Abstract
Products of the Steroid Receptor RNA Activator gene (SRA1) have the unusual property to modulate the activity of steroid receptors and other transcription factors both at the RNA (SRA) and the protein (SRAP) level. Balance between these two genetically linked entities is controlled by alternative splicing of intron-1, whose retention alters SRAP reading frame. We have previously found that both fully-spliced SRAP-coding and intron-1-containing non-coding SRA RNAs co-exist in breast cancer cell lines. Herein, we report a significant (Student's t-test, P < 0.003) higher SRA–intron-1 relative expression in breast tumors with higher progesterone receptor contents. Using an antisense oligoribonucleotide, we have successfully reprogrammed endogenous SRA splicing and increased SRA RNA–intron-1 relative level in T5 breast cancer cells. This increase is paralleled by significant changes in the expression of genes such as plasminogen urokinase activator and estrogen receptor beta. Estrogen regulation of other genes, including the anti-metastatic NME1 gene, is also altered. Overall, our results suggest that the balance coding/non-coding SRA transcripts not only characterizes particular tumor phenotypes but might also, through regulating the expression of specific genes, be involved in breast tumorigenesis and tumor progression.
INTRODUCTION
The Steroid Receptor RNA Activator (SRA) was first identified in 1999 as a non-coding RNA specifically co-activating steroid receptors (1). Through mutation analysis, the authors unequivocally showed in their original report that the core region, corresponding to sequences encompassing exons 2 to 5 (Figure 1A), was necessary and sufficient for SRA to act as co-activator (1). Several studies have since shed light on SRA's mechanisms of action [reviewed in (2)]. Briefly, it is believed that SRA acts embedded in ribonucleo-protein complexes recruited to the promoter of regulated genes. These complexes may contain positive regulators, such as the steroid receptor co-activator 1 (SRC-1), the DExD/H box family of RNA-helicase members p68 and p72, or the pseudouridine synthases Pus1p and Pus3p. Negative regulators, such as SMRT/HDAC1 Associated Repressor Protein (Sharp) and the recently identified SRA stem-loop interacting RNA binding protein (SLIRP), can also interact with SRA to decrease its activity (3,4).
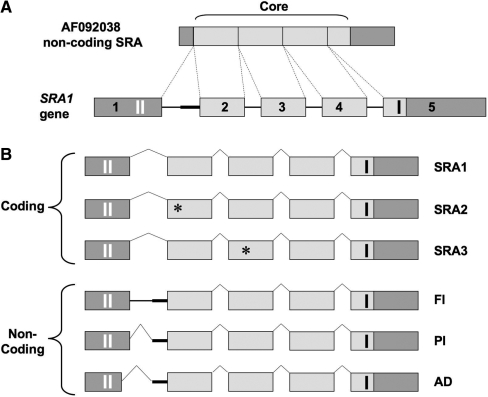
Figure 1.
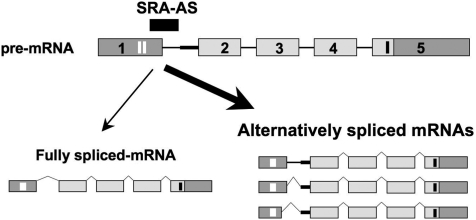
Genomic structure of SRA1, coding and non-coding SRA transcripts. (A) SRA1 gene, located on chromosome 5q31.3, consists of five exons (boxes) and four introns (plain lines). The originally described non-coding SRA sequence (AF092038) contains a core sequence (light gray), necessary and sufficient for SRA RNAs to act as co-activators. (B) Three coding isoforms have been identified (SRA1, SRA2, SRA3), which contain an extended 5′-extremity containing two AUG initiating codons (vertical white bars in exon 1). The stop codon of the resulting open reading frames (236/237 aa) is depicted by a black vertical bar in exon 5. Black stars in exons 2 and 3 correspond to a point mutation (position 98 of the core: U to C) and a point mutation followed by a full codon (position 271 of the core: G changed to CGAC), respectively (5). Three non-coding SRA isoforms containing a differentially-spliced intron-1 have been characterized FI, full intron-1 retention; PI, partial intron-1 retention; AD, alternative 5′ donor and partial intron retention (12). Thick straight line, 60 bp of intron 1 retained in PI.
If SRA was originally thought to specifically increase the activity of steroid receptors, further data have subsequently demonstrated that this RNA can also co-activate non-steroid nuclear receptors as well as other transcription factors such as MyoD (2). SRA has therefore a wider role than first anticipated and likely participates in signalling pathways still to be uncovered.
Additional SRA transcripts, almost identical to the original SRA and containing a full core sequence, have now been described (Figure 1B). As a result of gene polymorphism (5) they can however contain point mutations in exon-2 (SRA2) or an additional codon in exon-3 (SRA3). These transcripts can be divided in two categories: coding and non-coding SRAs. Coding isoforms have an extended exon-1, which contains two translation initiating methionine codons (Figure 1B). These coding SRAs have been proven to encode an endogenous SRA protein (SRAP) in several tissues including breast, prostate and muscle (5–8). Even though the exact functions of SRAP remain to be fully elucidated (2), independent reports suggest that this protein also regulate steroid receptor signalling, potentially as co-activator (8,9), or as a repressor (10,11).
Non-coding SRA transcripts, generated through alternative splicing of intron-1, have also been characterized (2,12). These transcripts contain either a full (FI) or a partial (PI) intron-1 sequence (Figure 1B). The partial intron retention results from the use of an alternative 3′ acceptor site located 60 bp upstream of exon-2 (12). Non-coding SRAs can also result from the concomitant use of an alternative 5′ donor site (located 15 bp upstream of the end of exon-1) and the alternative 3′ acceptor site (AD). All these different splicing events, which occur independently of the isoform considered (i.e. SRA1, SRA2 or SRA3), either shift SRAP open reading frame or introduce premature stop codons. The resulting alternatively spliced SRA RNAs are therefore unable to encode for SRAP.
It should be stressed that both fully- and alternatively-spliced SRA transcripts contain the functional core sequence. They can therefore act as transcriptional co-activators. The additional ability of fully-spliced SRA RNAs to encode for SRAP creates a peculiar level of functional complexity to the products of the SRA1 gene. We have shown that both coding and non-coding SRA coexist in breast cancer cells (12). Interestingly, their relative expression varies between breast cancer cells lines with different phenotypes (12). In particular, we have observed that invasive breast cancer cell lines (MDA-MB-231, MDA-MB-468) expressed higher relative levels of non-coding SRA than ‘closer to normal’ MCF-10A1 breast cells. This suggests that a balance ‘tipped’ toward the production of non-coding SRA RNA in breast cells might be associated with growth and/or invasion properties. This further raises the possibility that modifying the balance between these transcripts could trigger meaningful events in breast cancer cells.
To further explore the potential relevance of the balance coding/non-coding SRAs in breast cancer, we have herein investigated SRA–intron-1 relative expression in a small cohort of invasive breast tumors. Using an antisense oligoribonucleotide designed to target SRA exon-1/intron-1 junction, we have also assessed our ability to artificially alter the balance coding/non-coding SRA transcripts in T5 breast cancer cell line.
MATERIALS AND METHODS
Cells and tumor tissues
T5 breast cancer cells were kindly provided by Murphy (13). Cells were cultured in DMEM (GIBCO, Grand Island, NY) supplemented with 5% fetal bovine serum (CANSERA, Rexdale, ON), penicillin (100 units/ml), streptomycin (100 μg/ml) (GIBCO, Grand Island, NY) and 0.3% glucose. Cells were grown in a 37°C humidified incubator with 5% CO2.
Thirty-two tumor samples, with a wide range of estrogen receptor (ER, from 6.3 to 247 fmol/prot, median =82.5 fmol/prot) and progesterone receptor (PR, from 12.2 to 444 fmol/prot, median = 38.5 fmol/prot) levels, were selected from the Manitoba Breast Tumor Bank (14), which operates with the approval of the Faculty of Medicine, University of Manitoba, Research Ethics Board.
Oligonucleotide treatment
2′-O-Methyl-oligoribonucleotide phosphorothioate 20-mers anti-sense to the 5′-splice site of SRA intron-1 (SRA–AS, 5′-ACCCGGCUUCACGUACAGCU-3′) and to the 5′-splice site of a modified β-globin intron (βgl–AS, 5′-ACCUGCCCAGGGCCUCACCA-3′) were synthesized and purified by Trilink Biotechnologies, Inc. (San Diego, CA). Fluorophore (Indocarbocyanine-Cy3 and 5-Carboxyfluorescein_FAM) conjugated versions (SRA–AS–Cy3 and βgl–AS–FAM) were also obtained from Trilink Biotechnologies, Inc. Transfections were performed using DMRIE-C reagent (16 μg/ml; Invitrogen, Carlsbad, CA) according to the manufacturer's directions using concentrations ranging from 0 to 0.5 μM. It should be noted that when no oligonucleotide was added (mock), cells were also treated with DMRIE-C reagent. Five hours post-treatment, transfection medium was replaced with fresh medium, and cells were cultured for the indicated times. For oligonucleotide and minigene co-transfections, T5 cells were plated at 3.5 × 105 cells per well in 6-well plates and co-transfected with 1.65 μg of SRA minigene and 0.05, 0.1 or 0.5 μM oligoribonucleotides using DMRIE-C reagent (16 μg/ml, Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Medium containing the transfection complex was replaced by DMEM containing 5% FBS 5 h post transfection.
