Abstract
We have developed a high-throughput protein binding microarray (PBM) assay to systematically investigate transcription regulatory protein complexes binding to DNA with varied specificity and affinity. Our approach is based on the novel coupling of total internal reflectance fluorescence (TIRF) spectroscopy, swellable hydrogel double-stranded DNA microarrays and dye-labeled regulatory proteins, making it possible to determine both equilibrium binding specificities and kinetic rates for multiple protein:DNA interactions in a single experiment. DNA specificities and affinities for the general transcription factors TBP, TFIIA and IIB determined by TIRF–PBM are similar to those determined by traditional methods, while simultaneous measurement of the factors in binary and ternary protein complexes reveals preferred binding combinations. TIRF–PBM provides a novel and extendible platform for multi-protein transcription factor investigation.
INTRODUCTION
Transcriptional regulation is an essential gatekeeper in carcinogenesis (1), defining tissue and species identity, maintenance of cell function and cis-regulatory evolution (2). This regulation involves a myriad of proteins that assemble onto regulatory DNA elements to recruit RNA polymerases. Accurate determination of protein:DNA binding specificity is a key goal for understanding activity, and great effort has been devoted to generating databases of in vitro and in vivo determined sequence preferences for various factors [such as JASPAR (3) or TRANSFAC (4)]. Microarray analysis has allowed unprecedented access to high-throughput data on transcription factor binding locations by techniques such as chromatin immunoprecipitation on microarray (ChIP-chip) (5), and more recent, high-resolution techniques such as ChIP-seq (6), but few factors have been studied in high resolution, and the resulting data leave ambiguous whether the observed protein:DNA interaction is direct or indirect (7). One promising strategy for a better understanding of transcription factor binding specificities has been the use of DNA-based protein binding microarrays (PBMs), which enable the study of diverse DNA sequences through the equilibrium binding of individual epitope-tagged transcription factors to DNA microarrays (8,9), detected by subsequent antibody staining or SPR microscopy (10).
Organisms as diverse as human, rat, Drosophila and yeast use the same set of conserved general transcription factors (GTFs) to initiate mRNA synthesis (11). One of the central GTFs is TATA-binding protein (TBP), a sequence-specific DNA-binding protein that recognizes the TATA box sequence, typically located approximately 30 bp upstream of transcriptional start sites (12). Although TATA boxes are only found at ∼20% of all transcriptional start sites (13), TBP serves as a core protein to recruit RNA polymerase II to specific promoter regions. TBP forms large multi-protein complexes with TBP-associated factors (TAFs) and other GTFs, such as TFIIA and IIB, which alter its site specificity and aid in directing transcription (13). However, these assemblies may comprise more than 30 polypeptides, and the formation and DNA sequence preferences of distinct combinations of these GTFs remain poorly understood. Thus, an understanding of the combinatorial logic underlying genetic networks requires the ability to analyze multi-protein complexes, both at equilibrium and during binding, on a large number of DNA scaffolds.
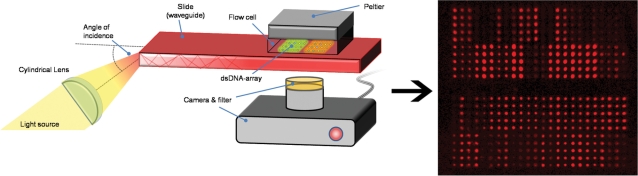
Here, we describe total internal reflectance fluorescence (TIRF)–PBM, a novel PBM assay that achieves multi-protein detection as well as enabling single experiment measurement of both thermodynamic equilibrium binding conditions and kinetic rates of association and dissociation. In TIRF–PBM (shown schematically in Figure 1), an array of hydrogel spots, each containing a unique double-stranded DNA (dsDNA), is synthesized on a slide and the slide is integrated into a flow cell chamber. The array slide is used as a waveguide to generate TIRF conditions in the flow-cell chamber, enabling the sensitive optical detection of fluorescently labeled proteins bound to the DNA in the array. This novel combination of TIRF, hydrogel arrays and fluorescently labeled proteins was used to analyze the binding of the GTFs TBP, TFIIA and IIB singly and in combination across an array of dsDNA containing binding site variants. The results of this analysis illustrate the relevance of multi-protein complexes as determinants of sequence specificity, validating TIRF–PBM and demonstrating its potential to elucidate multi-protein:DNA interactions, including general transcription complexes as well as multiple transcriptional activators.
Figure 1.
Schematic of TIRF–PBM. Amino-modified dsDNA oligonucleotides are linked via reaction with epoxide groups to polymer units and the DNA polymer is printed in a microarray on a slide, followed by treatment by UV to cross-link the polymer into stable, swellable hydrogel spots. This PBM is then probed by flowing fluorescently labeled protein/complex across the slide, with an evanescent excitation wave generated using the slide as an optical waveguide. Fluorescence in multiple excitation/emission pairs is scanned in real-time (shown is false-colored binding of TBP) across the arrays during the binding reaction, giving equilibrium and kinetic measurements for multiple proteins in complexes binding to the dsDNA features of the array.
MATERIALS AND METHODS
Oligonucleotide library generation
For the initial screening and proof of concept for our TIRF–PBM, a limited set of 96 dsDNA sequences were generated by primer extension (14) of a pool of synthetic 51-mer template strands (Integrated DNA Technologies), each with an invariant region recognized by a common amino-modified primer (primer sequence 5′-amino-C6-GGACCGATTGACTTGA-3′). Template strands, 5′ to 3′, are shown in Supplementary Table 1. Templates and primers were mixed in 96-well plates, annealed via slow cooling from 90°C and then extended by DeepVent exo-polymerase (New England BioLabs, see Supplementary Figure 1 for a representative gel of the extension reaction), manually purified by phenol:chloroform extraction and ethanol precipitation and transferred to 96-well plates for printing.
Hydrogel synthesis and arraying
Polyacrylamide-epoxide co-polymer hydrogel (poly dimethyl acrylamide co epoxy methacrylate co benzophenone methacrylate) was synthesized by a statistical radical polymerization at 65°C for 16 h in ethyl acetate using AIBN (azobisisobutyronitrile) as an initiator, followed by precipitation in ethyl ether. Polymer was reacted at 4°C overnight with amino-terminated dsDNA in 96-well plates, with a final concentration of 10 µM DNA and 2 mg/ml polymer. Poly(methyl methacrylate) (PMMA) microscope slides were spotted with the polymer–DNA mixture using a pin printing device (GMS 417 Arrayer, Affymetrix) at 50% humidity. After arraying, the polymer was UV-cross-linked into a hydrogel with 260 nm light with 1 J/cm2 energy. The average spot size was between 400 and 600 µm in diameter. SEM investigation of the surface in a dry state shows a regular array of well-formed polymeric dots with internal pore sizes compatible with protein complex access to DNA (Supplementary Figure 2).
