Abstract
Two epidemiologically unrelated carbapenem-resistant Acinetobacter baumannii isolates were investigated as representatives of the first Italian isolates producing the OXA-24 carbapenemase. Both isolates were of European clonal lineage II and carried an identical OXA-24-encoding plasmid, named pABVA01. Comparative analysis revealed that in pABVA01, blaOXA-24 was part of a DNA module flanked by conserved inverted repeats homologous to XerC/XerD binding sites, which in other Acinetobacter plasmids flank different DNA modules, suggesting mobilization by a novel site-specific recombination mechanism.
Acinetobacter baumannii is an opportunistic nosocomial pathogen of increasing clinical relevance (3, 4, 22). The species is naturally resistant to several antimicrobial agents and exhibits a remarkable propensity to acquire new resistances (4, 14, 22). Carbapenems are elective agents for treatment of A. baumannii infections, and the emergence of carbapenem-resistant strains is a matter of increasing clinical concern (22, 24). Acquired class D carbapenemases of the OXA-23, OXA-24 (also named OXA-40), and OXA-58 lineages are playing a major role as determinants of acquired carbapenem resistance in A. baumannii (24).
In Italy, production of OXA-58 is the dominant carbapenem resistance mechanism in A. baumannii, and several outbreaks caused by OXA-58-producing strains related to European clonal lineage II have been documented (11, 15, 17), while strains producing OXA-23 and OXA-24 have not been reported. Here we report the characterization of the first Italian isolates of carbapenem-resistant A. baumannii producing the OXA-24 carbapenemase.
(Part of this study was presented at the 18th European Congress of Clinical Microbiology and Infectious diseases, Barcelona, Spain, 2008. [M. M. D'Andrea, T. Giani, F. Luzzaro, and G. M. Rossolini, oral communication O300].)
Characterization of OXA-24-positive A. baumannii isolates.
In a survey of carbapenemase genes in carbapenem-resistant A. baumannii isolated from several Italian hospitals during the past decade, only two A. baumannii isolates, VA-566/00 and N50, tested positive for blaOXA-24-like genes by PCR analysis, while they lacked genes for other acquired class D carbapenemases (OXA-23- and OXA-58-like class D carbapenemases) and metallo-β-lactamases (IMP-, VIM-, or SIM-type metallo-β-lactamases). The primers and PCR conditions used for the detection of carbapenemase genes are described in Table 1. VA-566/00 was isolated from an inpatient at a tertiary care hospital in Varese (in northern Italy) in September 2000. No epidemiological links with areas of OXA-24 endemicity could be traced for this patient by analysis of clinical records. N50 was isolated from an inpatient at a general hospital in Rome (in central Italy) in January 2004. In this case, analysis of clinical records revealed a previous history of hospitalization (2 months earlier) in Spain, which is a country where OXA-24 is endemic (20, 24). Both isolates were from the respiratory tract and had apparently played the role of colonizers. Identification at the species level was performed by automatic identification systems (Phoenix; Becton Dickinson, Sparks, MD; and Vitek-2; bioMérieux, Marcy-l'Etoile, France) and confirmed by PCR detection of blaOXA-51-like alleles (33).
TABLE 1.
