Abstract
Human Ape2 protein has 3′ phosphodiesterase activity for processing 3′-damaged DNA termini, 3′–5′ exonuclease activity that supports removal of mismatched nucleotides from the 3′-end of DNA, and a somewhat weak AP-endonuclease activity. However, very little is known about the role of Ape2 in DNA repair processes. Here, we examine the effect of interaction of Ape2 with proliferating cell nuclear antigen (PCNA) on its enzymatic activities and on targeting Ape2 to oxidative DNA lesions. We show that PCNA strongly stimulates the 3′–5′ exonuclease and 3′ phosphodiesterase activities of Ape2, but has no effect on its AP-endonuclease activity. Moreover, we find that upon hydrogen-peroxide treatment Ape2 redistributes to nuclear foci where it colocalizes with PCNA. In concert with these results, we provide biochemical evidence that Ape2 can reduce the mutagenic consequences of attack by reactive oxygen species not only by repairing 3′-damaged termini but also by removing 3′-end adenine opposite from 8-oxoG. Based on these findings we suggest the involvement of Ape2 in repair of oxidative DNA damage and PCNA-dependent repair synthesis.
INTRODUCTION
Cellular DNA is continuously damaged by exogenous and endogenous agents. Reactive oxygen species (ROS) are a frequent source of DNA damage, and they can attack base and sugar moieties in DNA yielding a variety of lesions such as apurinic and apyrimidinic (AP) sites, DNA strand breaks, or oxidized bases (1–6). AP sites are one of the most common lesions in the cellular DNA which can also be formed spontaneously or by the action of DNA glycosylases on modified bases (7). An oxidized guanine, 7,8-dihydro-8-oxoguanine (8-oxoG), is also an abundant and highly mutagenic DNA lesion (8,9). Although with reduced efficiency replicative eukaryotic DNA polymerases (Pol), such as Polδ, can replicate through 8-oxoG; however, they incorporate A nucleotide opposite 8-oxoG more frequently than C yielding GC to TA transversions (10–12). ROS as well as the β-lyase activity of some DNA glycosylases can also induce strand breaks that have 3′-phosphate or 3′-phosphoglycolate (3′-PG) terminus which is inhibitory to DNA repair synthesis (3,13,14). Predictably thus, DNA repair mechanisms for removing 3′-damaged DNA-ends and reducing the mutagenic consequences of 8-oxoG and AP-site play a critical role in maintaining genomic stability (15–19).
In Saccharomyces cerevisiae, the base excision repair pathway constitutes the main defense against the mutagenic effect of 8-oxoG, which is initiated by Ogg1-catalysed excision of 8-oxoG from 8-oxoG/C pair (4,20–22). If escaped repair by Ogg1, during DNA replication 8-oxoG can be bypassed by replicative DNA polymerases, which dominantly insert an adenine across from the lesion generating 8-oxoG/A (10,23). Afterwards, 8-oxoG/A can be recognized by the mismatch repair system that excises the DNA fragment containing adenine opposite the lesion (24). The resulting DNA gap then can be filled in by DNA Polη, which inserts a cytosine opposite 8-oxoG resulting in 8-oxoG/C, which then can become accessible for Ogg1 (11,12). In humans, in addition to these mechanisms, the MutY protein that is missing from yeasts also plays an important role in preventing the mutagenic effect of 8-oxoG by catalyzing the excision of adenine residues from 8-oxoG/A (19,25). Recently, yeast Apn1 endonuclease has also been implicated in an alternative repair pathway to remove 3′ 8-oxoG from 3′ DNA-ends by its 3′–5′ exonuclease activity and thereby providing undamaged 3′-end to DNA synthesis (26).
Class II AP endonucleases are multifunctional enzymes that can remove AP-sites as well as 3′-blocking termini, and many of them also exhibit a 3′–5′ exonuclease activity of which function has not been well defined. In humans, the exonuclease III family of AP endonucleases has two members the Ape1 protein, the dominant AP endonuclease in human cells, and the Ape2 protein of which role is less clear. Nevertheless, the importance of Ape2 is indicated by the growth retardation and dyslymphopoiesis phenotype of Ape2 null mice (27). Whereas Ape1 exhibits a strong AP-endonuclease activity and accounts for more than 95% of the total AP endonuclease activity (28), the Ape2 protein has only a weak AP endonuclease activity but exhibits strong 3′-exonuclease and 3′-phosphodiesterase activities (29–31). In addition, Ape2 is able to remove preferentially mismatched 3′-nucleotides from DNA, which indicates a possible proofreading function for Ape2 in DNA repair synthesis (29).
Ape2 belongs to the Apn2-like protein subfamily, which in addition to the N-terminal catalytic exoIII protein domain also contains a C-terminal extension that is not present in the other members of the ExoIII family (30,32,33). This C-terminus of S. cerevisiae Apn2 contains a proliferating cell nuclear antigen (PCNA)-binding site by which it interacts with PCNA (34). PCNA is a homotrimeric protein that forms a sliding clamp around DNA and acts as a processivity factor for the replicative polymerase. PCNA also interacts with many other proteins such as translesion synthesis polymerases (35–39), which plays a crucial role in targeting them to the damage site and stimulating their enzymatic activity (40).
In the present study, we have explored further the function of human Ape2 and its interaction with PCNA in reducing the consequences of attack by ROS. We provide evidence for a role of interaction with PCNA in targeting Ape2 to hydrogen-peroxide-damaged DNA and in stimulating the 3′–5′ exonuclease and the 3′-phosphodiesterase activities of Ape2. Furthermore, we show that in addition to its role in removing 3′ blocking adducts, Ape2 is also able to remove preferentially 3′-end adenine opposite from 8-oxoG. These results suggest that Ape2 can function together with PCNA in the repair and bypass synthesis of ROS-damaged DNA.
