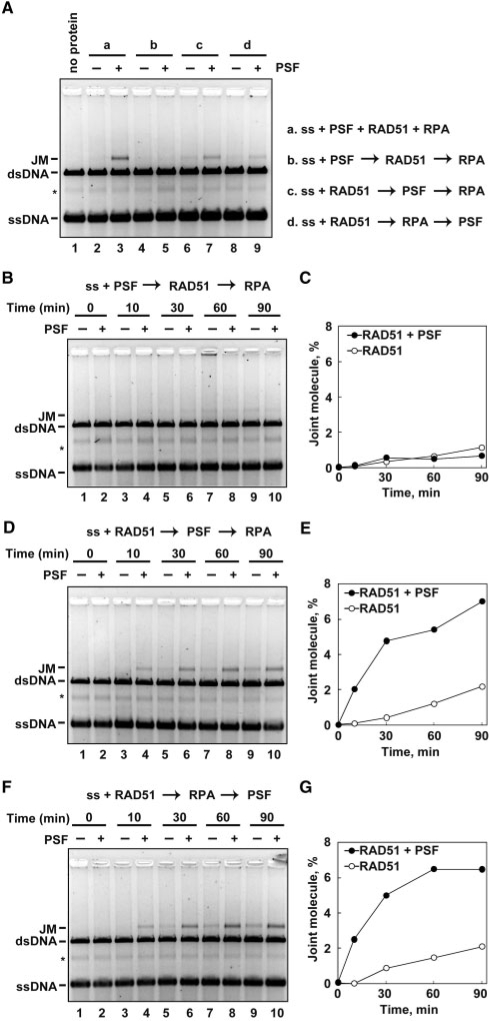
We then tested whether the order of the PSF addition affected the strand-exchange stimulation in the presence of the low RAD51 concentration (0.5 µM). As shown in Figure 3B and C, when PSF, RAD51, RPA and ssDNA were co-incubated together before initiating the reaction by the addition of dsDNA, the strand-exchange reaction was significantly enhanced (Figure 4A, lanes 2 and 3). In contrast, PSF did not enhance strand exchange, when it was incubated with ssDNA before RAD51 addition (Figure 4A, lanes 4 and 5, Figure 4B and C). Enhancement of strand exchange was also observed when PSF and RPA were added after the RAD51–ssDNA incubation (Figure 4A, lanes 6–9, and Figure 4D–G). These results suggested that, under low RAD51 conditions, PSF stimulates strand exchange by RAD51, when it was added after the RAD51–ssDNA filament formation. Since PSF inhibited strand exchange in the presence of an optimal amount of RAD51 (high RAD51 conditions), where RAD51 covered most of the ssDNA, PSF may bind to the RAD51-free ssDNA region under low RAD51 conditions and function as a RAD51 mediator.
Figure 4.
Effects of the PSF reaction order on RAD51-mediated strand exchange. The ϕX174 circular ssDNA (20 µM), RAD51 (0.5 µM), RPA (1.3 µM) and PSF (1.0 µM) were incubated at 37°C in various combinations. After this incubation, the reactions were then initiated by the addition of ϕX174 linear dsDNA (20 µM), and were continued at 37°C for the indicated times. The deproteinized products were separated by 1% agarose gel electrophoresis, and were visualized by SYBR Gold staining. (A) The proteins and ssDNA were incubated in the combinations represented on the right side of panel A. Lane 1 indicates a negative control experiment without proteins. Lanes 2, 4, 6 and 8 indicate control experiments without PSF, and lanes 3, 5, 7 and 9 indicate experiments with PSF. The reaction time was 60 min. (B) The ϕX174 circular ssDNA was incubated with PSF at 37°C for 10 min. After this incubation, RAD51 was added to the reaction mixture, which was incubated at 37°C for 5 min. RPA was then added, and the reactions were initiated by the addition of ϕX174 linear dsDNA. Reactions were continued for the indicated times. (C) Graphic representation of the strand-exchange experiments shown in panel B. The band intensities of the JM products were quantified. Closed and open circles represent the experiments with and without PSF, respectively. (D) The ϕX174 circular ssDNA was incubated with RAD51 at 37°C for 10 min. After this incubation, PSF was added to the reaction mixture, which was incubated at 37°C for 5 min. RPA was then added, and the reactions were initiated by the addition of ϕX174 linear dsDNA. Reactions were continued for the indicated times. (E) Graphic representation of the strand-exchange experiments shown in panel D. The band intensities of the JM products were quantified. Closed and open circles represent the experiments with and without PSF, respectively. (F) The ϕX174 circular ssDNA was incubated with RAD51 at 37°C for 10 min. After this incubation, RPA was added to the reaction mixture, which was incubated at 37°C for 5 min. PSF was then added, and the reactions were initiated by the addition of ϕX174 linear dsDNA. Reactions were continued for the indicated times. (G) Graphic representation of the strand-exchange experiments shown in panel F. The band intensities of the JM products were quantified. Closed and open circles represent the experiments with and without PSF, respectively.
