Abstract
The products of recombination activating genes (RAG) 1 and 2 mediate the assembly of antigen receptor genes during lymphocyte development in a process known as V(D)J recombination. Lack of structural information for the RAG proteins has hindered mechanistic studies of this reaction. We report here the crystal structure of an essential DNA-binding domain of the RAG1 catalytic core bound to its nonamer DNA recognition motif. The RAG1 nonamer-binding domain (NBD) forms a tightly interwoven dimer that binds and synapses two nonamer elements, with each NBD making contact with both DNA molecules. Biochemical and biophysical experiments confirm that the two nonamers are in close proximity in the RAG1/2-DNA synaptic complex and demonstrate the functional importance of the protein-DNA contacts revealed in the structure. These findings reveal a previously unsuspected function for the NBD in DNA synapsis and have implications for the regulation of DNA binding and cleavage by RAG1/2.
INTRODUCTION
V(D)J recombination assembles a diverse repertoire of immunoglobulin and T-cell receptor genes in developing B and T lymphocytes through rearrangement of different V (variable), in some cases D (diversity), and J (joining) gene segments. The first phase of the reaction, DNA cleavage, is catalyzed by the products of the recombination activating genes, RAG1 and RAG21,2, which form the core of the recombinase machinery. In vitro, the activity and specificity of this machinery are enhanced by the ubiquitous high-mobility-group proteins HMGB1/23,4. DNA binding and cleavage are targeted by recombination signal sequences (RSSs) that flank each gene segment (Fig. 1). Each RSS is composed of well-conserved heptamer (consensus 5'-CACAGTG) and nonamer (consensus 5'-ACAAAAACC) sequences separated by a spacer of either 12 bp or 23 bp (referred to as 12RSS or 23RSS, respectively). Efficient recombination occurs only between a 12RSS and a 23RSS, a phenomenon known as the 12/23 rule5.
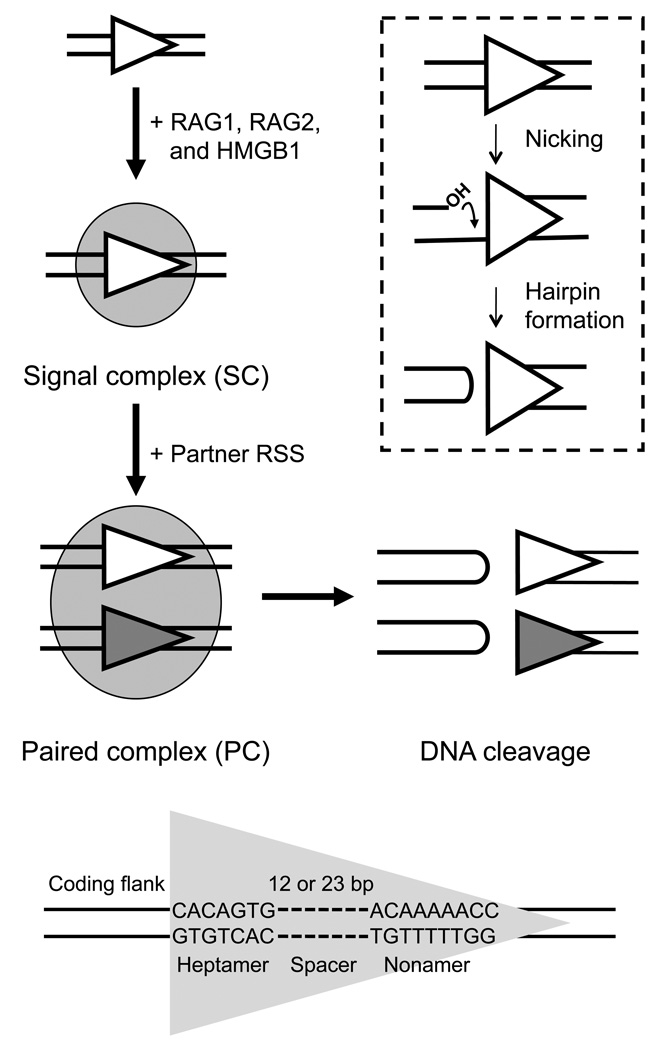
Figure 1.
Schematic of RAG-mediated DNA binding and cleavage. RAG1/RAG2/HMGB1 (shaded oval) bind to either a 12RSS or 23RSS (gray or white triangles) to form the SC and then capture a second, complementary RSS to form the PC, within which DNA cleavage is completed. DNA cleavage occurs in two steps, nicking and hairpin formation (inset). Nicking can occur in the SC or PC, while hairpin formation is restricted to the PC. The DNA sequence elements that make up the RSS DNA substrate are shown at bottom.
DNA cleavage occurs in two steps: nicking and transesterification (Fig. 1, inset). A nick is first introduced in the top strand immediately upstream of the heptamer, generating a 3'-hydroxyl group that can attack the phosphodiester bond on the opposite strand in a direct transesterification reaction, thereby creating four DNA ends: two hairpin coding ends and two blunt, 5'-phosphorylated signal ends6,7. According to the current model, the RAG complex first binds to one RSS to form a signal complex (SC), which can recruit a partner RSS to form the paired complex (PC), also called the synaptic complex, in which double-strand breaks are generated between the coding gene segments and the RSSs8–10. While nicking can occur in both the SC and PC, hairpin formation is largely restricted to the PC and is sensitive to 12/23 synapsis8,11–14. In the second phase of V(D)J recombination, the ends are processed and joined by proteins of the non-homologous end joining pathway15.
RSS-specific binding and cleavage activities are attributed primarily to RAG1, with RAG2 and HMGB1/2 functioning as its co-factors. RAG1 contains the catalytic triad DDE, a motif common to many recombinases and transposases, as well as multiple domains that interact with the RSS through base-specific and nonspecific DNA contacts16,17. Murine RAG1 core, the minimal region capable of supporting catalysis, includes amino acids 384–1008 of the total 1040 residues. Located at the N-terminus of the RAG1 core is the nonamer-binding domain (NBD), which recognizes the nonamer and is critical for anchoring the RAG proteins onto the RSS18,19. Footprinting and interference experiments mapped the primary site of RAG1-RSS interaction to the nonamer, while contacts in the heptamer and spacer are more dependent on RAG2 and HMGB1/217. Other domains in the RAG1 core contribute the DDE active site residues and interact with RSS elements proximal to the cleavage site and coding flank20. Less is known about the function of RAG2, but it is required for high affinity DNA binding and cleavage.
Although the basic biochemical steps in RAG-mediated DNA cleavage have been delineated, little is known about the protein-protein and protein-DNA interactions that govern the reaction. While several lines of evidence show that the dimeric form of RAG1 mediates binding of RAG1 alone to the RSS, the stoichiometry of RAG1 in the SC and PC is subject to debate10,20–24. To date, only the zinc dimerization domain and the plant homeodomain in RAG1 and RAG2, respectively, have yielded to structure determination25,26, but they offer little direct information about DNA recognition and cleavage as they are outside of the core regions. It has been proposed that the RAG1 NBD interacts with the nonamer through a helix-turn-helix (HTH) motif similar to the DNA-binding domain (DBD) of the Hin recombinase27, although a recent bioinformatic analysis has called this idea into question28. A lack of structural information for the RAG proteins has hindered efforts to understand the mechanism of V(D)J recombination.
Here, we report the crystal structure of the murine RAG1 NBD in complex with a 14-mer DNA containing the nonamer. The NBD adopts an intertwined dimer structure that mediates the synapsis of two DNA molecules and lacks the predicted HTH motif characteristic of Hin. Biochemical experiments with mutant RAG1 core proteins support the NBD structural findings and help to elucidate the mechanisms underlying RSS binding and synapsis, the differential regulation of nicking and hairpin formation, and the basis of some immunodeficiency diseases.
RESULTS
Structure determination and refinement
Based on previous studies that established the approximate boundaries of the RAG1 NBD18,19,29, we screened NBD proteins of varying lengths with several nonamer oligonucleotides for crystallization. His6-tagged RAG1 residues 389–464 in complex with a sticky ended 14-mer DNA duplex containing the consensus nonamer sequence (numbered 1–9) and four adjacent base pairs of the consensus 12RSS spacer (5'-CTTA, numbered S9-S12) yielded the best crystals (Fig. 2a). The structure of the NBD-DNA complex was determined at a resolution of 2.4 Å using multiple isomorphous replacement with anomalous scattering (MIRAS) from lead, mercury, and tungsten derivatives.
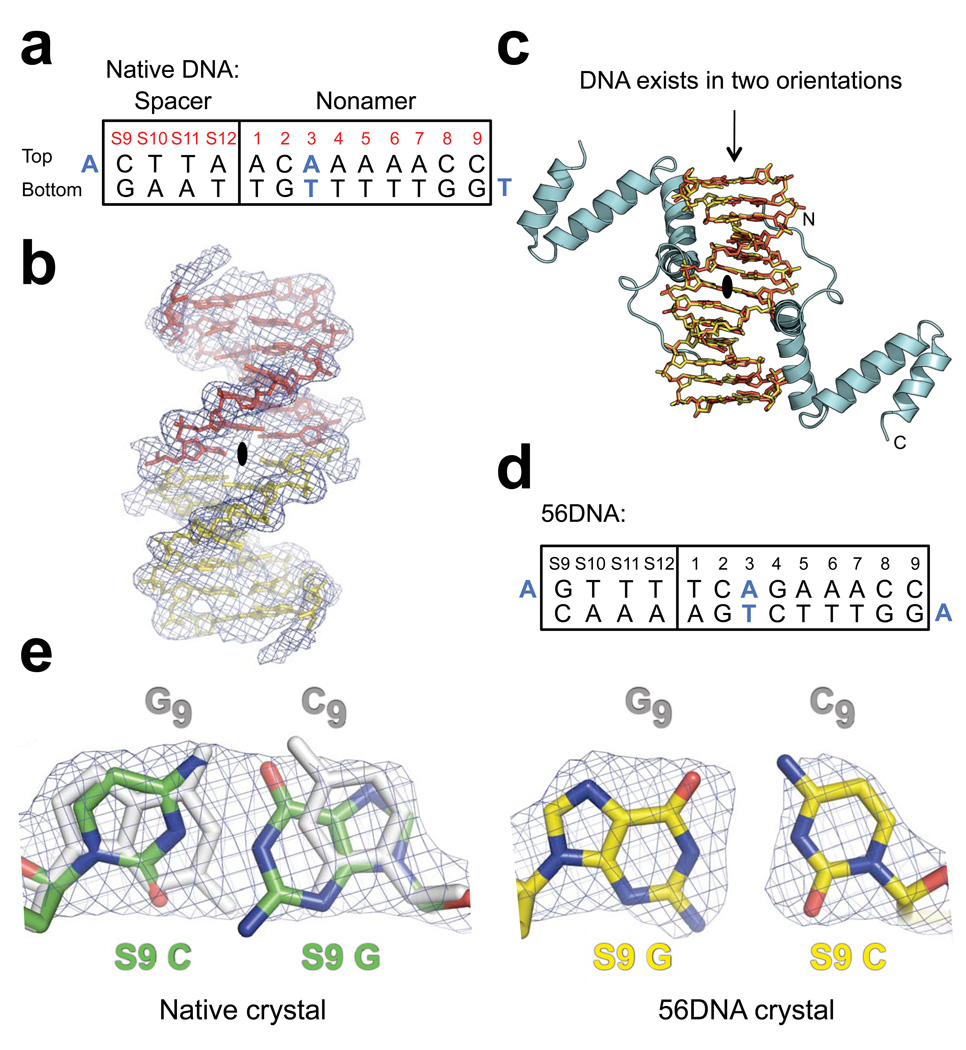
Figure 2.
