Abstract
The Stromelysin-1 gene promoter contains a palindrome of two Ets-binding sites (EBS) that bind the p51 and p42 isoforms of the human Ets-1-transcription factor. A previous study established that full gene transactivation is associated with a ternary complex consisting of two p51 bound to the two EBS on the promoter. p42, only able to bind one of the two EBS, induces only very weak activity. Here, we investigate the mechanism by which the Stromelysin-1 promoter discriminates between p51 and p42. The differential stoichiometry of the two Ets-1 isoforms arises from the Stromelysin-1 EBS palindrome. The ternary complex requires the presence of two inhibitory domains flanking the DNA-binding domain and the ability to form an intramolecular autoinhibition module. Most importantly, the p51-ternary and the p42-binary complexes induce DNA curvatures with opposite orientations. These results establish that differential DNA bending, via p51 and p42 differential binding, is correlated with the Stromelysin-1 promoter activation process.
INTRODUCTION
Ets-1 is the founding member of the E26 transforming specific (ETS) family of transcription factors characterized by a conserved DNA-binding domain (DBD), called the ETS domain. The DBD recognizes specific DNA elements called ETS-binding sites (EBS), that include the consensus 5′-GGAA/T-3′ core motif present in the promoter of target genes (1,2). By binding to these elements, Ets-1 activates the transcription of several genes implicated in various processes such as development, angiogenesis, tumor invasion and apoptosis (3). However, the mechanism of Ets-1-mediated transcriptional activation is not completely understood. The current model involves only interactions with co-activators that induce histone acetylation and allow contacts with components of the basal transcription machinery (3,4).
Among the gene promoters requiring Ets-1, the Stromelysin-1 (matrix metalloproteinase-3 or MMP-3) promoter is particularly relevant from a biological point of view. Stromelysin-1 is a matrix metalloproteinase involved in remodeling the extracellular matrix during certain physiological and pathological processes (5,6). Increased coexpression of both Ets-1 and Stromelysin-1 genes has been reported in rheumatoid arthritis (7), glomerulonephritis (8), angiogenesis (9) and tumor invasion (10).
Ets-1 binds to the Stromelysin-1 promoter at two head-to-head GGAA core motifs (i.e. the EBS) separated by four base pairs, the whole segment being palindromic. This palindrome is required for basal expression and for phorbol-12-myristate 13-acetate (PMA)- and interleukin-1β (IL-1β)-induced expression of the gene (11,12). Furthermore, our previous work demonstrated that the binding of Ets-1 to this EBS palindrome is essential for activating the Stromelysin-1 promoter and inducing protein expression in the cell (13,14).
The human protein Ets-1 physiologically exists in two isoforms: p51, the full-length isoform, and p42, a shorter isoform lacking exon VII due to alternative splicing (15,16). The lower DNA-binding activity of p51 compared to that of p42 (15,17) is most likely the consequence of the p51 allosteric mechanism of autoinhibition (18). In absence of the cognate DNA, the two inhibitory domains of p51 (the N-terminal-inhibitory domain, encoded by the exon VII and the C-terminal inhibitory domain) cooperatively interact to form an inhibitory module (19). The structure of this module, determined by NMR (18) and X-ray (20), shows that four structurally coupled inhibitory helices, HI-1, HI-2, H4 and H5, pack intramolecularly with each other and with the helix H1 of the DBD. However, this assembly has only limited stability, and changes between conformational states (18). These characteristics facilitate the structural reorganization of Ets-1 upon DNA binding (21), the optimization of the DNA/DBD interface involving a shift of the HI-2/H1 region and the unfolding of HI-1 (18,20). This non-trivial rearrangement is specific to p51; p42 lacks the N-terminal-inhibitory domain and therefore the inhibitory module.
Interactions with protein partners that counteract the autoinhibition mechanism allow p51 to increase its DNA-binding affinity (22). We established that p51 can act as its own partner when bound to the palindromic EBS in the Stromelysin-1 promoter (13). Thus, p51–p51 cooperative interaction allows the formation of a ternary p51–DNA–p51 complex (13). However, p42 only forms a binary p42–DNA complex despite the presence of two EBS. This intriguing difference of binding stoichiometry between p51 and p42 is of special interest since it correlates with distinct transcriptional activities on the Stromelysin-1 promoter: p51 induces full transactivation of the promoter, whereas p42 is much less efficient (13).
Here, we investigated the mechanistic and structural bases of the differential DNA-binding stoichiometry of p51 and p42 on the EBS palindrome, given its crucial role in the transcriptional control of Stromelysin-1. The role of the target DNA sequence and of the Ets-1 N-terminal and C-terminal-inhibitory domains was explored, by mutating both DNA and proteins. The structural DNA distortions induced upon the complexation of p51 and p42 were characterized. Overall, the results suggest a mechanism that accounts for the differential DNA-binding stoichiometry of p51 and p42. Furthermore, we propose that Stromelysin-1 promoter activation is correlated with the isoform-induced orientation of DNA bending.
MATERIALS AND METHODS
Luciferase-reporter gene
The Stromelysin-1 reporter plasmid pGL3 (Promega) constructs containing the wild-type (WT) EBS, or mutant forms M1, M2, M1M2 and inverted palindrome (IP) have been previously described (13). The WT + 4 reporter plasmid construct was generated by site-directed mutagenesis using the QuickChange Site-directed Mutagenesis kit (Stratagene®) with the WT Stromelysin-1 promoter construct as a template and the following oligonucleotides 5′-ACCAAGACAGGAAGCACGCACTTCCTGGAGATTAATC-3′ and 5′-GATTAATCTCCAGGAAGTGCGTGCTTCCTGTCTTGGT-3′.
Bacterial expression vector constructions
The construction of bacterial expression vectors containing the cDNA sequences of human Ets-1 isoforms p51 and p42, and of N-terminal deletion mutants ΔN301, and ΔN331 have been previously described (13). The cDNAs of the other deletion mutants were obtained by polymerase chain reaction (PCR) amplification using a unique reverse primer 5′-TTCAGGCGCCGATCCCCAGCAGGCTCTGCAG-3′ and the following forward primers 5′-TAGAATTCACATATGAAGGCGGCCGTCGATCTC-3′ for p51ΔC415 and p42ΔC329, 5′-AAGTATCATATGAAGGGCACCTTCAAGGAC-3′ for p51(301–415), and 5′-AAGTATCATATGGGCAGTGGACCAATCCAG-3′ for Ets-1 DBD. The human Ets-1 p51 or p42 cDNA inserted in a pSG5 vector was used as a template. The amplified fragments were digested with NdeI and SfoI and cloned into a pTYB2 plasmid (T7 Impact System, New England Biolabs) previously digested by NdeI and SmaI endonucleases.
Eukaryotic expression vector constructions
To construct the eukaryotic expression vectors for Ets-1 p51 and p42, the cDNAs of the two isoforms inserted in pSG5 plasmids were amplified by PCR using the following primers 5′-ATAAATAGATCTATGAAGGCGGCCGTCGATC-3′ and 5′-ATAAATAGATCTCACTCGTCGGCATCTGGC-3′. The amplified fragments were digested with BglII and inserted in a pcDNA3™ vector (Invitrogen) previously digested with BamHI. Correct insertion and orientation were checked by sequencing. The pcDNA3™–p51Y424A construct was generated by site-directed mutagenesis using the following oligonucleotides 5′-CAGAGCCTGCTGGGGGCCACCCCTGAGGAGC-3′ and 5′-GCTCCTCAGGGGTGGCCCCCAGCAGGCTCTG-3′ and the QuickChange Site-directed Mutagenesis kit (Stratagene®) with the pcDNA3™–p51 construct as a template.
Expression and purification of Ets-1 proteins
Expression and purification of Ets-1 proteins were carried out according to (13), by using the T7 Impact System (New England Biolabs). Briefly, a culture of Escherichia coli (ER2566) transformed with the appropriate recombinant plasmid was induced at cell density A595 nm = 0.7 with isopropyl-β-d-thiogalactopyranoside (IPTG) (0.3 mM final) in a Luria-Bertani medium. After 3–4 h of incubation at 30°C, the culture was pelleted, washed with phosphate-buffered saline (PBS), and suspended in 10 ml of lysis buffer [50 mM Tris, pH 8, 150 mM NaCl, 1 mM EDTA, 0.1% (v/v) Triton X-100, complete protease inhibitor mixture (CompleteTM, Roche Molecular Biochemical)]. Bacteria were lysed with a French press (1000 pounds/square inch pressure) and lysates were clarified by two successive centrifugations at 20 000 × g at 4°C (5 min and 15 min). Each clarified lysate was applied to a 5 ml chitin bead column (New England Biolabs), and columns were washed with 20 volumes of column buffer (lysis buffer without protease inhibitor) and rapidly flushed with 3 volumes of elution buffer (column buffer without Triton X-100) containing 50 mM dithiothreitol (DTT). Columns were stored 16 h at 4°C for peptidic cleavage and proteins were eluted by 15 ml of elution buffer in 1 ml fractions. Fractions were subjected to a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE) analysis and those containing the recombinant protein were quick-frozen by immersion in liquid nitrogen and stored at –80°C. Yields were measured by colorimetry (Bio-Rad protein assay).