Nucleotide primers used in this work for detection of β-lactamase genes and their linkages with insertion sequences in A. baumannii isolates
| Primer | Sequence (5′-3′)a | Target(s) | Expected amplicon size (bp) | Conditions for denaturation (°C/min)b | Conditions for annealing (°C/min)b | Conditions for extension (°C/min)b | Source or reference | Positive control (gene) [source or reference] |
|---|---|---|---|---|---|---|---|---|
| IMP-DIA-fwd | GGAATAGAGTGGCTTAATTCTC | blaIMP alleles | 361 | 94/1 | 50/1 | 72/1 | 13 | A. baumannii AC-54/97 (blaIMP-2) [25] |
| IMP-DIA-rev | GTGATGCGTCYCCAAYTTCACT | |||||||
| VIM-DIA-fwd | CAGATTGCCGATGGTGTTTGG | blaVIM alleles | 523 | 94/1 | 50/1 | 72/1 | 26 | P. aeruginosa VR143/97 (blaVIM-1) [18] |
| VIM-DIA-rev | AGGTGGGCCATTCAGCCAGA | |||||||
| SIM1-F | TACAAGGGATTCGGCATCG | blaSIM-1 | 570 | 94/1 | 50/1 | 72/1 | 19 | A. baumannii YMC 03/9/T104 (blaSIM-1) [19] |
| SIM1-R | TAATGGCCTGTTCCCATGTG | |||||||
| OXA23-F | GATGTGTCATAGTATTCGTCG | blaOXA-23 alleles | 748 | 94/1 | 50/1 | 72/1 | 2 | A. baumannii Ab13 (blaOXA-23) [10] |
| OXA23-R | TCACAACAACTAAAAGCACTG | |||||||
| OXA24-F | TTCCCCTAACATGAATTTGT | blaOXA-24 alleles | 582 | 94/1 | 50/1 | 72/1 | 2 | A. baumannii RYC 52763/97 (blaOXA-24-type) [6] |
| OXA24-R | GTACTAATCAAAGTTGTGAA | |||||||
| OXA-58_I_Fw | GCTGAGCATAGTATGAGTCG | blaOXA-58 alleles | 691 | 94/1 | 48/1 | 72/1 | This work | A. baumannii VA-900/05 (blaOXA-58) [this work] |
| OXA-58_I_Rev | AAGCAAATGCCACCACTTGC | |||||||
| OXA-69A | CTAATAATTGATCTACTCAAG | blaOXA-51 alleles | 975 or 2,168c | 94/1 | 48/1 | 72/2 | 16 | A. baumannii VA-804/03 (ISAba1 + blaOXA-90) [this work] |
| OXA-69B | CCAGTGGATGGATGGATAGATTATC | |||||||
| ISAba2_Fw | CCTTATCCTATCAGGGTTCTG | ISAba1 and blaADC alleles | 2,300 | 94/1 | 50/1 | 72/2 | This work | A. baumannii 16x46 (blaADC-type) [this work] |
| AmpCaba_Rev | GCATTCAGCACAGCATAAG |
For degenerate primers, the following code was used: Y, C/T.
All reactions included an initial denaturation step of 5 min at 94°C, 30 cycles of amplification, and a final extension step of 20 min at 72°C.
Primers give a larger amplicon in the presence of an ISAba1 upstream of the blaOXA-51 allele.
Susceptibility testing carried out by broth microdilution and interpreted according to CLSI guidelines (8, 9) revealed that both isolates were resistant to all β-lactams (including carbapenems), aminoglycosides, and fluoroquinolones. Variable susceptibilities toward colistin and tigecycline were observed (Table 2). PCR performed with the primers described in Table 1 revealed ISAba1 upstream of the resident blaADC gene but not upstream of the resident blaOXA-51-like gene in both isolates, suggesting that blaOXA-51-like gene overexpression was not involved in the carbapenem resistance phenotype.
TABLE 2.
Antibiotic susceptibility of the blaOXA-24-positive A. baumannii isolates investigated in this work and of the RUH134(pABVA01) transformanta
| Isolate | MIC (mg/liter) of the indicated antibioticb:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| CIP | GEN | AMK | CAZ | IPM | MEM | SAM | CST | TGC | |
| VA-566/00 | 128 | >256 | 64 | >256 | 256 | 256 | 64 | 0.5 | 4 |
| N50 | 128 | >256 | 256 | >256 | 128 | 128 | 32 | >8 | 1 |
| RUH134 | 1 | >256 | 4 | 16 | 0.25 | 1 | 64 | 0.5 | 1 |
| RUH134(pABVA01) | 1 | >256 | 4 | 16 | 64 | 128 | 64 | 0.5 | 1 |
The susceptibility of RUH134 is shown for comparison.
CIP, ciprofloxacin; GEN, gentamicin; AMK, amikacin; CAZ, ceftazidime; IPM, imipenem; MEM, meropenem; SAM, ampicillin/sulbactam; CST, colistin; TGC, tigecycline.