MATERIALS AND METHODS
Purification of Ape2 and PCNA proteins
Wild-type Ape2 and PCNA proteins were purified as described previously (29,41). PCNA-binding site mutant Ape2 Y396A F397A protein was generated on plasmid pIL1045 by the QuickChange site-directed mutagenesis kit (Stratagene) using oligonucleotides O1052 (5′-GAA AAA CCT GAA GAG CGC CGC TCA GTC CTC CCC TAG CTG-3′) and O1053 (5′-CAG CTA GGG GAG GAC TGA GCG GCG CTC TTC AGG TTT TTC-3′) resulting in plasmid pIL1278. For expressing Ape2 proteins we cloned the mutant Ape2 cDNA in fusion with the glutathione S-transferase (GST) gene under the control of the S. cerevisiae galactose-inducible phosphoglycerate promoter using the plasmid pBJ842, which resulted in plasmid pIL1285 expressing Ape2 Y396A F397A protein.
DNA substrates
DNA substrate (S1) used for the 3′–5′ exonuclease assay was generated by annealing a 75 nt oligomer template O21 (5′-AGC TAC CAT GCC TGC CTC AAG AAT TCG TAA CAT GCC TAC ACT GGA GTA CCG GAG CAT CGT CGT GAC TGG GAA AAC-3′) to the 31 nt 5′-32P-labelled primer O73 (5′-CGA CGA TGC TCC GGT ACT CCA GTG TAG GCA T-3′). DNA substrate (S2) used for the 3′-phosphodiesterase assay was generated by annealing a 70 nt template O105 (5′-AGC TGG ATT CGT GTC AGC CGC TAG CCA TTA GCT GGC GCA TTC CTC GAG AGA GTC CCG AGC ATC GTG ACT G-3′) to the 35 nt 5′-32P-labelled primer O102 (5′-CAG TCA CGA TGC TCG GGA CTC TCT CGA GGA ATG CG-PG-3′) containing a 3′-PG terminus. DNA substrate (S3) used for the AP endonuclease assay was generated by annealing a 75 nt template O82 (5′-GTT TTC CCA GTC ACG ACG ATG CTC CGG TAC TCC AGT GTA GGC ATA TTA CGA ATT CTT GAG GCA GGC ATG GTA GCT-3′) to the 75 nt 5′-32P-labelled oligomer O14 (5′-AGC TAC CAT GCC TGC CTC AAG AAT TCG TAA XAT GCC TAC ACT GGA GTA CCG GAG CAT CGT CGT GAC TGG GAA AAC-3′). In this oligomer X represents the place of a single tetrahydrofuran, an abasic site analogue. DNA substrates S4 and S5 used for assaying the Ape2 3′–5′ exonuclease activity opposite from the 8-oxoG were generated by annealing a 75 nt oligomer O20 (5′-AGC TAC CAT GCC TGC CTC AAG AAT TCG TAA 8-oxoGAT GCC TAC ACT GGA GTA CCG GAG CAT CGT CGT GAC TGG GAA AAC-3′) to the 32 nt 5′-32P-labelled oligomers O430 (5′-CGA CGA TGC TCC GGT ACT CCA GTG TAG GCA TA-3′) or O83 (5′-CGA CGA TGC TCC GGT ACT CCA GTG TAG GCA TC-3′), respectively. For control reactions shown in Figure 4C, the 8-oxoG residue of the O20 75 nt oligomer was changed to T or G.
Figure 4.
Preferential removal of A opposite from 8-oxoG by Ape2. (A) The 3′–5′ exonuclease activity of Ape2 (1 nM) was assayed on partial heteroduplexes (10 nM) containing an A (lanes 1–6) or a C (lanes 7–12) nucleotides at the 3′-primer ends opposite from 8-oxoG on the template strands. (B) Michaelis-Menten plots for calculating the efficiency of A and C removal opposite from 8-oxoG. Ape2 (1 mM) was incubated in the presence of increasing substrate concentration for 1 min at 37°C. (C) The steady-state parameters Vmax and Km values for removing A and C opposite from 8-oxoG and matched undamaged template residues represent the average from three independent determination and the standard deviations are indicated (±).
Ape2 enzyme reactions
A standard reaction mixture (10 µl) contained 40 mM Tris–HCl (pH 7.5), 8 mM MgCl2, 1 mM DTT, 100 µg/ml BSA, 10 nM DNA substrate and 1 nM Ape 2 protein. Where indicated, 5 nM PCNA was added together with Ape2 (Figure 2), the reaction buffer was supplemented with 150 mM NaCl (Figure 1D), and the MgCl2 was substituted with 0.5 mM MnCl2 (Figure 4). Reactions were assembled on ice, incubated at 37°C for 5 min, and stopped by the addition of loading buffer (20 µl) containing 20 mM EDTA, 95% formamide, 0.25% xylene cyanol and 0.25% bromophenol blue. The reaction products were resolved on 10% polyacrylamide gel containing 8 M urea followed by analyzing the band intensities with Molecular Dynamics STORM phosphorimager and ImageQuant 5.0 software.
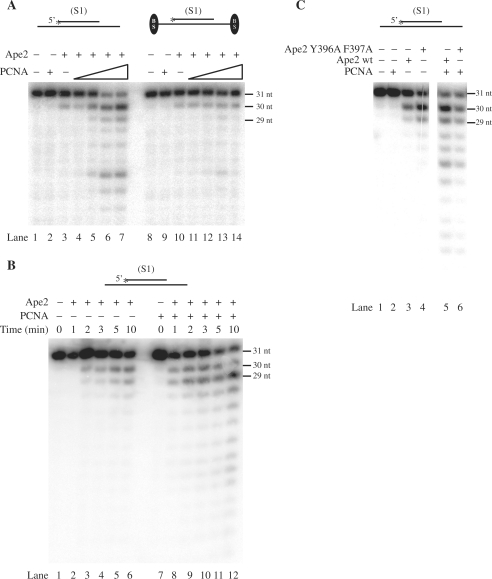
Figure 2.