Electron density maps of DNA in the native and 56DNA crystals. (a) Sequence of the DNA used in native crystals is shown with the consensus nonamer (numbered 1–9) and four base pairs from the consensus 12RSS spacer (numbered S9-S12). Bases coinciding with the crystallographic 2-fold symmetry axis are blue. (b) Solvent-flattened experimental electron density map (mesh) derived from heavy-atom phasing of the native crystal shows continuous electron density extending from the base pair at nonamer position 3 across the 2-fold symmetry axis (black oval; the axis is perpendicular to the page). Two halves of a single 14-mer DNA duplex minus the overhang bases are shown as yellow and red stick models. (c) Application of the crystallographic 2-fold symmetry operator generates a model consistent with the DNA existing in two possible orientations (overlap of yellow and red stick models), each at half occupancy, and the NBD (cyan) bound to the two A•T-rich sites in the DNA. The termini of one NBD are indicated with N and C. Crystals containing a DNA with additional symmetrized bases, 56DNA (d), in complex with the NBD were also obtained. (e) 2Fo-Fc omit electron density maps (mesh) of bases at S9, which were generated by performing simulated annealing refinement without the DNA coordinates, are shown for both the native (green) and 56DNA (yellow) crystals. The corresponding symmetry-mate bases at nonamer position 9 (subscript) are colored gray in both models, but are not visible in the case of 56DNA due to perfect overlap with the S9 base pair. Nitrogen and oxygen atoms are indicated with blue and red, respectively.
One molecule of the NBD could be unambiguously fit into the experimental electron density map in the asymmetric unit, while only half of the expected DNA could be fit. Inspection of the electron density map showed that continuous electron density extended from the base pair at nonamer position 3 across a crystallographic 2-fold symmetry axis (Fig. 2b). Applying this 2-fold symmetry operator to the partial model generated the entire DNA bound to two NBDs. This was possible because the spacer base pairs created a second A•T-rich region with which the NBD was able to interact in addition to the A•T tract in the nonamer. This suggested a model in which the 2-fold symmetry axis of the complex coincided with the crystallographic 2-fold axis, consistent with the DNA existing in two possible orientations in the lattice (Fig. 2c). Since the DNA sequence is not symmetric, the electron density map of the DNA represented the averaged density of the DNA in two orientations. When we accounted for the two DNA orientations in our model by setting each orientation at half occupancy, both Rwork and Rfree decreased substantially, suggesting that our modeling of the electron density was correct. The overhang bases at the ends of the DNA, which mediate DNA stacking in the lattice and do not contribute direct protein-DNA contacts, were excluded from the model due to poor electron density at these positions.
To further validate our model, we also grew crystals of the NBD in complex with a DNA containing additional symmetrized bases (referred to as 56DNA; Fig. 2d). As expected, when the model was refined against the 56DNA data, all of the symmetric base pairs could be fit unambiguously into the electron density map (Fig. 2e), confirming that our refinement of the model with the two DNA orientations is correct. Except for two solvent-exposed side chains, which show clearer electron densities in the 56DNA map, the electron density of the protein is identical in both the native and 56DNA crystals, indicating that the dual orientation of the DNA had minimal effect on the protein and its interactions with the DNA. Hereafter, we shall only refer to the DNA orientation that places the NBD in direct contact with the A•T tract in the nonamer, since this is consistent with existing biochemical data and the experiments described below. The final model, with the DNA at half occupancy, has an Rwork/Rfree of 23.6/27.1%.
Structure of the NBD dimer in complex with DNA
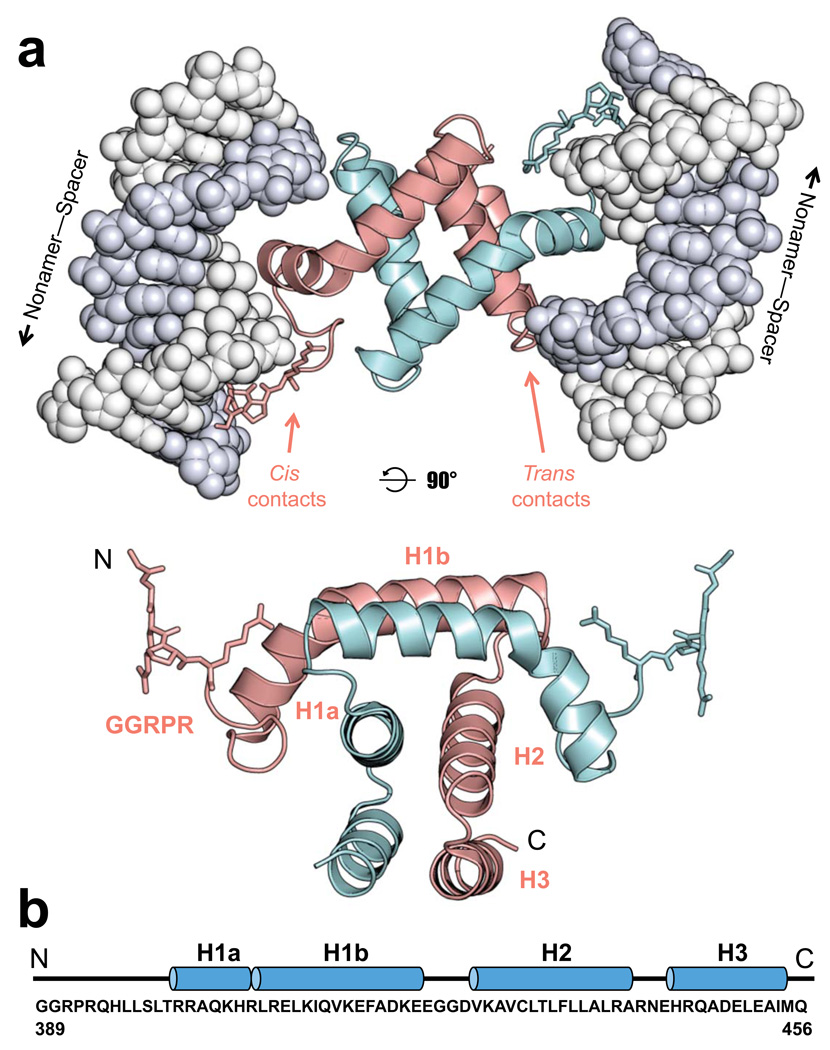
The NBD is a symmetrical homodimer and interacts with two molecules of DNA, which are bound to the outside of the protein core in a near anti-parallel configuration offset by 30 degrees (Fig. 3a). The DNA shows little deviation from the ideal B-form. Each NBD monomer is composed of three helices: H1 (residues 400–422), H2 (residues 426–441), and H3 (residues 444–454) (Fig. 3b). Helix 1 contains a kink that separates it into two smaller helices: H1a (residues 400–407) and H1b (residues 408–422). Helices H2 and H3 from each subunit form a 4-helix bundle through extensive hydrophobic interactions and constitute the bulk of the dimer interface, while the H1 helices from the two subunits wrap around one side of the 4-helix bundle with the N-terminal GGRPR motifs (residues 389–393) protruding from opposite sides of the dimer (Fig. 3a). The ability of the NBD dimer to simultaneously bind to and tether two DNA molecules is in agreement with its overall charge distribution, which shows two large positively charged surfaces on opposite sides of the dimer (Supplementary Fig. 1). Moreover, each NBD monomer interacts with both DNA molecules, with the resulting contacts being categorized as either cis or trans, depending on whether they involve the GGRPR motif and H1a or the loop between H2 and H3, respectively. A remarkable 1,900 Å2 surface area of each monomer becomes buried at the dimer interface. Given the extensive surface area of interaction and the interwrapped nature of the NBD structure, it is highly unlikely that the dimer interface is an artifact of crystal packing. Both gel filtration and multi-angle laser light scattering methods confirmed that the NBD exists as a dimer in solution in the absence of DNA (data not shown). Consistent with the structure, RAG1 core also forms a stable dimer that is resistant to dissociation20,23. Perturbations in the NBD dimer interface, such as alanine substitutions of residues 423–433, substantially reduced RAG1 core protein expression in vivo30, suggesting the importance of NBD dimerization in RAG1 protein folding and dimerization.
Figure 3.
Structure of the RAG1 NBD-DNA complex. (a) The NBD homodimer (subunits colored cyan and pink with the GGRPR motif shown as stick model) binds to two DNA molecules (gray space-filling model) and makes both cis and trans DNA contacts as indicated for the pink subunit. The lower panel shows the NBD dimer alone, rotated 90° from the view in the top panel. (b) The sequence of the NBD in the model (murine RAG1 residues 389–456) is shown below the corresponding secondary structures, with loops and helices represented as lines and blue cylinders, respectively.