Multiplex PCR for the definition of A. baumannii sequence groups, which has been proven to be a useful method for assigning isolates to major clonal lineages (31, 32), was performed using ompA, csuE, and blaOXA-51-like target genes as described previously (32). PCR amplicons were purified by using a Wizard PCR purification kit (Promega, Madison, WI) and were sequenced on both strands, using custom primers at an external sequencing facility (Macrogen, Seoul, Korea). Genotyping by macrorestriction analysis of ApaI-digested A. baumannii genomes was performed by pulsed-field gel electrophoresis (PFGE) as previously described (28), using a CHEF mapper (Bio-Rad, Hemel Hempstead, United Kingdom). PFGE profiles were interpreted according to the criteria proposed by Tenover et al. (30), with a difference of six bands or less used to define epidemiological relatedness, and were analyzed by BioNumerics software (Applied Maths, Kortrijk, Belgium). A. baumannii RUH875 and RUH134 were used as reference strains representative of European clonal lineages I and II, respectively (12, 21). A. baumannii isolates VA-900/05, N33, and N40, obtained from the same hospitals in which the OXA-24-producing isolates were detected, were used as representatives of the major carbapenem-resistant OXA-58-producing clone spreading in Italy. N33 and N40 showed close genetic relatedness to the OXA-58-producing prototypic Italian strain ACICU (11, 17).
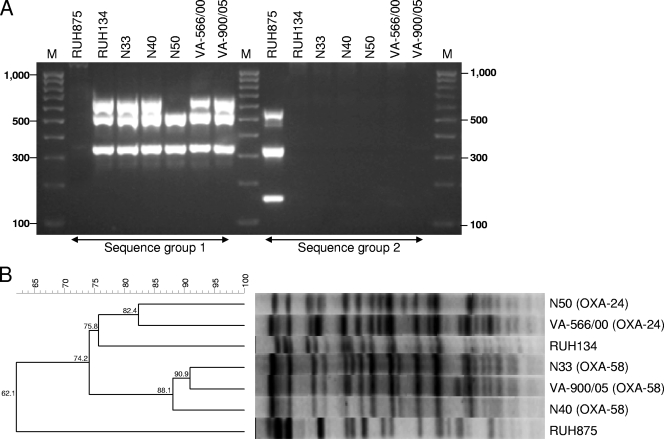
Sequencing of the ompA, csuE, and blaOXA-51-like alleles showed that VA-566/00 belonged in sequence group 1 (allelic profile 1-1-1), similar to RUH134 and the OXA-58 producers VA-900/05, N33, and N40 used for comparison, while N50 belonged in a variant of sequence group 1 called group 4, which does not yield an amplicon for the csuE allele (31) (Fig. 1A). As expected, RUH875 yielded results typical of sequence group 2, with an allelic profile of 2-2-2. The ApaI macrorestriction profiles of genomic DNA confirmed that the two OXA-24-positive isolates were related to each other (a difference of four bands) and, although to a lower extent, to RUH134 and the OXA-58-producing isolates from the same hospitals (differences of six to seven bands) (Fig. 1B). The topology of the dendrogram highlighted two major clusters at a similarity threshold of at least 82.4%, which differentiated the OXA-24- producing isolates from the OXA-58-producing isolates. Moreover, both clusters were related, having a 74.2% similarity with the prototypic strain RUH134 (Fig. 1B). Altogether, these results demonstrate a genetic correlation between the two OXA-24-producing isolates and that they belong to European clonal lineage II.
FIG. 1.
Results of genotyping of A. baumannii isolates investigated in this work and of the reference strains RUH875 and RUH134, representative of European clonal lineages I and II, respectively. (A) Sequence-based typing resulting from multiplex PCR targeting the ompA, csuE, and blaOXA-51-like genes. Lane M contains 100-bp molecular weight markers. (B) PFGE profiles. The dendrogram was generated with BioNumerics software (Applied Maths), using the unweighted-pair group method with arithmetic averages and the Dice coefficient. The percentage of similarity is shown at each node. Strain descriptions with the corresponding carbapenemases (in parentheses) are shown on the right.
Genetic support of blaOXA-24.
Plasmid extraction from VA-566/00 and N50, carried out with a Wizard Plus SV Minipreps DNA purification system (Promega), revealed in both cases the presence of a small plasmid, which in a Southern blotting experiment (27) hybridized to a blaOXA-24 probe (data not shown).