Mechanistic insight into PCNA-dependent stimulation of Ape2 exonuclease. (A) PCNA has to be loaded onto DNA in order to stimulate the 3′–5′ exonuclease activity of Ape2. The 3′–5′ exonuclease activity of Ape2 was assayed using a partial DNA duplex substrate (S1) without (lanes 1–7) or with (lanes 8–14) a biotin-streptavidin complex on the template ends. The DNA substrate (10 nM) was incubated with Ape2 (1 nM) in the presence of PCNA at increasing concentration (0.1 nM, 1 nM, 5 nM, 20 nM). Lane 1, no protein was added; lane 2, 20 nM PCNA was assayed. (B) The kinetics of the exonuclease activity of Ape2 (1 nM) was compared in the absence (lanes 1–6) or presence (lanes 7–12) of PCNA (5 nM) using a partial heteroduplex DNA (10 nM). (C) Effect of mutations in the PIP-box of Ape2 on its PCNA-dependent stimulation. The 3′–5′ exonuclease activity of wild type Ape2 and mutant Ape2 Y396A F397A proteins (1 nM) was assayed using a partial DNA heteroduplex substrate (10 nM) in the absence or presence of PCNA (5 nM) as indicated.
Figure 1.
PCNA-dependent stimulation of the 3′–5′ exonuclease and 3′ phosphodiesterase activities of Ape2. (A) Schematic representation of Ape2 protein structure. The N-terminus of Ape2 contains the conserved Exonuclease III domain (white box), characteristic of class II AP endonucleases, while the C-terminus is unique to Apn2/Ape2 subfamily and has a putative PCNA binding site (PIP) between 390–397 residues (black box). (B) The 3′-5′ exonuclease activity of Ape2 was assayed using a partial DNA heteroduplex substrate generated by annealing a 5′-labelled 31 nt oligonucleotide to the middle of a 75 nt oligonucleotide (S1). (C) The phosphodiesterase activity of Ape2 was tested on a DNA substrate containing a 5′-labelled 35 nt oligomer with a 3′-PG terminus annealed to a 70-nt oligomer (S2). The positions of the reaction product and the substrate are indicated by 3′-OH and 3′-PG, respectively. (D) The AP-endonuclease activity of Ape2 was assayed using a 75 nt double-stranded DNA substrate containing a single AP-site at position 31 on the 5′-labelled strand (S3). The position of the incision product at 30 nt is indicated. DNA substrates (10 nM) were incubated without any protein (lanes 1), with 5 nM PCNA (lanes 2), with 1 nM Ape2 (lanes 3), or with 1 nM Ape2 together with 5 nM PCNA (lanes 4) for 5 min at 37°C. The reactions (10 μl) were stopped by addition of loading buffer (20 μl) containing 20 mM EDTA, 95% formamide, 0.25% xylene cyanol and 0.25% bromophenol blue, and the reaction products were resolved on 10% polyacrylamide gel containing 8 M urea before autoradiography.
Localization study of Ape2
For localization studies we cloned the wild-type and PCNA-binding site mutant Ape2 cDNAs from plasmids pIL1045 and pIL1278 in fusion with N terminal Flag-tag into plasmid pRK2F, which resulted in plamids pIL1565, pIL1566, respectively. SV40-transformed human fibroblasts MRC5V1 were grown in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% FCS (Sigma). Transfection was carried out using Exgene transfection reagent (Fermentas) according to the instruction of the manufacturer. Cells cultivated and transfected on coverslips were washed once with PBS followed by incubation with 0.06 mM hydrogen-peroxide for 6 h or with 0.01% MMS for 1 h before processing for immunoflourescence.
Flag-Ape2 was stained using anti-Flag antibody (Sigma, M2 clone) diluted 1:200 and FITC-conjugated anti-mouse antibody (Calbiochem) diluted 1:10. To visualize endogenous PCNA, cells were incubated with anti-PCNA antibody (Santa Cruz, sc:7907) diluted 1:200 followed by incubation with Alexa568 conjugated anti-rabbit secondary antibody (Molecular probes) diluted 1:500. Samples were mounted in 25% glycerol in PBS containing 1 μg/ml DAPI. Microscopy was carried out using Olympus FV1000 confocal laser scanning microscope.
Steady-state kinetic analysis
Steady-state kinetic analysis for Ape2 DNA cleavage efficiency opposite from 8-oxoG was assayed on S4 and S5 DNA substrates. Ape2 was incubated with DNA substrates at increasing concentration (2–162 nM) for 1 min under standard reaction conditions. The gel bands intensity of the substrates and products were quantified with Molecular Dynamics STORM Phosphor Imager and the ImageQuant 5.0 software. The cleaved DNA substrate (nM) was plotted to the starting DNA substrate concentration. The data were fit by nonlinear regression, using SigmaPlot 5.0, to the Michaelis-Menten equation describing a hyperbola v = [Vmax × (DNA substrate)/Km + (DNA substrate)]. Apparent Vmax and Km steady-state parameters were obtained from the fit and used to calculate the efficiency of the 3′–5′ exonuclease activity.