GGRPR motif in the minor groove
The GGRPR motif is an example of an AT-hook, a structural element found in a variety of DNA-binding proteins31. This motif is the only region of strong sequence conservation between the RAG1 NBD and Hin DBD18 and adopts almost identical structures in the two proteins27. In the NBD, the GGRPR motif interacts predominantly with the bases at positions 5–7 of the nonamer (Fig. 4a), the most highly conserved nonamer positions in endogenous RSSs32.
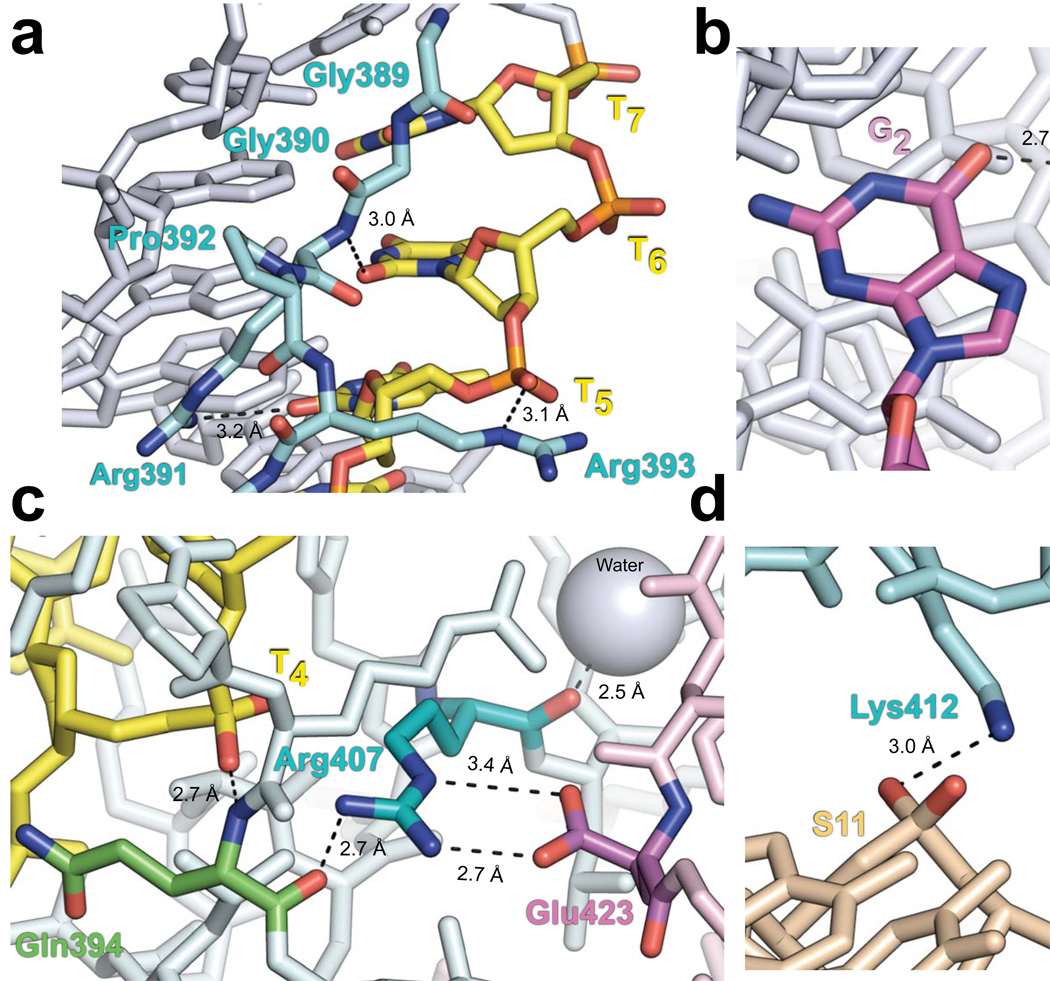
Figure 4.
Important protein-DNA and protein-protein contacts in the NBD-DNA crystal structure. Nitrogen, oxygen, and phosphate atoms are indicated with blue, red, and orange, respectively. Dashed lines represent bonds. Subscript denotes the nonamer position. (a) Structure of the GGRPR motif (cyan) in the minor groove of the nonamer is shown with thymines colored yellow, while all other DNA bases are gray. The Arg391 side chain amino group and main chain amide are hydrogen-bonded to the O2 atoms of T5 and T6, respectively. The Arg393 ε-imino group is hydrogen-bonded to the phosphate of T5. (b) Base-specific interaction between the Arg402 side chain (cyan) and the O6 atom of G2 (magenta). (c) The Arg407 guanidino group (cyan) is hydrogen-bonded to the carbonyl of Gln394 (green) in the same subunit (light cyan) and forms a salt bridge with Glu423 (magenta) of the other NBD subunit (light pink). The main chain amide of Gln394 is hydrogen-bonded to the phosphate of T4 (yellow). Additionally, the carbonyl of Arg407 is hydrogen-bonded to a nearby water molecule (gray sphere). (d) In the spacer, the side chains of Lys405 and Lys412 (cyan) form hydrogen bonds with the DNA backbone at positions S10 and S11 (beige), respectively. See also Supplementary Fig. 2.
Gly389 and Gly390 pack against the minor groove edges at nonamer positions 8 and 7, respectively. Arg391 makes two important contacts with the DNA; its main chain amide is hydrogen-bonded to the O2 atom of T6 (subscript denotes the position within the nonamer), while its side chain amino group forms another hydrogen bond with the O2 atom of T5. In addition, the Arg393 ε-imino group is hydrogen-bonded to the phosphate of T5 (Fig. 4a). The electrostatic interaction between the Arg393 guanidino group and the DNA backbone may be important in positioning the GGRPR motif in the minor groove. Although these interactions contribute to the binding of the NBD to the DNA, specificity is primarily achieved through conformational complementarity between the GGRPR motif and the minor groove of the nonamer (Supplementary Fig. 1c). Indeed, the structure of the motif is such that replacing the A•T tract with a stretch of G•C base pairs would cause the N2 atom of guanine at positions 5–7 to protrude into the minor groove, thereby resulting in steric clash with residues Gly390 and Arg391. This is consistent with the finding that G•C substitutions at nonamer positions 5–7 substantially reduced RSS-RAG binding in gel shift assays33. Interactions between the GGRPR residues and the thymines in the minor groove are also consistent with strong permanganate interference, hydroxy radical, and DNase I footprinting results in the nonamer of the RSS bound by RAG1 alone or by the entire RAG complex, suggesting that the NBD-DNA structure captured here is pertinent to complexes assembled with core RAG proteins17.
Major groove and spacer contacts
Although helix H1a in the major groove is functionally similar to the recognition helix in HTH motifs, the arrangement of the helices in the RAG1 NBD is unrelated to that of the HTH motif. The H1a helix axis is coplanar to the bases of the DNA, but only interacts marginally with base edges in the major groove. Of the five basic residues in H1a, only Arg402 makes base-specific contact with the DNA. The electron density of the Arg402 side chain in the native crystal appears to be an average of two possible rotamers. However, the most likely rotamer conformation places the arginine side chain amino group 2.7 Å away from the O6 atom of the guanine at nonamer position 2 (Fig. 4b), which is consistent with strong interference in RAG1-RSS binding upon methylation of G217 and 91% conservation of C•G at nonamer position 2 in endogenous RSSs34. Further validation of the position of this side chain was obtained from the 56DNA crystal in which the guanine is symmetric across the 2-fold symmetry axis. In this structure, we observed unambiguous electron density for the Arg402 side chain, confirming the indicated rotamer conformation and its base-specific contact with G2. Helix H1 also contacts the DNA through Arg407, whose guanidino group interacts electrostatically with the phosphate backbone (Fig. 4c). Arg407 also appears to play an important structural role, as evidenced by the multiple contacts it makes with surrounding protein residues. These include a hydrogen bond between the Arg407 side chain and the main chain carbonyl of Gln394, an interaction that helps define the conformation of the loop connecting the GGRPR motif to H1a and stabilizes the hydrogen bond between the Gln394 main chain amide and the phosphate of T4. The Arg407 guanidino group also forms a salt bridge with the carboxyl group of Glu423 from the neighboring NBD subunit. In addition, hydrogen bonding between the main chain carbonyl of Arg407 and a nearby water molecule stabilizes the kink in H1.
In addition to the nonamer contacts made by the GGRPR motif and H1a, which constitute the primary cis contacts, backbone contacts with the top strand in the spacer are contributed both in cis and in trans, by means of conformational and electrostatic complementarity between the NBD and DNA (Supplementary Fig. 2). Four residues of the cis-bound NBD subunit are observed to interact with the top strand in the spacer; Lys405 and Ly412 are hydrogen-bonded to the backbone phosphates at positions S10 and S11, respectively (Fig. 4d), while Arg401 and Arg409 are positioned near the backbone at positions S9 and S12, respectively, but are too far to form hydrogen bonds with the phosphate groups. Further shape complementarity is provided in trans by residues Asn443 and His445 of the other subunit, which are positioned near the backbone phosphates at S8 and S9 (Supplementary Fig. 2). Interactions with the backbone at positions S8-S12 in the spacer are consistent with published ethylation interference and DNase I footprinting results17.
Consistent with the relatively small number of sequence-specific interactions observed in the NBD-DNA structure, we were unable to detect sequence-specific interactions of the NBD with the RSS or isolated nonamer in gel shift, anisotropy, or FRET DNA binding assays (data not shown). Specific RSS recognition is therefore substantially facilitated by the heptamer, RAG2, HMGB1/2, and other domains of RAG1, consistent with many previous studies of DNA binding by the RAG proteins17,20.
Testing the NBD structure predictions in RAG1 core
We therefore investigated the functional significance of the DNA contacts observed in the crystal structure in the context of the RAG1 core protein. To accomplish this, we mutated basic residues within the NBD in RAG1 core and assayed for their effects on DNA binding and cleavage. Although some NBD mutants have been characterized previously18,35,36, our experiments here offer a more systematic comparison among individual mutants (Fig. 5 and Supplementary Fig. 3). To ensure that the mutations did not result in the misfolding of RAG1, we tested all of the mutants for their ability to interact with RAG2 using a GST-RAG2 pull-down assay and found that all of the RAG1 core mutants exhibited wild-type ability to interact with RAG2 (Supplementary Fig. 4).