PSF competes with RAD51 for ssDNA binding
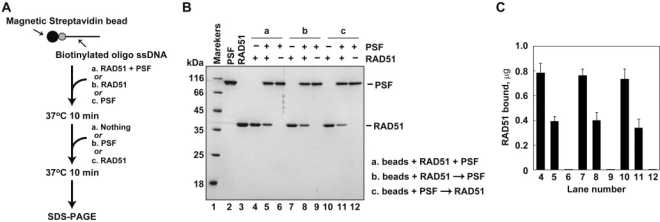
Under the high RAD51 conditions, PSF inhibited homologous pairing and strand exchange by RAD51, suggesting that PSF may compete with RAD51 for ssDNA binding. To test this possibility, we prepared streptavidin beads conjugated with a biotinylated ssDNA 80-mer, and performed a pull-down assay with these ssDNA beads (Figure 5A). As shown in Figure 5B (lanes 4 and 6) and 5C, RAD51 or PSF bound to the ssDNA beads. However, the amount of RAD51 bound to the ssDNA beads was significantly decreased, when both RAD51 and PSF were coincubated with the ssDNA beads (Figure 5B, lane 5, and Figure 5C). This inhibition of the RAD51–ssDNA binding by PSF was also observed when PSF was added to the ssDNA beads before and after RAD51 (Figure 5B, lanes 7–12, and Figure 5C). Therefore, we concluded that the PSF-dependent inhibition of homologous pairing and strand exchange, in the presence of an optimal amount of RAD51, may be due to RAD51 disassembly from the ssDNA by PSF. PSF may not disassemble RAD51 from ssDNA, when a RAD51-free ssDNA region is available for PSF–ssDNA binding, under the low RAD51 conditions.
Figure 5.
PSF competes with RAD51 for ssDNA binding. (A) A schematic representation of the ssDNA bead pull-down assay. (B) RAD51 (5 µM) and PSF (1 µM) were incubated with the ssDNA (16 µM) beads, and the proteins bound to the ssDNA were analyzed by 15% SDS–PAGE with Coomassie Brilliant Blue staining. Lane 1 indicates the molecular mass markers. Lane 5 indicates an experiment in which RAD51, PSF, and the ssDNA beads were simultaneously incubated. Lane 8 indicates an experiment in which RAD51 and the ssDNA beads were incubated before the addition of PSF. Lane 11 indicates an experiment in which PSF and the ssDNA beads were incubated before the addition of RAD51. Lanes 2 and 3 represent PSF and RAD51, respectively. Lanes 4, 7 and 10 indicate control experiments without PSF, and lanes 6, 9 and 12 indicate control experiments without RAD51. (C) Graphic representation of the ssDNA bead pull-down assay shown in panel B. The RAD51 band intensities were quantified, and the average values of three independent experiments are shown with the SD values.