RESULTS
PCNA stimulates the 3′-phosphodiesterase and 3′–5′ exonuclease activities of Ape2
Human Ape2 possesses a consensus PCNA-interacting protein (PIP) motif, after its conserved ExoIII domain at the C-terminal portion spanning amino-acids 390–397 (Figure 1A). A similar sequence motif is present in many proteins playing role in DNA metabolism, such as replicative and translesion synthesis DNA polymerases or Fen1 endonuclease, and via binding to this motif PCNA stimulates their enzymatic activity (42,43). We hypothesized that PCNA binding may modulate the 3′-phosphodiesterase, 3′–5′ exonuclease, or AP-endonuclease activities of Ape2, and to test this possibility we assayed Ape2 in the presence of PCNA. First, we examined the 3′–5′ exonuclease activity of purified Ape2 on a partial heteroduplex DNA substrate containing a 5′-32P-labelled strand with recessed 3′ terminus (Figure 1B). We incubated this DNA substrate in the presence of limiting Ape2 concentration which predominantly resulted in the removal of only one nucleotide from the recessed 3′ end (Figure 1B, lane 3). The addition of PCNA to this reaction greatly stimulated the 3′–5′ exonuclease activity of the Ape2, as there was greatly enhanced hydrolysis of the labelled strand as well as the generation of shorter digestion products. Next, we tested if PCNA can also stimulate the 3′-phosphodiesterase activity of Ape2 by monitoring the removal of 3′-PG group from the 5′ labelled strand of a double-stranded DNA substrate. Enzymatic removal of the 3′-PG group results in a 3′-hydroxyl terminus which confers slower electrophoretic mobility to DNA on a denaturing polyacrylamide gel. We found that addition of PCNA enhanced the removal of the 3′-PG by Ape2 (Figure 1C). Finally, we assayed if PCNA can enhance the AP endonuclease activity of Ape2 using a 75 nt duplex DNA substrate in which the 5′-labelled strand contained a single abasic site (Figure 1D). On such a substrate AP endonucleases can catalyse hydrolyses of the 5′-phosphodiester bound at the abasic site, generating a 30 nt labelled oligomer. We found that PCNA had no effect on the AP endonuclease activity of Ape2. In summary, we discovered that PCNA has a robust stimulatory effect on the 3′–5′ exonuclease and 3′-PG removal activities of Ape2.
Mechanistic insight into the functional interaction of Ape2 with PCNA
Having found that PCNA stimulates the 3′–5′ exonuclease activity but does not enhance the AP endonuclease activity of Ape2 indicated that PCNA may affect Ape2 only when it encircles DNA and only at a primer template junction. To prove this hypothesis, we compared the PCNA-dependent stimulation of Ape2 using two different DNA substrates having free or blocked ends on which PCNA can or can not slide on, respectively. As shown on Figure 2A, when we blocked PCNA loading by introducing biotin-streptavidin complex at both ends of the DNA, PCNA was not able to stimulate the exonuclease activity of Ape2 (compare Figure 2A, from lanes 1–7 to lanes 8–14). Thus, PCNA has to be loaded onto DNA in order to be able to stimulate the Ape2. The stimulation was proportional to PCNA concentration and reached saturation at PCNA concentration higher than the concentration of Ape2 as can be expected if PCNA acts via physically interacting with Ape2 (Figure 2A).
Next we asked if PCNA influences the processivity of Ape2 by comparing the kinetics of its exonuclease activity in the absence or presence of PCNA (Figure 2B). We found that in single hit reaction that dominated at early time points during which only a small percentage of the substrate DNA was processed, Ape2 alone took off mostly only a single nucleotide, while Ape2 together with PCNA removed several nucleotides. We concluded that PCNA not only increases the access of Ape2 to primer/template junction but also increases the processivity of Ape2 exonuclease.
It has been shown for a number of proteins that their PIP-motif autonomously is able to mediate their interaction with the interdomain connector loop (IDCL) of PCNA. For example, the crystal structure of FEN1 shows that its conserved PIP motif permits the docking of this region to the IDCL of PCNA via the insertion of the highly conserved hydrophobic residues into the hydrophobic pocket of IDCL (44). To gain further insight into the nature of the interaction between Ape2 and PCNA, we examined if the conserved PIP-motif encompassing 390–397 residues in Ape2 mediated the functional interaction of Ape2 with PCNA. We generated Ape2 Y396A F397A mutant, in which the highly conserved hydrophobic residues of the PIP-motif were changed to alanine residues (Figure 1A). The mutant protein was purified to apparent homogeneity and during purification it behaved similarly to the wild-type protein and displayed identical electrophoretic mobility on denaturing polyacrylamide gel (data not shown). The Y396A F397A point mutations did not affect the active centre of Ape2 exonuclease since the mutant Ape2 protein exhibited wild-type level of 3′–5′ exonuclease activity. We found that the PCNA-dependent stimulation of the exonuclease activity of the Ape2 Y396A F397A mutant protein was only reduced but not completely impaired as compared to that of wild-type Ape2 (Figure 2C). Therefore in addition to the conserved PIP motif located at 390–397 positions there should be other residues in the Ape2 protein sequence that mediate the stimulatory effect of PCNA.
Hydrogen-peroxide treatment induces colocalization of Ape2 with PCNA
To gain further insight into the functional significance of Ape2 interaction with PCNA, we studied the cellular localization of Ape2 and examined if it could colocalize with PCNA. In eukaryotic cells, many proteins playing role in DNA repair and replication form discrete nuclear foci. For example, PCNA forms nuclear foci that represent the sites of ongoing DNA replication and vary in morphology and number during S phase. To visualise Ape2, we generated a construct encoding a FLAG-tag fused in frame to the N-terminus of Ape2, and transfected it into SV40-transformed human fibroblasts MRC5. As revealed by fluorescence confocal microscopy using anti-FLAG monoclonal mouse antibody and FITC-conjugated secondary antibody, in most of the cells Ape2 distributed homogenously within the nucleus (Figure 3A), and only in a very few cases, Ape2 localized in discrete nuclear foci or showed cytoplasmic localization. As treatment of cells with DNA damaging agents can result in redistribution of several DNA repair proteins into nuclear foci formed at the lesion sites, we also tested if treatment by DNA damaging agents can redistribute Ape2. First, we examined the effect of MMS that alkylates bases in DNA; however, in MMS treated cells, the localization of Ape2 did not change and remained similar to untreated cells. Next, we chose an oxidative DNA-damaging agent, hydrogen-peroxide, which produces DNA strand breaks containing 3′-phosphate or 3′-phosphoglycolate termini as well as attacks bases in DNA, and 7,8-dihydro-8-oxoguanine (8-oxoG) is one of abundant adducts formed. Importantly, hydrogen-peroxide treatment induced the redistribution of Ape2 into discrete fluorescent spots within the nucleus suggesting the presence of high local concentration of Ape2 (Figure 3A). To examine if Ape2 colocalised with PCNA, we also stained endogenous PCNA with anti-PCNA antibody and found that hydrogen-peroxide treatment induced almost complete colocalization of Ape2 with PCNA (Figure 3B). The Ape2 Y396A F397A mutant retained only partial ability for colocalization with PCNA as revealed by calculating the percentage of colocalised Ape2–PCNA foci (Figure 3C). Thus we conclude that the 390–397 PIP motif of Ape2 has a role in mediating the targeting of Ape2 to PCNA and hydrogen-peroxide-induced DNA damage sites, but other motifs in Ape2 could partially substitute for this function.