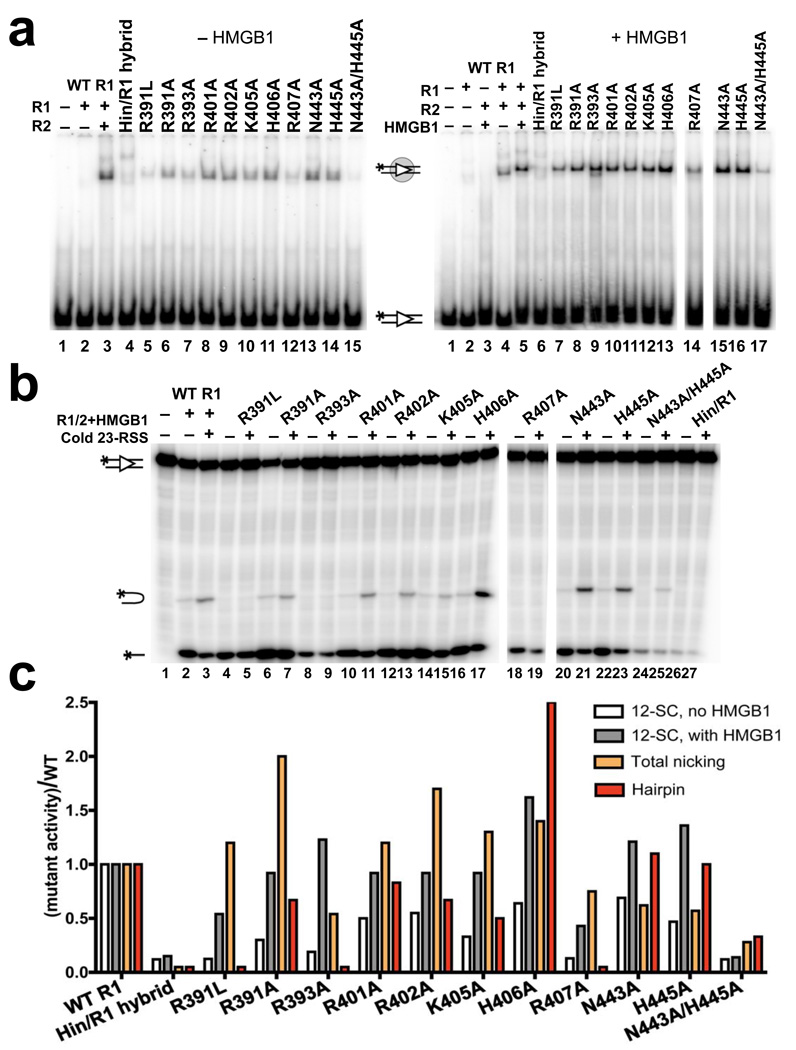
Figure 5.
NBD-DNA contacts are important for DNA binding and hairpin formation. (a) Gel shift analysis of RAG1/2 binding to a 32P-labeled consensus 12RSS substrate in the presence (right gel) or absence (left gel) of HMGB1 in a binding buffer containing 5 mM CaCl2. Proteins were added as indicated above the lanes. All reactions with mutant RAG1 proteins contained RAG2, and the positions of the free and bound substrates are indicated with diagrams. (b) DNA cleavage assay was performed with 32P-labeled consensus 12RSS and a 5-fold molar excess of unlabeled consensus 23RSS in the presence of 1.5 mM MgCl2. Proteins and 23RSS substrate were added as indicated above the lanes. All reactions with mutant RAG1 proteins contained RAG2 and HMGB1. The positions of the nicked product, hairpin product, and input substrate are indicated with diagrams. (c) Quantitation of binding and cleavage results from panels (a) and (b), with the activity of the mutants normalized to that of wild-type RAG1, whose activity was arbitrarily set to one. Similar results were obtained in repeat experiments (data not shown). See also Supplementary Fig. 3.
As expected, disruption of the NBD structure, as in the Hin/RAG1 core hybrid protein (see Methods), substantially reduced 12RSS binding by RAG1/RAG2 as assayed by gel shift (Fig. 5a,c). Consistent with previous mutagenesis studies and the structure, mutations in the GGRPR motif also resulted in appreciable defects in DNA binding18,35. Mutating the critical residue Arg407 also substantially reduced DNA binding. Alanine mutations of other residues in H1a caused only moderate defects in DNA binding, indicating that these basic residues individually make only small contributions to the binding interaction. In contrast, the clear binding defect of the N443A/H445A double mutant is consistent with the idea that these residues mediate important trans DNA contacts. Addition of HMGB1 to binding reactions stimulated DNA binding and partially suppressed the phenotype of several mutants, especially those in the GGRPR motif (compare Fig. 5a and 5b).
Next, we examined the effects of the RAG1 mutations on nicking and hairpin formation (Fig. 5b,c). Most mutants showed near wild-type levels and in some cases enhanced nicking activity. Even mutants that were severely impaired in DNA binding, such as the Hin/RAG1 hybrid and R407A mutants, exhibited reduced but detectable levels of nicking. In contrast, mutating residues that make specific contacts with the DNA, especially those in the minor groove, showed much stronger defects in hairpin formation (Fig. 5b,c). The R391L and R393A mutants exhibited dramatically reduced hairpin formation despite retaining substantial nicking activity. Although the R391A mutant supported detectable hairpin formation, it exhibited a substantially lower hairpin:nick ratio than wild-type RAG1 core, indicating a deficiency in the hairpin formation step.
Given the overall electropositive charge of the NBD and the lack of extensive base-specific contacts, it is not surprising that most single-residue substitutions in H1a had only minor effects on DNA cleavage. Mutation of Arg401 and His406, which do not make direct DNA contacts, allowed for robust hairpin activity (Fig. 5b,c). As with the R391A mutant, mutation of Arg402, whose side chain forms a hydrogen bond with G2 in the nonamer, resulted in a defect specifically in converting nicked products to hairpins. Mutating this guanine to adenine substantially reduced both nicking and hairpin formation by the wild-type RAG proteins (Supplementary Fig. 5). Mutation of Arg407 in H1a reduced hairpin formation to background levels despite a near wild-type level of nicking. The reduced hairpin activity of K405A and the severe binding and DNA cleavage defects of the N443/H445 double mutant highlight the importance of spacer contacts in RAG-mediated activities (Fig. 5b,c). This is consistent with previous studies that found that variations in the spacer sequence affect recombination frequency37. Since hairpin formation is largely restricted to the PC, we also performed a biotin pull-down assay to determine the effect of these NBD mutations on PC formation. As expected, mutants that were impaired in hairpin formation exhibited defective pull down of radiolabeled 12RSS with biotinylated 23RSS (data not shown).
Probing nonamer-mediated DNA synapsis in solution
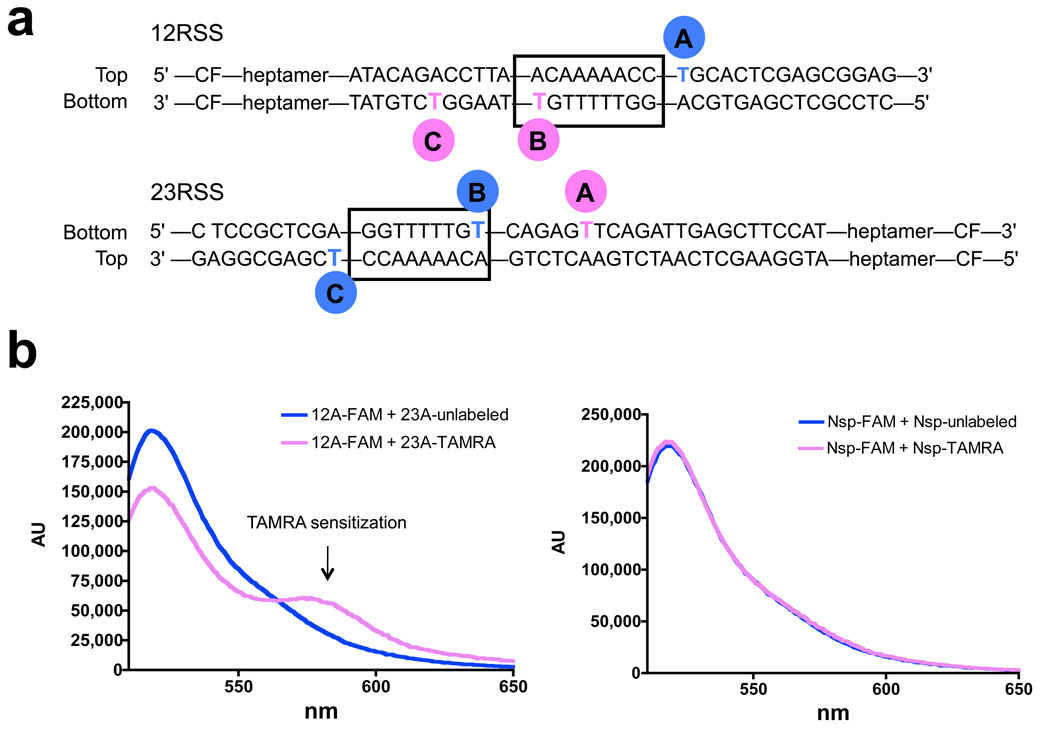
An important and surprising finding to emerge from the crystal structure is that a dimer of the NBD mediates a synaptic configuration in which the two nonamers are held in relatively close proximity (≈ 50 Å between the helix axes). The NBD has not been implicated in DNA synapsis, and previous models of RAG-RSS binding have invariably depicted a monomer of the NBD bound to a single nonamer. It was therefore important to test whether close approximation of the two nonamers occurs in the PC formed by RAG1/RAG2/HMGB1. To do so, we utilized fluorescence resonance energy transfer (FRET) to assess synapsis of fluorescently labeled RSSs. This method provides a sensitive assay for the distance between two fluorophores and has been used by us previously to study the structure of the PC38. Using the NBD structure as a model, we designed three 12/23 pairs of RSSs (A, B, and C; Fig. 6a) in which the donor (6-carboxyfluorescein, FAM) and acceptor (carboxytetramethylrhodamine, TAMRA) fluorophores were attached to bases predicted to be on the side of the helix facing the protein core and within the Förster radius of 55 Å (the distance at which energy transfer efficiency is 50%)38.