The plasmid from VA-566/00, named pABVA01, was transferred to RUH134 by electroporation, using a Gene Pulser apparatus (Bio-Rad). Competent cells were prepared as described for Escherichia coli (27). Transformants were selected on LB agar supplemented with imipenem (4 mg/liter). The carriage of blaOXA-24 in the transformants was confirmed by PCR. The RUH134(pABVA01) transformants showed high MICs of carbapenem (similar to those for parent strain VA-566/00), while their susceptibilities to other agents, including ceftazidime, were not affected (Table 2), confirming that OXA-24 is not active on this substrate (6). Ceftazidime resistance in the original host was likely related to overexpression of the blaADC cephalosporinase gene due to upstream insertion of ISAba1.
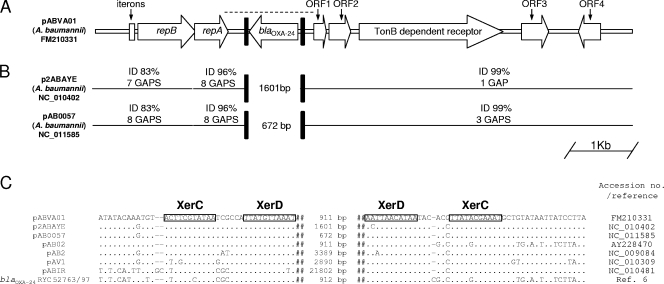
The nucleotide sequence of pABVA01 was determined on both strands by a primer-walking technique with a purified plasmid preparation. Plasmid pABVA01 is 8,963 bp long and carries eight open reading frames (ORFs) (Table 3, Fig. 2A). PCR mapping, followed by restriction analysis of amplicons and partial sequencing, showed that the plasmid obtained from N50 was indistinguishable from pABVA01 (data not shown). In particular, the nucleotide sequence of the region encompassing blaOXA-24 and its flanks (Fig. 2A) was identical.
TABLE 3.
ORFs and other features of plasmid pABVA01
| Nucleotide positiona | Strandb | Feature | Gene product (no. of amino acids)b | Properties and/or putative function |
|---|---|---|---|---|
| 319-519 | + | oriV | NA | Origin of DNA replication |
| 520-607 | NA | Repeat region | NA | Imperfect 4-repeat iterons; control of DNA replication |
| 663-1613 | + | repB | 316 | Replicase |
| 1606-2181 | + | repA | 191 | Replicase |
| 2481-2491 | + | Putative XerC binding site | NA | Protein binding site |
| 2498-2508 | + | Putative XerD binding site | NA | Protein binding site |
| 2519-3346 | − | blaOXA-24 | 275 | Carbapenem-hydrolyzing oxacillinase |
| 3420-3430 | − | Putative XerD binding site | NA | Protein binding site |
| 3437-3447 | − | Putative XerC binding site | NA | Protein binding site |
| 3617-3832 | + | ORF1 | 71 | Putative inner membrane protein |
| 3860-4243 | + | ORF2 | 127 | Putative cytoplasmic protein |
| 4371-6782 | + | TonB-dependent receptor | 803 | Putative TonB-dependent receptor |
| 7087-7548 | + | ORF3 | 153 | Hypothetical protein |
| 8044-8415 | − | ORF4 | 123 | Hypothetical protein |
Position no. 1 corresponds to the first nucleotide of p2ABAYE (NC_010402).
NA, not applicable.
FIG. 2.
(A) Linear map of plasmid pABVA01. Plasmid features are detailed in Table 3. The dashed line indicates the sequenced region of the plasmid from isolate N50. (B) Comparison of plasmid pABVA01 with plasmids p2ABAYE and pAB0057. For each homologous segment, the percentage of nucleotide identity and the number of gaps inserted into the alignments are shown. Filled boxes represent the putative recombination sites represented by the IRs homologous to the XerC/XerD binding sites. The sizes of the different DNA modules inserted between the IRs in place of the blaOXA-24 module are also shown. (C) Nucleotide sequences flanking different DNA modules (including that containing blaOXA-24) in different Acinetobacter plasmids. The IRs homologous to the XerC/XerD binding sites that resulted from comparison with sequences reported by Bui et al. (7) are boxed. The sizes of intervening DNA modules are also indicated. The regions flanking blaOXA-24 in the chromosome of strain RYC 52763/97 (6) are also shown. Accession numbers for each sequence and the reference for strain RYC 52763/97 are reported on the right.