Fluorescent microscopy
Cells were cultured on coverslips and transfected with SRA–AS–Cy3 or βgl–AS–FAM at the indicated concentrations and times post-treatment. Coverslips were then briefly washed with PBS and cells were fixed with 3.7% formaldehyde in PBS for 15 min at room temperature. Cover-slips were then rinsed with PBS and cells permeabilized with 0.2% Triton-X100 in PBS for 1–2 min prior to staining nuclei with 1 μg/ml 4′,6-diamidino-2-phenylindole-dihydrochloride (Dapi). Coverslips were mounted with FluorSave™Reagent (Calbiochem, La Jolla, CA). Fluorescent images were captured with an Eclipse E1000 epifluorescent microscope (Nikon), digitized with ACT-1 software (v.2.63; Nikon) and merged images were generated with Photoshop 6.0 (Adobe).
RT–PCR and triple-primer-PCR (TP-PCR)
Total RNA was extracted from cells or frozen tumor tissue sections using TrizolTM reagent (Gibco BRL, Grand Island, NY) and subjected to DNase treatment (Promega, Madison WI) as previously described (12,15). Half a microgram of total RNA was reverse transcribed in a final volume of 30 μl using Moloney Murine Leukemia Virus (M-MLV) reverse transcriptase and random hexamers as previously reported (5). One and a half microliters of reverse-transcription mixture was amplified in a final volume of 15 µl, in the presence of 60 mM Tris–HCl (pH 8.5), 15 mM [NH4]2SO4, 1.5 mM MgCl2, 0.2 mM of each dNTPs, 4 ng/µl of each primer (two or three primers for RT–PCR and TP-PCR, respectively), 1 unit of Platinum Taq DNA polymerase (Invitrogen Carlsbad, CA) and 10 nM α−32P dCTP. Each PCR consisted of a 5 min pre-incubation step at 94°C followed by 30 cycles of amplification (30 s at 94°C, 30 s at 60°C and 30 s at 72°C). The sequences of primers used are detailed in Supplementary Table S1.
Radio-labeled PCR products were then separated on denaturating poly-acrylamide/urea gels as previously described (15). Following electrophoresis, the gels were dried and exposed 30 min to a Molecular ImagerTM-FX Imaging screen (Bio-Rad, Hercules, CA). Exposed screen was then scanned using a Molecular ImagerTM-FX (Bio-Rad, Hercules, CA), which allows the subsequent quantification of each observed signal.
The identity of all PCR products obtained has been confirmed following subcloning and sequencing as described previously (12,16).
Quantification and normalization of PCR signals
PCR signals were quantified using a Molecular ImagerTM-FX (Bio-Rad, Hercules, CA) as previously described (15). For each TP-PCR experiment (Figures 2 and 4), the relative amount of fully spliced and intron-1 retained SRA of each sample was first expressed as the percentage of the total signal measured following amplification (12). For tumor cases (Figure 2), the percentage of intron-1 retained signal was then divided by the average intron-1 retained percentage observed in T5 cells always analyzed in parallel (arbitrary unit). For Figure 2C, each dot represents the average of three independent experiments. Differences between tumor subgroups were tested using the Student's t-test (two-tailed). For Figure 4, the average obtained in Mock transfected cells was used as reference (corresponding to 1 a.u.). For each time and experiment, differences with the signal obtained in mock cells were tested using the Student's t-test (two-sided).
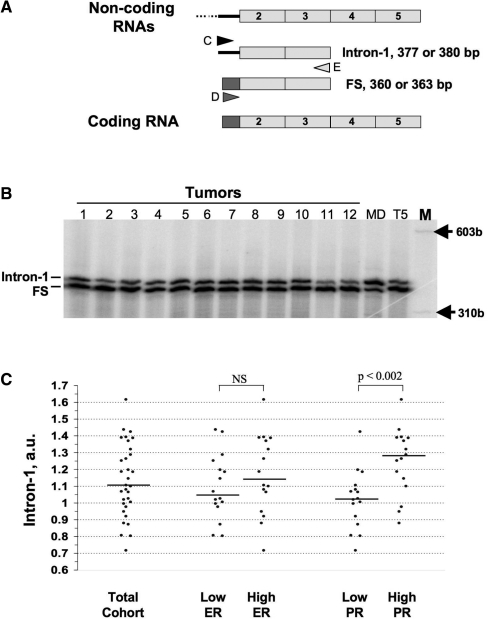
Figure 2.
TP-PCR amplification of coding and non-coding SRA mRNAs in breast cancers. (A) Principle: three primers are used during the PCR amplification. The lower primer (E) is common to the two sequences whereas, the upper D and C primers are specific for exon-1 (gray) and intron-1 (black), respectively (12). Triple-primer PCR will lead to an intron-1 or Fully spliced (FS) products, corresponding to non-coding and coding SRA RNAs, respectively. (B) RNA from frozen tissue sections (1–12) and from T5 as well as MDA-MB-468 (MD) cells was reverse transcribed and amplified by TP-PCR as described in ‘Materials and Methods’ section. radio-labeled PCR products, run on an acrylamide gel, are visualized using a Biorad Phosphorimager. (C) Signals from three experiments have been quantified and the average relative proportion of non-coding SRA RNA expressed in arbitrary unit (intron-1 a.u) as detailed in the ‘Materials and Methods’ section has been graphed for each tumor. For analysis purposes, tumors have been grouped according to their ER levels: lower and higher than the median; or according to their PR levels: lower and higher than the median. The horizontal bar indicates the median of intron-1 values for each subgroup. Differences have been tested using the Student's t-test two-tailed.
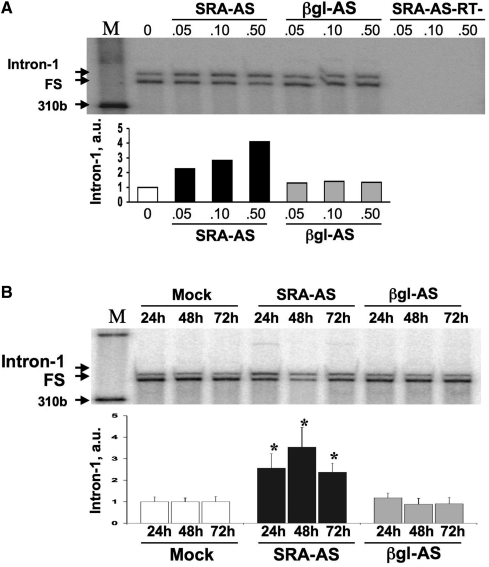
Figure 4.
Dose and time dependence analysis. (A) T5 cells were treated with increasing amounts (0, 0.05, 0.1, 0.5 μM) of SRA-AS or control oligos (βgl-AS). At t: 24 h, total RNA was extracted, reverse-transcribed and amplified by TP-PCR as described in ‘Material and methods’ section. Samples were separated on PAGE gel and visualized using a Molecular ImagerTM-FX. SRA-AS-RT- correspond to RNAs from SRA-AS amplified without reverse-transcription. Signals corresponding to intron-1 and FS were quantified and the proportion of intron-1 retention expressed in arbitrary unit (a.u) as described in ‘Materials and Methods’ section. (B) T5 cells were treated with no oligonucleotide (Mock), 0.5 μM of SRA–AS or 0.5 μM βgl–AS. At t: 24, 48 and 72 h, total RNA was extracted, reverse-transcribed and amplified by TP-PCR as described in ‘Material and methods’ section. Samples were separated on PAGE gel and visualized using a Molecular ImagerTM-FX. Signals corresponding to intron-1 and FS were quantified and the proportion of intron-1 retention expressed in arbitrary unit (a.u) as described in ‘Materials and Methods’ section. Bars represent the average value of five independent experiments normalized to values obtained for mock transfection at a given time point. Error bars represent standard deviation. *Correspond to P-values lower than 0.05 (Student's t-test).
Western blot analysis
Total cell lysates were analyzed and SRAP was detected using rabbit anti-SRAP 743 (Bethyl Laboratories Inc, Montgomery, TX) antibody in conjunction with a goat anti-rabbit HRP (Sigma, St Louis, MO) antibody at dilutions of 1/1000 and 1/3000, respectively, as described (6).
Real-time PCR
All PCR reactions were performed in an iCycler iQ™ Real-Time PCR Detection System (Bio-rad). For each PCR run, a master-mix was prepared on ice containing 2.5 mM MgCl2, 2.5 mM each dATP, dCTP, dGTP and dTTP, 0.1 μg each primer, 0.2x SYBR®Green1 (Molecular Probes, Inc.), 5 nM Fluorescein, 2.5% DMSO, 1× PCR Reaction Buffer (Invitrogen, Carlsbad, CA) and 1 unit of Platinum® Taq DNA polymerase (Invitrogen, Carlsbad, CA) (Figure 5). Two and a half microliters of each RT sample was added to 40 μl of PCR master-mix. The thermal cycling conditions comprised an initial denaturation step at 95°C for 10 min, then 40 cycles at 95°C for 30 s, 60°C for 45 s and 72°C for 30 s. Primers used are detailed in Supplementary Table S1. Three experiments were performed. For each treatment and experiment, the number of cycles needed to reach the defined cycle threshold for fully spliced and intron-1 alternatively spliced was first corrected for the number of cycles leading to the average 36B4 threshold cycle to obtain a normalized cycle number. The average number of normalized cycles for intron-1 retained and fully spliced RNA from the three experiments performed with no oligo (Mock) was then calculated to obtain intron-1 average mock and fully spliced average mock. For each treatment and experiment, the normalized cycle number of intron-1 retained and fully spliced SRA RNA was then subtracted from intron-1 average mock and fully spliced average mock, respectively, to get the number of cycle difference between treatments (ΔCt) and establish the standard deviations. A ΔCt of +1 represents a 21 = 2 fold increase in expression, whereas a ΔCt of −2 represents a 22 = 4-fold decrease in expression. Differences between Mock and oligo treatments were tested using the Student's t-test (two-sided).