Figure 3.
Localization of the wild type and PIP mutant Ape2. (A) MRC5 cells were transfected with FLAG-Ape2. Twenty-four hours after transfection, cells were treated with MMS (0.01%, 1 h) or H2O2 (0.06 mM, 6 h) before fixation. FLAG-Ape2 proteins were immunostained with anti-FLAG mAb. (Magnification: 600×). (B) Colocalization of Ape2 with PCNA. Cells were transfected with FLAG-Ape2 or FLAG-Ape2 Y396A F397A expressing constructs followed by H2O2-treatment before testing for colocalization of Ape2 with PCNA using anti-FLAG and anti-PCNA antibodies, respectively. (C) Quantitative analysis of Ape2 and PCNA colocalization. Nuclear spots of wild type or mutant Ape2 and PCNA were counted, and the percentage of colocalized Ape2–PCNA foci was calculated and graphed with error bars representing standard deviation.
Ape2 preferentially removes adenine opposite from 8-oxoG
The hydrogen-peroxide-induced nuclear foci formation of Ape2 and its colocalization with PCNA indicated that Ape2 may play roles in the repair of oxidatively damaged DNA. Attack by hydrogen-peroxide can cause DNA strand breaks containing modified 3′-termini like 3′-PG residues as well as oxidatively damaged bases such as 8-oxoG. Having discovered and characterized the 3′-PG processing activity of Ape2 our attention turned to its possible role in 8-oxoG repair. Opposite 8-oxoG, eukaryotic replicative DNA polymerases tend to insert an adenine; consequently, 8-oxoG is highly mutagenic and causes G:C to T:A transversions. Since Ape2 is able to remove mismatched nucleotides from the 3′-end, we investigated if Ape2 could also remove adenine opposite from 8oxo-G which would indicate that Ape2 has the enzymatic property to reduce the mutagenic consequences of 8-oxoG bypass. To test this hypothesis, we compared the 3′-5′ exonuclease activity of Ape2 on partial heteroduplex DNA substrates containing either a cytosine or an adenine on the 3′ recessed end opposite 8-oxoG (Figure 4A). We found that Ape2 removed A nucleotide more efficiently than C nucleotide opposite from 8-oxoG residue. For example, after 5 min incubation, Ape2 removed about 80% of A and <10% of C opposite from 8-oxoG (Figure 4A, compare lanes 5–11). PCNA had a robust stimulation effect on both A and C removal opposite from 8-oxoG, but it did not change the preference of removal since the ratio of A and C cleavage remained the same regardless of the presence of PCNA (data not shown). To confirm further the preference for A removal, we also determined the steady-state kinetic parameters Vmax and Km of the 3′–5′ exonuclease activity of the Ape2 for A and C removal opposite from 8-oxoG and undamaged template residues (Figure 4B and C). Comparing the Vmax/Km values describing the efficiency of the 3′–5′ exonuclease activity, we found that Ape2 removed A opposite from the 8-oxoG 8.5-fold more efficiently than it removed C opposite from this lesion (Figure 4C). Furthermore, Ape2 removed A opposite from 8-oxoG more efficiently than opposite from undamaged T (Figure 4C). Thus, Ape2 could play a proofreading role in PCNA dependent 8-oxoG bypass synthesis.
DISCUSSION
In this study we examined whether Ape2 can function together with PCNA in reducing the mutagenic consequences of attack by ROS. We found that PCNA is able to stimulate the 3′-phosphodiesterase and 3′-5′ exonuclease activities of Ape2. Furthermore, we showed that Ape2 can also remove 3′ adenine opposite 8-oxoG, which can arise if replicative polymerase inserts adenine opposite this lesion. In concert with the robust effect of PCNA on these biochemical activities of Ape2, we also demonstrated that hydrogen-peroxide treatment induced the redistribution of Ape2 into nuclear foci where it colocalized with PCNA.
Previously, we reported that purified Ape2 is a multifunctional enzyme, which has strong 3′-phosphodiesterase and 3′–5′exonuclease activities but a somewhat weak AP endonuclease activity (29). One can argue, however, that interacting protein partners could modify the intensity of these activities. Indeed, we found that PCNA strongly stimulated the 3′-phosphodiesterase and 3′–5′ exonuclease activities of Ape2, and for the stimulation to occur PCNA has to encircle the DNA. However, PCNA exhibited no effect on the AP-endonuclease activity of Ape2 despite that PCNA similarly to the exonuclease substrate could slide onto the substrate, as well. From these results we conclude that stimulation of Ape2 occurs only if PCNA encircles DNA and interacts with Ape2 at a primer-template junction.
To support the functional significance of interaction of Ape2 with PCNA, we asked whether Ape2 could colocalize with PCNA in human cells. We found that Ape2 distributed homogenously in the nucleus and did not colocalize with PCNA in untreated cells. We reasoned that treatment with DNA damaging agents may lead to redistribution of Ape2 to DNA lesions if, Ape2 was a DNA repair protein. However, alkylating agent, MMS, did not affect the distribution of Ape2. MMS can methylate bases in DNA, particularly adenine at the N3 position (3 MeA) and guanine at the N7 position (7 MeG). The alkylated bases can then be removed by the action of N-methylpurine DNA glycosylases and the resulting AP-sites could then be targeted by AP-endonucleases (45). Consequently, finding no evidence for the redistribution of Ape2 to the sites of alkylated bases or AP-sites may indicate that Ape2 has no role in their repair, and it is in concert with its very week AP endonuclease activity. Remarkably, however, treatment of cells with hydrogen-peroxide which can induce DNA strand breaks with 3′-modified termini as well as various types of oxidized bases such as 8-oxoG caused the redistribution of Ape2 into PCNA containing nuclear foci. Therefore, we conclude that Ape2 in cooperation with PCNA could have a role in the repair of DNA damaged by ROS.