Structure of plasmid pABVA01 and context of blaOXA-24.
Comparative analysis with other sequenced Acinetobacter plasmids revealed that the genetic organization of pABVA01 was overall very similar to that of p2ABAYE (35) and pAB0057 (1), two small Acinetobacter plasmids previously detected in French and American clinical isolates, respectively. The major differences between pABVA01 and these plasmids were found in the replicon region (the iteron and repB region of pABVA01 are more divergent; the 3′ end of the repA gene of pABVA01 carries a 7-bp deletion resulting in a premature stop codon) and in the region containing blaOXA-24, which in the other plasmids was replaced by unique regions of different size and composition (Fig. 2B).
A similar arrangement of intervening DNA modules inserted at the same position suggested the occurrence of a conserved recombination site where different DNA modules could be inserted by a site-specific recombination mechanism. Analysis of the sequences at the recombination junctions revealed, on both sides, the presence of conserved inverted repeats (IRs) separated by a 6-bp variable region (Fig. 2C). These structures, which partially overlap the previously described Re27-1 and Re27-2 regions associated with blaOXA-58 (23, 36), share high homology with dif-like binding sites that act as targets of the XerC and XerD recombinases that normally convert plasmid and chromosome dimers to monomers during cell division (29). XerC and XerD proteins and cognate recombination sites are also involved in other site-specific recombination mechanisms such as the integration of phage CTX-Φ at the dif1 site of the larger chromosome of Vibrio cholerae (34) and have also been exploited for artificial gene excision by site-specific recombination in E. coli and Bacillus subtilis (5). Interestingly, the conserved IRs homologous to the XerC/XerD binding sites were found to flank not only different DNA modules in related Acinetobacter plasmids but also the blaOXA-24 gene in the partially sequenced plasmid pAB02 and in the chromosome of strain RYC 52763/97 (6) (Fig. 2C). Altogether, these findings suggest that the XerC/XerD-like sites could act as site-specific recombination targets responsible for the mobilization of discrete DNA modules within Acinetobacter plasmids and chromosomes and that blaOXA-24 could be mobilized by a similar mechanism.
Concluding remarks.
To the best of our knowledge, this is the first description of carbapenem-resistant A. baumannii isolates carrying the blaOXA-24 determinant from Italy. The two isolates have a common ancestry and carry the same OXA-24-encoding plasmid, but they are not identical by PFGE profile and sequence type, which suggests a history of independent acquisition of the same resistance plasmid by the two related strains. Analysis of the genetic context of blaOXA-24 did not reveal structures typically involved in DNA mobilization (insertion sequences or other genes encoding known recombinases). However, blaOXA-24 was carried on a DNA module inserted between conserved inverted repeats homologous to XerC/XerD binding sites, which, in other plasmids, flank DNA modules of different sizes and compositions. This finding suggests the occurrence of a novel site-specific recombination mechanism that could play a role in the plasticity of Acinetobacter plasmids and in the mobilization of resistance genes.
Nucleotide sequence accession number.
The nucleotide sequence determined in this work has been submitted to the EMBL/DDBJ/GenBank database and assigned accession number FM210331.
Acknowledgments
This study was supported in part by grants from the European Commission (6th Framework, DRESP2 project) to G.M.R. and from Ricerca Corrente INMI 2007 to P.V.
We thank L. Poirel and P. Nordmann for providing reference strain Ab13, L. Dolzani for providing reference strains RUH134 and RUH875, G. Bou for providing the blaOXA-24-positive reference strain RYC 52763/97, and A. Dionisi for helping with BioNumerics analysis. Finally, we thank Maria Cristina Thaller for critical reading of the manuscript and helpful discussions.
Footnotes
Published ahead of print on 1 June 2009.