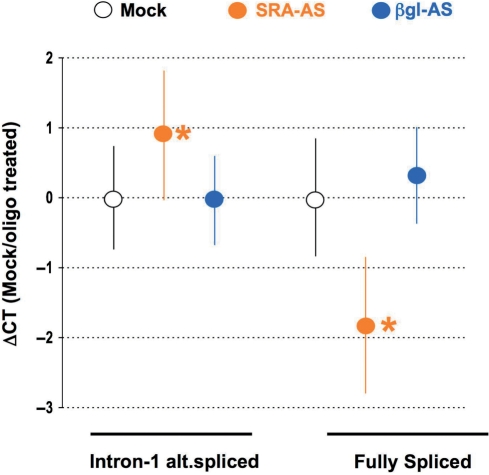
Figure 5.
Real-time PCR quantification of intron-1 retaining and fully spliced SRA RNAs expression following treatment of T5 cells. T5 cells were treated with no oligoribonucleotide (Mock, white circle), 0.5 μM of SRA–AS (orange circle) or 0.5 μM βgl–AS (blue circle). At t: 24 h, total RNA was extracted, reverse-transcribed and analyzed by real-time PCR as described in ‘Material and methods’ section. Three experiments were performed and for each treatment, ΔCt has been calculated as described in the ‘Materials and Methods’ section. Dots represent the average ΔCt for each treatment whereas bars correspond to standard deviations. Asterisk indicate a significant difference (Student's t-test, two-sided) in the corresponding SRA (intron-1 alternatively spliced or fully spliced) expression between mock and oligo-transfected cells.
Real-time PCR-array
Half a microgram of RNA was reverse transcribed using ReactionReady™ First Strand cDNA synthesis Kit (SuperArray Bioscience Corp., Frederick, MD) and applied to Breast Cancer and Estrogen Receptor RT2Profiler™ PCR arrays (SuperArray Bioscience Corp., Frederick, MD) as detailed by the manufacturer (Figures 7 and 8). For Figure 7, four experiments were performed. For each treatment (SRA–AS and βgl–AS), gene and experiment, the number of cycles needed to reach the defined threshold was normalized to the β-actin signal (ACTB) to obtain the corrected cycle numbers. For each gene the corrected cycle number per SRA–AS treatment was then subtracted from the reference gene–βgl–AS treatment to get the number of cycle difference between treatments (ΔCt) and establish the standard deviations. Differences between gene expression upon SRA–AS and βgl–AS treatments were tested using the Student's t-test (two-sided). Levels of expression presented in Table 1 were calculated as follow. If ΔCt was higher than or equal to 0, expression corresponds to 100 × 2ΔCt whereas when ΔCt is lower than 0 (i.e. negative), expression corresponds to 100/2−ΔCt.
Figure 7.
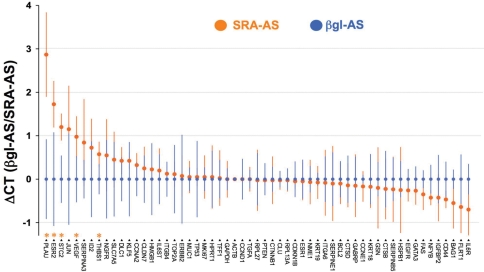
Change in gene expression in T5 cells following SRA–AS treatment. T5 cells were treated with 0.5 μM of SRA–AS or 0.5 μM βgl–AS as described above. At t: 24 h, RNA was extracted, reverse-transcribed, and used to assess, by real-time PCR, the expression of a series of 57 genes historically linked to breast cancer, as described in the ‘Materials and Methods’ section. Four independent experiments were performed. Blue dots represent, for each gene, the normalized expression upon βgl–AS treatment. Orange dots represent the average ΔCT increase or decrease in gene expression upon SRA–AS treatment. Bars represent the standard deviation for each gene and treatment. Genes whose expression is significantly modified (P < 0.05, Student's t-test, two-sided) upon SRA–AS treatment are indicated by orange stars.
Figure 8.
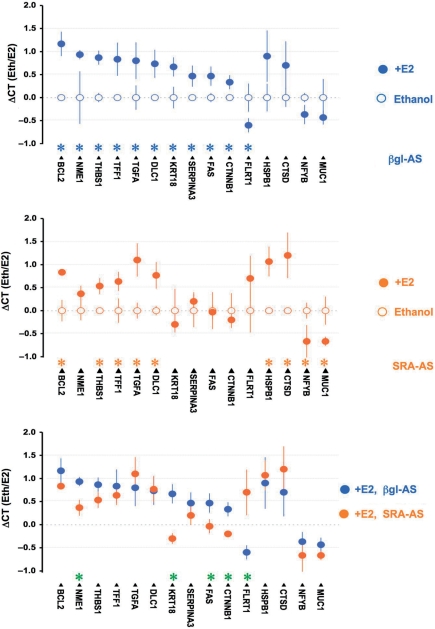
Differential estrogen regulation of gene expression in T5 cells following SRA–AS treatment. T5 cells were treated with 0.5 μM of βgl–AS or 0.5 μM SRA–AS as described earlier. Forty hours later, cells were treated for 4 h by E2 10−8M or vehicle (ethanol) alone, RNA was extracted, reverse-transcribed, and assessed for the expression of a series of 57 genes as described in the ‘Materials and Methods’ section. Three independent experiments were performed, and the expression of each gene normalized as detailed in the ‘Materials and Methods’ section. The average expression of 15 genes upon E2 treatment (filled circle) and ethanol (hollow circle) are depicted. Bars represent standard deviation. (A) Cells have been treated with 0.5 μM βgl–AS. Genes expression of which is significantly (p < 0.05, Student's t-test, two- sided) different between E2 treatment and Ethanol, are indicated by blue stars. (B) Cells have been treated with 0.5 μM SRA–AS. Genes expression of which is significantly (P < 0.05, Student's t-test, two-sided) different between E2 treatment and Ethanol are indicated by orange stars. (C) To visualize the differential effect of E2 in cells treated with SRA–AS (orange) and βgl–AS (blue), E2 corresponding dots from A and B have been plotted on the same graph. Genes whose expression is significantly (P < 0.05, Student's t-test, two-sided) different upon E2 treatment are indicated by green stars.
Table 1.
Change in gene expression at t: 24 h following treatment of T5 cells with SRA–AS
| Symbol | Description | ΔCT | Percentage expression | P-value |
|---|---|---|---|---|
| PLAU | Plasminogen activator, urokinase | 2.87 | 729 | 0.021 |
| ESR2 | Estrogen receptor 2 (ER beta) | 1.73 | 331 | 0.029 |
| STC2 | Stanniocalcin 2 | 1.20 | 230 | 0.009 |
| JUN | V-jun sarcoma virus 17 oncogene homolog (avian) | 1.15 | 222 | 0.164 |
| VEGF | Vascular endothelial growth factor | 0.98 | 197 | 0.034 |
| SERPINA3 | Serpin peptidase inhibitor, clade A, member 3 | 0.84 | 179 | 0.187 |
| ID2 | Inhibitor of DNA binding 2 | 0.72 | 165 | 0.234 |
| THBS1 | Thrombospondin 1 | 0.58 | 149 | 0.043 |
| NGFR | Nerve growth factor receptor | 0.55 | 146 | 0.404 |
| SLC7A5 | Solute carrier family 7, member 5 | 0.45 | 137 | 0.433 |
| DLC1 | Deleted in liver cancer 1 | 0.43 | 134 | 0.202 |
| KLF5 | Kruppel-like factor 5 (intestinal) | 0.43 | 134 | 0.369 |
| CCNA2 | Cyclin A2 | 0.33 | 125 | 0.223 |
| CLDN7 | Claudin 7 | 0.25 | 119 | 0.523 |
| HMGB1 | High-mobility group box 1 | 0.23 | 117 | 0.637 |
| IL6ST | Interleukin 6 signal transducer (gp130, oncostatin M receptor) | 0.20 | 115 | 0.620 |
| ITGB4 | Integrin, beta 4 | 0.12 | 109 | 0.642 |
| TOP2A | Topoisomerase (DNA) II alpha 170 kDa | 0.12 | 108 | 0.819 |
| ERBB2 | V-erb-b2 erythroblastic leukemia viral oncogene homolog 2 | 0.07 | 105 | 0.898 |
| MUC1 | Mucin 1, transmembrane | 0.05 | 104 | 0.834 |
| TP53 | Tumor protein p53 (Li-Fraumeni syndrome) | 0.05 | 104 | 0.915 |
| MKI67 | Antigen identified by monoclonal antibody Ki-67 | 0.05 | 104 | 0.867 |
| HPRT1 | Hypoxanthine phosphoribosyltransferase 1 | 0.05 | 104 | 0.922 |
| TFF1 | Trefoil factor 1 | 0.03 | 102 | 0.951 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 0.00 | 100 | 1.000 |
| ACTB | Actin, beta | – | 100 | |
| CCND1 | Cyclin D1 | 0.00 | 100 | 1.000 |
| TGFA | Transforming growth factor, alpha | 0.00 | 100 | 1.000 |
| RPL27 | Ribosomal protein L27 | −0.03 | 98 | 0.959 |
| PTEN | Phosphatase and tensin homolog | −0.03 | 98 | 0.902 |
| CTNNB1 | Catenin (cadherin-associated protein), beta 1, 88 kDa | −0.03 | 98 | 0.938 |
| CLU | Clusterin rotein J) | −0.03 | 98 | 0.868 |
| RPL13A | Ribosomal protein L13a | −0.03 | 98 | 0.894 |
| CDKN1B | Cyclin-dependent kinase inhibitor 1B (p27, Kip1) | −0.05 | 97 | 0.830 |
| ESR1 | Estrogen receptor 1 | −0.05 | 97 | 0.878 |
| NME1 | Non-metastatic cells 1, protein (NM23A) expressed in | −0.07 | 95 | 0.850 |
| KRT19 | Keratin 19 | −0.08 | 95 | 0.725 |
| ITGA6 | Integrin, alpha 6 | −0.08 | 94 | 0.869 |
| SERPINE1 | Serpin peptidase inhibitor, clade E, member 1 | −0.10 | 93 | 0.857 |
| BCL2 | B-cell CLL/lymphoma 2 | −0.10 | 93 | 0.797 |
| CTSD | Cathepsin D (lysosomal aspartyl peptidase) | −0.14 | 91 | 0.740 |
| GABRP | Gamma-aminobutyric acid (GABA) A receptor, pi | −0.15 | 90 | 0.760 |
| CCNE1 | Cyclin E1 | −0.17 | 89 | 0.473 |
| KRT18 | Keratin 18 | −0.18 | 89 | 0.660 |
| GSN | Gelsolin (amyloidosis, Finnish type) | −0.20 | 87 | 0.706 |
| CTSB | Cathepsin B | –0.22 | 86 | 0.654 |
| SERPINB5 | Serpin peptidase inhibitor, clade B (ovalbumin), member 5 | –0.23 | 85 | 0.677 |
| HSPB1 | Heat shock 27kDa protein 1 | –0.25 | 84 | 0.658 |
| EGFR | Epidermal growth factor receptor | –0.26 | 84 | 0.222 |
| GATA3 | GATA-binding protein 3 | –0.27 | 83 | 0.305 |
| FAS | Fas (TNF receptor superfamily, member 6) | –0.35 | 78 | 0.142 |
| NFYB | Nuclear transcription factor Y, beta | –0.43 | 74 | 0.102 |
| IGFBP2 | Insulin-like growth factor binding protein 2, 36 kDa | –0.43 | 74 | 0.357 |
| CD44 | CD44 antigen (Indian blood group) | –0.47 | 72 | 0.148 |
| BAG1 | BCL2-associated athanogene | –0.55 | 68 | 0.129 |
| FLRT1 | Fibronectin Leucine-rich transmembrane protein 1 | –0.64 | 65 | 0.610 |
| IL6R | Interleukin 6 receptor | –0.70 | 62 | 0.088 |
*Boldface depicts significant P-values.