Electrophoretic mobility shift assay (EMSA)
Double-stranded oligonucleotides corresponding to the WT, M1, M2, IP and WT + 4 mutants of the Stromelysin-1 (−223/−194) promoter region (Table 1) were end-labeled using T4 polynucleotide kinase and [γ-32P]ATP and were subsequently purified by electrophoresis on a 20% polyacrylamide (acrylamide:bisacrylamide 19:1, Euromedex) non-denaturing gel in Tris borate EDTA (TBE) buffer (90 mM Tris borate, 1 mM EDTA). Recombinant proteins (4 pmol) were incubated with 0.5 ng of each probe in 20 μl of binding reaction buffer (20 mM Tris, pH 8, 80 mM NaCl, 1 mM EDTA, 2 mM DTT, 10% glycerol) for 20 min on ice. Complexes formed were resolved on a 5% or 8% polyacrylamide (acylamide/bisacrylamide 29:1, Euromedex) non-denaturing gel in 0.25× TBE buffer at 11.7 V cm−1 at room temperature. Gels were dried and autoradiographed at −80°C.
Table 1.
Wild type (WT) and mutant sequences of the Stromelysin-1 promoter
| Name | Sequences | EBS topology |
|---|---|---|
| WT | 5′-ACCAAGACAGGAAGCACTTCCTGGAGATTA-3′ | → ←a |
| M1 | 5′-ACCAAGACAAAAAGCACTTCCTGGAGATTA-3′ | × ← |
| M2 | 5′-ACCAAGACAGGAAGCACTTTTTGGAGATTA-3′ | → ×b |
| IP | 5′-ACCAAGAGCTTCCTGCAGGAAGTGAGATTA-3′ | ← → |
| WT+4 | 5′-ACCAAGACAGGAAGCACGCACTTCCTGGAGATTA-3′ | → ← |
→ and ← represent the orientation of the EBS.
X mutated EBS.
Several modifications were done for the circular permutation assay and phasing analysis. EMSA experiments were calibrated to determine optimal polyacrylamide concentration, voltage, temperature and migration time. Probes corresponded to digestion fragments of circular permutated or phasing plasmids amplified by PCR as described below (see ‘Circular permutation assay’ section and ‘Phasing analysis’ section). Recombinant proteins (4 pmol) were incubated with 20 ng of each probe in 20 µl of binding reaction buffer containing 1 µg of bovine serum albumin (BSA). Samples were applied to a 5% polyacrylamide gel (acylamide/bisacrylamide 29:1, Euromedex) in 0.25× TBE buffer at 20.6 V cm−1 at 4°C.
Moreover, in all EMSA experiments, p42 gave rise to small, but detectable amounts of an additional binary complex which behaved like the main binary complex, but with higher mobility. This supplementary complex, already detected in previous studies (13), is most likely due to a degradation product of p42 that is still able to bind DNA. We considered this complex to be non-relevant.
Circular permutation assay
The circular permutated plasmids containing WT or M1 mutant fragment of the Stromelysin-1 (−223/−194) promoter region were constructed using a pBend2 plasmid (23) digested with SalI and XbaI and inserting the following annealed oligonucleotides:
WT, 5′-CTAGAACCAAGACAGGAAGCACTTCCTGGAGATTAG-3′ and 5′-TCGACTAATCTCCAGGAAGTGCTTCCTGTCTTGGTT-3′;
M1, 5′-CTAGAACCAAGACAAAAAGCACTTCCTGGAGATTAG-3′ and 5′-TCGACTAATCTCCAGGAAGTGCTTTTTGTCTTGGTT-3′.
The pBend2-WT and -M1 constructs were digested with MluI, ClaI, SpeI, EcoRV, NruI, KpnI and BamHI. The resulting 147–155 bp fragments (containing the oligonucleotides) were amplified by PCR and 2 μg of each fragment was end-labeled with [γ-32P]ATP (50 μCi) using T4 polynucleotide kinase. The probes were purified using the QIAquick® gel extraction kit (Qiagen®) according to the manufacturer's instructions and used for EMSA as described above (see ‘electrophoretic mobility shift assay’).
Migration of both the protein–DNA complexes (Rbound) and the probes (Rfree) were analyzed using a PhosphorImage Analyser (Molecular Dynamics) and relative mobility (Rbound/Rfree) of each complex was calculated from several independent experiments. The relative mobility of each complex was normalized to the average relative mobility of all the complexes and fitted to a second order polynomial to determine a, b and c coefficients (GraphPad Prism) of the following equation:
| 1 |
where L represents the length of the probe and D, the distance of the center of the two or single EBS (WT or M1 respectively) from the 5′ end of the DNA molecule (24). The angle, θ, obtained from the following equation:
| 2 |
gave the bending angle α = 180° – θ as described (24).
Phasing analysis
The phasing plasmid constructs were obtained by inserting into the pBend2-WT plasmid, previously digested with SalI, the following annealed oligonucleotides containing A:T tracts: +34, 5′-TCGACAAAAACGGGCAAAAACGGGCAAAAAG-3′ and 5′-TCGACTTTTTGCCCGTTTTTGCCCGTTTTTG-3′; +36, 5′-TCGACGCAAAAACGGGCAAAAACGGGCAAAAACGG-3′ and 5′-TCGACCGTTTTTGCCCGTTTTTGCCCGTTTTTGCG-3′; +38, 5′-TCGACACGCAAAAACGGGCAAAAACGGGCAAAAACGCAG-3′ and 5′-TCGACTGCGTTTTTGCCCGTTTTTGCCCGTTTTTGCGTG-3′; +40, 5′-TCGACACACGCAAAAACGGGCAAAAACGGGCAAAAACGCACAG-3′ and 5′-TCGACTGTGCGTTTTTGCCCGTTTTTGCCCGTTTTTGCGTGTG-3′; +42, 5′-TCGACCGACACGCAAAAACGGGCAAAAACGGGCAAAAACGCACAGCG-3′ and 5′-TCGACGCTGTGCGTTTTTGCCCGTTTTTGCCCGTTTTTGCGTGTCGG-3′; +44, 5′TCGACGACGACACGCAAAAACGGGCAAAAACGGGCAAAAACGCACAGCAGG-3′ and 5′-TCGACCTGCTGTGCGTTTTTGCCCGTTTTTGCCCGTTTTTGCGTGTCGTCG-3′.
The plasmids pBend2-WT +34, +36, +38, +40, +42, +44 were digested with EcoRV and the 181–199 bp fragments underwent PCR amplification. Two micrograms of each amplified fragment were end-labeled with [γ-32P]ATP (50 μCi) using T4 polynucleotide kinase. The probes were purified using the QIAquick® gel extraction kit (Qiagen®) according to the manufacturer's instructions and used for EMSA as described above (see ‘Electrophoretic mobility shift assay’ section).
Relative mobility of each protein–DNA complex (Rbound/Rfree) was determined as described above (see ‘Circular permutation assay’ section) from several independent experiments and normalized to the average relative mobility of all the complexes. The normalized relative mobilities were plotted as a function of linker length, which is defined as the distance (bp) between the center of the intrinsic bend (A-tract) and the center of the EBS palindrome. The best fit to a cosine function (phasing function) was determined using GraphPad Prism (25). The DNA bend orientation was determined from the minima of the phasing functions (26) and expressed as the angle separating this bend from the intrinsic bend in a plane perpendicular to the DNA helix axis. The intrinsic DNA bend is directed toward the minor groove at the center of the A-tract (27). The helical periodicity was assumed to be 10.5 bp per turn of the DNA helix.