Previous data indicated that Ape2 can be co-immunoprecipitated with PCNA from undamaged cells (46). In concert, using purified Ape2 and PCNA and testing for a functional interaction, we found evidence for a direct interaction between Ape2 and PCNA. However, we were not able to isolate a stable Ape2–PCNA complex using purified proteins neither in the presence nor in the absence of DNA (data not shown) and we found no evidence for colocalization of Ape2 with PCNA in undamaged cells since this became apparent only after treating cells with hydrogen-peroxide. Therefore, we suggest a quite dynamic interaction for Ape2 and PCNA which is triggered by ROS-damaged DNA. Our finding that mutational inactivation of the highly conserved PIP motif of Ape2 encompassing residues 390–397 impairs only partially the functional interaction of Ape2 with PCNA suggests that other PCNA-binding motifs in Ape2 could also play role in mediating their interaction. In this regard we note that Ape2 exhibits other PIP like motifs such as the one between its 486–496 amino acid residues, and that PCNA interaction can also be mediated by non-canonical PCNA-binding boxes, as well (47). There are also examples of proteins that bind PCNA via two separate PCNA-binding sites. For example, recent finding indicates that human traslesion synthesis DNA polymerase η contains two functional PCNA interacting boxes which can substitute for one another and that the mutational alteration of both of these domains causes a complete loss in hPolη's ability to function in translesion synthesis (48). Thus further studies could reveal interesting details about the mechanism and regulation of Ape2 and PCNA interaction.
One of the main points of our study is that the enzymatic activity of Ape2 is suitable for decreasing the mutagenic consequences of ROS by various ways. In addition to cleansing 3′ damaged DNA termini, we found that Ape2 can preferentially remove 3′adenine opposite from 8-oxoG. Since replicative DNA polymerases synthesize through 8-oxoG by incorporating dAMP, the occurrence of 3′ adenine opposite 8-oxoG in the template is quite frequent. The molecular rationale for this is that the formation of an 8-oxoG(anti) •C(anti) base pair during replication induces template and polymerase distortions similar to those seen when the active site of DNA polymerase encounters mismatches. By contrast, inaccurate bypass of 8-oxoG results from the ability of the 8-oxoG(syn)•A (anti) base pair to mimic a normal base pair, which can explain the lack of this error detection by replicative DNA polymerases. This could lead to G:C → T:A transversion during the second round of DNA synthesis (49). Further studies could reveal if simultaneous binding of PCNA homotrimer to Ape2 and to a DNA polymerase plays a role in promoting error-free bypass of 8-oxoG as well as efficient repair of 3′ oxidative DNA damage by coordinating the degradative and polymerizing activities of two different proteins.
Although here we provide only biochemical evidence for a possible role of Ape2 in decreasing the mutagenic consequences of 8-oxoG, we note that yeast genetic evidence have already indicated an alternative role for an AP endonuclease, Apn1, in 8-oxoG repair (26). Thus, using the same genetic approach, it would be interesting to examine if the yeast Apn2, the homologue of human Ape2, can increase the fidelity of 8-oxoG bypass synthesis. In concert with our findings that Ape2 has a role in the repair of H2O2-induced DNA damage yeast genetic studies have indicated the involvement of APN2 in a pathway alternate to APN1 in repairing strand breaks arising from the reaction of DNA with reactive oxygen species (50).
In summary, based on the colocalization of Ape2 with PCNA upon DNA damage by ROS, their functional interaction, and the 3′ damaged termini removal and proofreading activity of Ape2 opposite 8-oxoG we suggest a role for Ape2 in PCNA mediated repair of oxidatively damaged DNA.
FUNDING
This work was supported by the Wellcome Trust International Senior Research Fellowship, Howard Hughes Medical Institute grant 55005612, Hungarian Science Foundation grant OTKA 077495, Bolyai János fellowships, the Hungarian National Office for Research and Technology grant KFKT-1-2006-0010. Funding for open access charge: Hungarian Science Foundation grant, OTKA.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Katalin Illesne Kovacs and Ildiko Kravjar for technical assistance.