For Figure 8, cells were treated with SRA–AS or βgl–AS as indicated earlier. Twenty-four hours post treatment, medium was changed to medium without serum for an additional 24 h. Cells were then treated with E2 10−8 M or vehicle alone (ethanol 1/1000) for 4 h before extraction of total RNA and SuperArray analysis. Three experiments were performed. For all treatment (SRA–AS and βgl–AS with Ethanol or E2), gene and experiment, the number of cycles needed to reach the defined threshold was first normalized to the β-actin signal (ACTB) to obtain the corrected cycle numbers. For each gene, the corrected cycle number following E2 treatment was then subtracted from the Ethanol treatment to get an arbitrary measurement of E2 action: A ΔCt(Eth/E2) = 1 corresponds to 21 = 2-fold increase in expression, whereas a ΔCt(Eth/E2) = −1 represents a 21 = 2-fold decrease in expression. Differences between gene expressions upon treatments were tested using the Student's t-test (two-sided).
Cell proliferation assay
Cell viability/proliferation was measured using CellTiter 96®Aqueous One Solution Reagent (Promega Co., Madison, WS) as previously described (17). Briefly, T5 cells were treated as described earlier with anti-sense oligos or not (mock). Twenty-four hours later, cells (3 × 103 cells) were re-seeded in 96 wells dishes in 5%FBS–DMEM. Twenty-four hours after re-seeding (48 h post oligo treatment), T0 measurements were taken. Media was changed every two days after T0. All time-point measurements were taken by adding, at various times (T0, 1, 3, 5 days), 40 µl of CellTiter 96®AQueous One Solution Reagent to each well containing 200 µl medium, followed by incubation for 3 h at 37°C. The absorbance was then recorded at 490 nm with 96-well plate reader (SpectraMax 190, Molecular Devices, Sunnyvale, CA.) All time point measurements were first blanked to the appropriate media not exposed to cells. For each experiment (performed in quadruplicate) the average of absorbance at T0 was calculated and deduced from each experimental points. For each point, the resulting mean of two independent experiments (expressed as Viability, a.u) and standard deviations were plotted using Prism 5 (GraphPad software, Inc, La Jolla, CA). Differences were tested using the Student's t-test (two-sided).
RESULTS
Higher SRA–intron-1 relative expression in breast tumors with higher progesterone receptor contents
Thirty-two different human breast tumors were selected from the Manitoba Breast Tumor Bank. These tumors spanned a wide range of estrogen receptor alpha (ER) and progesterone receptor (PR) levels, as assessed by ligand binding assay. Total RNA was extracted from frozen tumor tissue sections, reverse-transcribed and the relative expression of non-coding SRA transcripts containing intron-1 analyzed by radioactive TP-PCR, as described in the ‘Materials and Methods’ section. TP-PCR, which relies on the use of three primers during the PCR reaction, has been previously validated to assess the relative proportion of transcripts sharing a common extremity but differing in the other (12,16,18). As illustrated Figure 2A, the lower primer (primer E) recognizes the common exon 3 sequence, whereas the upper primers (C and D) are designed to recognize intron-1 and exon-1 sequence, specific of non-coding and coding SRA transcripts, respectively. Two PCR products (377–380 and 360–363 bp), corresponding respectively, to SRA transcripts containing intron-1 sequences (FI, PI and AD in Figure 1B) or fully spliced (FS), are expected. The potential 3 bp size difference corresponds to the additional codon specific for SRA3 isoform and located in the amplified part of exon-3 analyzed (Figure 1B). PCR products corresponding to both coding and non-coding transcripts were detected in all samples analyzed (Figure 2B). For each sample, signals were quantified and the relative proportion of SRA intron-1 containing RNA was expressed in arbitrary unit (a.u, Figure 2C). Relative level of this transcript varies from one case to another (min: 0.71 a.u, max: 1.61 a.u, median 1.10 a.u). No difference was found in the relative expression of non-coding SRA when comparing cases with low ER (n = 16, ER < 82.5 fmol/mg prot., intron-1 median: 1.04 a.u) and high ER (n = 16, ER > 82.5 fmol/mg prot., intron-1 median: 1.14 a.u.).
In contrast, a significant (Student's t-test, two-sided, P < 0.003) higher intron-1 retention was observed in samples with high PR (n = 16, PR > 38.5 fmol/mg prot., intron-1 median: 1.27 a.u.), than with low PR (n = 16, PR < 38.5 fmol/mg prot., intron-1 median: 1.02 a.u.).
Altering intron-1 splicing events using a modified anti-sense oligoribonucleotide
Reprogramming splicing events through transfection of modified antisense oligoribonucleotides targeting exon–intron junctions to interfer with donor or acceptor splicing sites has been successfully achieved in different systems (19,20). We hypothesized that an oligonucleotide complementary to the junction exon-1–intron-1 could reprogram the fate of pre-messenger RNA and lead to the production of more alternatively spliced non-coding SRA transcripts Figure 3.
Figure 3.
Principle: an oligonucleotide complementary to the junction exon-1/intron-1 is designed to interfere with the proper splicing of SRA pre-messenger. As a result, a higher proportion of alternatively spliced transcripts (containing Full—FI- or partial—PI-intron-1 retention, or resulting from the use of an alternative donor site (AD) as defined Figure 1) relative to the fully spliced (FS) should be observed.
To establish proof of principle that such an approach might be suitable to block SRA intron-1 donor site and ultimately alter SRA intron-1 splicing events, we initially used a previously described artificial SRA–β-globin minigene model (12). Two different 2′-O-methyl-modified anti-sense oligoribonucleotide phosphorothioate 20-mers were designed to recognize the splice donor sites of β-globin (βgl–AS) and SRA intron-1 (SRA–AS), present on this mini-gene (Supplementary Figure S1). We first established that both SRA–AS and βgl–AS similarly entered and remained in T5 cells (Supplementary Figure S2). We then demonstrated that SRA–AS oligoribonucleotide, as opposed to βgl–AS, had the ability to interfere with SRA intron-1 donor site and promoted SRA intron-1 retention within transcripts originating from our artificial SRA minigene (Supplementary Figure S3).
The proportion of endogenous SRA transcripts retaining intron-1 is increased following treatment of T5 cells with SRA–AS
To further determine if the introduction of SRA–AS could also alter the balance of endogenous coding and non-coding SRA RNAs, dose and time dependence experiments were performed and the relative proportion of SRA intron-1 retention assessed by triple-primer PCR (TP-PCR). T5 cells were transfected with increasing concentrations of either SRA-AS or βgl-AS oligoribonucleotides and total RNA analyzed by TP-PCR as described earlier. Co-amplification of two products, migrating at an apparent size of 377 and 360 bp, occurred (Figure 4A). These products were found, following sequencing, to correspond to the expected non-coding (containing intron-1) and coding (fully spliced) SRA transcripts. No products were amplified when RNA was not reverse-transcribed, confirming that no DNA could possibly be amplified in this assay. Signals were quantified and SRA intron-1 relative levels expressed in arbitrary unit as described in the ‘Materials and Methods’ section. A dose-dependent shift towards more intron-1 retention was observed in T5 cells when treated with SRA–AS. At the highest concentration tested of 0.5 μM, which corresponded to a 100% transfection efficiency (Supplementary Figure S2), SRA–AS led to an approximate 4-fold increase in the relative amount of intron-1 retention (Figure 4A, 0.5 μM SRA–AS), as compared to mock transfected control (Figure 4A, 0). As expected, no effect was observed on the proportion of endogenous intron-1 retaining SRA RNAs when cells were transfected with equivalent amounts of βgl–AS oligoribonucleotides (Figure 4A, 0.05 μM βgl–AS). βgl–AS oligonucleotides have been used as negative controls in subsequent experiments.