Transient transfection and reporter assay
Human Embryonic Kidney (HEK) 293 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum in 12-well plates (2 ml per well) to reach 40–60% confluence at time of transfection. Before transfection, ExGen 500 Transfection Reagent (2 μl per well, Euromedex) was incubated with 250 ng of each respective reporter (pGL3) and expression vector (pcDNA3), and 1.25 ng of a control reporter plasmid (pRL-null, Promega) for 15 min at room temperature in a volume of 50 μl of a 150 mM NaCl solution. Cell medium was changed for 500 μl of Opti-modified Eagle's medium (Invitrogen), and DNA-ExGen 500 (Euromedex) mixture was added. After 6 h, DMEM was added (1.5 ml per well). Cells were harvested 48 h after transfection with 250 μl of cell lysis buffer (1% Triton X-100, 25 mM glycylglycine, pH 7.8, 15 mM MgSO4, 4 mM EGTA, 1 mM DTT) per well. Twenty-microliter aliquots of each supernatant were sequentially tested for firefly and Renilla luciferases (Dual-Luciferase® Reporter Assay System, Promega) using a Lumat LB 9501 (Berthold). For each transfection condition, firefly luciferase activity (pGL-3 constructs) was normalized to Renilla luciferase activity (pRL-null) to correct for variation in the number of transfected cells.
Molecular modeling
The ternary protein–DNA complexes were constructed by docking two p51ΔN331 proteins on the following DNA sequences: (i) 5′-N6GGAAN4TTCCN6-3′ (WT) (ii) 5′-N5GGAAN4 + xTTCCN5-3′ (WT + x) (iii) 5′-N6TTCCN4GGAAN6-3′ (IP). These sequences differ by the number of base pairs composing the spacers N4 or N4 + x, with x from 1 to 3, and by the relative orientation of the two target sites (IP). The models were based on the crystallographic structure of the binary complex with p51ΔN331 bound to the 5′-N7GGAAN4-3′ target DNA [(28); PDB code 1K79], p51ΔN331 being simply duplicated on the two EBS of different sequences by superimposing the EBS core motifs.
Calculations of DNA deformation were performed using the JUMNA program (29) which represents nucleic acid flexibility using a combination of helicoidal and internal variables. Solvent damping of electrostatic interactions was included using a sigmoidal distance-dependent dielectric function and counter-ion screening was dealt with by reducing total charges on each phosphate group to −0.5e. Although rather rudimentary, this solvent representation has proved useful in modeling DNA solution structure (30,31). The force field Parm98 (32) was applied to the WT sequence in a canonical B-DNA conformation. This structure was then minimized under graduated, simple quadratic distance restraints with steps of 1 Å, in order to move the two GGAA-binding sites away from each other. Analyses of DNA structure were carried out using CURVES (33).
RESULTS
EBS configuration and binding stoichiometry of p51 and p42 Ets-1 isoforms
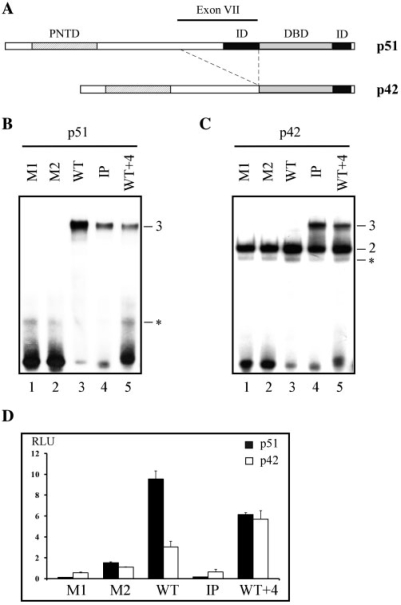
To determine if the configuration of the Stromelysin-1 EBS palindrome (WT, two head-to-head core consensus motifs separated by four base pairs, Table 1) is required for differential binding stoichiometry between both human isoforms, p51 and p42 (Figure 1A), we used the EBS variants shown in Table 1. Two mutated sequences with only one EBS (M1 and M2) were used for monitoring the binary complexes. We designed two other sequences containing two EBS in which (i) the orientation of the two EBS were inverted (IP, two tail-to-tail core consensus motifs separated by 4 bp) and (ii) the spacer length was increased by 4 bp (WT + 4), keeping the original orientation of the EBS (Table 1). EMSA experiments were performed with these EBS sequences and either p51 or p42.
Figure 1.
EBS configuration in the Stromelysin-1 promoter: differential binding stoichiometry and transactivation properties of p51 and p42 Ets-1 isoforms. (A) Schematic organization of the human Ets-1 p51 and p42 isoforms. DBD, DNA-binding domain; PNTD, pointed domain; ID, inhibitory domain. (B, C) Gel shift assays. Equal amounts of p51 (B) or p42 (C) were incubated with M1, M2, WT, IP and WT + 4 variants, as defined in Table 1. Binary (2) and ternary (3) complexes are indicated. Non-relevant complexes (see ‘Materials and Methods’ section) are indicated by an asterisk. (D) Transient transfection assays with reporter plasmids containing WT or variant EBS defined in Table 1 and either p51 or p42 eukaryotic expression vector. Luciferase activity was normalized and expressed as relative luciferase activity (RLU).
As expected (13), no complex could be detected with the single EBS sequences M1 and M2 and p51, due to its autoinhibitory module (Figure 1B, lanes 1 and 2). Indeed, relatively high dissociation constants (Kd) of ∼100 nM have previously been measured on p51–M1 and p51–M2 complexes using a surface plasmon resonance kinetic assay (13), which also thereby provided an estimation of EMSA sensitivity for detecting protein/DNA binding. M1 and M2 formed binary complexes with the non-autoinhibited p42 (Figure 1C, lanes 1 and 2). p51 and p42 bound to WT (containing two EBS core motifs) with distinct stoichiometries. The existence of the ternary complex p51–WT–p51 was indicated by a slow migrating band of high intensity (Figure 1B, lane 3), reflecting a Kd of ∼3 nM measured when p51 cooperatively bound to the second EBS (13). This kind of ternary complex cannot be observed with p42, which was only able to generate a p42–WT binary complex (Figure 1C, lane 3). In fact, p42 recognizes M1, M2 and WT with the same affinity (Kd ∼20 nM, unpublished data). When the two EBS motifs were either inverted (IP) or separated by a spacer of 8 bp (WT + 4), ternary complexes were detected with p42 and, to a much lesser extent, with p51 (Figure 1B and C, lanes 4 and 5).
Changes in the EBS configuration encountered in the Stromelysin-1 promoter tended to eliminate the differences in binding stoichiometry between p51 and p42 and thus the discrimination between p51 and p42 Ets-1 isoforms.
EBS configuration and transactivation activities of p51 and p42 Ets-1 isoforms
We then used transfection experiments to determine whether the Stromelysin-1 promoter containing the WT variant is able to induce differential transactivation properties with p51 and p42, compared to the M1, M2, IP and WT + 4 variants.
As previously shown (13), p51 and p42 induced distinct levels of promoter activity with WT. In contrast to p42, p51 fully transactivated the promoter in a synergistic manner, the promoter activity being more than three times higher compared to promoter activity induced by p42 (Figure 1D). Transactivation drastically decreased with p51 and p42 bound to M1, M2 and IP (Figure 1D). In contrast, WT + 4 recovered substantial activity with p51 and p42 (Figure 1D), although clearly lower than the activity obtained with p51 with the WT promoter configuration.
Only the WT configuration generated distinct levels of promoter activity in response to p51 and p42, paralleling the differential binding stoichiometry described above. The full transactivation of the Stromelysin-1 promoter was associated with the ternary complex p51–WT–p51; we therefore investigated which p51 domains are required for the formation of this complex.
Role of the inhibitory domains and DBD in the ternary complex formation
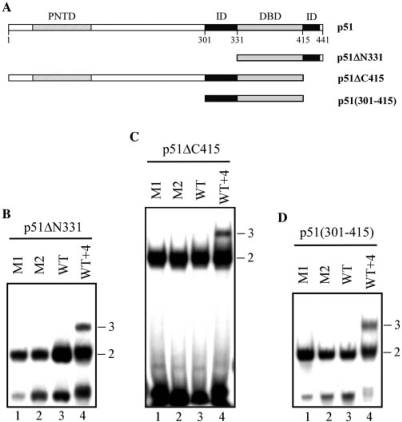
Previous work showed that deleting the N-terminal region of p51 spanning from amino acids 1–300 does not alter the binding stoichiometry for the EBS palindrome found in the Stromelysin-1 promoter (13,34). The p51ΔN301 mutant, which contains the two inhibitory domains in addition to the DBD, was the shortest Ets-1 protein able to form a ternary complex with the WT promoter configuration. Thus, we focused on the role of each inhibitory domain in p51ΔN301 on Ets-1-binding stoichiometry.