REFERENCES
- 1.Adams, M. D., K. Goglin, N. Molyneaux, K. M. Hujer, H. Lavender, J. J. Jamison, I. J. MacDonald, K. M. Martin, T. Russo, A. A. Campagnari, A. M. Hujer, R. A. Bonomo, and S. R. Gill. 2008. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J. Bacteriol. 190:8053-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afzal-Shah, M., N. Woodford, and D. M. Livermore. 2001. Characterization of OXA-25, OXA-26, and OXA-27, molecular class D β-lactamases associated with carbapenem resistance in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 45:583-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergogne-Bérézin, E. 2001. The increasing role of Acinetobacter species as nosocomial pathogens. Curr. Infect. Dis. Rep. 3:440-444. [PubMed] [Google Scholar]
- 4.Bergogne-Bérézin, E., and K. J. Towner. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9:148-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloor, A. E., and R. M. Cranenburgh. 2006. An efficient method of selectable marker gene excision by Xer recombination for gene replacement in bacterial chromosomes. Appl. Environ. Microbiol. 72:2520-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bou, G., A. Oliver, and J. Martinez-Beltran. 2000. OXA-24, a novel class D β-lactamase with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob. Agents Chemother. 44:1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bui, D., J. Ramiscal, S. Trigueros, J. S. Newmark, A. Do, D. J. Sherratt, and M. E. Tolmasky. 2006. Differences in resolution of mwr-containing plasmid dimers mediated by the Klebsiella pneumoniae and Escherichia coli XerC recombinases: potential implications in dissemination of antibiotic resistance genes. J. Bacteriol. 188:2812-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial susceptibility testing; 17th informational supplement. M100-S19. Clinical and Laboratory Standards Institute, Wayne, PA.
- 9.Clinical and Laboratory Standards Institute. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M07-A8. Clinical and Laboratory Standards Institute, Wayne, PA.
- 10.Corvec, S., L. Poirel, T. Naas, H. Drugeon, and P. Nordmann. 2007. Genetics and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-23 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:1530-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Arezzo, S., A. Capone, N. Petrosillo, and P. Visca. 2009. Epidemic multidrug-resistant Acinetobacter baumannii related to European clonal types I and II in Rome (Italy). Clin. Microbiol. Infect. 15:347-357. [DOI] [PubMed] [Google Scholar]
- 12.Dijkshoorn, L., H. Aucken, P. Gerner-Smidt, P. Janssen, M. E. Kaufmann, J. Garaizar, J. Ursing, and T. L. Pitt. 1996. Comparison of outbreak and nonoutbreak Acinetobacter baumannii strains by genotypic and phenotypic methods. J. Clin. Microbiol. 34:1519-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Docquier, J. D., M. L. Riccio, C. Mugnaioli, F. Luzzaro, A. Endimiani, A. Toniolo, G. Amicosante, and G. M. Rossolini. 2003. IMP-12, a new plasmid-encoded metallo-β-lactamase from a Pseudomonas putida clinical isolate. Antimicrob. Agents Chemother. 47:1522-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fournier, P. E., D. Vallenet, V. Barbe, S. Audic, H. Ogata, L. Poirel, H. Richet, C. Robert, S. Mangenot, C. Abergel, P. Nordmann, J. Weissenbach, D. Raoult, and J. M. Claverie. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS. Genet. 2:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giordano, A., P. Varesi, A. Bertini, L. Villa, A. M. Dionisi, M. Venditti, P. Carfagna, I. Luzzi, C. Mancini, and A. Carattoli. 2007. Outbreak of Acinetobacter baumannii producing the carbapenem-hydrolyzing oxacillinase OXA-58 in Rome, Italy. Microb. Drug Resist. 13:37-43. [DOI] [PubMed] [Google Scholar]