Protein expression, purification and labeling
His-tagged constructs for yeast TBP, TFIIA and IIB were generously donated by Laurie Stargell (CSU). All constructs were transformed into BL21 host cells, grown to OD600 0.6 in Luria-Bertani media, induced with 0.2% Isopropyl β-d-1-thiogalactopyranoside at 37°C for 4 h and the cells were harvested by centrifugation. For TBP and TFIIB, cells were lysed via French press under native conditions (50 mM sodium phosphate, pH 8.0, 300 mM NaCl, 0.1 mM phenylmethylsulphonyl fluoride (PMSF)) and proteins were purified on NiNTA agarose (Qiagen) with 80 mM imidazole wash and 300 mM imidazole elution. This was followed by dialysis and cation exchange chromatography using BioRex70 resin (BioRad) with a linear gradient from 100 mM NaCl to 1 M NaCl in 50 mM sodium phosphate, pH 8.0, 0.1 mM PMSF, 10% glycerol. Cells containing the subunits of TFIIA, Toa1 and Toa2, were lysed under denaturing conditions (8 M urea, 100 mM sodium phosphate, 10 mM Tris–HCl, pH 8.0) via French press and the two cell lysates were combined. Urea was dialyzed out of the solution to refold the subunits together (15). The refolded TFIIA was then purified by NiNTA agarose as for TBP and TFIIB as explained above (Supplementary Figure 3, sodium docecyl sulfate (SDS) gel of purified proteins). Purified protein was then covalently conjugated to a variety of commercial dye labels on solvent-exposed lysine residues using N-hydroxysuccinimide ester (NHS) attachment chemistry under careful stoichiometric control (Pierce). Briefly, purified protein was dialyzed into a buffer lacking primary amines (1× PBS: 10 mM sodium phosphate, pH 7.4, 137 mM NaCl, 2.7 mM KCl, with 10% glycerol) and concentrated to >20 µM. NHS-DyLight conjugate (DyLight 549, DyLight 649 or DyLight 488) was suspended in dimethyl formamide and added to the solution. Care must be taken to avoid excess labeling or reaction with lysines important for protein function, and to ensure minimal disruption, the labeling reaction was carried out substoichiometrically (<0.5 moles NHS per mole of protein) for 1 h at 4°C. The labeling reaction was halted by introduction of 1 mM ethanolamine and 10 mM Tris–HCl, pH 7.5, and excess dye was removed via dialysis into 1× PBS with 10% glycerol. The function of the labeled proteins was assessed by gel electrophoretic mobility shift assays (EMSAs), which were performed as described previously (16,17). This step ensures that the random labeling does not significantly disrupt function or activity, and we find that labeled TBP, TFIIA and IIB formed complexes with expected DNA affinity (data not shown).
TIRF instrumentation
A high-power light-emitting diode (LED) (Luxeon II) serves as the TIRF light source. The beam is focused with a cylindrical lens, and intersects the edge of the microscope slides at a range of angles 70° ± 10°, all greater than the critical angle of 60° for total internal reflectance conditions in our setup. This range of angles leads to a uniform evanescent field across the array (18) (Figure 1). The microscope slide is fitted into a reaction chamber with a peltier temperature control. Emitted light from the array is collected through the microscope slide by a peltier-cooled (−18°C) charge-coupled device camera. The following excitation sources, excitation filters and emission filters were used: for DyLight 649 and Cy5, a Luxeon III 620-645 nm LED source (Phillips) with a 650 nm lowpass emission filter and a 700BP40 excitation filter (Thorlabs). For Dylight 549, a Luxeon V 520-500 nm LED source with a 550nm lowpass emission filter and a 600BP40 excitation filter was used. For DyLight 488, a Luxeon V 460–490 nm LED source with a 500 nm lowpass emission filter and a 550BP20 excitation filter was used. For SYBR Gold staining, a Luxeon 460–490 nm LED source with a 500 nm lowpass excitation filter and a 600BP40 excitation filter was used.
Reaction conditions
The reaction chamber was first blocked at 30°C for 15 min with amine buffer (10 mM Tris–HCl pH 8, 10 mM ethanolamine, 0.1% SDS solution) to react with any remaining epoxide groups, then with phosphate buffered saline (1× PBS) supplemented with 5% BSA and 1% Tween 20. Proteins were suspended in a running buffer of 1× PBS, 0.5% BSA and 0.01% Tween 20. To facilitate TBP binding in the absence of cofactors, 5 mM MgCl2 was included in trials with TBP alone. The chamber was then briefly washed with running buffer, and protein samples in running buffer were introduced at 25°C using a syringe pump with a constant flow at 50 ul/min. Ten seconds integrations of fluorescent signal were collected, typically for 1 h. To stimulate protein:DNA dissociation, running buffer without protein was then introduced at the same flow rate, and signal capture was continued. The arrays were regenerated by washing with a regeneration buffer (1× PBS with 1 M NaCl and 1% Tween 20).
Data normalization and analysis
Data were collected for each sequence across five replicate spots using ATR Reader 1.0 (Imtek). Measured spot intensities were quantified using Signalyse 2.0 (from Holgar Klapproth). Spots with high error (SD/signal >0.5) were flagged and excluded from further analysis. For each batch of arrays, one array was stained with SYBR Gold and analyzed to establish DNA intensity for each spotted sequence. Any spots displaying DNA signal more than 2 times or less than 0.5 times, the average DNA signals were flagged and excluded from further analysis. Normalized binding intensity was generated by dividing raw protein signal fluorescent intensity by normalized DNA intensity for that sequence. These normalized values were used for all subsequent analysis. Data were processed with Excel 2000 (Microsoft), Mathematica 6 (Wolfram) and Graphpad Prism 5.01 (Graphpad). Position weight matrixes (PWMs) were generated and analyzed by Multiple Em for Motif Elicitation (MEME) (19), using the correction for small sample size. Clusters of sequences were compared using pair wise two-tailed Student's t-test with Welsh's correction. Comparison to existing consensus motifs was performed using Hamming distance of the degenerated motif to the known motif, as previously described (20).