We used three mutants that contained the DBD and (i) the complete C-terminal region (p51ΔN331), (ii) the complete N-terminal region (p51ΔC415), or (iii) only the N-terminal-inhibitory domain [p51(301–415)] (Figure 2A). These C-terminal and N-terminal mutants, submitted to EMSA, formed binary complexes with M1 and M2 (Figure 2B, C and D, lanes 1 and 2), due to the loss of their potential to autoinhibit DNA binding. When assessed on WT, they only formed binary complexes (Figure 2B, C and D, lane 3). The ternary complex was however restored with WT + 4, whatever the mutant (Figures 2B, C and D, lane 4).
Figure 2.
Role of both inhibitory domains of p51 in DNA-binding stoichiometry to the EBS palindrome. (A) Map of the p51 isoform and p51ΔN331, p51ΔC415 and p51(301–415) deletion mutants. (B, C, D) Gel shift assays. Equal amounts of p51ΔN331 (B), p51ΔC415 (C), or p51(301–415) (D) were incubated with M1, M2, WT and WT + 4 variants, as defined in Table 1. Binary (2) and ternary (3) complexes are indicated.
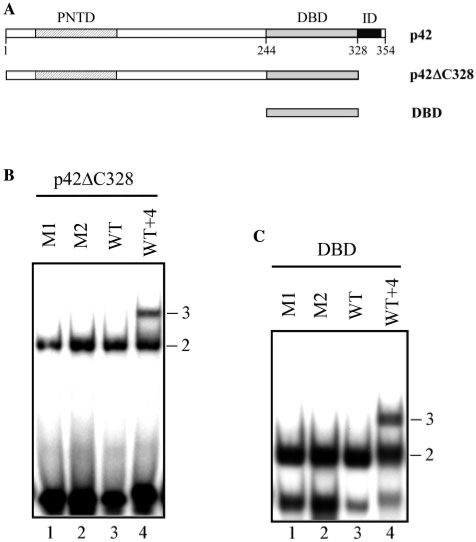
We then investigated the ability of DBD to form a ternary complex in the absence of any inhibitory domain, using a p42 deletion mutant lacking the C-terminal region (p42ΔC328) or possessing only the DBD region (Figure 3A). Only binary complexes were observed with M1, M2 and WT (Figure 3B and C, lanes 1, 2 and 3), while ternary complexes appeared with WT + 4 (Figure 3B and C, lane 4).
Figure 3.
Role of the N- and C-terminal parts of p42 in DNA-binding stoichiometry to the EBS palindrome. (A) Map of p42 isoform and p42ΔC328 and DBD deletion mutants. (B, C) Gel shift assays. Equal amounts of p42ΔC328 (B) or DBD (C) were incubated with M1, M2, WT and WT + 4 variants, as defined in Table 1. Binary (2) and ternary (3) complexes are indicated.
Therefore, proteins containing the DBD but amputated of one or both inhibitory regions were unable to generate ternary complexes with WT. Our hypothesis is that, in the WT promoter variant, the neighboring DBDs of the two Ets-1 molecules cause steric hindrances that can be overcome by structural reorganization in the promoter sequence or by the inhibitory regions. Next, we examine the possible structural distortions of the promoter sequence induced in the binary and ternary complexes before considering the role of the autoinhibitory module.
DNA bending induced by binding of p51 and p42 Ets-1 isoforms to the WT EBS palindrome
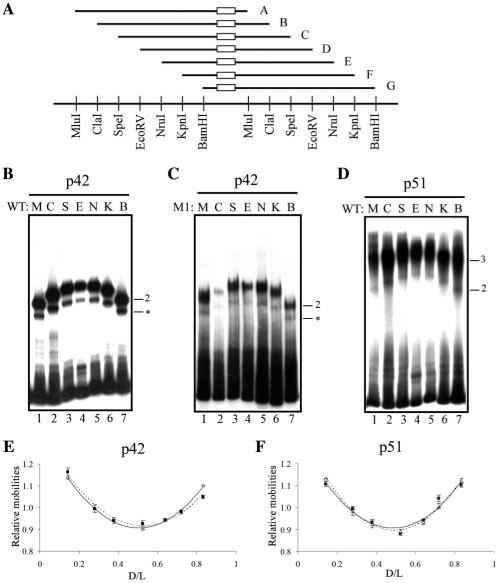
We used circular permutation assays for detecting potential DNA bending (23). The circular permutated DNA probes consisted of seven DNA fragments containing either WT or M1 differently positioned along the pBend vector (Figure 4A).
Figure 4.
Determination of DNA bending amplitude induced by p51 and p42 Ets-1 isoforms. (A) Schematic representation of DNA probes used for the circular permutation assays. WT or M1 are symbolized by an open rectangle in probes A-G that correspond to M, C, S, E, N, K and B nomenclature according to the restriction sites. Restriction enzyme sites are given below the probe representations. (B, C, D) Gel shift assays performed with p42 on WT probe (B), p42 on M1 probe (C) and p51 on WT probe (D), as described in A. Binary (2) and ternary (3) complexes are indicated. Non-relevant complexes are indicated by an asterisk. (E, F) Relative mobility as a function of D/L; D is the number of base pairs separating the 5′ end of the probe and the EBS centers (center of M1 or center of WT palindrome); L is the length of the probe, expressed in base pairs. The lines correspond to the best second-order polynomial fit. In E, open diamond: p42-WT and filled square: p42-M1. In F, unfilled triangle: p51–WT and filled square: p51–WT–p51.
The binary complexes formed by p42 and either WT or M1 located near the vector center (Figure 4B and C, probes E and N) migrated at slower rates than those in which the EBS sites were positioned near the ends (Figure 4B and C, probes M and B). Analyzing the normalized relative mobilities (24) shows that the DNA molecules noticeably bend in p42–WT or p42–M1 complexes (Figure 4E), with the value of DNA curvatures being assessed at around 70° in our experimental conditions (Table 2).
Table 2.
Amplitude and orientation of DNA bend induced by p51 and p42 Ets-1 isoforms
| Protein | Site | Complex type | Circular permutation analysis |
Phasing analysis |
||
|---|---|---|---|---|---|---|
| Bend angle (°) | R2a | Bend orientation (°) | R2b | |||
| P42 | WT | Binary | 70 | 0.99 | 269 | 0.98 |
| M1 | Binary | 66 | 0.97 | ND | ND | |
| P51 | WT | Binary | 73 | 0.99 | ND | ND |
| WT | Ternary | 70 | 0.94 | 84 | 0.99 | |
| P51Y424A | WT | Binary | ND | ND | 259 | 0.98 |
R2 is the coefficient of determination of the fit to a second order polynomial.
bR2 is the coefficient of determination of the fit to a cosine function.
p51 bound to the WT probes (Figure 4D) showed the same relative mobility patterns (Figure 4F) as p42 (Figure 4E). Employing specific migration conditions (i.e. at low temperature, see ‘Materials and Methods’ section) made it possible to detect a p51–WT binary complex (Figure 4D), previously observed and characterized by cross-linking experiments (13). This binary complex, not observed with M1, showed that p51 induced similar DNA bending intensity in both binary and ternary complexes (Table 2), the curvature amplitude being estimated around 70°. Thus, the binding of a second molecule of p51 to the EBS palindrome had no dramatic impact on the DNA curvature amplitude induced by the first molecule.
Both p42 and p51 induced DNA bending and their curvature amplitudes were comparable across the different types of binary and ternary complexes.
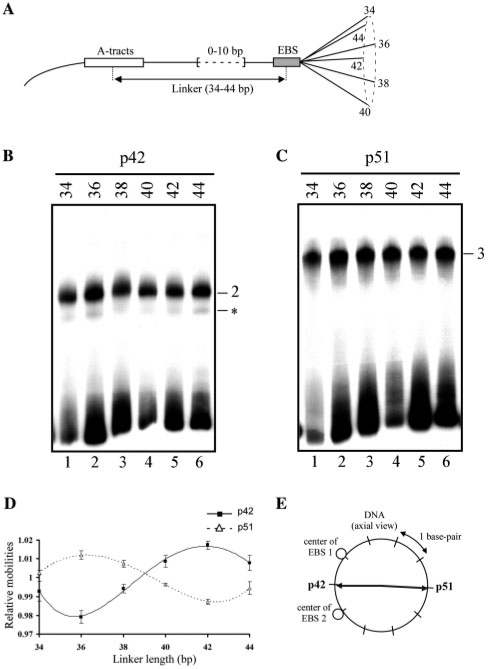
DNA curvature is defined by both amplitude and orientation. Given the above, phasing analyses were performed to determine the orientation of the DNA curvatures generated by p51 and p42. A set of DNA probes containing WT located near to or far from a sequence-directed A-tract bend (Figure 5A) was used for EMSA in the presence of p51 and p42. The linker length between WT and the A-tract varied through one helical turn with 2 bp increments. Two bends in the same phase (in phase) cooperate and decrease electrophoretic mobility, while two bends in opposite phase (out of phase) cancel each other and enhance electrophoretic mobility.