Time course experiments similarly performed with no oligoribonucleotides (Mock), 0.5 μM SRA–AS or 0.5 μM βgl–AS oligoribonucleotides, revealed a significant (n = 5, p < 0.05, Student's t-test) increase (2.5 ± 0.6 SD fold) in relative intron-1 retention at t: 24 h upon SRA–AS treatment (Figure 4B, 24 h SRA–AS). A maximal 3.5-fold average increase in the relative level of intron-1 retention was observed at t: 48 h (Figure 4B, 48 h SRA–AS) over that observed for time-matched mock transfected cells. This effect is maintained, albeit decreased at t: 72 h when levels of intron-1 retention have returned to that of samples at t: 24 h. As expected, βgl–AS oligoribonucleotide transfection had no effect in promoting endogenous SRA intron-1 retention in T5 cells.
Increased relative expression of non-coding SRA transcripts corresponds to both an increase in intron-1 containing SRA RNAs and a decrease in the levels of fully spliced SRA RNA
To clarify whether the relative increase in non-coding SRA RNAs expression observed by TP-PCR resulted from an increase in intron-1 retaining RNA, a decrease in fully spliced RNA or both, we assessed the expression of these SRA RNAs by real-time PCR. T5 cells were treated with no oligoribonucleotides (Mock), SRA–AS or βgl–AS oligoribonucleotides and RNA extracted and reverse-transcribed 24 h post-treatment. Following amplification with a lower primer annealing to exon-3 and upper primers annealing to either intron-1 or exon-1, the percentage change in the level of intron-1 retained or fully spliced was quantified in three independent experiments, as described in the ‘Materials and Methods’ section (Figure 5). A significant (P < 0.05, Student's t-test) average increase of 90% (corresponding to a ΔCt of 0.92) in the expression of SRA intron-1 retaining RNA was observed in cells treated with SRA–AS as compared to mock transfection. Conversely, a significant decrease of 70% (corresponding to a ΔCt of −1.82) in fully spliced SRA RNA expression was observed. As expected, no effect of βgl–AS oligoribonucleotides on either intron-1 retaining or fully spliced SRA levels was observed.
Decrease of endogenous SRAP in T5 cells treated with 0.5 μM SRA–AS
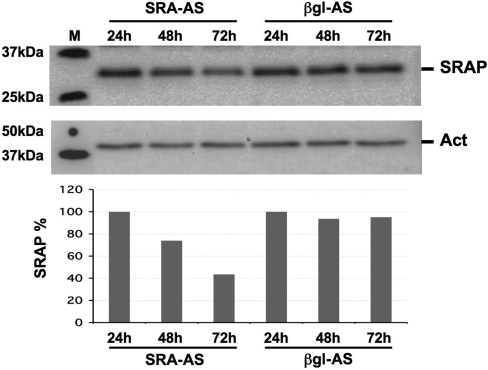
T5 cells were treated with either SRA–AS or βgl–AS for 24, 48 and 72 h and SRAP expression assessed by Western blot as detailed in the ‘Materials and Methods’ section. No apparent difference between SRA–AS and βgl–AS treated cells can be seen after 24 h (Figure 6). In contrast, lower levels of SRAP protein are seen after 48 and 72 h of SRA–AS oligoribonucleotide treatment. As expected, βgl–AS oligoribonucleotide does not appear to have any effect on SRAP levels.
Figure 6.
Western blot analysis of SRAP in T5 cells treated with 0.5 μM SRA–AS. T5 cells were treated with 0.5 μM of SRA–AS or 0.5 μM βgl–AS. At t: 24, 48 and 72 h, total were extracted and analyzed by Western using anti-SRAP antibodies as described in ‘Materials and Methods’ section. This Figure is representative of results obtained from three independent experiments.
Modulation of the balance coding/non-coding endogenous SRA RNAs alters gene expression in T5 breast cancer cells
To establish whether this artificial alteration of the balance between coding/non coding SRA1 RNAs impacts on gene expression, total RNA from T5 cells transfected with SRA–AS or βgl–AS oligoribonucleotides was extracted and analyzed 24 h post-transfection by real-time quantitative PCR using a Breast Cancer and Estrogen Receptor Signaling RT2 Profiler™ PCR Array (SuperArray Biosciences USA) as described in the ‘Materials and Methods’ section. This technology consists of ready-to-use PCR plates with each well containing an optimized pair of primers corresponding to a series of known genes. It provides the fastest way to interrogate the effect of a given treatment on genes historically linked to different pathways or pathologies (21). The expression of 57 genes was evaluated in cells treated with SRA–AS and βgl–AS oligoribonucleotides and differences in expression assessed using the Student's t-test (see Table 1 for a full description of the genes assessed and the results obtained). The expression of 51 genes remained constant upon SRA–AS treatment (Figure 7). The expression of five genes was however significantly increased. A strong increase (ΔCt = 2.87, corresponding to a 729% increase in the expression compared to the control) in the expression of the urokinase plasminogen activator PLAU, gene intimately linked to invasion mechanisms (22), was observed. Similarly, enhanced levels (ΔCt = 1.73, corresponding to a 331% increase in the expression compared to the control) of estrogen receptor beta mRNA (ESR2) was also found. Expression of Stanniocalcin 2 (STC2), vascular endothelial growth factor (VEGF) and Thrombospondin 1 (THBS1) was significantly increased, although to a lesser extent.
Modulation of the balance coding/non-coding endogenous SRA RNAs alters the response to estrogen of T5 breast cancer cells
The modification of the expression of estrogen receptor beta together with the known importance of estrogen signaling in breast cancer led us to investigate the effect of altering the balance coding/non-coding SRA RNAs on the response of T5 breast cancer cells to estrogen. T5 cells were transfected with βgl–AS or SRA–AS oligoribonucleotides and treated 48 h later with vehicle (ethanol) or E2 (10−8 M) for a period of 4 h, as described in the ‘Materials and Methods’ section. The expression of the 57 genes listed in Table 1 has been assessed as described earlier. Estradiol treatment of control cells pre-treated with βgl–AS oligoribonucleotide significantly altered the expression of 10 genes (Figure 8A). Nine genes (B-cell lymphoma 2: BCL2, Non-metastatic cells protein: NME1, Thrombospondin 1: THBS1, Trefoil factor 1: TFF1, Transforming growth factor alpha: TGFA, Deleted in liver cancer 1: DLC1, Keratin 18: KRT18, Serpin peptidase inhibitor 3: SERPINA3, Fas, Catenin beta 1: CTNNB1) are up-regulated whereas a single gene (Fibronectin Leucine-rich transmembrane protein 1: FLRT1) is downregulated. Expression of only five of the up-regulated genes (BCL2, THBS1, TFF1, TGFA and DLC1) remain significantly increased when cells are pre-treated with SRA–AS oligoribonucleotide (Figure 8B). In these latter conditions, estradiol treatment also led to a significantly increase in Heat shock 27 kDa protein 1 (HSPB1) and Cathepsin D (CTSD) expression and a decrease in Nuclear transcription factor Y-beta (NFYB) and Mucin 1 (MUC1) mRNA levels. Interestingly, a significant difference in estrogen effect when comparing directly SRA–AS or βgl–AS oligoribonucleotides pre-treatment is observed for NME1, KRT18, FAS, CTNNB1 and FLRT1 (Figure 8 C).
Modulation of the balance coding/non-coding endogenous SRA RNAs alters T5 breast cancer cells growth
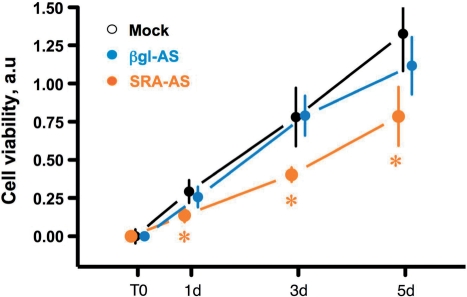
To further assess the relevance of the balance coding/non-coding SRA transcripts, we have compared the viability of cells treated with SRA–AS, βgl–AS or without oligonucleotides (Mock). Cells were treated as described earlier, re-seeded after 24 h and allowed to grow in normal medium for an additional 24 h. Viability of cells was measured at this time (T0, corresponding to 48 h after transfection) and following 1, 3 or 5 days, as described in the ‘Materials and Methods’ section. It should be noted that we have confirmed, using fluorescent oligonucleotides, that more than 70% of re-seeded cells still contained SRA–AS and βgl–AS (data not shown). As shown in Figure 9, the number of viable cells remains identical in cells treated without (Mock) or with a control oligonucleotide (βgl–AS). In contrast, significantly less viable cells are present at 1, 3 and 5 d, which corresponds to 72, 120 and 168 h following SRA–AS treatment, respectively.
Figure 9.
Differential viability of T5 cells following SRA–AS treatment. Cells were treated with SRA–AS (orange), βgl–AS (blue) or without (Mock, Black) oligonucleotides, re-seeded in 96 wells plates after 24 h and allowed to grow in normal medium for an additional 24 h. At this time (T0), or 1 (1d), 3 (3d) and 5 (5d) days later, the amount of live cells was determined and expressed in arbitrary unit as described in the ‘Materials and Methods’ section. For each point, the mean and the standard deviation corresponding to two independent experiments, each performed in quadruplicate, are indicated. Stars indicate a significant (P < 0.05) difference between SRA–AS and both negative controls (βgl–AS and Mock), as assessed by the Student's t-test, two-tailed.