RESULTS
TIRF as a tool for PBM analysis
TIRF has been used to great effect in investigations of fluorescent analytes within a confined area, such as in studies of membrane-bound proteins in cells (21). In this study, TIRF is coupled to a microarray to enable real-time detection of GTF binding across a microarray. Our TIRF instrument (Figure 1) has a temperature and flow-rate controlled reaction chamber positioned on top of the printed hydrogel–DNA dot array region on a microscope slide. A cylindrical lens spreads the incident light to multiple, similar angles of incidence all greater than the critical angle (18) on the edge of the microscope slide (waveguide), creating an evanescent field of uniform intensity which excites the fluorescently labeled proteins. The exponential decrease of the field with distance results in low background signal, precluding time-consuming washing steps and makes TIRF–PBM sensitive to detection of only molecules located within the hydrogel, as only a few tenths of a percent of molecules in solution generate signal (22). The high sensitivity of TIRF, coupled with this real-time detection capability, expands PBM analysis and provides a route for studying protein complex formation on DNA.
dsDNA hydrogel synthesis and characterization
The attachment of DNA to silane-coated glass (23) is poorly suited to the study of DNA-binding proteins due to surface irregularities, roughness and high levels of nonspecific protein binding (24). To overcome this challenge, TIRF–PBM uses arrays of swellable hydrogel dots impregnated with dsDNA oligonucleotides, which are more favorable for unperturbed protein binding (25), especially for kinetic measurements. In this initial study, a minimal set of 96 sequences was used, but the technique is compatible with much larger microarrays. dsDNA is made by primer extension (14) and covalently linked to the hydrogel to enable reuse of each slide and prevent signal irregularities from leeching processes. This process generates spots with a uniform quantity of dsDNA, retained even under stringent wash conditions (Supplementary Figure 4), and shows greater uniformity and more robust signal than coupling to epoxy-silane glass (data not shown). The accessibility of protein to the dsDNA was confirmed using Cy5-labeled streptavidin and biotin-modified dsDNA (Supplementary Figure 5).
GTF equilibrium binding specificities
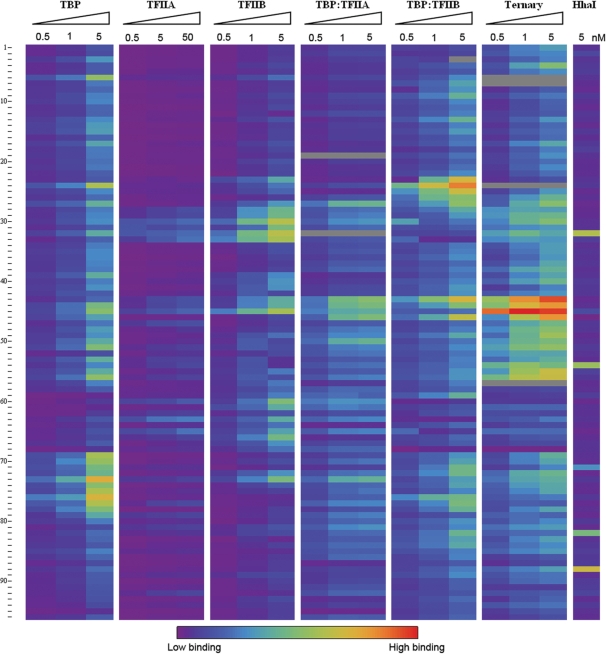
We examined the interactions of several GTFs (TBP, and TFIIA and IIB from Saccharomyces cerevisiae) to study how protein–protein and protein:DNA interactions influence consensus DNA site specificity and affinity. A deeper understanding of how these interactions drive the specificity and affinity of the preinitiation complex has profound biological implications. Our initial study focuses on 96 DNA sequences that contain variants of recognition elements for the GTFs; these are derived from genomic promoters (26,27), previous investigations (12,17,28,29) and single-base substitutions. Purified GTFs were covalently tagged using N-hydroxy succinimide cross-linking with the fluorescent dyes DyLight 549, DyLight 649 and DyLight 488; at substoichiometric ratios this resulted in ∼10% labeling (16). Although NHS cross-linking may not be an applicable technique for the labeling of all recombinant proteins, we verified that the protein labeling procedure did neither alter DNA specificity (Supplementary Figure 6) nor DNA affinity [as described previously (16), data not shown] in the proteins used here. TIRF–PBM analysis was conducted for each factor at multiple protein concentrations under equilibrium binding conditions. TBP, TFIIA and IIB display fluorescent binding intensity differences that vary in sequence-specific and protein concentration-dependent fashion, revealing significant binding specificity preferences to variant DNA sequences (Figure 2, sequences clustered by DNA preferences across all GTF binding conditions). GTF concentrations in excess of the KconsensusD show uniformly less specific binding.
Figure 2.
TIRF–PBM data for 96 DNA sequences and several protein conditions. The labeled proteins DyLight 649-TBP, DyLight 649-TFIIA and DyLight 649-TFIIB, as well as the binary complexes generated by the addition of DyLight 488-TFIIA or DyLight 549-TFIIB to a fixed concentration (0.5 nM) of DyLight 649-TBP and the ternary complex of DyLight 649-TBP: DyLight 549-TFIIB: DyLight 488-TFIIA (TBP and TFIIB held at 0.5 nM) at varied concentrations were flowed across a PBM with 480 features (96 unique sequences) and allowed to reach equilibrium binding. Equilibrium binding fluorescence intensities of DyLight 649 for each condition are shown in a spectrum from lowest (violet) to highest (red) signal, with inconsistent values removed (gray). A control reaction with the DyLight 649-labeled methyltransferase M.HhaI is included. DNA sequences are clustered by similarity of binding across all conditions tested, and form 12 distinct clusters (P < 0.05). For observed patterns and correlation to existing sequence binding annotation, see Results and Discussion section.
TBP binding specificities confirm previous results
We focused initially on the binding of TBP and its modulation by TFIIA and IIB, as TBP is an essential and well-conserved factor used for transcription in all eukaryotes, with well-established binding properties and a known consensus of TATA(A/T)A(A/T)(A/G). ‘Good’ or ‘poor’ TBP binding predictions for 58 of the 96 sequences in our array were identified using existing databases (26,27) (Supplementary Table 1). The observed fluorescent binding intensities of TBP varied in a DNA sequence-dependent manner (Supplementary Figure 7), and we define protein sequence specificity as the normalized fluorescent intensity at equilibrium for a given sequence, as compared across the intensities of the complete dataset (kinetic association and dissociation constants were also obtained and used to determine thermodynamic constants, see below and Supplementary Table 2).