REFERENCES
- 1.Epe B, Hegler J. Oxidative DNA damage: endonuclease fingerprinting. Methods Enzymol. 1994;234:122–131. doi: 10.1016/0076-6879(94)34083-8. [DOI] [PubMed] [Google Scholar]
- 2.Bacsi A, Chodaczek G, Hazra TK, Konkel D, Boldogh I. Increased ROS generation in subsets of OGG1 knockout fibroblast cells. Mech. Ageing Dev. 2007;128:637–649. doi: 10.1016/j.mad.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Demple B, Harrison L. Repair of Oxidative Damage to Dna – Enzymology and Biology. Annu. Rev. Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 4.Girard PM, Boiteux S. Repair of oxidized DNA bases in the yeast Saccharomyces cerevisiae. Biochimie. 1997;79:559–566. doi: 10.1016/s0300-9084(97)82004-4. [DOI] [PubMed] [Google Scholar]
- 5.Henle ES, Linn S. Formation, prevention, and repair of DNA damage by iron hydrogen peroxide. J. Biol. Chem. 1997;272:19095–19098. doi: 10.1074/jbc.272.31.19095. [DOI] [PubMed] [Google Scholar]
- 6.Guillet M, Boiteux S. Origin of endogenous DNA abasic sites in Saccharomyces cerevisiae. Mol. Cell Biol. 2003;23:8386–8394. doi: 10.1128/MCB.23.22.8386-8394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boiteux S, Guillet M. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair. 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Boiteux S, Gellon L, Guibourt N. Repair of 8-oxoguanine in Saccharomyces cerevisiae: interplay of DNA repair and replication mechanisms. Free Radic. Biol. Med. 2002;32:1244–1253. doi: 10.1016/s0891-5849(02)00822-5. [DOI] [PubMed] [Google Scholar]
- 9.Kozmin S, Slezak G, Reynaud-Angelin A, Elie C, de RY, Boiteux S, Sage E. UVA radiation is highly mutagenic in cells that are unable to repair 7,8-dihydro-8-oxoguanine in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2005;102:13538–13543. doi: 10.1073/pnas.0504497102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moriya M. Single-stranded shuttle phagemid for mutagenesis studies in mammalian cells: 8-oxoguanine in DNA induces targeted G.C–>T.A transversions in simian kidney cells. Proc. Natl Acad. Sci. USA. 1993;90:1122–1126. doi: 10.1073/pnas.90.3.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de PM, Slezak G, uffret van Der KP, Boiteux S. The post-replication repair RAD18 and RAD6 genes are involved in the prevention of spontaneous mutations caused by 7,8-dihydro-8-oxoguanine in Saccharomyces cerevisiae. Nucleic Acids Res. 2004;32:5003–5010. doi: 10.1093/nar/gkh831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase eta. Nat. Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 13.Mitra S, Hazra TK, Roy R, Ikeda S, Biswas T, Lock J, Boldogh I, Izumi T. Complexities of DNA base excision repair in mammalian cells. Mol. Cells. 1997;7:305–312. [PubMed] [Google Scholar]
- 14.Seeberg E, Eide L, Bjoras M. The base excision-repair pathway. Trends Biochem. Sci. 1995;20:391–397. doi: 10.1016/s0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- 15.Epe B. Role of endogenous oxidative DNA damage in carcinogenesis: what can we learn from repair-deficient mice? Biol. Chem. 2002;383:467–475. doi: 10.1515/BC.2002.049. [DOI] [PubMed] [Google Scholar]
- 16.Eiberger W, Volkmer B, Amouroux R, Dherin C, Radicella JP, Epe B. Oxidative stress impairs the repair of oxidative DNA base modifications in human skin fibroblasts and melanoma cells. DNA Repair. 2008;7:912–921. doi: 10.1016/j.dnarep.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 17.Khobta A, Kitsera N, Speckmann B, Epe B. 8-Oxoguanine DNA glycosylase (Ogg1) causes a transcriptional inactivation of damaged DNA in the absence of functional Cockayne syndrome B (Csb) protein. DNA Repair. 2009;8:309–317. doi: 10.1016/j.dnarep.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, Hodges AK, Davies DR, David SS, Sampson JR, et al. Inherited variants of MYH associated with somatic G:C–>T:A mutations in colorectal tumors. Nat. Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 19.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Girard PM, Guibourt N, Boiteux S. The Ogg1 protein of Saccharomyces cerevisiae: a 7,8-dihydro-8-oxoguanine DNA glycosylase/AP lyase whose lysine 241 is a critical residue for catalytic activity. Nucleic Acids Res. 1997;25:3204–3211. doi: 10.1093/nar/25.16.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Girard PM, D'Ham C, Cadet J, Boiteux S. Opposite base-dependent excision of 7,8-dihydro-8-oxoadenine by the Ogg1 protein of Saccharomyces cerevisiae. Carcinogenesis. 1998;19:1299–1305. doi: 10.1093/carcin/19.7.1299. [DOI] [PubMed] [Google Scholar]
- 22.Thomas D, Scot AD, Barbey R, Padula M, Boiteux S. Inactivation of OGG1 increases the incidence of G. C–>T. A transversions in Saccharomyces cerevisiae: evidence for endogenous oxidative damage to DNA in eukaryotic cells. Mol. Gen. Genet. 1997;254:171–178. doi: 10.1007/s004380050405. [DOI] [PubMed] [Google Scholar]
- 23.Osterod M, Hollenbach S, Hengstler JG, Barnes DE, Lindahl T, Epe B. Age-related and tissue-specific accumulation of oxidative DNA base damage in 7,8-dihydro-8-oxoguanine-DNA glycosylase (Ogg1) deficient mice. Carcinogenesis. 2001;22:1459–1463. doi: 10.1093/carcin/22.9.1459. [DOI] [PubMed] [Google Scholar]
- 24.Ni TT, Marsischky GT, Kolodner RD. MSH2 and MSH6 are required for removal of adenine misincorporated opposite 8-oxo-guanine in S. cerevisiae. Mol. Cell. 1999;4:439–444. doi: 10.1016/s1097-2765(00)80346-9. [DOI] [PubMed] [Google Scholar]
- 25.Noll DM, Gogos A, Granek JA, Clarke ND. The C-terminal domain of the adenine-DNA glycosylase MutY confers specificity for 8-oxoguanine.adenine mispairs and may have evolved from MutT, an 8-oxo-dGTPase. Biochemistry. 1999;38:6374–6379. doi: 10.1021/bi990335x. [DOI] [PubMed] [Google Scholar]
- 26.Ishchenko AA, Yang X, Ramotar D, Saparbaev M. The 3′->5′ exonuclease of Apn1 provides an alternative pathway to repair 7,8-dihydro-8-oxodeoxyguanosine in Saccharomyces cerevisiae. Mol. Cell Biol. 2005;25:6380–6390. doi: 10.1128/MCB.25.15.6380-6390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ide Y, Tsuchimoto D, Tominaga Y, Nakashima M, Watanabe T, Sakumi K, Ohno M, Nakabeppu Y. Growth retardation and dyslymphopoiesis accompanied by G2/M arrest in APEX2-null mice. Blood. 2004;104:4097–4103. doi: 10.1182/blood-2004-04-1476. [DOI] [PubMed] [Google Scholar]