DISCUSSION
The full sequencing of the human genome has led to a growing interest in alternative splicing events. Indeed, originally thought to possibly result from ‘hiccups’ of the splicing machinery, it became evident that these events are highly controlled and define a new level of complexity in gene expression (23–26). The critical participation of alternative splicing in regulating normal major biological events, such as sex determination or tuning of brain receptor sensitivity, has been underlined in many different systems (27–29). It is now also suggested that alteration of splicing can also be involved in many pathological situations including but not limited to skin diseases (30), neurodegeneration (31), and cancer (32–37). Interestingly, it has even been proposed that specific splicing events may be associated to tissue specific cancer (38).
We have previously reported that different breast cancer cell lines grown in culture have different relative levels of coding/non coding SRA transcripts (12). This balance ultimately controls two functional entities (SRA RNA and SRAP) modulating the action of ER and PR, major players in human breast tumorigenesis and tumor progression (39). It was critical to establish whether the relative proportion of these transcripts differed in vivo within tumor tissues. The selected cohort consisted of tumors with ER and PR values higher than 3 and 10 fmol/mg of total prot, respectively (40), therefore belonging to the clinically established ER+/PR+ subgroup. This subgroup has been selected as it represents the main subtype of cases contained in the Manitoba Breast Tumor Bank (MBTB). Indeed, within more than 5000 specimens collected in the MBTB, 48% of tumors are ER+/PR+, whereas 25%, 5% and 22% are ER+/PR−, ER−/PR+ and ER−/PR−, respectively.
Observed relative levels of non-coding intron-1-containing RNA varied from one sample to another, ranging from 0.71 to 1.61 a.u. (Figure 2C). Frozen tissue sections analyzed are intrinsically heterogeneous, containing cancer cells but also normal epithelial, fibroblastic and blood cells. These values could therefore represent an average of the relative SRA transcripts expression within all these cell types. These numbers are however fully consistent with the ones obtained on isolated cancer cell lines (between 0.80 and 2 a.u. for MCF-10A1 cells and MDA-MB-468 cells, respectively) (12). We have also shown that SRA expression was higher in breast cancer tissue than in normal breast tissue (41). The relative expression of SRA transcripts assessed on tumor tissues therefore likely reflects the balance coding/non coding SRAs in breast cancer cells rather than in normal other cells contained in these sections. However further in-situ experiments, specifically highlighting individual SRA splicing variants, are needed to confirm this hypothesis.
Surprisingly, with such a small tumor subset, a significant higher relative intron-1-retention in tumors with higher PR levels was identified. Indeed this suggests that within these ER+/PR+ cases, the balance coding/non-coding SRA transcript might characterize particular subgroups. It is well established that patients with ER+/PR+ tumors have a higher chance to respond to endocrine therapy than other patients (42). It is also known that within this group, higher ER and PR level are associated with a better response to tamoxifen (40). Unfortunately some patients, even though defined through ER and PR levels as most likely to benefit, will not respond to endocrine therapy. This reflects the heterogeneous biology of tumors belonging to the currently established ER+/PR+ subgroup and underscores the need to further characterize these lesions to better predict their behaviour upon treatment. It is believed that understanding the exact connections between traditional prognostic/predictive factors, gene expression signatures and patient outcome will allow the establishment of more adequately targeted treatment (43). Our observation that SRA intron-1 retention potentially characterizes a particular tumor subgroup fits with the current idea that assessing alternative splicing events might help profiling cancer sub-types (38,44–46).
We have herein successfully shifted the relative levels of endogenous coding/non coding SRA transcripts in the estrogen receptor alpha positive breast cancer cell line T5. Our approach, consisted in masking a splicing donor site with a modified oligonucleotide, and has been used successfully by many different groups to redirect splicing events involving specific target RNAs (19,47–49). Such an approach presents several advantages. As an anti-sense method, it benefits from the fact that 2′-O-modified (either methyl or methoxy-ethyl) oligoribonucleotide phosphorothioates bind to pre-mRNA target sequences in vivo with high specificity but do not form RNase H competent RNA-oligonucleotide hybrid complexes (50). Therefore, unlike more classical anti-sense oligonucleotide techniques that result in mRNA degradation, these modified anti-sense oligoribonucleotides are not expected to significantly affect RNA stability but rather serve as negative regulators that mask splice recognition sequences thereby preventing recruitment of splicing factors. It should however be noted that if a modification of splicing events is likely, a potential change in the stability of the RNAs considered cannot be fully excluded. Applied to the endogenous SRA RNA population, which ultimately consists of fully-spliced coding, and intron-1 alternatively spliced non-coding species, this strategy allows reprogramming the fate of immature RNAs toward the production of more non-coding RNAs. Reprogramming endogenous RNAs, as opposed to introducing exogenous non-coding RNAs under the control of artificial promoters such as the Cytomegalovirus promoter (CMV), presents an obvious advantage to interrogate physiological balance modifications. Treatment of cells with SRA–AS oligoribonucleotides allowed the relative intron-1 retention levels to increase by a factor of 2.5-fold. This increase is very similar to the difference previously observed (2-fold) between MDA-MB-468 invasive breast cancer cells and T5 non-invasive breast cancer cells (12). This confirms the suitability of the approach to increase the relative proportion of non-coding SRA RNA up to levels naturally occurring in the various cell lines previously characterized.
We herein showed that the increase (2.5-fold) in the relative amount of intron-1 retained SRA RNA as assessed by TP-PCR corresponded, using real-time PCR, to an absolute increase of ∼90% of this transcript and a ∼70% decrease of fully spliced SRA RNA (Figure 5). As expected, the decrease in coding SRA RNA resulted in a decrease in endogenous SRAP levels detected by Western blot (Figure 6). This underscores that whatever effects of SRA–AS oligonucleotide treatment might be, they may result from either the increase in non-coding functional SRA transcript, or a decrease in protein function or both.
PLAU (plasminogen urokinase activator also called uPA), is known to play a critical role in the development of metastases through the activation of several metalloproteinase (22,51) and was among the genes whose expression was modified after SRA–AS oligonucleotide treatment. Interestingly, we have previously reported that invasive MDA-MB-231 and MDA-MB-468 breast cancer cells expressed significant more SRA RNAs retaining intron-1 than non-invasive MCF-7, T47D or BT20 cells, whereas the more ‘normal’ MCF-10A1 breast cell line expressed the lowest relative level of SRA intron-1 RNA (12). This suggests that a balance ‘tipped’ toward the production of non-coding SRA1 RNA in breast cells might affect growth and/or invasion properties. Altogether, this observation suggests that non-coding SRA RNA, through the over-expression of genes such as PLAU, might directly participate in the establishment of an invasive phenotype in breast cancer cells. Further studies are needed to corroborate this hypothesis.
A significant increase in ESR2, and to a lesser extend in Stanniocalcin 2, Vascular endothelial growth factor and Thrombospondin 1 expression were observed upon SRA–AS treatment. All these genes have previously been shown to be up-regulated by estrogens in breast cancer cells (52–54). Their altered expression following the artificial shift in endogenous coding/non-coding SRA transcripts outline the relevance of this balance in mediating estrogen effects in breast cancer cells. Interestingly, increasing the relative levels of non-coding SRA had distinct potential effects on estrogen mediated gene regulation. For genes such as Bcl2, THSB1, TFF1, TGFA, DLC1 the significant and previously described estrogen induced over-expression (55–58) remains significant (Figure 8A and B). This could result from an absence of effect of SRA RNA and SRAP on the estrogen regulation of these particular genes. As outlined earlier, our approach modifies both functional RNA and protein levels. This lack of effect could therefore also reflect a neutralization of opposite individual actions. For genes like CTSD or MUC1, the respective trend toward an increase or a decrease in expression upon estrogen treatment becomes significant (Figure 8A and B). This amplification of estrogen action fits with the concept that increasing the relative levels of the functional non-coding RNA co-activator enhances estrogen receptor activity. It should however be noted that for these genes, no significant differences in estrogen effect is observed upon modifying coding/non-coding SRA transcripts (Figure 8C). Additional time-course experiments as well as experiments designed to investigate the respective actions of non-coding SRA and SRAP are needed to further characterize the effect the balance of coding/non-coding transcripts plays on the estrogen regulation of these genes.
Most interestingly, altering the balance of coding/non-coding SRA significantly disrupts estrogen's effect on NME1, CTNNB1, KRT18, Fas and FLRT1 genes (Figure 8C). CTNNB1 or beta catenin, an inherent component of the Wnt/β-Catenin canonical signalling pathway, is a critical player in development as well as cell–cell contact and migration processes (59–61). NME1 (non-metastatic cells 1), whose expression is regulated by estradiol in breast cancer cells (62), is also believed to itself modulate estrogen receptor alpha action (63). These results overall suggest that relative levels of SRA transcripts still containing intron-1 have the potential to control selectively estrogen effect on specific genes highly relevant to development and cancer. Further experiments are warranted to establish the exact mechanisms behind these effects and to assess the full extent of the impact of the balance of SRA transcripts on estrogen action.
We have herein used a colorimetric method for determining the number of viable cells following modification of the balance of endogenous SRA towards the production of more non-coding transcripts. It should be stressed that the number of viable cells at a given time is a direct result of cell proliferation and cell death. Our experiments strongly suggest that the balance of coding/non-coding transcripts participates to the growth of breast cancer cells. Indeed, compared to both our controls, less viable cells are present following SRA–AS treatment. We cannot however, at the present stage, establish whether enhancing the relative amount of non-coding SRA RNA increases apoptosis or decreased cell proliferation. Additional experiments are needed to address this issue.
Herein, we have shown that the balance of coding/non-coding SRA RNAs could be altered through the use of modified oligoribonucleotides in breast cancer cells. This led to a change in expression of genes likely to have an important impact on two critical aspects of breast cancer cell phenotype, namely invasion and response to estrogen. It also led to a decrease in cell growth/viability. It has been previously hypothesized that strategies aimed at favouring the production of a given splice variant could be developed and proposed as new therapeutic tools (64–66). We propose that modifying SRA splicing events might lead to establishing potential new breast cancer treatments.