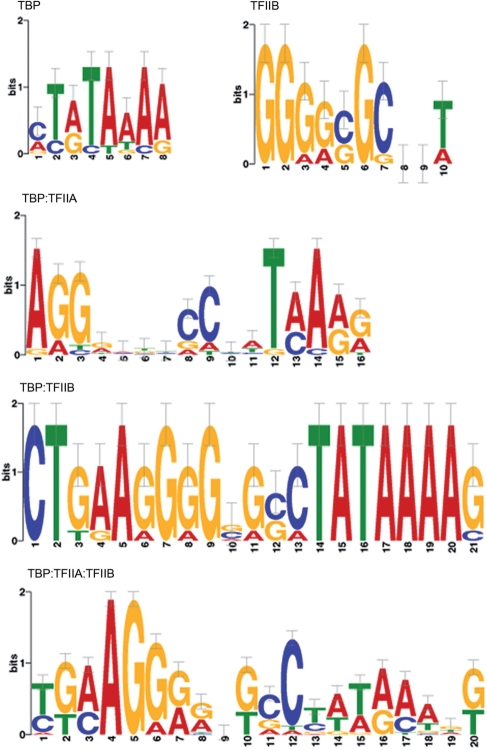
These normalized binding intensities (specificities) are highly correlated with the database-predicted binding behavior, with 51 in agreement and 7 (sequences 2, 15, 22, 27, 52, 63 and 86) showing deviations from database prediction (as seen below, at least one of these seven sequences agreed with prior ChIP data). PWMs were generated from TIRF–PBM data by alignment of the highest specificity sequences using MEME (19) (Figure 3). The TBP motif agrees well with the known TATA box consensus (P < 2.44 × 10−4). Disruptions of this core TATA box sequence cause the predicted changes in TBP binding, as do changes involving the flanking sequences (Figure 2, TBP column). For example, sequence 13 (core site TATAAAG) displays the expected, high TBP binding specificity and systematic replacement of individual bases with cytosine (1, 7, 16, 23, 27 and 34) or guanine (2, 17, 28, 43, 47 and 48) results in modest changes in TBP specificity, as previously reported (17). Specificity for sequences derived from genomic promoter regions is highly correlated with previous studies. For example, sequences derived from coding sections of the S. cerevisiae genome (sequences 16, 35, 38, 40, 48, 83 and 84) show low TBP specificity in our assay, as well as low specificity for the TBP:TFIIA:TFIIB ternary complex (Figure 2, TBP:TFIIA:TFIIB column). Sequence 16 is an exception, and displayed high specificity for the TBP:TFIIB and TBP:TFIIA:TFIIB complexes, consistent with the presence of the consensus TFIIB recognition element (BRE). Sequences with known TBP preferences (sequences 9, 11, 12, 14, 18, 25, 26–28, 30, 34, 36, 41, 42, 46, 50, 53, 63, 71, 72, 91 and 93) display the expected specificity for TBP and the TBP:TFIIA:TFIIB complex (27,28). Ten of these sequences (9, 18, 26–28, 30, 53, 71, 72 and 93) were previously investigated by ChIP-chip studies (30). Eight of these displayed consistent TBP specificity [including sequence 27, which had different behavior predicted from in vitro work (17)], while two (18 and 53) showed TBP and TBP:TFIIA:TFIIB complex specificity contrary to the previous analysis (30).
Figure 3.
The binding of different GTFs favors different sequence motifs. Binding logograms for different proteins and complexes were generated by aligning the sequences of the highest intensity scores on the array (n > 9 for each condition). These logograms demonstrate that the labeled proteins TBP and TFIIB in the TIRF–PBM exhibit DNA sequence specificity in agreement with known consensus sequences. Additionally, the binary and ternary complexes exhibit differential binding preference, reflecting the role of TFIIA and IIB in organizing TBP on the correct sequence.
Gel shifts were performed to validate our observed specificities on sequences 15, 63 and 86 (which were inconsistent with database predictions) as well as on sequences 1, 6, 44, 46, 75 and 76 (which agree with database predictions and span a range of observed affinities). We find dissociation constants that confirm TIRF–PBM observations of both tight and weak binding specificities (Table 1), suggesting that the small degree of disagreement with prediction stems largely from the effects of flanking sequence [e.g. sequence 86 is extremely guanine-rich, consistent with reduced TBP binding even in the presence of a TATA box (31)] and DNA structural effects (e.g. we predict sequence 15 to form extensive hairpin structures based on sequence analysis).
Table 1.
Comparison of TIRF–PBM and EMSA dissociation constants
| Sequence | KappD EMSA (nM) | KappD TIRF (nM) | EMSA/TIRF ratio |
|---|---|---|---|
| 1 | 25.2 ± 2.6 | 19.4 ± 1.6 | 1.30 |
| 6 | 5.5 ± 1.5 | 1.6 ± 0.1 | 3.43 |
| 15 | 5.9 ± 2.6 | 3.5 ± 0.3 | 1.69 |
| 44 | 4.2 ± 1.6 | 3.8 ± 0.4 | 1.11 |
| 91 | 37 ± 14 | 21.0 ± 2.0 | 1.76 |
| 63 | 37 ± 21 | 39 ± 51 | 0.95 |
| 75 | 3.1 ± 1.3 | 1.2 ± 0.1 | 2.58 |
| 46 | 4.5 ± 0.9 | 4.2 ± 0.3 | 1.07 |
| 76 | 1.6 ± 0.5 | 1.8 ± 0.1 | 0.89 |
| 86 | 170 ± 90 | 122 ± 17 | 1.39 |
EMSA KDs were determined under standard gel electrophoretic mobility shift conditions using dsDNA prepared for the array and labeled on the 5′-terminus of the template strand with P32. TIRF KDs was determined by fitting normalized fluorescence data to an association, followed by dissociation kinetic model. EMSA- and TIRF-derived KDs presented have an average ratio of 1.3 ± 0.6.
TFIIA and IIB binding and alteration of TBP specificity
TIRF–PBMs for TFIIA and IIB correlate well with the limited previous studies of the individual proteins. TFIIA has negligible affinity for DNA but cross-linking studies have shown localization to regions surrounding the TATA box (32,33) and DNA contacts may contribute to TFIIA's modulation of TBP binding (32,34). The small degree of binding shown with even a 10-fold excess of TFIIA (Figure 2, TFIIA column) may result from indirect readout of the overall DNA structure (33). In contrast, TFIIB interacts significantly with basepairs on both sides of the TATA box; the consensus for the upstream BRE is (G/C)(G/C)(A/G)CGCC (35). TFIIB specificity varies less than what was observed with TBP, but varies among sequences with a consensus BRE (e.g. sequences 3, 6, 21, 23 and 58), indicating that flanking interactions and DNA structural changes influence TFIIB binding. In our assay, TFIIB displays a PWM similar to the consensus BRE (Figure 3, P-values for sequences containing this motif were ≤6.1 × 10−4) (19,36).
TBP binding specificity with either TFIIA or IIB was examined with TBP held at 0.5 nM, while TFIIA or IIB were varied from 0.5 nM to 5 or 50 nM, respectively. The binding of these binary complexes (measured by the TBP binding intensity) showed patterns of specificity different from when any of the three proteins were incubated separately (Figure 2, TBP:TFIIA and TBP:TFIIB columns), and the pattern was maintained whether or not the interacting factor was dye labeled (Supplementary Figure 6). The modulation of TBP site preference, dependent on the presence of either TFIIA or IIB, is consistent with prior work identifying TFIIA and IIB as regulators of TBP specificity and affinity to DNA binding sites (17,31–33,37–42). We generated PWMs for the highest binding binary and ternary complexes of our GTFs (Figure 3), although comparison is complicated by the lack of multi-factor motifs present in the literature. The motif for TBP:TFIIB shows a preference for both a BRE and TATA box (P < 6.6 × 10−11), as predicted from structural studies (43). Interestingly, the highest specificity sequences for the TBP:TFIIA:TFIIB complex displays a weak TATA box and an upstream BRE-like region (P < 2.3 × 10−5). The TATA box motif is often found in ChIP-chip studies of TBP (30,44), but TBP is also known to extensively act at TATA-less promoters (45,46).