PSF promotes strand exchange between ssDNA and an RNA/DNA hybrid
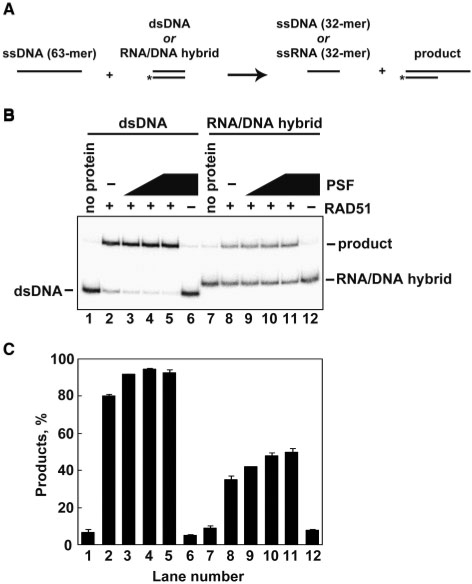
PSF contains RNA-binding motifs, and is known to be involved in transcription (20–24), suggesting that PSF may be involved in a transcription-associated recombination event. Therefore, we tested whether PSF affects strand exchange between ssDNA and an RNA/DNA hybrid, which may be formed when transcription stalls. To do so, we performed a strand exchange assay with short oligonucleotides (Figure 6A). In this assay, the 32-mer dsDNA and RNA/DNA hybrid were purified by polyacrylamide gel electrophoresis. Consistent with the results from the D-loop formation assay and the strand-exchange assay with long DNA substrates, PSF stimulated the RAD51-mediated strand exchange with oligonucleotides under the suboptimal RAD51 conditions. We found that RAD51 promoted strand exchange between ssDNA and an RNA/DNA hybrid (Figure 6B and C). Intriguingly, PSF also stimulated this reaction (Figure 6B and C). Therefore, PSF has the potential to function in transcription-associated recombination.
Figure 6.
The strand-exchange assay between the ssDNA 63-mer and either the dsDNA 32-mer or the RNA/DNA hybrid 32-mer. (A) A schematic representation of the strand-exchange assay with oligonucleotides. Asterisks indicate the 32P-labeled end of the ssDNA 32-mer, which is complementary to the ssDNA 63-mer. The dsDNA and RNA/DNA hybrid 32-mers were prepared by anealing the 32P-labeled ssDNA strand with the complementary ssDNA 32-mer or the complementary RNA 32-mer, and were purified by native polyacrylamide gel electrophoresis. (B) The strand-exchange assay. The ssDNA 63-mer (15 µM) was incubated with RAD51 (3 µM) in the presence of the indicated amounts of PSF. The reactions were initiated by adding dsDNA (lanes 1–6) or the RNA/DNA hybrid (lanes 7–12). The strand-exchange products were analyzed by 15% native polyacrylamide gel electrophoresis. Lanes 1 and 7 indicate negative control experiments without proteins. Lanes 6 and 12 indicate control experiments without RAD51 in the presence of 2 µM PSF. The PSF concentrations were 0 µM (lanes 2 and 8), 0.5 µM (lanes 3 and 9), 1 µM (lanes 4 and 10) and 2 µM (lanes 5 and 11). (C) Graphic representation of the strand-exchange assay shown in panel B. The average values of three independent experiments are shown with the SD values. Lane numbers correspond to those in panel B.
Domain analysis of PSF
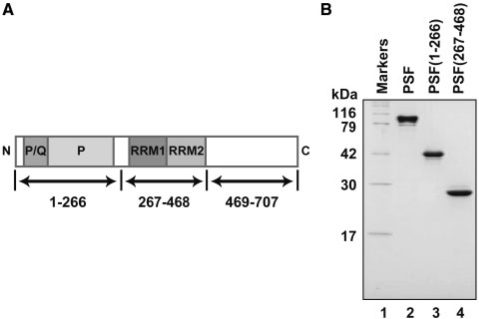
To identify the functional domains of PSF, we constructed three PSF deletion mutants, PSF(1–266), PSF(267–468) and PSF(469–707), which were composed of amino acid residues 1–266, 267–468 and 469–707, respectively (Figure 7A). These PSF deletion mutants were expressed in E. coli cells, and PSF(1–266) and PSF(267–468) were successfully obtained in the soluble fractions. However, PSF(469–707) was only found in the insoluble fraction, suggesting that it did not fold properly. Therefore, we purified PSF(1–266) and PSF(267–468) as recombinant proteins (Figure 7B). The purified PSF deletion mutants did not contain the His6 tag. The predicted molecular masses of PSF, PSF(1–266), and PSF(267–468) are 76, 26 and 23 kDa, respectively. However, PSF, PSF(1–266) and PSF(267–468) migrated on an SDS-denaturing polyacrylamide gel with distances corresponding to about 100, 42 and 25 kDa, respectively (Figure 7B). A mass spectrometric analysis revealed that the molecular masses of purified PSF, PSF(1–266), and PSF(267–468) were 76, 26 and 23 kDa, respectively. Therefore, the abnormal migrations of PSF and PSF(1–266) on the SDS-denaturing polyacrylamide gel may be a consequence of their basic pI values, 9.45 and 12.01, respectively.