Figure 6.
Detection of nonamer synapsis using FRET. (a) Diagram showing the positions of fluorophore labeling in the A, B, and C pairs of 12/23 RSS FRET substrates. TAMRA (pink circle); FAM (blue circle); CF, coding flank. (b) Representative corrected emission spectra obtained with the A substrate pair (left panel) or nonspecific (nsp) substrates, which are identical to the A substrates except that the nonamer and heptamer elements have been mutated in both the 12 and 23 RSSs (right panel). In each case, the FAM-labeled substrate was incubated with a 4-fold molar excess of either a TAMRA-labeled (pink spectrum) or unlabeled (blue, control spectrum) partner substrate together with RAG1/RAG2/HMGB1 in a buffer containing 5 mM MgCl2. Background intrinsic TAMRA fluorescence has been subtracted from both pink spectra. Donor quenching and TAMRA sensitization are observed only with the consensus A substrates.
Fluorophores were attached to thymines with a 6-carbon linker of about 10 Å in length. Control experiments demonstrated that binding of the proteins caused substantial quenching of the fluorescence of FAM in all three donor substrates (Supplementary Fig. 6), indicative of protein-fluorophore interactions that could influence the trajectories of the 6-carbon linkers and constrain fluorophore rotation. The distances between the fluorophores were therefore difficult to predict accurately, ranging from ≈ 30–70 Å depending on how the linkers are oriented relative to one another in the PC (calculated inter-base distance is ≈ 48 Å for the B pair).
Coupled 12/23 RSS cleavage experiments comparing fluorophore-labeled and control substrates demonstrated that the fluorophores do not alter DNA cleavage efficiency (data not shown), indicating that they do not interfere with critical protein-DNA interactions. FRET experiments were then performed with labeled substrates containing intact or mutant heptamer and/or nonamer elements (representative corrected emission spectra shown in Fig. 6b). Percent energy transfer efficiency was calculated from acceptor sensitization as measured by TAMRA emission. When the entire ensemble of proteins was added to the RSSs to allow PC formation, all three RSS pairs supported substantial energy transfer (Fig. 7a). From the measured energy transfer efficiencies, distances between fluorophores in the A, B, and C pairs were calculated to be 59 Å, 66 Å, and 66 Å, respectively, which are within the predicted range. As noted above, the flexibility of the 6-carbon linker and protein-fluorophore interactions make it difficult to accurately determine the distances between the fluorophores in solution. Furthermore, the distances derived from FRET will be overestimates if not all FAM-labeled substrates synapse with a TAMRA-labeled substrate. However, qualitatively, these data support the notion that the two nonamers are brought together in the PC in a manner consistent with the NBD structure.
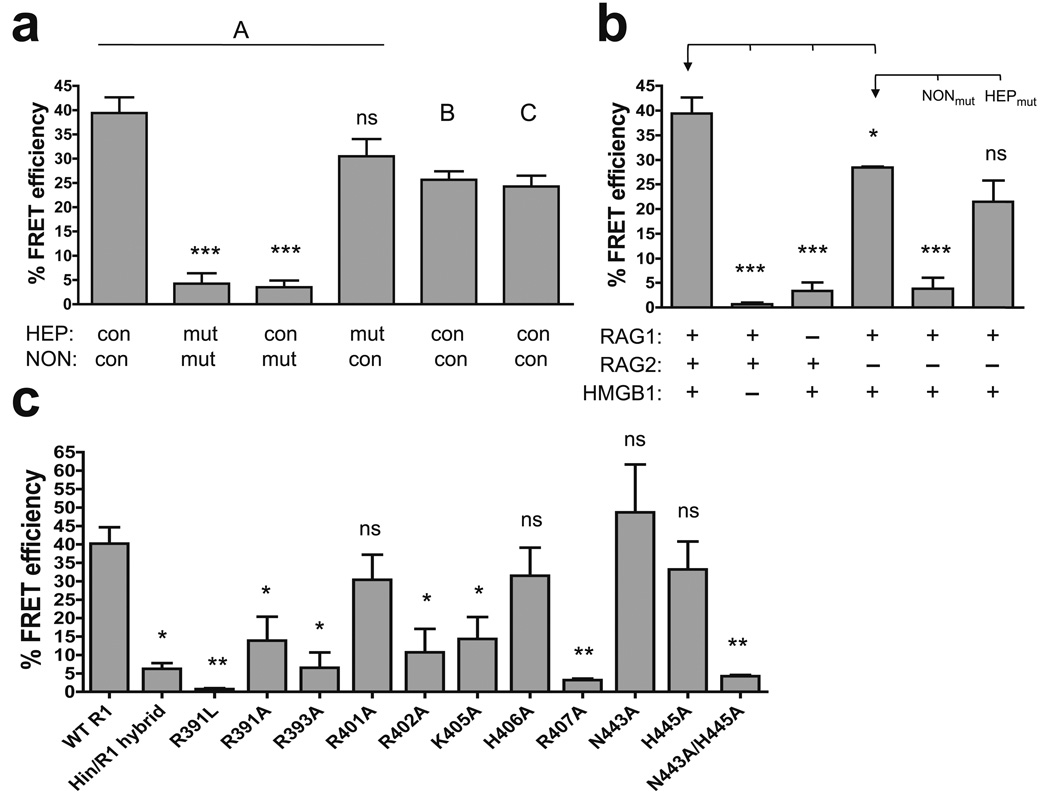
Figure 7.
FRET analysis of the DNA and protein requirements for nonamer synapsis. Average FRET efficiencies of at least two experiments are plotted with error bars indicating the s.e.m. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant (unpaired two-tailed t-test). (a) Energy transfer was assessed with the A, B, and C substrate pairs (as indicated above the bars), and with A substrates in which the heptamer (HEP) and/or nonamer (NON) were mutated in both the 12 and 23 RSSs, as indicated below the bars. mut, mutant; con, consensus. Statistical comparisons were made with the data obtained with the consensus A substrate pair. (b) Energy transfer was assessed using various combinations of RAG1, RAG2, and HMGB1, as indicated below the bars, with the A substrate pair, or the A substrate pair with mutant nonamer (NONmut), or mutant heptamer (HEPmut) sequences. Statistical comparisons (brackets) were made with the data indicated with an arrow. (c) Energy transfer was assessed with the A substrate pair using WT and mutant RAG1 core proteins, as indicated below the bars. Statistical comparisons were made with the data obtained with WT RAG1 core.
Since the A pair yielded the strongest energy transfer, we chose it for subsequent analyses. Experiments performed with substrates containing mutant heptamer and/or nonamer elements demonstrated that energy transfer is strongly nonamer-dependent and only weakly dependent on the heptamer (Fig. 7a). As expected, omitting RAG1 resulted in a dramatic reduction in energy transfer, and a similar effect was observed when HMGB1 was omitted from the reaction (Fig. 7b). Interestingly, RAG1 and HMGB1 in the absence of RAG2 supported substantial energy transfer, which again was strongly dependent on the nonamer but not the heptamer. Together, these data indicate that the full complement of proteins, or RAG1 and HMGB1 alone, are capable of mediating close juxtaposition of two nonamers without requiring heptamer interactions, a conclusion consistent with a number of previous findings17. Using the FRET assay, we also analyzed the RAG1 mutants for their ability to support nonamer-mediated DNA synapsis (Fig. 7c). The efficiencies with which these mutants supported energy transfer were strikingly similar to their phenotypes in hairpin formation (compare with Fig. 5c), emphasizing the importance of these nonamer contacts in coupling PC formation and DNA cleavage.
DISCUSSION
The crystal structure of the RAG1 NBD in complex with the nonamer and proximal spacer sequences reveals a unique intertwined homodimer bound to two DNA molecules. Dimerization is accomplished by three helices that form an extensive hydrophobic interface, while DNA binding is mediated by three distinct elements: a GGRPR AT-hook that is inserted in the minor groove, a short helix that interacts with the major groove, and spacer contacts that include two basic residues contributed in trans by the other subunit. Unlike most site-specific recombinases, the RAG recombinase must tolerate substantial sequence variation in its DNA substrates. To this end, instead of forming extensive base-specific contacts, nonamer recognition relies more on structural and electrostatic complementarity between the NBD and DNA and cooperative binding with HMGB1.
Biochemical and FRET experiments demonstrate that NBD residues predicted to engage in direct DNA contacts are important for DNA binding, synapsis, and hairpin formation by RAG1/RAG2/HMGB1. This suggests that the NBD-DNA structure captured here represents an important piece of the physiological synaptic complex, although it is clear that the structure reflects only a small portion of the protein-DNA interactions responsible for RSS recognition by the RAG proteins. RAG1 exists as a tight dimer in solution20,22,24, and our data indicate that this dimer, together with HMGB1, can support the synapsis of two nonamers. The simplest interpretation of this is that the PC contains a single RAG1 dimer22,24, although this point remains controversial10,21. The interwrapped nature of the NBD dimer provides an appealing structural basis for the finding that the RAG1 active site and nonamer-binding domain GGRPR/H1a regions that engage a particular RSS are donated in trans by different RAG1 protomers (see model by Swanson)39.