To test our prediction that the ternary complex binding reveals a binding motif reflective of in vivo behavior discovered by ChIP, we used MAST (47) to find the set of open reading frame (ORFs) in the yeast genome that contain the ternary motif in their upstream promoter regions, and compared these to the set of ORFs with promoter regions bound by TBP, as determined in a recent, high-resolution ChIP study (44). The ORFs for each set were sorted by cellular process, and there is a significant correlation between the ChIP-derived TBP motif and the TBP:TFIIA:TFIIB motif we find in TIRF–PBM (Pearson correlation coefficient of 0.92); whereas the single protein TBP motif we discover in TIRF–PBM displays significantly less correlation (Pearson coefficient 0.65, Supplementary Figure 8). TBP:TFIIA exhibited more selective binding than TBP alone, suggesting that TFIIA may reduce the specificity of TBP for improper sites, consistent with the known in vivo role of TFIIA (32,39). TBP alone displays less than 103 preference for the TATA box, while most TFs display approximately 106 preference for their cognate sites over nonspecific DNA, highlighting the role of additional GTFs in directing TBP binding. The preferred TBP:TFIIA and TBP:TFIIB motifs that we observe are similar, with a degenerate TATA box with a G-rich 5′-region (P < 1.4 × 10−3). The affinity of TFIIA for BRE sites is established (48), and this motif is biologically relevant; for example, it appears in sequence 46, which is derived from the adenovirus major later promoter (AdMLP), shows high specificity by TBP:TFIIA and is well-known by highly preferred by GTFs (49). TBP:TFIIB bind more tightly than TBP:TFIIA to TATA box sites (Figure 2, TBP:TFIIB column), consistent with previous in vitro studies (31,32,39,40). The TBP:TFIIB complex PWM (Figure 3, P < 6.6 × 10−11) is consistent with binding contributions from both proteins, along with enhanced specificity for a 3′-region of the core TATA box rich in G. The preferred sequences for the ternary TBP:TFIIA:TFIIB complex have a degenerate TATA box and G-rich 5′-BRE motif (Figure 3, P < 2.3 × 10−5) and the promoters containing this motif correspond well with the observed in vivo behavior of the GTFs in the transcriptional complex as shown through TBP ChIP studies (44). Notably, the most preferred sequence for the ternary complex is the AdMLP sequence (46), which has been exhaustively studied with these and other GTFs (49).
Our observations are consistent with the known roles of sequence content on GTF binding. For example, guanine-rich tracts flanking the TATA box (such as in sequence 90) are known to reduce TBP binding (31), which is recovered when TBP is in complex with TFIIB. Sequences with overall low specificity for TBP alone have an increased level of binding for TBP:TFIIB (e.g. 16, 28 and 93), consistent with TFIIB's role in increasing TBP's ability to bind off-consensus sites (31,39). TFIIB can also increase the specificity of TBP for favored sites, such as sequence 68 (which contains a TATA box and BRE), which displays high binding of TBP alone, which further increases when bound by the TBP:TFIIB complex (Figure 2) (39). In our system, we also observed the known ability of TFIIB to direct the TBP:TFIIB complex away from sites that lack G-rich features (31), seen in a sequence that contains a TATA box surrounded by primarily A and C repeats (sequence 18). This sequence binds TBP alone, but shows reduced specificity in the TBP:TFIIB complex.
Multiplex detection of DNA-binding proteins
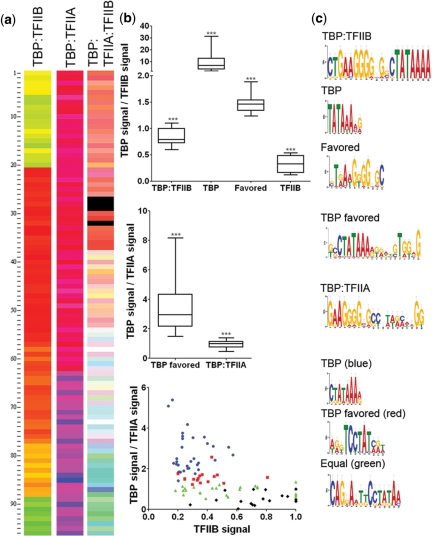
The instrument's excitation and emission filters are easily changed, allowing the simultaneous detection of multiple uniquely labeled GTFs across the array (Figure 4a). Four statistically distinct clusters of sequences (pair wise two-tailed Student's t-test with Welsh's correction, P < 0.01) are apparent for the binding of co-incubated TBP and TFIIB (Figure 4b). One sequence cluster with an average TBP to TFIIB ratio of 9.5 ± 1.0 reflects sequences where TBP binds essentially to the exclusion of TFIIB and generates a conserved TATA box PWM (P < 2 × 10−4). A second cluster displays a TBP:TFIIB ratio of 0.83 ± 0.03 and the PWM shows both a TATA box and a G-rich 5′-region similar to a BRE (P < 4 × 10−9). This equal binding cluster correlates with the sequences that display the highest specificity of the ternary complex (shown in Figure 2). A third cluster displays a 3-fold preference for TFIIB (ratio of 0.33 ± 0.06), but displayed little sequence conservation. The fourth cluster, composed almost exclusively of low-specificity sequences, displays a weak TATA box motif (P < 8 × 10−5). This cluster shows that with nonpreferred sites, TBP displays a low specificity and may associate with TFIIB despite the lack of a clear TFIIB binding site. No clear patterns of regulation were apparent when the Gene Ontology of the motifs generated from each cluster was compared, which is not wholly surprising in light of the diverse involvement of the GTFs in gene regulation (44,46).
Figure 4.