Figure 7.
Purification of the PSF deletion mutants. (A) Schematic representation of human PSF. P/Q and P indicate the proline/glutamine-rich and proline-rich domains, respectively. RRM1 and RRM2 indicate the RNA-recognition motifs 1 and 2, respectively. Arrows represent regions corresponding to the deletion mutants. (B) The purified PSF, PSF(1–266), and PSF(267–468) proteins were analyzed by 15% SDS–PAGE with Coomassie Brilliant Blue staining. These proteins did not contain a His6 tag.
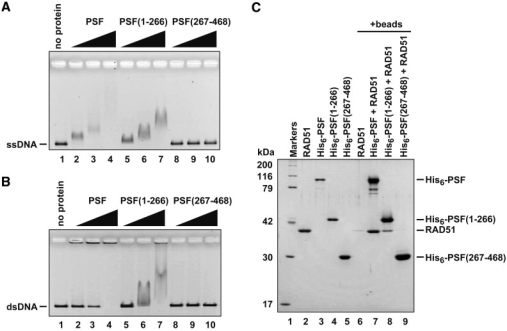
As previously reported by Akhmedov and Lopez (28), PSF bound to ssDNA and dsDNA (Figure 8A and B, lanes 2–4). PSF(1–266) also bound to both ssDNA and dsDNA (Figure 8A and B, lanes 5–7). In contrast, PSF(267–468), which contains RNA recognition motifs, did not bind either ssDNA or dsDNA under these reaction conditions (Figure 8A and B, lanes 8–10). Therefore, the DNA- and RNA-binding domains are separately located in the N-terminal and central domains of PSF, respectively.
Figure 8.
DNA-binding and RAD51-binding activities of the PSF domains. ϕX174 ssDNA (20 µM) (A) or ϕX174 linear dsDNA (20 µM) (B) was incubated with PSF, PSF(1–266) or PSF(267–468) at 37°C for 10 min. The samples were then separated by 0.8% agarose gel electrophoresis in TAE buffer and were visualized by ethidium bromide staining. The protein concentrations for panel A were 0 µM (lane 1), 0.15 µM (lanes 2, 5 and 8), 0.3 µM (lanes 3, 6 and 9) and 0.6 µM (lanes 4, 7 and 10). The protein concentrations for panel B were 0 µM (lane 1), 0.1 µM (lanes 2, 5 and 8), 0.2 µM (lanes 3, 6 and 9) and 0.4 µM (lanes 4, 7 and 10). (C) The pull-down assay with Ni–NTA beads. Lanes 2–5 represent purified RAD51, His6-tagged PSF, His6-tagged PSF(1–266) and His6-tagged PSF(267–468), respectively. His6-tagged PSF, His6-tagged PSF(1–266) or His6-tagged PSF(267–468) (3.8 µg) was mixed with RAD51 (7.4 µg). The RAD51 bound to the His6-tagged proteins was pulled down by the Ni–NTA agarose beads, and was analyzed by 12% SDS–PAGE. Bands were visualized by Coomassie Brilliant Blue staining.
We next tested the RAD51-binding activity of PSF(1–266) and PSF(267–468). To do so, we prepared the His6-tagged PSF, PSF(1–266) and PSF(267–468) proteins (Figure 8C, lanes 3–5), and performed a pull-down assay with the Ni–NTA agarose beads. Consistent with the results from the pull-down assay by the PSF-conjugated beads (Figure 1C), His6-tagged PSF also bound to RAD51 in the Ni–NTA pull-down assay (Figure 8C, lane 7). A substantial amount of RAD51 was pulled down with His6-tagged PSF(1–266) (Figure 8C, lane 8), but not with His6-tagged PSF(267–468) (Figure 8C, lane 9). Therefore, the N-terminal 1–266 region of PSF contains both the DNA- and RAD51-binding sites.
DISCUSSION
Previous studies have suggested that PSF is a multifunctional nuclear protein, which functions in RNA synthesis, RNA processing, RNA transport, and transcription (40). In addition to these functions, interestingly, PSF was purified from a HeLa cell extract as a protein that promotes homologous pairing between ssDNA and dsDNA in vitro (28). This finding suggested that PSF may also function in homologous recombination, and prompted us to test whether PSF affects the recombination activity of RAD51, which is a central protein for homologous recombination in eukaryotes.