- 16.Héritier, C., L. Poirel, P. E. Fournier, J. M. Claverie, D. Raoult, and P. Nordmann. 2005. Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:4174-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iacono, M., L. Villa, D. Fortini, R. Bordoni, F. Imperi, R. J. Bonnal, T. Sicheritz-Ponten, G. De Bellis, P. Visca, A. Cassone, and A. Carattoli. 2008. Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob. Agents Chemother. 52:2616-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lauretti, L., M. L. Riccio, A. Mazzariol, G. Cornaglia, G. Amicosante, R. Fontana, and G. M. Rossolini. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43:1584-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, K., J. H. Yum, D. Yong, H. M. Lee, H. D. Kim, J. D. Docquier, G. M. Rossolini, and Y. Chong. 2005. Novel acquired metallo-β-lactamase gene, blaSIM-1, in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrob. Agents Chemother. 49:4485-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez-Otsoa, F., L. Gallego, K. J. Towner, L. Tysall, N. Woodford, and D. M. Livermore. 2002. Endemic carbapenem resistance associated with OXA-40 carbapenemase among Acinetobacter baumannii isolates from a hospital in northern Spain. J. Clin. Microbiol. 40:4741-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nemec, A., L. Dijkshoorn, and T. J. van der Reijden. 2004. Long-term predominance of two pan-European clones among multi-resistant Acinetobacter baumannii strains in the Czech Republic. J. Med. Microbiol. 53:147-153. [DOI] [PubMed] [Google Scholar]
- 22.Perez, F., A. M. Hujer, K. M. Hujer, B. K. Decker, P. N. Rather, and R. A. Bonomo. 2007. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:3471-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poirel, L., and P. Nordmann. 2006. Genetic structures at the origin of acquisition and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-58 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 50:1442-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poirel, L., and P. Nordmann. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12:826-836. [DOI] [PubMed] [Google Scholar]
- 25.Riccio, M. L., N. Franceschini, L. Boschi, B. Caravelli, G. Cornaglia, R. Fontana, G. Amicosante, and G. M. Rossolini. 2000. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of blaIMP allelic variants carried by gene cassettes of different phylogeny. Antimicrob. Agents Chemother. 44:1229-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riccio, M. L., L. Pallecchi, J. D. Docquier, S. Cresti, M. R. Catania, L. Pagani, C. Lagatolla, G. Cornaglia, R. Fontana, and G. M. Rossolini. 2005. Clonal relatedness and conserved integron structures in epidemiologically unrelated Pseudomonas aeruginosa strains producing the VIM-1 metallo-β-lactamase from different Italian hospitals. Antimicrob. Agents Chemother. 49:104-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.Seifert, H., L. Dolzani, R. Bressan, T. van der Reijden, B. van Strijen, D. Stefanik, H. Heersma, and L. Dijkshoorn. 2005. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J. Clin. Microbiol. 43:4328-4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Summers, D. K., and D. J. Sherratt. 1988. Resolution of ColE1 dimers requires a DNA sequence implicated in the three-dimensional organization of the cer site. EMBO J. 7:851-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Towner, K. J., K. Levi, and M. Vlassiadi. 2008. Genetic diversity of carbapenem-resistant isolates of Acinetobacter baumannii in Europe. Clin. Microbiol. Infect. 14:161-167. [DOI] [PubMed] [Google Scholar]
- 32.Turton, J. F., S. N. Gabriel, C. Valderrey, M. E. Kaufmann, and T. L. Pitt. 2007. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin. Microbiol. Infect. 13:807-815. [DOI] [PubMed] [Google Scholar]
- 33.Turton, J. F., N. Woodford, J. Glover, S. Yarde, M. E. Kaufmann, and T. L. Pitt. 2006. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 44:2974-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Val, M. E., M. Bouvier, J. Campos, D. Sherratt, F. Cornet, D. Mazel, and F. X. Barre. 2005. The single-stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae. Mol. Cell 19:559-566. [DOI] [PubMed] [Google Scholar]
- 35.Vallenet, D., P. Nordmann, V. Barbe, L. Poirel, S. Mangenot, E. Bataille, C. Dossat, S. Gas, A. Kreimeyer, P. Lenoble, S. Oztas, J. Poulain, B. Segurens, C. Robert, C. Abergel, J. M. Claverie, D. Raoult, C. Medigue, J. Weissenbach, and S. Cruveiller. 2008. Comparative analysis of Acinetobacters: three genomes for three lifestyles. PLoS One 3:e1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zarrilli, R., D. Vitale, A. Di Popolo, M. Bagattini, Z. Daoud, A. U. Khan, C. Afif, and M. Triassi. 2008. A plasmid-borne blaOXA-58 gene confers imipenem resistance to Acinetobacter baumannii isolates from a Lebanese hospital. Antimicrob. Agents Chemother. 52:4115-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]