Figure 5.
Determination of DNA bend orientation induced by p51 and p42 Ets-1 isoforms. (A) Schematic representation of DNA probes used for phasing analysis. The probes contain A-tracts (open rectangle) and the Stromelysin-1 EBS palindrome (shaded rectangle). They are designated by linker length, i.e. the number of base pairs separating the A-tract and the EBS centers, which varied by 2 bp increments (0–10 bp), through one helical turn. (B, C) Gel shift assays performed with p42 (B) or p51 (C) in the presence of the probes described in A. Binary (2) and ternary (3) complexes are indicated. Non relevant complexes are indicated by an asterisk. (D) Relative mobility averaged from several independent EMSA experiments as a function of linker length and fitted to a sinusoidal function (lines). (E) DNA bend directions induced by p51 and p42 are represented by arrows in a plane perpendicular to the long DNA axis, positioned at the EBS palindrome center. EBS 1 and 2 are the centers of the two core motifs in the WT EBS palindrome.
The mobilities of the free probes vary with two parameters, their length and the presence of intrinsically bent A-tracts; they regularly decreased from lanes 1–6 (Figure 5B and C). These variations were not reproduced when the probes were bound to p42 or p51 (Figure 5B and C). Analysis of the normalized relative migrations of the complexes (Rbound probe/Rfree probe) showed that p42–WT and p51–WT–p51 followed sinusoidal patterns typical of DNA bending (Figure 5D). Despite their unusually low amplitudes, these patterns were faithfully reproduced across several independent experiments. They are in full agreement with the results obtained by circular permutation assays, and, in addition, indicate that p42 and p51 induced DNA curvatures with distinct orientations. The DNA bend orientations were then determined by calculating the angle separating the A-tract-directed bend and the bend induced by protein binding (‘Materials and Methods’ section). The values found for p42 and p51 (Table 2), separated spatially by about 180°, indicate that these proteins generate opposite DNA bends.
Replacing the center of each EBS related to bend orientation showed that p42 causes the DNA to bend towards the minor groove, i.e. towards the protein–DNA interface (Figure 5E). Conversely, the DNA curvature induced by p51 ternary complex is directed away from the protein–DNA interface, compressing the DNA major groove (major groove bending) (Figure 5E).
The DNA bending observed in p42-WT is in agreement with the binary complex structures [(28), PDB code 1K79, (35), PDB codes 2STT and 2STW], which revealed a noticeable DNA minor groove curvature induced by p51ΔN331. Conversely, DNA bending observed here with the p51–WT–p51 complex does not correspond to its crystallographic counterpart, two p51Δ280–436 proteins bound to WT [(34), PDB code 2NNY]. In this symmetric ternary complex, the minor groove curvature detected for the EBS core motif in the X-ray binary complex disappeared and the EBS palindrome was globally straight, contrary to our results. However, this X-ray ternary complex also raises another problem concerning protein homo-dimerization. In 2NNY, the dimerization of p51Δ280–436 proteins was ensured by patches of amino acids belonging to the N-terminal-inhibitory region adjacent to the DBD (amino acids 379–382) and the DBD itself (amino acids 327–335), with the most crucial contacts involving amino acids 332–335 and 379–382. The C-terminal-inhibitory region was not involved either in the protein–protein or in the DNA–protein interfaces. The first intriguing point is that p42 and p51ΔN331 both contain the key amino acids 332–335 and 379–382; however, they do not form any observable ternary complexes with WT (Figures 1C and 2B). We could nevertheless hypothesize that the 331–335 loop adopts a shape conducive to the protein–protein interface owing to a fold involving the outermost N-terminal region. For instance, the X-ray structures of Ets-1 dimers (20), as well as 2NNY, show that, in this part of the protein, the pairs of amino acids 332–327, 343–323 and 346–321 are hydrogen bonded. However, the second point that is more difficult to explain is that 2NNY cannot account for the incapacity of p51ΔC415 and p51(301–415) to form a ternary complex (Figure 2C and D). If the N-terminal-inhibitory region and the DBD were sufficient to stabilize the ternary complex, these mutants should have been able to dimerize in the presence of WT. Discrepancies between solid-state and solution data may be due to the complex–complex contacts generated by the crystal lattice (34). In 2NNY, the ternary complexes were in fact associated in pairs, the structural unit forming a hexameric assemblage. This particular crystal packing mobilized the 308–322 section of the N-terminal-inhibitory regions and consequently prohibited any interaction between the N- and C-terminal domains, as our results suggest. Given that our migration patterns ruled out any durable complex-complex interaction, we thus hypothesize that lattice packing modifies the ternary complex structure so that it cannot be directly extrapolated to interactions that occur in solution.
To further examine the role of the N- and C-terminal domains in the sharp DNA curvature induced by p51, we addressed the necessity of a pre-existing inhibitory module in the interaction process.
Role of the inhibitory module in ternary complex formation and in Stromelysin-1 promoter transactivation
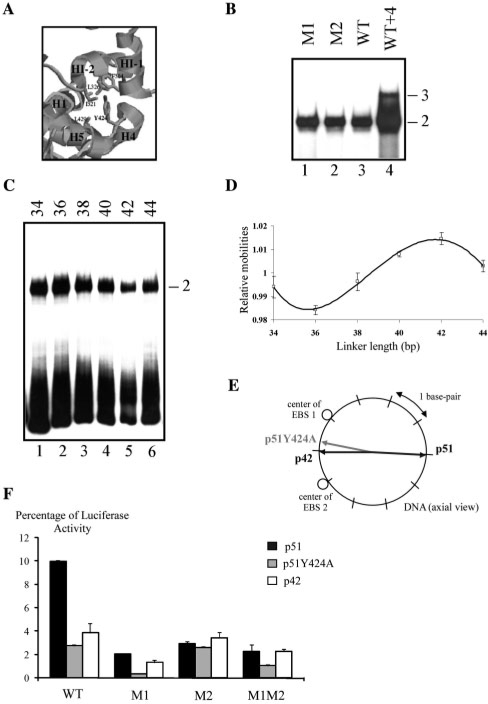
Tyrosine 424 is crucial for the formation of the p51-inhibitory module (18,20) (Figure 6A). Indeed, a single mutation of this tyrosine to alanine in p51 disrupts DNA-binding autoinhibition due to the unfolding of the HI-1 helix (20). This mutant, p51Y424A, is thus useful for testing the role of a pre-existing inhibitory module on ternary complex formation and on Stromelysin-1 transactivation.
Figure 6.
Importance of DNA-binding autoinhibition and DNA bend orientation for Stromelysin-1 promoter transactivation. (A) Detail of the structure of the Ets-1 inhibitory module around Tyr 424 (20). (B) Gel shift assays. p51Y424A was incubated with M1, M2, WT and WT + 4 variants, as defined in Table 1. Binary (2) and ternary (3) complexes are indicated. (C) Gel shift assays were performed with p51Y424A in the presence of the different probes as described in Figure 5A. (D) Relative mobility as a function of linker length expressed in base pairs and fitted to a sinusoidal function. (E) DNA-bending directions induced by p51Y424A, p42 and p51 are represented by arrows in a plane perpendicular to the long DNA axis, positioned at the EBS palindrome center. EBS 1 and 2 are the centers of the two core motifs in the WT EBS palindrome. (F) Transient transfection assays with reporter plasmids containing M1, M2, WT and WT + 4, defined in Table 1, and either p51Y424A, p51 or p42 eukaryotic expression vector. Luciferase activities were normalized and expressed as relative luciferase activity (RLU).