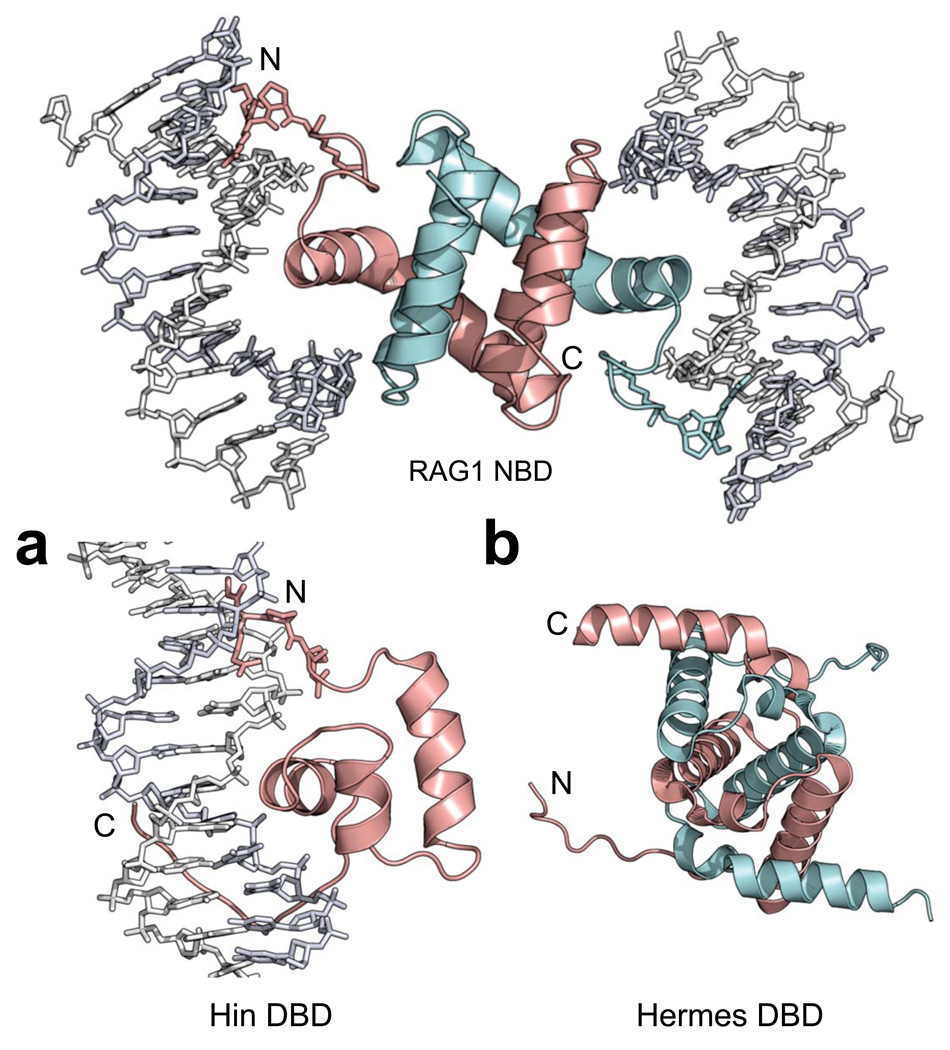
Despite some similarities in their modes of DNA recognition, the RAG1 NBD and the Hin DBD27 have very different arrangements of their structural elements (Fig. 8a). The strongest functional parallels to RAG1/2 are provided by the hAT family transposase Hermes, which like RAG1/2 contains a DDE active site motif and generates hairpins on the flanking DNA during DNA cleavage40. The structure of the Hermes transposase has been helpful in elucidating the function of residues in RAG1, suggesting that there are structural parallels between the two proteins41–43. Interestingly, the RAG1 NBD and the Hermes DBD are both intertwined dimers formed by domain-swapped helices held together by extensive hydrophobic interactions (Fig. 8b). However, the absence of DNA or any obvious DNA-binding motifs in the Hermes structure41 makes it difficult to align the structures for a more detailed analysis.
Figure 8.
Structural comparisons of the RAG1 NBD with the DNA-binding domains (DBDs) of Hin and Hermes. The NBD dimer (subunits colored cyan and pink) with the GGRPR motif (stick model) is shown. (a) The Hin DBD (pink) is a monomer that uses a HTH motif to interact with DNA in the major groove, while a GGRPR motif (stick model) and a C-terminal tail interact with the minor grooves. Hin PDB: 1HCR (b) The Hermes DBD (residues 76–159) also has an intertwined dimer structure (subunits colored cyan and pink). Hermes PDB: 2BW3.
The structure of the NBD provides insight into the mechanism by which mutations in the NBD cause Omenn syndrome (OS) (Supplementary Fig. 7), an immunodeficiency disease characterized by severe defects in B and T lymphocyte development due to inefficient V(D)J recombination44. Mutation of Arg393 (residue numbering as in murine RAG1) to Cys or His, as observed in OS, is predicted to perturb DNA contacts made by the GGRPR motif (Fig. 4a) and to severely reduce hairpin formation (Fig. 5b,c). The S398P mutation in OS is predicted to alter the structure of the helical turn in the loop connecting the GGRPR motif and H1a, thereby perturbing DNA interactions in both the minor and major grooves. Mutation of Asp426 to Gly in OS could destabilize the dimer core by perturbing the positioning of H2 within the 4-helix bundle. Also, mutation of Asp426 abolishes a salt bridge with His395 from the other subunit, leading to destabilized DNA interaction by the neighboring GGRPR motif. Finally, substitution of the bulkier Val for Ala441 in the turn connecting H2 and H3, as seen in OS, could interfere with the trans DNA contacts made by nearby residues Asn443 and His445.
Our findings provide insight into the structural basis for differential regulation of nicking and hairpin formation. Most NBD mutations did not substantially compromise nicking, consistent with previous results showing that nicking can occur independent of the nonamer and a partner RSS13,45–47. The deleterious effects of NBD mutations on stable 12RSS binding could be partially overcome by HMGB1, consistent with the ability of HMGB1 to interact directly with the NBD and enhance RAG-RSS binding, in part by inducing additional DNA contacts near the nonamer17,20,29,48. In contrast, NBD interactions with DNA are critical for efficient PC formation and hairpin activity. Our data indicate that RAG1 and HMGB1 are sufficient for nonamer-dependent RSS synapsis, while full engagement of the heptamer has been shown to be RAG2-dependent49. These findings suggest a model in which the dimeric NBD-nonamer module provides an anchor point for HMGB1-, RAG2-, and heptamer-dependent conformational changes, such as DNA bending29,38,50,51, that underlie assembly of the PC. Such structural changes are not captured in the NBD-nonamer crystal and could cause the structure of the NBD and nonamer in the PC to deviate from our model. The intertwined NBD-nonamer module also provides a potential means of communication between two RAG1 protomers, allowing the binding of an appropriate 12/23 RSS pair to be sensed and to activate the RAG complex for hairpin formation.
DNA-binding domains that form obligate homodimers, as in RAG1 and Hermes, provide an interesting contrast with the Sin serine recombinase, in which pairs of HTH DNA-binding domains from different dimers mediate the formation of a synaptic complex52, as well as recombinases like Hin, whose DBD does not contribute to synapsis27. In the case of RAG1, the relatively rigid NBD dimer imposes its geometry on the two nonamers and directly couples DNA binding with synapsis. In the case of Sin, the disposition of binding sites on each target DNA duplex determines the geometry of the DBDs and hence their ability to synapse, leaving DNA binding and synapsis uncoupled and allowing Sin the flexibility to interact with different configurations of binding sites52.
In summary, our findings indicate that the RAG1 NBD plays an important role in the synapsis of two DNA molecules. It will now be a priority to determine how the NBD-nonamer module is used in building the 12/23 RSS synaptic complex.
METHODS
Proteins
We cloned DNA encoding His6-tagged NBD (murine RAG1 residues 389–464) into the pET28A vector, which added 21 amino acids at the N-terminus, and expressed the recombinant protein in E. coli BL21(DE3). The soluble fraction of the cell lysate was then purified using nickel-chelating resin followed by heparin and size-exclusion columns in buffers similar to the final storage buffer (1 M NaCl, 30 mM Tris-HCl pH 7.5, and 5% (v/v) glycerol). The NaCl concentration was reduced to about 100 mM prior to the heparin column. The NBD protein was soluble through all steps of the purification. Except for the Hin/RAG1 hybrid mutant, all other mutations were introduced into the wild-type RAG1 construct, which encodes MBP-RAG1 core-(residues 384–1008)-His6, using the QuikChange XL site-directed mutagenesis kit from Stratagene. The Hin/RAG1 hybrid mutant is similar to the previously published mutant in which RAG1 residues 389–444 have been replaced with Hin DBD residues 138–19018, except the substitution was done in the context of the above mentioned MBP-RAG1 core construct. Wild-type and mutant MBP-RAG1 core proteins were purified from bacteria using nickel-chelating resin, amylose beads, and gel filtration as described previously23, with the dimer peak fractions isolated and used in all experiments. GST-tagged murine RAG2 core (residues 1–383) expressed in 293T cells and His-tagged full-length human HMGB1 expressed in bacteria were purified as described previously18,53.
DNA oligonucleotides
Oligonucleotides used for crystallography, FRET, and biochemical assays were synthesized by Integrated DNA Technologies (IDT). Primers used for cloning were synthesized by Invitrogen. The sequences of the 14-mer oligonucleotides used in crystallography can be found in Fig. 2a and 2d. The sequences of the top strands of the consensus 12RSS and 23RSS are: 5'-GATCTGGCCTGTCTTACACAGTGATACAGACCTTAACAAAAACCTGCACTCGAGCGG AG and 5'-GATCTGGCCTGTCTTACACAGTGATGGAAGCTCAATCTGAACTCTG ACAAAAACCTCGAGCGGAG, respectively. In nonamer-mutant RSSs, the consensus nonamer was replaced with 5'-CTCTGGCTG. Heptamer-mutant RSSs contained 5'-GCTCAGT in place of the consensus heptamer. Radiolabeled substrates were 5' end labeled on the top strand using γ-32 P-ATP and T4 polynucleotide kinase, followed by annealing with the complementary cold strand and gel purification. For FRET substrates, fluorophores were attached to position 5 of the thymine ring via a 6-carbon spacer arm during oligonucleotide synthesis by IDT. A complete list of all the oligonucleotides can be found in Supplementary Table 1 online.
