Abstract
Interactions between host factors and the viral replication complex play important roles in host adaptation and regulation of influenza virus replication. A cellular protein, nuclear factor 90 (NF90), was copurified with H5N1 viral nucleoprotein (NP) from human cells in which NP was transiently expressed and identified by matrix-assisted laser desorption ionization-time of flight mass spectrometry analysis. In vitro coimmunoprecipitation of NF90 and NP coexpressed in HEK 293T cells or individually expressed in bacterial and HEK 293T cells, respectively, confirmed a direct interaction between NF90 and NP, independent of other subunits of the ribonucleoprotein complex. This interaction was prevented by a mutation, F412A, in the C-terminal region of the NP, indicating that the C-terminal of NP is required for NF90 binding. RNase V treatment did not prevent coprecipitation of NP and NF90, which demonstrates that the interaction is RNA binding independent. After small interfering RNA knockdown of NF90 expression in A549 and HeLa cells, viral polymerase complex activity and virus replication were significantly increased, suggesting that NF90 negatively affects viral replication. Both NP and NF90 colocalized in the nucleus of virus-infected cells during the early phase of infection, suggesting that the interaction between NF90 and NP is an early event in virus replication. Quantitative reverse transcription-PCR showed that NF90 downregulates both viral genome replication and mRNA transcription in infected cells. These results suggest that NF90 inhibits influenza virus replication during the early phase of infection through direct interaction with viral NP.
Sixteen hemagglutinin (HA) and nine neuraminidase (NA) subtypes of influenza A virus have been identified in aquatic birds, the recognized natural reservoir for all avian influenza A viruses (62). Host range restriction has largely limited the cross-species transmission of avian and other animal influenza viruses to humans. Historically, only three subtypes of influenza virus, namely, H1N1, H2N2, and H3N2, have been found to cross the host barrier to become human influenza viruses; this was achieved by reassortment and/or a yet-to-be-confirmed direct adaptation mechanism and in each case triggered an influenza pandemic (19, 57). Of these, only H1N1 and H3N2 are currently circulating in humans; however, in recent years sporadic human transmissions of three other avian influenza virus subtypes—H5N1, H7N7, and H9N2—have occurred (10, 16, 47, 65). Of note, more than 400 cases of human infection with avian H5N1 virus have been observed since it was first documented in Hong Kong in 1997 (1, 61). The replication and transmission abilities of these viruses in mammalian hosts are therefore of great interest, since another influenza pandemic may occur if an animal influenza virus crosses the species barrier and becomes adapted to humans. The April 2009 emergence of a swine-origin H1N1 virus, which infected humans first in Mexico and the United States and has since spread to more than 40 countries in less than 1 month, confirmed another successful cross-species transmission of influenza A virus that may be leading to another pandemic (19).
Evolutionary fitness of influenza virus involves establishment of compatibility of viral functional elements and adaptation to host interacting factors so that the virus may gain cellular entry, replicate, and assemble and release new virions. Infection by influenza viruses may be restricted by receptor specificity; human influenza viruses preferentially bind to cell surface sialic acids linked by α2,6 bonds to galactose (SA α2,6 Gal), whereas avian and equine viruses prefer SA α2,3 Gal (55). However, receptor specificity may not exclusively account for the inefficient human infections by current H5N1 viruses, which are still of the avian type with respect to receptor specificity. An engineered reassortant influenza virus containing genes for the two viral surface glycoproteins, HA and NA, of a human H3N2 virus, A/Victoria/3/75, and the remaining six internal genes from an H5N1 virus, A/Hong Kong/486/97, was unable to transmit efficiently in ferrets (34). In addition, despite avian H9N2 virus exhibiting a certain degree of SA α2,6 Gal receptor binding affinity and being endemic in chickens in Asia, human transmissions are still rare (7, 35, 47). These observations support the notion that as-yet undetermined virus-host interactions limit the replication cycles of these viruses, thus reducing the efficiency of transmission to humans. Host restricting factors have been found to block viral replication in retroviruses (63). Therefore, identification of host factors and a deeper understanding of the mechanism by which avian influenza virus replication is inhibited in mammalian cells may provide insights into the host adaptation of this virus.