Overall, p51Y424A behaves as p42. Binary complexes were observed with M1 or M2 (Figure 6B, lanes 1 and 2), confirming the loss of DNA-binding autoinhibition. This binary complex was also observed with WT (Figure 6B, lane 3), the ternary complex only being detected with WT + 4 (Figure 6B, lane 4). Using the approach detailed above to measure DNA bending, we found that the patterns obtained with p51Y424A (Figure 6C and D) exactly parallel those obtained with p42 (Figure 5B and D). Both proteins induced the same curvature in terms of both amplitude and direction (Table 2, Figures 5E and 6E). Finally, the Stromelysin-1 promoter activity associated with p51Y424A was similar to activity obtained with p42 (Figure 6F). Moreover, these results emphasize the problem posed by the 2NNY X-ray structure that fails to explain the absence of observable p51Y424A–WT–p51Y424A ternary complex in our experiments since this mutant contains all the amino acids belonging to the X-ray Ets1-Ets1 interface.
Therefore, the inability of p51Y424A to reproduce the properties of p51 with WT implies that the integrity of the inhibitory module of p51 is required to form the ternary complex. The results confirm the correlation between binding stoichiometry, induced DNA bend orientation and transactivation of the Stromelysin-1 promoter.
Steric protein hindrances on WT and DNA curvature
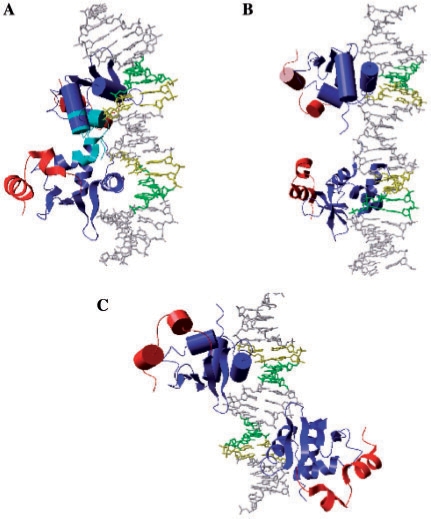
To better understand why the binding of two p42 was prevented with WT and not with IP and WT + 4, modeling investigations were undertaken. Given the results described above, we chose to investigate the capacity of DBD in generating ternary complexes using a very simple rigid body approach where only steric clashes could be detected. We focused on the DBD because its structure, identical across the available X-ray (20,28,34) and NMR (18,35) structures, is not controversial and widely accepted. In contrast, the N-terminal region of the 2NNY ternary complex (34) is likely to suffer from crystallographic artifacts, as explained above. Several structures of the N-terminal are available but they correspond to Ets-1/Ets-1 or Pax/Ets-1/DNA complexes (20) and are not relevant in the context of Ets-1/DNA/Ets-1 interactions. Thus, we constructed models based on the X-ray structure of the binary complex [(28), PDB code 1K79]. The protein is p51ΔN331. The DNA structures consisted of 1K79 EBS core motifs bent towards the minor groove and straight B-canonical spacers. Despite their simplicity, the models gave a good estimate of the relative position of the DBD linked to the C-terminal domain and their steric hindrance.
With the WT configuration, two DBD bind in the two successive major grooves on the same DNA face and strongly overlap (Figure 7A). The C-terminal domains do not participate in the steric hindrances because they are far from each other. The N-terminal-inhibitory regions, absent in our model, would increase the area of the steric hindrance zone, the N-terminal DBD parts being face-to-face and tightly tangled. It is clear that this ternary complex cannot exist without drastically distorting the DNA molecule.
Figure 7.
Model of p51ΔN331–DNA–p51ΔN331 ternary complex. Models of ternary complex of p51ΔN331 proteins bound to WT (A), WT + 3 (B) or IP (C). The DBD monomers are in blue (one in ‘ribbon’, one in ‘cartoon’), and the C-terminal regions in red. The clash zones in A are in cyan. The EBS core motifs are in green for GG.CC and yellow for AA.TT bases.
Introducing at least three base pairs in the spacer (WT + 3) sufficiently increases the distance between the geometric centers of the two recognition sites to circumvent the steric hindrance between the two DBD (Figure 7B) since two p42 molecules are able to bind to this type of site (Figure 1C). In this model, the N-terminal and C-terminal regions, while rather far from each other, remain face-to-face. These relative positions could allow for some level of cooperativity between two p51 molecules, leading to small amount of active ternary complexes and in agreement with the experimental data (Figure 1B and D). Changing the relative orientation of the two EBS and keeping the 4 bp spacer intact (IP) corresponds to a configuration that also comfortably supports two proteins (Figure 7C). The two DBD are now located on opposite sides of the DNA and, in addition, the N-terminal and C-terminal regions are not in position to interact. This disposition allows p42 to form ternary complexes (Figure 1C). However, the minor, observable amount of p51–IP–p51 observed in Figure 1B cannot be explained by the model. Since the related transactivation activity associated with p51–IP–p51 was not detectable (Figure 1D), this ternary complex may differ from the native one. In any case, further investigations are needed to fully comprehend the nature of this ternary complex.
We then investigated how the WT spacer reacts to constraints that push the two EBS core motifs away and what sort of distortions are needed to remove the steric hindrances between two DBD. This strategy was successfully used to account for the DNA deformations induced by the TATA box-binding protein (30).
The progressive distance constraints applied between the two EBS geometrical centers moved away from the EBS, generating an under-twisting and a major groove curvature on the spacer (Table 3). Systematic dockings of two p51ΔN331 proteins extracted from 1K79 on these constrained DNA models showed that a total under-twisting of around 20° associated with a major groove curvature of 42° were the minimal changes required to circumvent DBD clashes (Table 3). Despite of the reductionism of this approach, the predicted major groove curvature perfectly matches the bending direction inferred from the experimental data (Figure 6E).
Table 3.
Spacer elongation in WT restrained simulations
| Δda (Å) | ΔEb (Kcal/mol) | Icc (°) | Twavd(°) | Protein–protein steric clashes |
|---|---|---|---|---|
| 0 | 0 | 5 | 37 | ++ |
| 2.5 | 4 | 22 | 34 | + |
| 4.5 | 15 | 42 | 30 | ∼0 |
| 7.5 | 37 | 67 | 23 | – |
| 8.5 | 44 | 82 | 20 | – |
Δd: increase of the distance separating two EBS core motifs in reference to the starting structure.
ΔE: difference between total energies of constained and starting structures.
Ic: curvative intensity.
Twav: average twist of the 4 bp spacer.
These theoretical investigations completely corroborate the experimental results. They help understand why both IP and WT + 4 are able to form ternary complexes with p51 and p42 (Figure 1B and C). The fact that, in contrast to IP, WT + 4 seemed able to accommodate an interface between two neighboring proteins could account for Stromelysin-1 transactivations, which were substantial with WT + 4 but negligible with IP (Figure 1D). Concerning the curvature, its cost, around 15 kcal mol−1 for a bending magnitude of 42° (Table 3), is as low or lower than the lowest deformation energies calculated on DNA extracted from 71 X-ray complexes using the same modeling program (36). This modest energetic cost should be easily compensated by the gain resulting from the homo-dimerization of the two neighboring proteins.
The nature of the intermolecular protein-protein interface cannot be deduced from our investigations and requires further experiments in conjunction with sophisticated modeling. The extended conformation of the N-terminal-inhibitory region observed in the X-ray ternary complex appears unrelated to the assembly mechanism (34). Nevertheless, active participation of this N-terminal-inhibitory region seems more than likely. Indeed, 15N relaxation and hydrogen exchange NMR measurements (18) demonstrated that the inhibitory module consists of a stable scaffold made up of the H1, H4 and H5 helices and a mobile N-terminal-inhibitory region subject to conformational equilibria. The binding to DNA is accompanied by drastic changes in the inhibitory module. Without completely obliterating the inhibitory module folding, the N-terminal-inhibitory domain unpacks through the unfolding of HI-1, inducing a global sliding motion that results in the optimal fit between H1 and the DNA (18,20). Given that the ternary complex requires the amino acid residues 333 and 334 (34) close to H1, and a complete, intact and functional-inhibitory module, this kind of N-terminal-inhibitory domain rearrangement guided by the C-terminal region acting as a scaffold, is a good candidate for overriding the autoinhibition mechanism and, at the same time, mediating the p51–p51 interface within the ternary complex.
DISCUSSION AND CONCLUSION
The main aim of this work was to better understand the differential DNA-binding stoichiometry of the two human Ets-1 isoforms p51 and p42 on the EBS palindrome found in the Stromelysin-1 promoter. Indeed, the formation of a ternary complex with p51 and a binary complex with p42 allows for distinctive controls of Stromelysin-1 promoter activity (13), the full transactivation being associated with p51.