Crystallization and data collection
At least a 3-fold molar excess of the 14-mer DNA duplex was added to purified NBD (final protein concentration ≥ 5 mg ml−1 ). The complex was then dialyzed against a buffer containing 30 mM HEPES pH 7.5 and 200 mM NaCl for at least 4 hours prior to setting up crystal trays. Crystals were grown at 4 °C using hanging-drop method and precipitant solution containing 0.1 M HEPES pH 7.5, 0.2 M NaCl, and 30% (v/v) PEG 400. Crystals grew to 0.1–0.2 mm in about one week and were soaked in a cryo-protectant solution (mother liquor supplemented with 35% (v/v) PEG 400) for at least 30 minutes before flash-freezing in liquid propane. Lead, mercury, and tungsten derivatives were obtained by soaking native crystals in the cryo-protectant solution containing 50 mM trimethyl lead acetate (TMLA), 100 mM lead (II) acetate, 50 mM mercury (II) acetate, or 1 mM sodium tungstate for at least 1 hour before freezing. Diffraction data were collected using the 24ID beamline at Argonne Photon Source (Argonne, IL) and the X25 beamline at Brookhaven National Laboratory (Upton, NY).
Structure determination and refinement
Data were indexed and scaled using the HKL2000 program suite54. General handling of the scaled data was done in CCP455. SHELX was used to find lead sites in the TMLA derivative crystal first56. The phases derived from these lead sites were then used to calculate the difference Fourier map and to find the heavy-atom sites in the other derivatives (Table 1). All phases were calculated using SOLVE and improved by solvent-flattening in RESOLVE57,58. Model building was performed in COOT59. The 7-mer DNA and one NBD monomer, which constitute the asymmetric unit, were built first. Then, the crystallographic 2-fold symmetry operator was applied to generate the remaining half of the 14-mer DNA, which was used to model in the correct DNA sequence with each base set at half occupancy. The overhang nitrogenous bases were excluded because they showed poor electron densities. This structure of the DNA with the NBD monomer was then refined through iterative cycles of model building in COOT and refinement in REFMAC with NCS restraints and DNA backbone (phosphate and d-ribose atoms) constraints about the 2-fold symmetry axis60. The final model contains RAG1 residues 389–456 and the 14-mer DNA minus the overhang bases. The N-terminal 21 residues added by the tag and the C-terminal 8 residues in the NBD construct were disordered. The Ramachandran plot of the final model showed 96.9% of all residues in the favored regions and none in the disallowed region. All figures were generated using PyMOL (http://pymol.sourceforge.net).
Table 1.
Data collection, phasing, and refinement statistics (MIRAS)
| Native | 56DNA | Heavy-atom derivatives | |||||||
|---|---|---|---|---|---|---|---|---|---|
| TMLA | Pb(CH3COO)2 | Hg(CH3COO)2 | Na2WO4 | ||||||
| Data collection | |||||||||
| Space group | I422 | I422 | I422 | ||||||
| Cell dimensions | |||||||||
| a, b, c (Å) | 96.7, 96.7, 71.5 | 96.6, 96.6, 71.5 | 98.2, 98.2, 70.4 | 98.1, 98.1, 70.6 | 98.3, 98.3, 70.6 | 98.0, 98.0, 69.9 | |||
| abg (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | ||||
| Wavelength (nm) | 1.255 | 0.979 | 0.949 | 0.944 | 1.004 | 1.211 | |||
| Resolution (Å)* | 50–2.4 | 50–3.0 | 50–3.8 | 50–3.2 | 50–3.8 | 50–3.2 | |||
| (2.5–2.4) | (3.1–3.0) | (3.3–3.2) | (3.2–3.1) | (3.9–3.8) | (3.3–3.2) | ||||
| Rmerge | 0.048 (0.717) | 0.097 (0.209) | 0.084 (0.458) | 0.091 (0.767) | 0.064 (0.642) | 0.066 (0.614) | |||
| I / σI | 30.1 (2) | 27.8 (9) | 23.2 (3.3) | 18.1 (2.2) | 23.6 (2.6) | 25.1 (2.7) | |||
| Completeness (%) | 99.9 (100) | 99.3 (100) | 99.8 (100) | 99.9 (99.3) | 99.8 (99.4) | 99.7 (100) | |||
| Redundancy | 23.0 (17.8) | 8.7 (9.0) | 7.5 (6.4) | 7.3 (6.3) | 7.4 (6.5) | 7.7 (7.7) | |||
| Refinement | Number of heavy-atom sites | ||||||||
| Resolution (Å) | 20.0–2.4 | 20.0–3.0 | 5 | 1 | 1 | 2 | |||
| No. reflections | 6,475 | 3,382 | Phasing powera (centric/acentric) | ||||||
| Rwork / Rfree | 23.6/27.1% | 24.2/26.7% | 1.17/1.58 | 1.05/1.62 | 1.27/1.34 | 0.72/1.30 | |||
| No. atoms | 1,113 | 1,112 | FOM | ||||||
| Protein | 556 | 556 | 0.44 | 0.55 | 0.52 | 0.31 | |||
| Ligand/ion | 549 | 548 | |||||||
| Water | 8 | 8 | |||||||
| B-factors | |||||||||
| Protein | 92.94 | 53.69 | |||||||
| Ligand/ion | 93.12 | 53.60 | |||||||
| Water | 82.33 | 57.40 | |||||||
| R.m.s. deviations | |||||||||
| Bond lengths (Å) | 0.006 | 0.005 | |||||||
| Bond angles (°) | 1.204 | 1.071 | |||||||
Values in parentheses refer to the highest-resolution shell.
R.m.s.(FH)/r.m.s.(E).
12-SC binding and coupled cleavage assays
For the 12-SC gel shift assay, 68 nM MBP-RAG1 core, 107 nM GST-RAG2 core, and 150 nM His-HMGB1 were incubated with 120 fmol of radiolabeled consensus 12RSS in 20 µL reactions containing 10 mM Tris-HCl pH 7.5, 0.1 mg ml−1 BSA, 33 mM KCl, 5 mM CaCl2, 2 mM DTT, 500 nM nonspecific single-strand oligonucleotide (5'-TTTGGTCGATATCCATATGGGGGGAC), and 6% (v/v) glycerol. Samples were incubated at 30 °C for 15 minutes before electrophoresis on 6% (w/v) 80:1 native polyacrylamide gels in 0.5x TBE buffer at room temperature. For coupled cleavage experiments, 91 nM of MBP-RAG1 core, 143 nM GST-RAG2 core, and 200 nM His-HMGB1 were incubated with 120 fmol of radiolabeled 12RSS and a 5-fold molar excess of unlabeled 23RSS in 20 µL reactions containing 10 mM Tris-HCl pH 7.5, 0.1 mg ml−1 BSA, 50 mM KCl, 1.5 mM MgCl2, 2 mM DTT, and 6% (v/v) glycerol at 37 °C for 1 hour unless noted otherwise. Each cleavage reaction was stopped by adding 65 µL of a buffer containing 0.2% (w/v) SDS, 100 mM NaCl, 20 mM EDTA, 20 mM HEPES pH 7.5, and 60 µg ml−1 proteinase K and incubation at 55 ° C for at least 2 hours before ethanol precipitation of the DNA. DNA samples were then electrophoresed in 1x TBE buffer on 10% (w/v) denaturing polyacrylamide gels. For both assays, gels were dried before imaging with a PhosphorImager (Molecular Dynamics) and quantitation using ImageQuant 5.2 software.
Fluorescence resonance energy transfer assay
FRET experiments and data analysis were performed as previously described with a few modifications38. Final concentrations of DNA substrates were 20 nM TAMRA-labeled RSS and 5 nM FAM-labeled RSS, and final protein concentrations were 76 nM MBP-RAG1 core, 120 nM GST-RAG2 core, and 160 nM His-HMGB1. Samples were incubated at 30 °C for 15 minutes before excitation at 492 nm followed by emission scan from 510–650 nm at room temperature using a Photon Technology International scanning spectrofluorometer and FeliX32 software. Emission spectra were corrected as described previously, including correction for intrinsic TAMRA fluorescence in the absence of donor fluorophore38. Acceptor sensitization (emission from 567–620 nm) was used to calculate the FRET efficiency as described previously38. GraphPad Prism 4 was used to calculate the peak areas in all spectra, and Maple 7.0 software was used to calculate the fluorophore separation distance as described previously38.
Supplementary Material
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
ACKNOWLEDGMENTS
We thank the staff at APS beamline 24ID and NSLS beamline X25. We are grateful to N. Grindley, L. Regan, and A. Miranker for the generous use of their spectrofluorometers. We also thank W. Eliason and J. Kavran of the Steitz lab, S. Unniraman of the Schatz lab, and the staff of the Center for Structural Biology Core Facility at Yale for their help. This work was supported by US National Institutes of Health grant AI32524 to D.G.S., training grant T32 GM08283, and a Gershon Fellowship to F.F.Y. D.G.S. and T.A.S. are investigators and C.A.I. and S.B. are research associates of the Howard Hughes Medical Institute.