While SRA remains one the best characterized bi-functional RNA to date, other studies have highlighted the existence of similar other cases (67–69). Such systems, even though they challenge the paradigm classifying RNA as either strictly coding or as non–coding, provide a unique opportunity to further explore the mechanisms used by nature to control gene expression. There is an urgent need to design new experimental approaches to address the respective functions of these peculiar bi-faceted RNAs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institute of Health Research, the Manitoba Health Research Council, CancerCare Manitoba Foundation and a Trilink Research Award; USA-MR/MC pre-doctoral training grant (to S.C.-K. and Y.Y.); National Science and Engineering Research Council Canada Graduate Scholarship (to S.C.-K.); Le cancer du sein, parlons-en!, prix Ruban Rose 2008 (to F.H.). Funding for open access charge: Canadian Breast Cancer Foundation.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, Tsai MJ, O'Malley BW. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 2.Leygue E. Steroid receptor RNA activator (SRA1): unusual bi-faceted gene products with suspected relevance to breast cancer. Nuclear Recep. Signal. 2007;5:e006. doi: 10.1621/nrs.05006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi YH, Downes M, Xie W, Kao HY, Ordentlich P, Tsai CC, Hon M, Evans RM. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 2001;15:1140–1151. doi: 10.1101/gad.871201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatchell EC, Colley SM, Beveridge DJ, Epis MR, Stuart LM, Giles KM, Redfern AD, Miles LEC, Barker A, MacDonald LM, et al. SLIRP, a small SRA binding protein, is a nuclear receptor corepressor. Mol. Cell. 2006;22:657–668. doi: 10.1016/j.molcel.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 5.Emberley E, Huang GJ, Hamedani MK, Czosnek A, Ali D, Grolla A, Lu B, Watson PH, Murphy LC, Leygue E. Identification of new human coding steroid receptor RNA activator isoforms. Biochem. Biophys. Res. Commun. 2003;301:509–515. doi: 10.1016/s0006-291x(02)03070-x. [DOI] [PubMed] [Google Scholar]
- 6.Chooniedass-Kothari S, Emberley E, Hamedani MK, Troup S, Wang X, Czosnek A, Hube F, Mutawe M, Watson PH, Leygue E. The steroid receptor RNA activator is the first functional RNA encoding a protein. FEBS Lett. 2004;566:43–47. doi: 10.1016/j.febslet.2004.03.104. [DOI] [PubMed] [Google Scholar]
- 7.Chooniedass-Kothari SHM, Caracossa S, Jalaguier S, Cavailles V, Leygue E. Montreal, Canada: 2006. Canadian Breast Cancer Research Alliance (CBCRA) Vol. Oral communication. [Google Scholar]
- 8.Kurisu T, Tanaka T, Ishii J, Matsumura K, Sugimura K, Nakatani T, Kawashima H. Expression and function of human steroid receptor RNA activator in prostate cancer cells: role of endogenous hSRA protein in androgen receptormediated transcription. Prost. Cancer Prost. Dis. 2006;9:173–178. doi: 10.1038/sj.pcan.4500867. [DOI] [PubMed] [Google Scholar]
- 9.Kawashima H, Takano H, Sugita S, Takahara Y, Sugimura K, Nakatani T. A novel steroid receptor co-activator protein (SRAP) as an alternative form of steroid receptor RNA-activator gene: expression in prostate cancer cells and enhancement of androgen receptor activity. Biochem. J. 2003;369:163–171. doi: 10.1042/BJ20020743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chooniedass-Kothari S, Hamedani MK, Troup S, Hube F, Leygue E. The steroid receptor RNA activator protein is expressed in breast tumor tissues. Int. J. Cancer. 2006;118:1054–1059. doi: 10.1002/ijc.21425. [DOI] [PubMed] [Google Scholar]
- 11.Chooniedass-Kothari SHM, Caracossa S, Jalaguier S, Cavailles V, Leygue E. Keystone Symposia: Steroid Sisters. Banff, Alberta: 2006. Vol. Abstract 124, pp. 144. [Google Scholar]
- 12.Hube F, Guo JM, Chooniedass-Kothari S, Cooper C, Hamedani MK, Dibrov AA, Blanchard AAA, Wang XM, Deng G, Myal Y, et al. Alternative splicing of the first intron of the steroid receptor RNA activator (SRA) participates in the generation of coding and noncoding RNA isoforms in breast cancer cell lines. DNA Cell Biol. 2006;25:418–428. doi: 10.1089/dna.2006.25.418. [DOI] [PubMed] [Google Scholar]
- 13.Coutts AS, Davie JR, Dotzlaw H, Murphy LC. Estrogen regulation of nuclear matrix-intermediate filament proteins in human breast cancer cells. J. Cell Biochem. 1996;63:174–184. doi: 10.1002/(sici)1097-4644(19961101)63:2<174::aid-jcb5>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 14.Watson PH, Snell L, Parisien M. The NCIC-Manitoba Breast Tumor Bank: a resource for applied cancer research. CMAJ. 1996;155:281–283. [PMC free article] [PubMed] [Google Scholar]
- 15.Leygue E, Dotzlaw H, Watson PH, Murphy LC. Expression of the steroid receptor RNA activator in human breast tumors. Cancer Res. 1999;59:4190–4193. [PubMed] [Google Scholar]
- 16.Leygue E, Murphy L, Kuttenn F, Watson P. Triple primer polymerase chain reaction. A new way to quantify truncated mRNA expression. Am. J. Pathol. 1996;148:1097–1103. [PMC free article] [PubMed] [Google Scholar]
- 17.Wan J, Sazani P, Kole R. Modification of HER2 pre-mRNA alternative splicing and its effects on breast cancer cells. Int. J. Cancer. 2009;124:772–777. doi: 10.1002/ijc.24052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leygue E, Dotzlaw H, Watson PH, Murphy LC. Expression of estrogen receptor beta1, beta2, and beta5 messenger RNAs in human breast tissue. Cancer Res. 1999;59:1175–1179. [PubMed] [Google Scholar]
- 19.Mercatante DR, Kole R. Control of alternative splicing by antisense oligonucleotides as a potential chemotherapy: effects on gene expression. Biochim. Biophys. Acta. 2002;1587:126–132. doi: 10.1016/s0925-4439(02)00075-3. [DOI] [PubMed] [Google Scholar]
- 20.Khoo B, Roca X, Chew SL, Krainer AR. Antisense oligonucleotide-induced alternative splicing of the APOB mRNA generates a novel isoform of APOB. BMC Mol. Biol. 2007;8:3. doi: 10.1186/1471-2199-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arikawa E, Sun Y, Wang J, Zhou Q, Ning B, Dial S, Guo L, Yang J. Cross-platform comparison of SYBR(R) Green real-time PCR with TaqMan PCR, microarrays and other gene expression measurement technologies evaluated in the MicroArray Quality Control (MAQC) study. BMC Genomics. 2008;9:328. doi: 10.1186/1471-2164-9-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han B, Nakamura M, Mori I, Nakamura Y, Kakudo K. Urokinase-type plasminogen activator system and breast cancer (Review) Oncol. Rep. 2005;14:105–112. [PubMed] [Google Scholar]
- 23.Brett D, Pospisil H, Valcarcel J, Reich J, Bork P. Alternative splicing and genome complexity. Nat. Genet. 2002;30:29–30. doi: 10.1038/ng803. [DOI] [PubMed] [Google Scholar]
- 24.Lemischka IR, Pritsker M. Alternative splicing increases complexity of stem cell transcriptome. Cell Cycle. 2006;5:347–351. doi: 10.4161/cc.5.4.2424. [DOI] [PubMed] [Google Scholar]
- 25.Modrek B, Lee C. A genomic view of alternative splicing. Nat. Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 26.Roberts GC, Smith CW. Alternative splicing: combinatorial output from the genome. Curr. Opin. Chem. Biol. 2002;6:375–383. doi: 10.1016/s1367-5931(02)00320-4. [DOI] [PubMed] [Google Scholar]
- 27.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 28.Black DL, Grabowski PJ. Alternative pre-mRNA splicing and neuronal function. Progr. Mol. Subcell. Biol. 2003;31:187–216. doi: 10.1007/978-3-662-09728-1_7. [DOI] [PubMed] [Google Scholar]
- 29.Lee CJ, Irizarry K. Alternative splicing in the nervous system: an emerging source of diversity and regulation. Biol. Psychiatry. 2003;54:771–776. doi: 10.1016/s0006-3223(03)00375-5. [DOI] [PubMed] [Google Scholar]
- 30.Wessagowit V, Nalla VK, Rogan PK, McGrath JA. Normal and abnormal mechanisms of gene splicing and relevance to inherited skin diseases. J. Dermatol. Sci. 2005;40:73–84. doi: 10.1016/j.jdermsci.2005.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallo JM, Jin P, Thornton CA, Lin H, Robertson J, D'Souza I, Schlaepfer WW. The role of RNA and RNA processing in neurodegeneration. J. Neurosci. 2005;25:10372–10375. doi: 10.1523/JNEUROSCI.3453-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall PA, Russell SH. New perspectives on neoplasia and the RNA world. Hematol. Oncol. 2005;23:49–53. doi: 10.1002/hon.748. [DOI] [PubMed] [Google Scholar]
- 33.Kalnina Z, Zayakin P, Silina K, Line A. Alterations of pre-mRNA splicing in cancer. Genes Chromosomes Cancer. 2005;42:342–357. doi: 10.1002/gcc.20156. [DOI] [PubMed] [Google Scholar]