In the present study, we purified human PSF as a recombinant protein, and found that PSF directly binds to RAD51. Interestingly, PSF stimulates RAD51-mediated homologous pairing and strand exchange at low RAD51 concentrations, but inhibits these reactions at high RAD51 concentrations, which are optimal for the reactions without PSF. These findings suggest that PSF may be a novel recombination factor, which has both activator and suppressor functions for the RAD51 recombinase activity. During the initial period of homologous recombination in cells, PSF may function as activator, perhaps because the RAD51 accumulation at the recombination sites is not yet sufficient. The RAD51 concentration at the recombination sites is supposed to increase during the homologous recombination processes. Excess RAD51 accumulation on chromosomes at later stages of homologous recombination may induce undesired recombination, which may cause the chromosomal aberrations frequently found in tumor cells. PSF may then function as an inhibitor of RAD51, to suppress such inappropriate recombination reactions by removing RAD51 from ssDNA during homologous recombination. We found that PSF competes with RAD51 for ssDNA binding, and removes RAD51 from ssDNA. This fact indicated that PSF does not form a three-component complex with RAD51 and ssDNA, although the PSF–RAD51 complex is formed in the absence of ssDNA. Therefore, PSF may be recruited to the recombination sites as a complex form with RAD51, and may be released from RAD51 to free the ssDNA region when RAD51 is assembled on the recombination site.
We and others found that PSF alone (i) binds to ssDNA and dsDNA, (ii) promotes D-loop formation between ssDNA and dsDNA, and (iii) anneals complementary ssDNAs (28). In this study, we also found that (iv) the N-terminal domain of PSF directly binds to the central ATPase domain of RAD51, and (v) PSF modulates the RAD51-mediated homologous pairing and strand exchange reactions. These biochemical characteristics of PSF partially overlap with those of RAD52, which is an important activator for RAD51 (7–10). In the yeast Saccharomyces cerevisiae, the rad52 mutation reportedly exhibits significant sensitivity to DSB inducing agents, such as ionizing radiation (41,42). However, the RAD52-knockout chicken DT40 cells did not show significant defects in DSB repair (43). This discrepancy may explain the functional redundancy of RAD52 with other factors in higher eukaryotes (44). PSF may share a common function with RAD52 in vertebrates.
PSF was also purified from a nuclear extract of HeLa cells, as a complex form with p54(nrb). Intriguingly, the PSF–p54(nrb) complex stimulated a non-homologous DNA end joining (NHEJ) reaction by Ku70/80, DNA ligase IV and XRCC4 (27). NHEJ is a rapid, error-prone DSB repair pathway. On the other hand, homologous-recombinational repair (HRR) is an accurate, error-free DSB repair pathway. The present study has provided new evidence that PSF physically and functionally interacts with RAD51, which is the central enzyme for the homologous recombination reactions. Therefore, PSF may be involved in both DSB repair pathways, NHEJ and HRR, with different partners, p54(nrb) and RAD51, respectively.
FUNDING
Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology, Japan. Funding for open access charge: Waseda University.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENT
We thank Mr K. Kuribayashi (Waseda University) for technical assistance.