TIRF–PBM serves as a robust platform for multiplex protein detection. (a) Fluorescence binding intensity data for each of multiple proteins in a complex incubated on the PBM. Shown are false colors merges (TBP in red, TFIIB in green and TFIIA in blue) for the complexes formed with 0.5 nM of: TBP:TFIIB, TBP:TFIIA and TBP:TFIIB:TFIIA (each condition probed on a separate PBM run). Sequences are individually clustered for each merge by ratio of GTFs. (b) TBP:TFIIB, the ratio of binding of TBP and TFIIB, when co-incubated, clusters into four distinct behavior patterns (P < 0.01). Each of these clusters reflects a bias in those sequence for binding by TBP alone, TFIIB alone, the TBP:TFIIB complex or those that can bind either TBP or TBP:TFIIB. TBP:TFIIA, under the same analysis, the binding of TBP and TFIIA simultaneously forms two distinct clusters (P < 0.01): sequences that bind the TBP:TFIIA complex, and sequences that exclusively bind TBP. TBP:TFIIA:TFIIB, the binding of the three GTFs is plotted by compared normalized TFIIB binding to the ratio of TBP/TFIIA binding, and analysis of the binding for all three proteins indicates four clusters (P < 0.01). These clusters reveal sequences where TBP binds much higher than the other GTFs (blue), where the GTFs bind equally in complex (green) and two additional clusters which favor TBP (red) or TFIIB (black). (c) PWM motifs for each condition were generated by MEME and reflect sequence specificity of each cluster.
A similar analysis was performed for TBP and TFIIA, and the sequences form two distinct clusters (P < 0.01): sequences that bind both TBP and TFIIA (binding ratio 0.97 ± 0.04, P < 2.9 × 10−11) and sequences that favor TBP binding (3.4 ± 0.2 ratio, P < 4 × 10−7). As TFIIA has minimal affinity for DNA by itself, this clustering is consistent with expectations (32,40). The sites that favor binding of both GTFs display a motif with a TATA box flanked on both ends by G-rich regions, and this motif is significantly enriched in the yeast genome in the upstream promoter regions of tRNA genes and translation involved ORFs (33.3% of ORFs bound by the motif, as compared with <7.9% translation involved across all yeast genes). Previous studies have shown the involvement of TFIIA specifically in the expression of tRNA (50,51), suggesting that our 1:1 TBP:TFIIA binding motif coincides with tRNA gene regulation, and Gene Ontology searches using our TBP:TFIIA motif find tRNA gene promoter regions over-represented [15 of the 51 (29.4%) high confidence matches].
The simultaneous binding signal for all three GTFs was compared and found to form four distinct clusters of sequences (P < 0.01). When the normalized intensity of TFIIB binding is plotted against the ratio of TBP to TFIIA binding, these clusters are visible as separate regions in Figure 4b, TBP:TFIIA:TFIIB. One cluster (blue) displays TBP specificity far higher than seen for the other proteins, and its PWM shows a conserved TATA box (P < 2.2 × 10−4). A second cluster (red) shows favored TBP binding, but this cluster is distinguished from the first cluster by different specificity for TFIIA and IIB, although it retains a TATA box PWM (P < 2.2 × 10−4). The overlap seen between the red and blue clusters is due to different ranges of values for TBP and TFIIA, which result in similar TBP/TFIIA ratios. A third cluster (green) has all three GTFs binding at equivalent levels (TBP/TFIIA ratio 0.94 ± 0.2, TBP:TFIIB ratio 0.94 ± 0.2) and the PWM shows a very poorly conserved TATA box with a purine-rich 5′-region (P < 2 × 10−6). The remaining cluster (black) corresponds to slightly favored TFIIB binding and does not show significant sequence conservation.
Kinetic and thermodynamic investigation of TBP binding affinity
Although the binding intensity analysis described in Figures 2–4 does not allow a quantitative assessment of kinetics and thermodynamics, the TIRF–PBM assay is fully capable of providing this. We determined the kinetic on (kon) and off (koff) rates of the binding of the GTFs to the oligonucleotide sequences in our array using a continuous buffer flow through the flow cell of our TIRF–PBM instrument, following the methodology of SPR sensor techniques (10). The fluorescent intensity for each protein was observed to follow first-order association kinetics; and removal of the GTFs from solution results in a drop in fluorescent intensity consistent with predicted dissociation kinetics (e.g. see TBP kinetic traces in Figure 5). For each binding condition, kon and koff were determined and used to generate the apparent thermodynamic dissociation constants (KappD, Supplementary Table 2). The TIRF–PBM KDS's for TBP span a more than 600-fold range in affinity and agree with known values for specific and nonspecific TBP binding sequences. The range of dissociation constants captured in our initial experiments was limited (e.g. low signal prevented measurement of many KDs for TFIIA), but illustrate the potential for TIRF–PBM to capture KDs over a wide range, comparable with SPR kinetic determinations (52). Dissociation constants determined by traditional gel EMSAs show accurate agreement with dissociation constants derived from TIRF–PBM, with approximately 2-fold variance (Table 1). These results demonstrate that TIRF–PBM captures both equilibrium binding intensity and kinetic rate measurements in a single experiment.
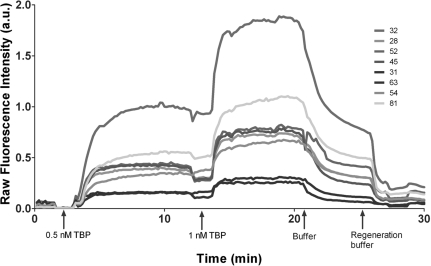
Figure 5.
TIRF–PBMs collect kinetic association and dissociation traces from every feature. Time course of raw fluorescence intensity across eight features on the array, illustrating association when protein is present in the flowing buffer, followed by dissociation when protein is removed, and regeneration of the surface when buffer containing 1 M NaCl is introduced.
DISCUSSION
It is well appreciated that the transcription factors in a eukaryotic cell assemble in transient and promoter-specific fashion to direct the transcription of particular genes (53), but there is still a pressing need for techniques to probe these transient complexes. TIRF–PBM provides a robust method to elucidate the intricate, combinatorial binding behavior of multi-transcription factor complexes, which is most evident in our multiplex analysis of individually labeled factors in complex with one another (Figure 4). To establish the validity of TIRF–PBM measurements, we performed an extensive comparison and validation of TIRF–PBM-derived specificity for TBP (Figure 2), compared with prior in vitro and in vivo measurements (27,28,30,44,54). These measurements indicate broad agreement with studies of sequence effects on TBP binding (17,27,28,31). On sequences exhibiting differential binding behavior between our work and prior in vitro measurements (17) relying on single proteins, our observations were largely shown to be consistent with in vivo experiments (30,44). Since our method and the in vivo work both reflect the preferences of multi-protein complexes, we suggest that the use of in vitro multi-protein approaches better recapitulate the in vivo behavior of GTFs.