We first summarize the key results presented in this work. The configuration of the Stromelysin-1 EBS palindrome is crucial for generating either ternary complexes with p51 or binary complexes with p42. Changes in the orientation of the two EBS core motifs or in spacer length cancel the differences between the two Ets-1 isoforms. Indeed, these types of EBS variants form ternary complexes regardless of the Ets-1 isoform, but do not result in efficient transactivation. Operational ternary complexes require both the N-terminal and C-terminal-inhibitory domains that also require a complete, intact and functional-inhibitory module. Deleting one or both inhibitory domains or preventing the formation of the inhibitory module, only generated binary complexes. This implies that the two inhibitory regions of Ets-1 are intimately, structurally as well as functionally, coupled to binding to the Stromelysin-1 EBS palindrome. Lastly, the functional ternary complex induces noticeable DNA major groove curvature, whereas the binary complex bends the DNA in the opposite direction.
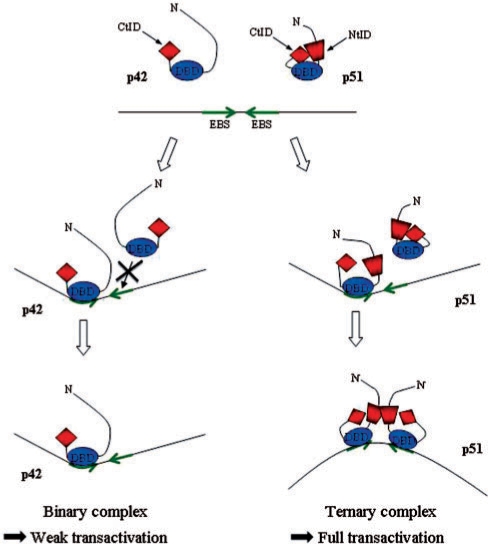
These experimental results associated with modeling investigations made it possible to reveal the mechanism that underlies the formation of p42 binary and p51 ternary complexes on the WT EBS palindrome (Figure 8). Our simulations showed that the formation of a binary complex prohibited the binding of a second protein because of severe clashes between two neighboring DBD, aggravated by the N-terminal regions. Inverting the respective orientation of the two EBS, increasing the spacer length or generating a curvature towards the major groove on the EBS palindrome center avoided the DBD steric hindrance, in agreement with the experimental results. Furthermore, our experimental data demonstrated that the cooperative binding of a second p51 on the EBS palindrome occurred via the simultaneous creation of a major groove DNA curvature and a p51–p51 interaction. Most likely, the intermolecular protein-protein interface corresponds to drastic structural rearrangements of the inhibitory module primarily implicating the N-terminal inhibitory domains, mimicking and amplifying the displacement detected in the formation of the binary complex (18). The strict prerequisite of the presence of two, interacting C- and N-terminal inhibitory domains explains why p42, lacking the N-terminal inhibitory domain, does not form a ternary complex.
Figure 8.
Model for the differential regulation of the Stromelysin-1 promoter by p51 and p42 Ets-1 isoforms. The binding of one p42 molecule on the EBS palindrome (core consensus motifs in green) induced DNA minor groove bending. Two p42 molecules are unable to bind the EBS palindrome due to steric clashes between the two DBD domains (DBD, in blue). In contrast, two p51 are able to overcome these steric hindrances through a cooperative interaction leading to (i) a drastic rearrangement of the inhibitory module (CtID, red diamond and NtID, red trapezoid) and the creation of an intermolecular interface between the N-terminal inhibitory domains (NtID, red trapezoid) (ii) the induction of major DNA bending (opposite to the protein–DNA interface). This ternary complex, via the induced specific DNA curvature, is responsible for the full transactivation of the promoter.
In addition, our results strongly suggest that the different capacities of p51 and p42 for activating the Stromelysin-1 promoter were associated with the induced DNA bending. p51 induces a bend directed away from the protein–DNA interface and fully transactivates the Stromelysin-1 promoter, while p42 generates an opposite bend and activates the promoter only weakly. The p42-induced DNA curvature detected in this work has previously been established at the molecular level (28,35). Precise characterization of p51–induced DNA curvature requires further investigations using techniques specifically dedicated to bending measurements. However, the global coherency of our results, together with their reproducibility, indicates that p51 and p42 produce different DNA distortions, most likely related to the curvature direction.
This relationship between DNA bending and transcription activation has only been reported for Thyroid Receptor (TR) (37) and the Sox2 (38) systems. However, DNA bending is associated with a large number of eukaryotic-transcription factors (39,40). In particular, many pairs of homo- or heterodimers induce DNA curvatures of opposite orientation (Fos-Jun/Jun-Jun (25), Myc-Myc/Max-Max (41), and TR-TR/TR-Retinoid X receptor dimers (42)). In addition, we found other similar palindromic EBS (i.e. two core motifs separated by 4 base pairs) in various promoter data banks (data not shown). We recently showed that Ets-1 behaves identically with the EBS palindromes of the human p53 and Stromelysin-1 promoters, in terms of both cooperative binding and transcriptional transactivation (43). Taken together, these observations suggest that the mechanism revealed in this study could be a common system used to specifically activate a repertoire of various promoters. Hence, characterizing induced DNA bending should be a prerequisite to a complete description and understanding of the subtle structural dialog between DNA and transcription factors that guarantees the fine tuning of transcription regulation.
FUNDING
This work was supported by the Centre National de la Recherche Scientifique (CNRS). The Association de Recherche contre le Cancer and the Ligue Nationale Contre le Cancer that provided student fellowships (to G.L. and D.B.). Funding for open access charge: La Ligue contre le Cancer-Comité du Pas-de-Calais
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr S. Adhya for generously providing pBend2; Dr J. Coll for stimulating discussions and F. Guillot and I. Roland for technical assistance.
REFERENCES
- 1.Graves BJ, Petersen JM. Specificity within the Ets family of transcription factors. Adv. Cancer Res. 1998;75:1–55. doi: 10.1016/s0065-230x(08)60738-1. [DOI] [PubMed] [Google Scholar]
- 2.Sharrocks AD. The ETS-domain transcription factor family. Nat. Rev. Mol. Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 3.Dittmer J. The biology of the Ets1 proto-oncogene. Mol. Cancer. 2003;2:29. doi: 10.1186/1476-4598-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Myers E, Hill AD, Kelly G, McDermott EW, O'Higgins NJ, Buggy Y, Young LS. Associations and interactions between Ets-1 and Ets-2 and coregulatory proteins, SRC-1, AIB1, and NCoR in breast cancer. Clin. Cancer Res. 2005;11:2111–2122. doi: 10.1158/1078-0432.CCR-04-1192. [DOI] [PubMed] [Google Scholar]
- 5.Sternlicht MD, Werb Z. How matrix metalloproteinases regulate cell behavior. Annu. Rev. Cell Dev. Biol. 2001;17:463–516. doi: 10.1146/annurev.cellbio.17.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vu TH, Werb Z. Matrix metalloproteinases: effectors of development and normal physiology. Genes Dev. 2000;14:2123–2133. doi: 10.1101/gad.815400. [DOI] [PubMed] [Google Scholar]
- 7.Sun HB, Yokota H. Messenger-RNA expression of matrix metalloproteinases, tissue inhibitors of metalloproteinases, and transcription factors in rheumatic synovial cells under mechanical stimuli. Bone. 2001;28:303–309. doi: 10.1016/s8756-3282(00)00454-3. [DOI] [PubMed] [Google Scholar]
- 8.Naito T, Razzaque MS, Nazneen A, Liu D, Nihei H, Koji T, Taguchi T. Renal expression of the Ets-1 proto-oncogene during progression of rat crescentic glomerulonephritis. J. Am. Soc. Nephrol. 2000;11:2243–2255. doi: 10.1681/ASN.V11122243. [DOI] [PubMed] [Google Scholar]
- 9.Oda N, Abe M, Sato Y. ETS-1 converts endothelial cells to the angiogenic phenotype by inducing the expression of matrix metalloproteinases and integrin beta3. J. Cell Physiol. 1999;178:121–132. doi: 10.1002/(SICI)1097-4652(199902)178:2<121::AID-JCP1>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 10.Wernert N, Gilles F, Fafeur V, Bouali F, Raes MB, Pyke C, Dupressoir T, Seitz G, Vandenbunder B, Stehelin D. Stromal expression of c-Ets1 transcription factor correlates with tumor invasion. Cancer Res. 1994;54:5683–5688. [PubMed] [Google Scholar]