Footnotes
Accession codes. Protein Data Bank: Coordinates for the NBD native and 56DNA structures have been deposited under accession codes 3GNA and 3GNB, respectively.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
REFERENCES
- 1.Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 2.Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 3.Sawchuk DJ, et al. V(D)J recombination: modulation of RAG1 and RAG2 cleavage activity on 12/23 substrates by whole cell extract and DNA-bending proteins. J. Exp. Med. 1997;185:2025–2032. doi: 10.1084/jem.185.11.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Gent DC, Hiom K, Paull TT, Gellert M. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 1997;16:2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 6.McBlane JF, et al. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 7.van Gent DC, Mizuuchi K, Gellert M. Similarities between initiation of V(D)J recombination and retroviral integration. Science. 1996;271:1592–1594. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- 8.Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol. Cell. 1998;1:1011–1019. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- 9.Jones JM, Gellert M. Ordered assembly of the V(D)J synaptic complex ensures accurate recombination. EMBO J. 2002;21:4162–4171. doi: 10.1093/emboj/cdf394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mundy CL, Patenge N, Matthews AG, Oettinger MA. Assembly of the RAG1/RAG2 synaptic complex. Mol. Cell. Biol. 2002;22:69–77. doi: 10.1128/MCB.22.1.69-77.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Gent DC, Ramsden DA, Gellert M. The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell. 1996;85:107–113. doi: 10.1016/s0092-8674(00)81086-7. [DOI] [PubMed] [Google Scholar]
- 12.Eastman QM, Leu TM, Schatz DG. Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature. 1996;380:85–88. doi: 10.1038/380085a0. [DOI] [PubMed] [Google Scholar]
- 13.West RB, Lieber MR. The RAG-HMG1 complex enforces the 12/23 rule of V(D)J recombination specifically at the double-hairpin formation step. Mol. Cell. Biol. 1998;18:6408–6415. doi: 10.1128/mcb.18.11.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steen SB, Gomelsky L, Roth DB. The 12/23 rule is enforced at the cleavage step of V(D)J recombination in vivo. Genes Cells. 1996;1:543–553. doi: 10.1046/j.1365-2443.1996.d01-259.x. [DOI] [PubMed] [Google Scholar]
- 15.Critchlow SE, Jackson SP. DNA end-joining: from yeast to man. Trends Biochem. Sci. 1998;23:394–398. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- 16.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 17.Swanson PC. The bounty of RAGs: recombination signal complexes and reaction outcomes. Immunol. Rev. 2004;200:90–114. doi: 10.1111/j.0105-2896.2004.00159.x. [DOI] [PubMed] [Google Scholar]
- 18.Spanopoulou E, et al. The homeodomain region of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell. 1996;87:263–276. doi: 10.1016/s0092-8674(00)81344-6. [DOI] [PubMed] [Google Scholar]
- 19.Difilippantonio MJ, McMahan CJ, Eastman QM, Spanopoulou E, Schatz DG. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell. 1996;87:253–262. doi: 10.1016/s0092-8674(00)81343-4. [DOI] [PubMed] [Google Scholar]
- 20.De P, Rodgers KK. Putting the pieces together: identification and characterization of structural domains in the V(D)J recombination protein RAG1. Immunol. Rev. 2004;200:70–82. doi: 10.1111/j.0105-2896.2004.00154.x. [DOI] [PubMed] [Google Scholar]
- 21.Landree MA, Kale SB, Roth DB. Functional organization of single and paired V(D)J cleavage complexes. Mol. Cell. Biol. 2001;21:4256–4264. doi: 10.1128/MCB.21.13.4256-4264.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swanson PC. A RAG-1/RAG-2 tetramer supports 12/23-regulated synapsis, cleavage, and transposition of V(D)J recombination signals. Mol. Cell. Biol. 2002;22:7790–7801. doi: 10.1128/MCB.22.22.7790-7801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ciubotaru M, et al. RAG1-DNA binding in V(D)J recombination. Specificity and DNA-induced conformational changes revealed by fluorescence and CD spectroscopy. J. Biol. Chem. 2003;278:5584–5596. doi: 10.1074/jbc.M209758200. [DOI] [PubMed] [Google Scholar]
- 24.Bailin T, Mo X, Sadofsky MJ. A RAG1 and RAG2 tetramer complex is active in cleavage in V(D)J recombination. Mol. Cell. Biol. 1999;19:4664–4671. doi: 10.1128/mcb.19.7.4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bellon SF, Rodgers KK, Schatz DG, Coleman JE, Steitz TA. Crystal structure of the RAG1 dimerization domain reveals multiple zinc-binding motifs including a novel zinc binuclear cluster. Nat. Struct. Biol. 1997;4:586–591. doi: 10.1038/nsb0797-586. [DOI] [PubMed] [Google Scholar]
- 26.Matthews AG, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng JA, Johnson RC, Dickerson RE. Hin recombinase bound to DNA: the origin of specificity in major and minor groove interactions. Science. 1994;263:348–355. doi: 10.1126/science.8278807. [DOI] [PubMed] [Google Scholar]
- 28.Banerjee-Basu S, Baxevanis AD. The DNA-binding region of RAG 1 is not a homeodomain. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-8-interactions1004. INTERACTIONS1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aidinis V, et al. The RAG1 homeodomain recruits HMG1 and HMG2 to facilitate recombination signal sequence binding and to enhance the intrinsic DNA-bending activity of RAG1-RAG2. Mol. Cell. Biol. 1999;19:6532–6542. doi: 10.1128/mcb.19.10.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swanson PC, Desiderio S. V(D)J recombination signal recognition: distinct, overlapping DNA-protein contacts in complexes containing RAG1 with and without RAG2. Immunity. 1998;9:115–125. doi: 10.1016/s1074-7613(00)80593-2. [DOI] [PubMed] [Google Scholar]
- 31.Aravind L, Landsman D. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 1998;26:4413–4421. doi: 10.1093/nar/26.19.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowell LG, Davila M, Yang K, Kepler TB, Kelsoe G. Prospective estimation of recombination signal efficiency and identification of functional cryptic signals in the genome by statistical modeling. J. Exp. Med. 2003;197:207–220. doi: 10.1084/jem.20020250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagawa F, et al. Footprint analysis of the RAG protein recombination signal sequence complex for V(D)J type recombination. Mol. Cell. Biol. 1998;18:655–663. doi: 10.1128/mcb.18.1.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramsden DA, Baetz K, Wu GE. Conservation of sequence in recombination signal sequence spacers. Nucleic Acids Res. 1994;22:1785–1796. doi: 10.1093/nar/22.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akamatsu Y, Oettinger MA. Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences. Mol. Cell. Biol. 1998;18:4670–4678. doi: 10.1128/mcb.18.8.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huye LE, Purugganan MM, Jiang MM, Roth DB. Mutational analysis of all conserved basic amino acids in RAG-1 reveals catalytic, step arrest, and joining-deficient mutants in the V(D)J recombinase. Mol. Cell. Biol. 2002;22:3460–3473. doi: 10.1128/MCB.22.10.3460-3473.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feeney AJ, Goebel P, Espinoza CR. Many levels of control of V gene rearrangement frequency. Immunol. Rev. 2004;200:44–56. doi: 10.1111/j.0105-2896.2004.00163.x. [DOI] [PubMed] [Google Scholar]
- 38.Ciubotaru M, Kriatchko AN, Swanson PC, Bright FV, Schatz DG. Fluorescence resonance energy transfer analysis of recombination signal sequence configuration in the RAG1/2 synaptic complex. Mol. Cell. Biol. 2007;27:4745–4758. doi: 10.1128/MCB.00177-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swanson PC. The DDE motif in RAG-1 is contributed in trans to a single active site that catalyzes the nicking and transesterification steps of V(D)J recombination. Mol. Cell. Biol. 2001;21:449–458. doi: 10.1128/MCB.21.2.449-458.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou L, et al. Transposition of hAT elements links transposable elements and V(D)J recombination. Nature. 2004;432:995–1001. doi: 10.1038/nature03157. [DOI] [PubMed] [Google Scholar]
- 41.Hickman AB, et al. Molecular architecture of a eukaryotic DNA transposase. Nat. Struct. Mol. Biol. 2005;12:715–721. doi: 10.1038/nsmb970. [DOI] [PubMed] [Google Scholar]
- 42.Grundy GJ, Hesse JE, Gellert M. Requirements for DNA hairpin formation by RAG1/2. Proc. Natl. Acad. Sci. U S A. 2007;104:3078–3083. doi: 10.1073/pnas.0611293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu CP, Sandoval H, Brandt VL, Rice PA, Roth DB. Amino acid residues in Rag1 crucial for DNA hairpin formation. Nat. Struct. Mol. Biol. 2006;13:1010–1015. doi: 10.1038/nsmb1154. [DOI] [PubMed] [Google Scholar]
- 44.Santagata S, Villa A, Sobacchi C, Cortes P, Vezzoni P. The genetic and biochemical basis of Omenn syndrome. Immunol. Rev. 2000;178:64–74. doi: 10.1034/j.1600-065x.2000.17818.x. [DOI] [PubMed] [Google Scholar]
- 45.Eastman QM, Schatz DG. Nicking is asynchronous and stimulated by synapsis in 12/23 rule-regulated V(D)J cleavage. Nucleic Acids Res. 1997;25:4370–4378. doi: 10.1093/nar/25.21.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu K, Lieber MR. The nicking step in V(D)J recombination is independent of synapsis: implications for the immune repertoire. Mol. Cell. Biol. 2000;20:7914–7921. doi: 10.1128/mcb.20.21.7914-7921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramsden DA, McBlane JF, van Gent DC, Gellert M. Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J. 1996;15:3197–3206. [PMC free article] [PubMed] [Google Scholar]
- 48.Swanson PC. Fine structure and activity of discrete RAG-HMG complexes on V(D)J recombination signals. Mol. Cell. Biol. 2002;22:1340–1351. doi: 10.1128/mcb.22.5.1340-1351.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swanson PC, Desiderio S. RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol. Cell. Biol. 1999;19:3674–3683. doi: 10.1128/mcb.19.5.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bergeron S, Madathiparambil T, Swanson PC. Both High Mobility Group (HMG)-boxes and the Acidic Tail of HMGB1 Regulate Recombination-activating Gene (RAG)-mediated Recombination Signal Synapsis and Cleavage in Vitro. J. Biol. Chem. 2005;280:31314–31324. doi: 10.1074/jbc.M503063200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dai Y, et al. Determinants of HMGB proteins required to promote RAG1/2-recombination signal sequence complex assembly and catalysis during V(D)J recombination. Mol. Cell. Biol. 2005;25:4413–4425. doi: 10.1128/MCB.25.11.4413-4425.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mouw KW, et al. Architecture of a serine recombinase-DNA regulatory complex. Mol. Cell. 2008;30:145–155. doi: 10.1016/j.molcel.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bergeron S, Anderson DK, Swanson PC. RAG and HMGB1 proteins: purification and biochemical analysis of recombination signal complexes. Methods Enzymol. 2006;408:511–528. doi: 10.1016/S0076-6879(06)08032-3. [DOI] [PubMed] [Google Scholar]
- 54.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. Macromol. Crystallogr. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 55.The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 56.Sheldrick GM. A short history of SHELX. Acta Crystallogr. A. 2008:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 57.Terwilliger T. SOLVE and RESOLVE: automated structure solution, density modification and model building. J. Synchrotron. Radiat. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- 58.Terwilliger TC. Maximum likelihood density modification. Acta Crystallogr. 2000;D56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 60.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.