We investigated the specificity of TFIIA and IIB (Figure 2), and their affect on TBP specificity, to illustrate a strategy for addressing inquiries into multi-protein effects on binding behavior. Comparison of our multi-protein GTF motifs (Figure 3) to ChIP studies (44) shows much greater agreement than is seen from measurements of the specificity of TBP alone in our assay (Supplementary Figure 8) or based on the reported TBP consensus sequence (3). This provides evidence for the expectation that multi-factor complexes will display binding more consistent with their in vivo functionality. Additionally, the reduced specificity for the TATA box and raised specificity for other elements that we observe in our TBP:TFIIA:TFIIB multi-protein motif (Figure 3) is consistent with the observation that in vivo, the GTF complex binds a larger number of sites than the number of possible TATA boxes (13,29). Our data clearly shows that the ternary complex of TBP:TFIIA:TFIIB binds to a different set of sequences than any individual GTF (Figures 2 and 3), illustrating the novel information available to multi-protein studies. We also collected kinetic binding information (Figure 5) for each GTF and combinations of GTFs, and the observed values for TBP binding (Supplementary Table 2) closely match previous observations (31,32). Altogether, these results agree with the known roles of TFIIA and TFIIB in driving promoter site specificity and affinity for TBP in vivo (17,31–34,38,39,42), and suggest that there is untapped information in the great diversity of sequences that bind at a lower, but still significant, level. These novel observations of binary and ternary multi-protein complexes on DNA (Figure 4) cannot be obtained from single protein studies, and suggest a dynamic interplay of different, functional complexes that may exist in vivo.
The application of this technology to more complex transcription factor assemblies, to the study of human transcription factors and to the study of how mutations of individual proteins alter such complex assemblies present intriguing ongoing opportunities. Additionally, this initial study investigated a comparatively small set of DNA sequences, but the TIRF–PBM technique can be directly applied to microarrays of any feature density and additional sequence complexity could better elucidate complex binding specificity. Further, the identification of small molecules which can disrupt the interactions of specific protein:protein interactions in such complexes provides a potentially powerful new route to the development of highly focused drugs with cancer therapeutic applications (1). TIRF–PBM can, in principle, be applied to the analysis of any complex of DNA- or RNA-binding proteins. We have shown TIRF–PBM to be a sensitive, effective and high-throughput means to obtain detailed equilibrium and kinetic data on the process of transcription factor binding, without complicating wash steps, antibodies or other detection reagents.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Institute for Collaborative Biotechnologies (Grant DAAD19-03-D-0004 from the U.S. Army Research Office). Funding for open access charge: Institute for Collaborative Biotechnologies (Grant DAAD19-03-D-0004).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Laurie Stargell, Colorado State University, for her generous donation of protein constructs used in this study; Teisha Rowland (UCSB) for critical reading of the manuscript; and Gary Braun (UCSB) for helpful discussion.
REFERENCES
- 1.Darnell JE. Transcription factors as targets for cancer therapy. Nat. Rev. Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 2.Prabhakar S, Visel A, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Morrison H, FitzPatrick DR, Afzal V, et al. Human-specific gain of function in a developmental enhancer. Science. 2008;321:1346–1350. doi: 10.1126/science.1159974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sandelin A, Alkema W, Engstrom P, Wasserman W, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucliec Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, Meinhardt T, Pruss M, Reuter I, Schacherer F. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horak CE, Snyder M. ChIP-chip: a genomic approach for identifying transcription factor binding sites. Methods Enzymol. 2002;350:469–483. doi: 10.1016/s0076-6879(02)50979-4. [DOI] [PubMed] [Google Scholar]
- 6.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 7.Zhu C, Byers K, McCord R, Shi Z, Berger M, Newburger D, Saulrieta K, Smith Z, Shah M, Radhakrishnan M, et al. High-resolution DNA binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mukherjee S, Berger MF, Jona G, Wang XS, Muzzey D, Snyder M, Young RA, Bulyk ML. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat. Genet. 2004;36:1331–1339. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Peña-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell CT, Kim G. SPR microscopy and its applications to high-throughput analyses of biomolecular binding events and their kinetics. Biomaterials. 2007;28:2380–2392. doi: 10.1016/j.biomaterials.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 11.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003;72:449. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 12.Wobbe CR, Struhl K. Yeast and human TATA-binding proteins have nearly identical DNA-sequence requirements for transcription in vitro. Mol. Cell. Biol. 1990;10:3859–3867. doi: 10.1128/mcb.10.8.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat. Rev. Genet. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- 14.Bulyk ML, Gentalen E, Lockhart DJ, Church GM. Quantifying DNA-protein interactions by double-stranded DNA arrays. Nat. Biotechnol. 1999;17:573–577. doi: 10.1038/9878. [DOI] [PubMed] [Google Scholar]
- 15.Ranish JA, Hahn S. The yeast general transcription factor TFIIa is composed of 2 polypeptide subunits. J. Biol. Chem. 1991;266:19320–19327. [PubMed] [Google Scholar]
- 16.Bonham AJ, Braun G, Pavel I, Moskovits M, Reich NO. Detection of sequence-specific protein-DNA interactions via surface enhanced resonance Raman scattering. J. Am. Chem. Soc. 2007;129:14572–14573. doi: 10.1021/ja0767837. [DOI] [PubMed] [Google Scholar]
- 17.Stewart JJ, Fischbeck JA, Chen X, Stargell LA. Non-optimal TATA elements exhibit diverse mechanistic consequences. J. Biol. Chem. 2006;281:22665–22673. doi: 10.1074/jbc.M603237200. [DOI] [PubMed] [Google Scholar]
- 18.Lehr HP, Reimann M, Brandenburg A, Sulz G, Klapproth H. Real-time detection of nucleic acid interactions by total internal reflection fluorescence. Anal. Chem. 2003;75:2414–2420. doi: 10.1021/ac0206519. [DOI] [PubMed] [Google Scholar]
- 19.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chua G, Morris QD, Sopko R, Robinson MD, Ryan O, Chan ET, Frey BJ, Andrews BJ, Boone C, Hughes TR. Identifying transcription factor functions and targets by phenotypic activation. Proc. Natl Acad. Sci. USA. 2006;103:12045–12050. doi: 10.1073/pnas.0605140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Axelrod D. Total internal reflection fluorescence microscopy in cell biology. Traffic. 2001;2:764–774. doi: 10.1034/j.1600-0854.2001.21104.x. [DOI] [PubMed] [Google Scholar]