- 11.Sirum-Connolly K, Brinckerhoff CE. Interleukin-1 or phorbol induction of the stromelysin promoter requires an element that cooperates with AP-1. Nucleic Acids Res. 1991;19:335–341. doi: 10.1093/nar/19.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wasylyk B, Wasylyk C, Flores P, Begue A, Leprince D, Stehelin D. The c-ets proto-oncogenes encode transcription factors that cooperate with c-Fos and c-Jun for transcriptional activation. Nature. 1990;346:191–193. doi: 10.1038/346191a0. [DOI] [PubMed] [Google Scholar]
- 13.Baillat D, Begue A, Stehelin D, Aumercier M. ETS-1 transcription factor binds cooperatively to the palindromic head to head ETS-binding sites of the stromelysin-1 promoter by counteracting autoinhibition. J. Biol. Chem. 2002;277:29386–29398. doi: 10.1074/jbc.M200088200. [DOI] [PubMed] [Google Scholar]
- 14.Baillat D, Leprivier G, Regnier D, Vintonenko N, Begue A, Stehelin D, Aumercier M. Stromelysin-1 expression is activated in vivo by Ets-1 through palindromic head-to-head Ets binding sites present in the promoter. Oncogene. 2006;25:5764–5776. doi: 10.1038/sj.onc.1209583. [DOI] [PubMed] [Google Scholar]
- 15.Fisher RJ, Fivash M, Casas-Finet J, Erickson JW, Kondoh A, Bladen SV, Fisher C, Watson DK, Papas T. Real-time DNA binding measurements of the ETS1 recombinant oncoproteins reveal significant kinetic differences between the p42 and p51 isoforms. Protein Sci. 1994;3:257–266. doi: 10.1002/pro.5560030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koizumi S, Fisher RJ, Fujiwara S, Jorcyk C, Bhat NK, Seth A, Papas TS. Isoforms of the human ets-1 protein: generation by alternative splicing and differential phosphorylation. Oncogene. 1990;5:675–681. [PubMed] [Google Scholar]
- 17.Lionneton F, Lelievre E, Baillat D, Stehelin D, Soncin F. Characterization and functional analysis of the p42Ets-1 variant of the mouse Ets-1 transcription factor. Oncogene. 2003;22:9156–9164. doi: 10.1038/sj.onc.1207241. [DOI] [PubMed] [Google Scholar]
- 18.Lee GM, Donaldson LW, Pufall MA, Kang HS, Pot I, Graves BJ, McIntosh LP. The structural and dynamic basis of Ets-1 DNA binding autoinhibition. J. Biol. Chem. 2005;280:7088–7099. doi: 10.1074/jbc.M410722200. [DOI] [PubMed] [Google Scholar]
- 19.Skalicky JJ, Donaldson LW, Petersen JM, Graves BJ, McIntosh LP. Structural coupling of the inhibitory regions flanking the ETS domain of murine Ets-1. Protein Sci. 1996;5:296–309. doi: 10.1002/pro.5560050214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garvie CW, Pufall MA, Graves BJ, Wolberger C. Structural analysis of the autoinhibition of Ets-1 and its role in protein partnerships. J. Biol. Chem. 2002;277:45529–45536. doi: 10.1074/jbc.M206327200. [DOI] [PubMed] [Google Scholar]
- 21.Jonsen MD, Petersen JM, Xu QP, Graves BJ. Characterization of the cooperative function of inhibitory sequences in Ets-1. Mol. Cell Biol. 1996;16:2065–2073. doi: 10.1128/mcb.16.5.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goetz TL, Gu TL, Speck NA, Graves BJ. Auto-inhibition of Ets-1 is counteracted by DNA binding cooperativity with core-binding factor alpha2. Mol. Cell Biol. 2000;20:81–90. doi: 10.1128/mcb.20.1.81-90.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J, Zwieb C, Wu C, Adhya S. Bending of DNA by gene-regulatory proteins: construction and use of a DNA bending vector. Gene. 1989;85:15–23. doi: 10.1016/0378-1119(89)90459-9. [DOI] [PubMed] [Google Scholar]
- 24.Ferrari S, Harley VR, Pontiggia A, Goodfellow PN, Lovell-Badge R, Bianchi ME. SRY, like HMG1, recognizes sharp angles in DNA. EMBO J. 1992;11:4497–4506. doi: 10.1002/j.1460-2075.1992.tb05551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerppola TK, Curran T. Fos-Jun heterodimers and Jun homodimers bend DNA in opposite orientations: implications for transcription factor cooperativity. Cell. 1991;66:317–326. doi: 10.1016/0092-8674(91)90621-5. [DOI] [PubMed] [Google Scholar]
- 26.Leonard DA, Rajaram N, Kerppola TK. Structural basis of DNA bending and oriented heterodimer binding by the basic leucine zipper domains of Fos and Jun. Proc. Natl Acad. Sci. USA. 1997;94:4913–4918. doi: 10.1073/pnas.94.10.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zinkel SS, Crothers DM. DNA bend direction by phase sensitive detection. Nature. 1987;328:178–181. doi: 10.1038/328178a0. [DOI] [PubMed] [Google Scholar]
- 28.Garvie CW, Hagman J, Wolberger C. Structural studies of Ets-1/Pax5 complex formation on DNA. Mol. Cell. 2001;8:1267–1276. doi: 10.1016/s1097-2765(01)00410-5. [DOI] [PubMed] [Google Scholar]
- 29.Lavery R, Zakrzewska K, Sklenar H. JUMNA (junction minimisation of nucleic acids) Comp. Phy. Commun. 1995;91:135–158. [Google Scholar]
- 30.Lebrun A, Shakked Z, Lavery R. Local DNA stretching mimics the distortion caused by the TATA box-binding protein. Proc. Natl Acad. Sci. USA. 1997;94:2993–2998. doi: 10.1073/pnas.94.7.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varnai P, Djuranovic D, Lavery R, Hartmann B. Alpha/gamma transitions in the B-DNA backbone. Nucleic Acids Res. 2002;30:5398–5406. doi: 10.1093/nar/gkf680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheatham TE, 3rd, Cieplak P, Kollman PA. A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. J. Biomol. Struct. Dyn. 1999;16:845–862. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- 33.Lavery R, Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 34.Lamber EP, Vanhille L, Textor LC, Kachalova GS, Sieweke MH, Wilmanns M. Regulation of the transcription factor Ets-1 by DNA-mediated homo-dimerization. EMBO J. 2008;27:2006–2017. doi: 10.1038/emboj.2008.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Werner MH, Clore GM, Fisher CL, Fisher RJ, Trinh L, Shiloach J, Gronenborn AM. Correction of the NMR structure of the ETS1/DNA complex. J. Biomol. NMR. 1997;10:317–328. doi: 10.1023/a:1018399711996. [DOI] [PubMed] [Google Scholar]
- 36.Zakrzewska K. DNA deformation energetics and protein binding. Biopolymers. 2003;70:414–423. doi: 10.1002/bip.10476. [DOI] [PubMed] [Google Scholar]
- 37.Leidig F, Shepard AR, Zhang WG, Stelter A, Cattini PA, Baxter JD, Eberhardt NL. Thyroid hormone responsiveness in human growth hormone-related genes. Possible correlation with receptor-induced DNA conformational changes. J. Biol. Chem. 1992;267:913–921. [PubMed] [Google Scholar]
- 38.Scaffidi P, Bianchi ME. Spatially precise DNA bending is an essential activity of the sox2 transcription factor. J. Biol. Chem. 2001;276:47296–47302. doi: 10.1074/jbc.M107619200. [DOI] [PubMed] [Google Scholar]
- 39.Dickerson RE, Chiu TK. Helix bending as a factor in protein/DNA recognition. Biopolymers. 1997;44:361–403. doi: 10.1002/(SICI)1097-0282(1997)44:4<361::AID-BIP4>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 40.Luscombe NM, Austin SE, Berman HM, Thornton JM. An overview of the structures of protein-DNA complexes. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-1-reviews001. REVIEWS001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wechsler DS, Dang CV. Opposite orientations of DNA bending by c-Myc and Max. Proc. Natl Acad. Sci. USA. 1992;89:7635–7639. doi: 10.1073/pnas.89.16.7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shulemovich K, Dimaculangan DD, Katz D, Lazar MA. DNA bending by thyroid hormone receptor: influence of half-site spacing and RXR. Nucleic Acids Res. 1995;23:811–818. doi: 10.1093/nar/23.5.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baillat D, Laitem C, Leprivier G, Margerin C, Aumercier M. Ets-1 binds cooperatively to the palindromic Ets-binding sites in the p53 promoter. Biochem. Biophy. Res. Commun. 2009;378:213–217. doi: 10.1016/j.bbrc.2008.11.035. [DOI] [PubMed] [Google Scholar]