REFERENCES
- 1.Petronczki M, Siomos MF, Nasmyth K. Un ménage à quatre: the molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–440. doi: 10.1016/s0092-8674(03)00083-7. [DOI] [PubMed] [Google Scholar]
- 2.Neale MJ, Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pittman DL, Cobb J, Schimenti KJ, Wilson LA, Cooper DM, Brignull E, Handel MA, Schimenti JC. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol. Cell. 1998;1:697–705. doi: 10.1016/s1097-2765(00)80069-6. [DOI] [PubMed] [Google Scholar]
- 4.Yoshida K, Kondoh G, Matsuda Y, Habu T, Nishimune Y, Morita T. The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol. Cell. 1998;1:707–718. doi: 10.1016/s1097-2765(00)80070-2. [DOI] [PubMed] [Google Scholar]
- 5.Bannister AA, Pezza RJ, Donaldson JR, de Rooij DG, Schimenti KJ, Camerini-Otero RD, Schimenti JC. A dominant, recombination-defective allele of Dmc1 causing male-specific sterility. PLoS Biol. 2007;5:1–10. doi: 10.1371/journal.pbio.0050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 7.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.West SC. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 9.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Ann. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 10.Sung P, Krejci L, Van Komen S, Sehorn MG. Rad51 recombinase and recombination mediators. J. Biol. Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- 11.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 12.Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 13.Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 14.Maeshima K, Morimatsu K, Horii T. Purification and characterization of XRad51.1 protein, Xenopus RAD51 homologue: recombinant XRad51.1 promotes strand exchange reaction. Genes Cells. 1996;1:1057–1068. doi: 10.1046/j.1365-2443.1996.d01-224.x. [DOI] [PubMed] [Google Scholar]
- 15.Gupta RC, Bazemore LR, Golub EI, Radding CM. Activities of human recombination protein Rad51. Proc. Natl Acad. Sci. USA. 1997;94:463–468. doi: 10.1073/pnas.94.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wesoly J, Agarwal S, Sigurdsson S, Bussen W, Van Komen S, Qin J, van Steeg H, van Benthem J, Wassenaar E, Baarends WM, et al. Differential contributions of mammalian Rad54 paralogs to recombination, DNA damage repair, and meiosis. Mol. Cell Biol. 2006;26:976–989. doi: 10.1128/MCB.26.3.976-989.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Modesti M, Budzowska M, Baldeyron C, Demmers JA, Ghirlando R, Kanaar R. RAD51AP1 is a structure-specific DNA binding protein that stimulates joint molecule formation during RAD51-mediated homologous recombination. Mol. Cell. 2007;28:468–481. doi: 10.1016/j.molcel.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 18.Wiese C, Dray E, Groesser T, San Filippo J, Shi I, Collins DW, Tsai MS, Williams GJ, Rydberg B, Sung P, et al. Promotion of homologous recombination and genomic stability by RAD51AP1 via RAD51 recombinase enhancement. Mol. Cell. 2007;28:482–490. doi: 10.1016/j.molcel.2007.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patton JG, Porro EB, Galceran J, Tempst P, Nadal-Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993;7:393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- 20.Urban RJ, Bodenburg Y, Kurosky A, Wood TG, Gasic S. Polypyrimidine tract-binding protein-associated splicing factor is a negative regulator of transcriptional activity of the porcine p450scc insulin-like growth factor response element. Mol. Endocrinol. 2000;14:774–782. doi: 10.1210/mend.14.6.0485. [DOI] [PubMed] [Google Scholar]
- 21.Urban RJ, Bodenburg YH, Wood TG. NH2 terminus of PTB-associated splicing factor binds to the porcine P450scc IGF-I response element. Am. J. Physiol. Endocrinol. Metab. 2002;283:E423–E427. doi: 10.1152/ajpendo.00057.2002. [DOI] [PubMed] [Google Scholar]
- 22.Urban RJ, Bodenburg Y. PTB-associated splicing factor regulates growth factor-stimulated gene expression in mammalian cells. Am. J. Physiol. Endocrinol. Metab. 2002;283:E794–E798. doi: 10.1152/ajpendo.00174.2002. [DOI] [PubMed] [Google Scholar]