- 22.Budach W, Abel AP, Bruno AE, Neuschafer D. Planar waveguides as high-performance sensing platforms for fluorescence-based multiplexed oligonucleotide hybridization assays. Anal. Chem. 1999;71:3347–3355. [Google Scholar]
- 23.Conzone SD, Pantano CG. Glass slides to DNA microarrays. Mater. Today. 2004;7:20–26. [Google Scholar]
- 24.Guilleaume B, Bune A, Schmidt C, Wiemann S, Poustka A. Systematic comparison of surface coatings for protein microarrays. Proteomics. 2005;5:4705–4712. doi: 10.1002/pmic.200401324. [DOI] [PubMed] [Google Scholar]
- 25.Linnell J, Mott R, Field S, Kwiatkowski DP, Ragoussis J, Udalova IA. Quantitative high-throughput analysis of transcription factor binding specificities. Nucleic Acids Res. 2004;32:e44. doi: 10.1093/nar/gnh042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wakaguri H, Yamashita R, Suzuki Y, Sugano S, Nakai K. DBTSS: database of transcription start sites, progress report 2008. Nucleic Acids Res. 2008;36:D97–D101. doi: 10.1093/nar/gkm901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu J, Zhang MQ. SCPD: a promoter database of the yeast Saccharomyces cerevisiae. Bioinformatics. 1999;15:607–611. doi: 10.1093/bioinformatics/15.7.607. [DOI] [PubMed] [Google Scholar]
- 28.Juo ZS, Chiu TK, Leiberman PM, Baikalov I, Berk AJ, Dickerson RE. How proteins recognize the TATA box. J. Mol. Biol. 1996;261:239–254. doi: 10.1006/jmbi.1996.0456. [DOI] [PubMed] [Google Scholar]
- 29.Kim TH, Barrera LO, Zheng M, Qu CX, Singer MA, Richmond TA, Wu YN, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- 31.Wolner BS, Gralla JD. TATA-flanking sequences influence the rate and stability of TATA-binding protein and TFIIB binding. J. Biol. Chem. 2001;276:6260–6266. doi: 10.1074/jbc.M008273200. [DOI] [PubMed] [Google Scholar]
- 32.Weideman CA, Netter RC, Benjamin LR, McAllister JJ, Schmiedekamp LA, Coleman RA, Pugh BF. Dynamic interplay of TFIIA, TBP and TATA DNA. J. Mol. Biol. 1997;271:61–75. doi: 10.1006/jmbi.1997.1152. [DOI] [PubMed] [Google Scholar]
- 33.Lagrange T, Kim TK, Orphanides G, Ebright YW, Ebright RH, Reinberg D. High-resolution mapping of nucleoprotein complexes by site-specific protein-DNA photocrosslinking: organization of the human TBP-TFIIA-TFIIB-DNA quaternary complex. Proc. Natl Acad. Sci. USA. 1996;93:10620–10625. doi: 10.1073/pnas.93.20.10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hieb AR, Halsey WA, Betterton MD, Perkins TT, Kugel JF, Goodrich JA. TFIIA changes the conformation of the DNA in TBP/TATA complexes and increases their kinetic stability. J. Mol. Biol. 2007;372:619–632. doi: 10.1016/j.jmb.2007.06.061. [DOI] [PubMed] [Google Scholar]
- 35.Deng WS, Roberts SGE. TFIIB and the regulation of transcription by RNA polymerase II. Chromosoma. 2007;116:417–429. doi: 10.1007/s00412-007-0113-9. [DOI] [PubMed] [Google Scholar]
- 36.Lee CY, Li XY, Hechmer A, Eisen M, Biggin MD, Venters BJ, Jiang CZ, Li J, Pugh BF, Gilmour DS. NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol. Cell Biol. 2008;28:3290–3300. doi: 10.1128/MCB.02224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dion V, Coulombe B. Interactions of a DNA-bound transcriptional activator with the TBP-TFIIA-TFIIB-promoter quaternary complex. J. Biol. Chem. 2003;278:11495–11501. doi: 10.1074/jbc.M211938200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buratowski RM, Downs J, Buratowski S. Interdependent interactions between TFIIB, TATA binding protein, and DNA. Mol. Cell Biol. 2002;22:8735–8743. doi: 10.1128/MCB.22.24.8735-8743.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imbalzano AN, Zaret KS, Kingston RE. Transcription factor (TF) IIB and TFIIA can independently increase the affinity of the TATA-binding protein for DNA. J. Biol. Chem. 1994;269:8280–8286. [PubMed] [Google Scholar]
- 40.Stewart JJ, Stargell LA. The stability of the TFIIA-TBP-DNA complex is dependent on the sequence of the TATAAA element. J. Biol. Chem. 2001;276:30078–30084. doi: 10.1074/jbc.M105276200. [DOI] [PubMed] [Google Scholar]
- 41.Stargell LA, Moqtaderi Z, Dorris DR, Ogg RC, Struhl K. TFIIA has activator-dependent and core promoter functions in vivo. J. Biol. Chem. 2000;275:12374–12380. doi: 10.1074/jbc.275.17.12374. [DOI] [PubMed] [Google Scholar]
- 42.Mousson F, Kolkman A, Pijnappel WWMP, Timmers HTM, Heck AJR. Quantitative proteomics reveals regulation of dynamic components within TATA-binding protein (TBP) transcription complexes. Mol. Cell. Proteomics. 2008;7:845–852. doi: 10.1074/mcp.M700306-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Nikolov DB, Chen H, Halay ED, Usheva AA, Hisatake K, Lee DK, Roeder RG, Burley SK. Crystal-structure of a TFIIb-TBP-TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- 44.Venters BJ, Pugh BF. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 2009;19:360–371. doi: 10.1101/gr.084970.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aso T, Conaway JW, Conaway RC. Role of core promoter structure in assembly of the RNA-polymerase II preinitiation complex. A common pathway for formation of preinitiation intermediates at many TATA and TATA-less promoters. J. Biol. Chem. 1994;269:26575–26583. [PubMed] [Google Scholar]
- 46.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CAM, Taylor MS, Engstrom PG, Frith MC, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 47.Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- 48.Deng W, Malecova B, Oelgeschlager T, Roberts SGE. TFIIB recognition elements control the TFIIA-NC2 axis in transcriptional regulation. Mol. Cell Biol. 2009;29:1389–1400. doi: 10.1128/MCB.01346-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Young CSH. The structure and function of the adenovirus major late promoter. Curr. Top Microbiol. Immunol. 2003;272:213–249. doi: 10.1007/978-3-662-05597-7_8. [DOI] [PubMed] [Google Scholar]
- 50.Meissner W, Wanandi I, Carbon P, Krol A, Seifart KH. Transcription factors required for the expression of Xenopus-Laevis selenocysteine transfer-RNA in-vitro. Nucleic Acids Res. 1994;22:553–559. doi: 10.1093/nar/22.4.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meissner W, Holland R, Waldschmidt R, Seifart KH. Transcription factor-IIa stimulates the expression of classical poIII-genes. Nucleic Acids Res. 1993;21:1013–1018. doi: 10.1093/nar/21.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith EA, Erickson MG, Ulijasz AT, Weisblum B, Corn RM. Surface plasmon resonance imaging of transcription factor proteins: interactions of bacterial response regulators with DNA arrays on gold filmsΓÇá. Langmuir. 2003;19:1486–1492. [Google Scholar]
- 53.Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- 54.Kraemer SM, Goldstrohm DA, Berger A, Hankey S, Rovinsky SA, Moye-Rowley WS, Stargell LA. TFIIA plays a role in the response to oxidative stress. Eukaryotic Cell. 2006;5:1081–1090. doi: 10.1128/EC.00071-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.