- 34.Cork DM, Lennard TW, Tyson-Capper AJ. Alternative splicing and the progesterone receptor in breast cancer. Breast Cancer Res. 2008;10:207. doi: 10.1186/bcr2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayes NV, Gullick WJ. The neuregulin family of genes and their multiple splice variants in breast cancer. J. Mammary Gland Biol. Neoplasia. 2008;13:205–214. doi: 10.1007/s10911-008-9078-4. [DOI] [PubMed] [Google Scholar]
- 36.Shelton BP, Misso NL, Shaw OM, Arthaningtyas E, Bhoola KD. Epigenetic regulation of human epithelial cell cancers. Curr. Opin. Mol. Ther. 2008;10:568–578. [PubMed] [Google Scholar]
- 37.Sundvall M, Iljin K, Kilpinen S, Sara H, Kallioniemi OP, Elenius K. Role of ErbB4 in breast cancer. J. Mammary Gland Biol. Neoplasia. 2008;13:259–268. doi: 10.1007/s10911-008-9079-3. [DOI] [PubMed] [Google Scholar]
- 38.He C, Zhou F, Zuo Z, Cheng H, Zhou R. A global view of cancer-specific transcript variants by subtractive transcriptome-wide analysis. PLoS ONE. 2009;4:e4732. doi: 10.1371/journal.pone.0004732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cordera F, Jordan VC. Steroid receptors and their role in the biology and control of breast cancer growth. Semin. Oncol. 2006;33:631–641. doi: 10.1053/j.seminoncol.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 40.Elledge RM, Green S, Pugh R, Allred DC, Clark GM, Hill J, Ravdin P, Martino S, Osborne CK. Estrogen receptor (ER) and progesterone receptor (PgR), by ligand-binding assay compared with ER, PgR and pS2, by immuno-histochemistry in predicting response to tamoxifen in metastatic breast cancer: a southwest oncology group study. Int. J. Cancer. 2000;89:111–117. [PubMed] [Google Scholar]
- 41.Murphy LC, Simon SL, Parkes A, Leygue E, Dotzlaw H, Snell L, Troup S, Adeyinka A, Watson PH. Altered expression of estrogen receptor coregulators during human breast tumorigenesis. Cancer Res. 2000;60:6266–6271. [PubMed] [Google Scholar]
- 42.Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 43.Wirapati P, Sotiriou C, Kunkel S, Farmer P, Pradervand S, Haibe-Kains B, Desmedt C, Ignatiadis M, Sengstag T, Schutz F, et al. Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 2008;10:R65. doi: 10.1186/bcr2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xi L, Feber A, Gupta V, Wu M, Bergemann AD, Landreneau RJ, Litle VR, Pennathur A, Luketich JD, Godfrey TE. Whole genome exon arrays identify differential expression of alternatively spliced, cancer-related genes in lung cancer. Nucleic Acids Res. 2008;36:6535–6547. doi: 10.1093/nar/gkn697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klinck R, Bramard A, Inkel L, Dufresne-Martin G, Gervais-Bird J, Madden R, Paquet ER, Koh C, Venables JP, Prinos P, et al. Multiple alternative splicing markers for ovarian cancer. Cancer Res. 2008;68:657–663. doi: 10.1158/0008-5472.CAN-07-2580. [DOI] [PubMed] [Google Scholar]
- 46.Zhang CL, Li HR, Fan JB, Wang-Rodriguez J, Downs T, Fu XD, Zhang MQ. Profiling alternatively spliced mRNA isoforms for prostate cancer classification. Bmc Bioinformatics. 2006;7:202. doi: 10.1186/1471-2105-7-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khoo B, Roca X, Chew SL, Krainer AR. Antisense oligonucleotide-induced alternative splicing of the APOB mRNA generates a novel isoform of APOB. Bmc Mol. Biol. 2007;8:3. doi: 10.1186/1471-2199-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mercatante DR, Bortner CD, Cidlowski JA, Kole R. Modification of alternative splicing of Bcl-x pre-mRNA in prostate and breast cancer cells. analysis of apoptosis and cell death. J. Biol. Chem. 2001;276:16411–16417. doi: 10.1074/jbc.M009256200. [DOI] [PubMed] [Google Scholar]
- 49.Mercatante DR, Sazani P, Kole R. Modification of alternative splicing by antisense oligonucleotides as a potential chemotherapy for cancer and other diseases. Curr. Cancer Drug Targets. 2001;1:211–230. doi: 10.2174/1568009013334124. [DOI] [PubMed] [Google Scholar]
- 50.Stein CA. Controversies in the cellular pharmacology of oligodeoxynucleotides. Antisense Nucleic Acid Drug Develop. 1997;7:207–209. doi: 10.1089/oli.1.1997.7.207. [DOI] [PubMed] [Google Scholar]
- 51.Duffy MJ. The urokinase plasminogen activator system: role in malignancy. Curr. Pharm. Des. 2004;10:39–49. doi: 10.2174/1381612043453559. [DOI] [PubMed] [Google Scholar]
- 52.Cappelletti V, Saturno G, Miodini P, Korner W, Daidone MG. Selective modulation of ER-beta by estradiol and xenoestrogens in human breast cancer cell lines. Cell Mol. Life Sci. 2003;60:567–576. doi: 10.1007/s000180300048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003;144:4562–4574. doi: 10.1210/en.2003-0567. [DOI] [PubMed] [Google Scholar]
- 54.Ghosh MG, Thompson DA, Weigel RJ. PDZK1 and GREB1 are estrogen-regulated genes expressed in hormone-responsive breast cancer. Cancer Res. 2000;60:6367–6375. [PubMed] [Google Scholar]
- 55.Perillo B, Sasso A, Abbondanza C, Palumbo G. 17beta -estradiol inhibits apoptosis in MCF-7 cells, inducing bcl-2 expression via two estrogen-responsive elements present in the coding sequence. Mol. Cell Biol. 2000;20:2890–2901. doi: 10.1128/mcb.20.8.2890-2901.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reid G, Metivier R, Lin CY, Denger S, Ibberson D, Ivacevic T, Brand H, Benes V, Liu ET, Gannon F. Multiple mechanisms induce transcriptional silencing of a subset of genes, including oestrogen receptor alpha, in response to deacetylase inhibition by valproic acid and trichostatin A. Oncogene. 2005;24:4894–4907. doi: 10.1038/sj.onc.1208662. [DOI] [PubMed] [Google Scholar]
- 57.Coser KR, Chesnes J, Hur J, Ray S, Isselbacher KJ, Shioda T. Global analysis of ligand sensitivity of estrogen inducible and suppressible genes in MCF7/BUS breast cancer cells by DNA microarray. Proc. Natl Acad. Sci. USA. 2003;100:13994–13999. doi: 10.1073/pnas.2235866100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rae JM, Johnson MD, Scheys JO, Cordero KE, Larios JM, Lippman ME. GREB 1 is a critical regulator of hormone dependent breast cancer growth. Breast Cancer Res. Treat. 2005;92:141–149. doi: 10.1007/s10549-005-1483-4. [DOI] [PubMed] [Google Scholar]
- 59.Fodde R, Brabletz T. Wnt/beta-catenin signaling in cancer stemness and malignant behavior. Curr. Opin. Cell Biol. 2007;19:150–158. doi: 10.1016/j.ceb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 60.Jeanes A, Gottardi CJ, Yap AS. Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene. 2008;27:6920–6929. doi: 10.1038/onc.2008.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mulholland DJ, Dedhar S, Coetzee GA, Nelson CC. Interaction of nuclear receptors with the Wnt/beta-catenin/Tcf signaling axis: what you like to know? Endocr. Rev. 2005;26:898–915. doi: 10.1210/er.2003-0034. [DOI] [PubMed] [Google Scholar]
- 62.Lin K-H, Wang W-J, Wu Y-H, Cheng S-Y. Activation of antimetastatic Nm23-H1 gene expression by estrogen and its {alpha}-receptor. Endocrinology. 2002;143:467–475. doi: 10.1210/endo.143.2.8620. [DOI] [PubMed] [Google Scholar]
- 63.Curtis CD, Likhite VS, McLeod IX, Yates JR, Nardulli AM. Interaction of the tumor metastasis suppressor nonmetastatic protein 23 homologue H1 and estrogen receptor {alpha} alters estrogen-responsive gene expression. Cancer Res. 2007;67:10600–10607. doi: 10.1158/0008-5472.CAN-07-0055. [DOI] [PubMed] [Google Scholar]
- 64.Wilton SD, Fletcher S. RNA splicing manipulation: strategies to modify gene expression for a variety of therapeutic outcomes. Curr. Gene Ther. 2005;5:467–483. doi: 10.2174/156652305774329249. [DOI] [PubMed] [Google Scholar]
- 65.Puttaraju M, Jamison SF, Mansfield SG, Garcia-Blanco MA, Mitchell LG. Spliceosome-mediated RNA trans-splicing as a tool for gene therapy. Nat. Biotechnol. 1999;17:246–252. doi: 10.1038/6986. [DOI] [PubMed] [Google Scholar]
- 66.Hayes GM, Carrigan PE, Beck AM, Miller LJ. Targeting the RNA splicing machinery as a novel treatment strategy for pancreatic carcinoma. Cancer Res. 2006;66:3819–3827. doi: 10.1158/0008-5472.CAN-05-4065. [DOI] [PubMed] [Google Scholar]
- 67.Dinger ME, Pang KC, Mercer TR, Mattick JS. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput. Biol. 2008;4 doi: 10.1371/journal.pcbi.1000176. e1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kloc M, Wilk K, Vargas D, Shirato Y, Bilinski S, Etkin LD. Potential structural role of non-coding and coding RNAs in the organization of the cytoskeleton at the vegetal cortex of Xenopus oocytes. Development. 2005;132:3445–3457. doi: 10.1242/dev.01919. [DOI] [PubMed] [Google Scholar]
- 69.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.