- 23.Song X, Sui A, Garen A. Binding of mouse VL30 retrotransposon RNA to PSF protein induces genes repressed by PSF: effects on steroidogenesis and oncogenesis. Proc. Natl Acad. Sci. USA. 2004;101:621–626. doi: 10.1073/pnas.0307794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song X, Sun Y, Garen A. Roles of PSF protein and VL30 RNA in reversible gene regulation. Proc. Natl Acad. Sci. USA. 2005;102:12189–12193. doi: 10.1073/pnas.0505179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-m-RNA 3′ processing and transcription termination. Genes Dev. 2007;21:1779–1789. doi: 10.1101/gad.1565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buxadé M, Morrice N, Krebs DL, Proud CG. The PSF.p54nrb complex is a novel Mnk substrate that binds the mRNA for tumor necrosis factor α. J. Biol. Chem. 2008;283:57–65. doi: 10.1074/jbc.M705286200. [DOI] [PubMed] [Google Scholar]
- 27.Bladen CL, Udayakumar D, Takeda Y, Dynan WS. Identification of the polypyrimidine tract binding protein-associated splicing factor.p54(nrb) complex as a candidate DNA double-strand break rejoining factor. J. Biol. Chem. 2005;280:5205–5210. doi: 10.1074/jbc.M412758200. [DOI] [PubMed] [Google Scholar]
- 28.Akhmedov AT, Lopez BS. Human 100-kDa homologous DNA-pairing protein is the splicing factor PSF and promotes DNA strand invasion. Nucleic Acids Res. 2000;28:3022–3030. doi: 10.1093/nar/28.16.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurumizaka H, Aihara H, Kagawa W, Shibata T, Yokoyama S. Human Rad51 amino acid residues required for Rad52 binding. J. Mol. Biol. 1999;291:537–548. doi: 10.1006/jmbi.1999.2950. [DOI] [PubMed] [Google Scholar]
- 30.Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 31.Kagawa W, Kurumizaka H, Ikawa S, Yokoyama S, Shibata T. Homologous pairing promoted by the human Rad52 protein. J. Biol. Chem. 2001;276:35201–35208. doi: 10.1074/jbc.M104938200. [DOI] [PubMed] [Google Scholar]
- 32.Matsuo Y, Sakane I, Takizawa Y, Takahashi M, Kurumizaka H. Roles of the human RAD51 L1 and L2 loops in DNA binding. FEBS J. 2006;273:3148–3159. doi: 10.1111/j.1742-4658.2006.05323.x. [DOI] [PubMed] [Google Scholar]
- 33.Ishida T, Takizawa Y, Sakane I, Kurumizaka H. The Lys313 residue of the human Rad51 protein negatively regulates the strand-exchange activity. Genes Cells. 2008;13:91–103. doi: 10.1111/j.1365-2443.2007.01143.x. [DOI] [PubMed] [Google Scholar]
- 34.Aihara H, Ito Y, Kurumizaka H, Yokoyama S, Shibata T. The N-terminal domain of the human Rad51 protein binds DNA: structure and a DNA binding surface as revealed by NMR. J. Mol. Biol. 1999;290:495–504. doi: 10.1006/jmbi.1999.2904. [DOI] [PubMed] [Google Scholar]
- 35.Pellegrini L, Yu DS, Lo T, Anand S, Lee M, Blundell TL, Venkitaraman AR. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 36.Shin DS, Pellegrini L, Daniels DS, Yelent B, Craig L, Bates D, Yu DS, Shivji MK, Hitomi C, Arvai AS, et al. Full-length archaeal Rad51 structure and mutants: mechanisms for RAD51 assembly and control by BRCA2. EMBO J. 2003;22:4566–4576. doi: 10.1093/emboj/cdg429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu Y, He Y, Moya IA, Qian X, Luo Y. Crystal structure of archaeal recombinase RadA: a snapshot of its extended conformation. Mol. Cell. 2004;15:423–435. doi: 10.1016/j.molcel.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 38.Conway AB, Lynch TW, Zhang Y, Fortin GS, Fung CW, Symington LS, Rice PA. Crystal structure of a Rad51 filament. Nat. Struct. Mol. Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- 39.Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl Acad. Sci. USA. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shav-Tal Y, Zipori D. PSF and p54nrb/NonO- multi-functional nuclear proteins. FEBS Lett. 2002;531:109–114. doi: 10.1016/s0014-5793(02)03447-6. [DOI] [PubMed] [Google Scholar]
- 41.Ho KS. The gene dosage effect of the rad52 mutation on X-ray survival curves of tetraploid yeast strains. Mutat. Res. 1975;33:165–172. doi: 10.1016/0027-5107(75)90191-8. [DOI] [PubMed] [Google Scholar]
- 42.Prakash S, Prakash L, Burke W, Montelone BA. Effects of the RAD52 Gene on Recombination in Saccharomyces cerevisiae. Genetics. 1980;94:31–50. doi: 10.1093/genetics/94.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamaguchi-Iwai Y, Sonoda E, Buerstedde JM, Bezzubova O, Morrison C, Takata M, Shinohara A, Takeda S. Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in RAD52. Mol. Cell Biol. 1998;18:6430–6435. doi: 10.1128/mcb.18.11.6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujimori A, Tachiiri S, Sonoda E, Thompson LH, Dhar PK, Hiraoka M, Takeda S, Zhang Y, Reth M, Takata M. Rad52 partially substitutes for the Rad51 paralog XRCC3 in maintaining chromosomal integrity in vertebrate cells. EMBO J. 2001;20:5513–5520. doi: 10.1093/emboj/20.19.5513. [DOI] [PMC free article] [PubMed] [Google Scholar]