- 28.Wilson DM, Takeshita M, Grollman AP, Demple B. Incision Activity of Human Apurinic Endonuclease (Ape) at Abasic Site Analogs in Dna. J. Biol. Chem. 1995;270:16002–16007. doi: 10.1074/jbc.270.27.16002. [DOI] [PubMed] [Google Scholar]
- 29.Burkovics P, Szukacsov V, Unk I, Haracska L. Human Ape2 protein has a 3′-5′ exonuclease activity that acts preferentially on mismatched base pairs. Nucleic Acids Res. 2006;34:2508–2515. doi: 10.1093/nar/gkl259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hadi MZ, Wilson D.M., III Second human protein with homology to the Escherichia coli abasic endonuclease exonuclease III. Environ. Mol. Mutagen. 2000;36:312–324. [PubMed] [Google Scholar]
- 31.Hadi MZ, Ginalski K, Nguyen LH, Wilson D.M., III Determinants in nuclease specificity of Ape1 and Ape2, human homologues of Escherichia coli exonuclease III. J. Mol. Biol. 2002;316:853–866. doi: 10.1006/jmbi.2001.5382. [DOI] [PubMed] [Google Scholar]
- 32.Johnson RE, Torres-Ramos CA, Izumi T, Mitra S, Prakash S, Prakash L. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev. 1998;12:3137–3143. doi: 10.1101/gad.12.19.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Unk I, Haracska L, Johnson RE, Prakash S, Prakash L. Apurinic endonuclease activity of yeast Apn2 protein. J. Biol. Chem. 2000;275:22427–22434. doi: 10.1074/jbc.M002845200. [DOI] [PubMed] [Google Scholar]
- 34.Unk I, Haracska L, Gomes XV, Burgers PMJ, Prakash L, Prakash S. Stimulation of 3′–> 5′ exonuclease and 3′-phosphodiesterase activities of yeast Apn2 by proliferating cell nuclear antigen. Mol. Cell Biol. 2002;22:6480–6486. doi: 10.1128/MCB.22.18.6480-6486.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haracska L, Kondratick CM, Unk I, Prakash S, Prakash L. Interaction with PCNA is essential for yeast DNA polymerase eta function. Mol. Cell. 2001;8:407–415. doi: 10.1016/s1097-2765(01)00319-7. [DOI] [PubMed] [Google Scholar]
- 36.Haracska L, Johnson RE, Unk I, Phillips B, Hurwitz J, Prakash L, Prakash S. Physical and functional interactions of human DNA polymerase eta with PCNA. Mol. Cell Biol. 2001;21:7199–7206. doi: 10.1128/MCB.21.21.7199-7206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haracska L, Johnson RE, Unk I, Phillips BB, Hurwitz J, Prakash L, Prakash S. Targeting of human DNA polymerase iota to the replication machinery via interaction with PCNA. Proc. Natl Acad. Sci. USA. 2001;98:14256–14261. doi: 10.1073/pnas.261560798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haracska L, Kondratick CM, Unk I, Prakash S, Prakash L. Interaction with PCNA is essential for yeast DNA polymerase eta function. Mol. Cell. 2001;8:407–415. doi: 10.1016/s1097-2765(01)00319-7. [DOI] [PubMed] [Google Scholar]
- 39.Haracska L, Unk I, Johnson RE, Phillips BB, Hurwitz J, Prakash L, Prakash S. Stimulation of DNA synthesis activity of human DNA polymerase kappa by PCNA. Mol. Cell Biol. 2002;22:784–791. doi: 10.1128/MCB.22.3.784-791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boldogh I, Milligan D, Lee MS, Bassett H, Lloyd RS, McCullough AK. hMYH cell cycle-dependent expression, subcellular localization and association with replication foci: evidence suggesting replication-coupled repair of adenine:8-oxoguanine mispairs. Nucleic Acids Res. 2001;29:2802–2809. doi: 10.1093/nar/29.13.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Unk I, Hajdu I, Fatyol K, Szakal B, Blastyak A, Bermudez V, Hurwitz J, Prakash L, Prakash S, Haracska L. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc. Natl Acad. Sci. USA. 2006;103:18107–18112. doi: 10.1073/pnas.0608595103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gomes XV, Burgers PM. Two modes of FEN1 binding to PCNA regulated by DNA. EMBO J. 2000;19:3811–3821. doi: 10.1093/emboj/19.14.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haracska L, Acharya N, Unk I, Johnson RE, Hurwitz J, Prakash L, Prakash S. A single domain in human DNA polymerase iota mediates interaction with PCNA: implications for translesion DNA synthesis. Mol. Cell Biol. 2005;25:1183–1190. doi: 10.1128/MCB.25.3.1183-1190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hosfield DJ, Mol CD, Shen B, Tainer JA. Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell. 1998;95:135–146. doi: 10.1016/s0092-8674(00)81789-4. [DOI] [PubMed] [Google Scholar]
- 45.Roy R, Brooks C, Mitra S. Purification and biochemical characterization of recombinant N-methylpurine-DNA glycosylase of the mouse. Biochemistry. 1994;33:15131–15140. doi: 10.1021/bi00254a024. [DOI] [PubMed] [Google Scholar]
- 46.Tsuchimoto D, Sakai Y, Sakumi K, Nishioka K, Sasaki M, Fujiwara T, Nakabeppu Y. Human APE2 protein is mostly localized in the nuclei and to some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen. Nucleic Acids Res. 2001;29:2349–2360. doi: 10.1093/nar/29.11.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu H, Zhang P, Liu L, Lee M. A Novel PCNA-Binding Motif Identified by the Panning of a Random Peptide Display Library. Biochemistry. 2001;40:4512–4520. doi: 10.1021/bi010103+. [DOI] [PubMed] [Google Scholar]
- 48.Acharya N, Yoon JH, Gali H, Unk I, Haracska L, Johnson RE, Hurwitz J, Prakash L, Prakash S. Roles of PCNA-binding and ubiquitin-binding domains in human DNA polymerase eta in translesion DNA synthesis. Proc. Natl Acad. Sci. USA. 2008;105:17724–17729. doi: 10.1073/pnas.0809844105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 50.Unk I, Haracska L, Prakash S, Prakash L. 3′-phosphodiesterase and 3′–> 5′ exonuclease activities of yeast Apn2 protein and requirement of these activities for repair of oxidative DNA damage. Mol. Cell Biol. 2001;21:1656–1661. doi: 10.1128/MCB.21.5.1656-1661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]