There is increasing evidence that the viral polymerase complex plays a critical role in host adaptation and pathogenesis (8, 17, 18, 40, 60). Influenza virus relies on its own polymerase complex, consisting of PA, PB1, and PB2 subunits, together with nucleoprotein (NP), to initiate the replication of viral genome in the nucleus during infection of host cells. However, viral gene transcription, translation, and protein trafficking are critically dependent on host machinery; host barriers may dwell mainly in host factors associated with these processes (11, 14). Some cellular factors interacting with human influenza viruses have been identified by using yeast two-hybrid system and proteomics approaches (41). In addition to those associated with innate immunity and interacting with NS1, many other host factors are associated with viral replication and transcription processes (12, 15, 24, 28, 36, 39, 59). Unlike most other RNA viruses, influenza virus replicates in the cell nucleus, rather than in the cytoplasm, and may thus encounter some distinct antiviral host factors. Some host factors remain unidentified, and some may also interact differently with avian, as opposed to human, influenza viruses. In a recent study, an inhibitory activity associated with the function of avian-like influenza virus polymerase PB2 627E was found in human cells; however, the specific host factor remains unknown (31, 37). Viruses may escape these restrictions by adaptive mutations, as described for avian H5N1 virus and some mouse-adapted viruses (6, 17, 18).
NP plays a critical role, interacting with the polymerase complex to form the viral ribonucleoprotein (RNP) complex, that is necessary for replication and transcription of the viral genome during influenza virus infection. Since it has been hypothesized that NP plays a role in determining host range, it is important to identify and understand host factors that interact with NP (4, 54, 56, 62). In the present study, nuclear factor 90 (NF90), a double-stranded RNA (dsRNA)-binding protein (29, 53), was identified to interact with NP. Biological assays showed that NF90 inhibits the activity of the viral polymerase complex and replication of influenza virus during the early phase of infection, suggesting that NF90 is a host antiviral/restricting factor that targets influenza virus NP function in the nucleus.
MATERIALS AND METHODS
Cells and viruses.
Human embryonic kidney (HEK) 293T cells and HeLa cells were maintained in Dulbecco minimal essential medium, and Madin-Darby canine kidney (MDCK) and human lung carcinoma A549 cells were cultured in Eagle minimal essential medium (MEM), in all instances supplemented with 10% fetal bovine serum, 100 IU of penicillin G/ml and 100 μl of streptomycin sulfate/ml and cultured at 37°C with 5% CO2. For transfection studies, subconfluent (70%) monolayers were transiently transfected or cotransfected with plasmids by using TransIT-LT1 transfection reagent (Mirus). Transfected cultures were incubated for an additional 48 h for pulldown or coimmunoprecipitation (Co-IP) assay.
Influenza virus strains A/PR/8/34 (PR8; H1N1) and A/VNM/1194/04 (VNM1194; H5N1) were propagated in 10-day-embryonated specific-pathogen-free chicken eggs at 35°C for 48 and 30 h, respectively. Allantoic fluid was harvested and virus titrated in MDCK cells by plaque assay. Briefly, virus stocks were serially diluted in phosphate-buffered saline (PBS) and adsorbed onto confluent MDCK cells for 30 min at 37°C. The inoculum was removed; the cells were washed twice with PBS and covered with 2 ml of an agar medium (1% agarose, 1 μg of TPCK [tolylsulfonyl phenylalanyl chloromethyl ketone]-trypsin/ml in MEM). After 3 days of incubation, plaques were counted, and the virus concentration in PFU/ml was calculated.
Plasmid construction.
Expression vectors containing a Flag epitope (5′-GATTACAAGGATGACGACGATAAG-3′) or a V5 tag (5′-GATTACAAGGATGACGACGATAAG-3′) were constructed by using a pCMV-script vector (Stratagene) and designated pCMV-Flag and pCMV-V5, respectively. The NP gene of the H5N1 strain A/VNM/1194/04 (GenBank accession no. AY651498) was cloned into the HindIII/SalI site of pCMV-Flag to create pCMV-VNM1194NP-Flag. An N-terminal truncated NP deletion mutant (Δ1-180) was constructed by PCR from the full-length cDNA encoding NP. A collection of nine point-mutated versions of NP—S314A, R267A, R8A, R199A, Q405A, R342A, R338A, F412A, and R416A—was constructed by using the QuikChange II site-directed mutagenesis kit (Stratagene) and cloned into pCMV-Flag. Plasmid pCMV-NF90-V5 (NF90-V5) was generated by inserting NF90 (GenBank accession no. NM_004516) into pCMV-V5 at the BamHI/EcoRI site. All of the plasmids used in the present study contained C-terminal-tagged epitopes and a Kozak consensus sequence (CCACC) to optimize expression of the fusion proteins.
Pulldown and coimmunoprecipitation.
HEK 293T cells were transfected as described above and incubated for 48 h, and then pull-down or Co-IP assays were performed. Transfected cells were lysed in a buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Triton X-100, and protease inhibitor cocktail (Roche). Cell lysates were precleared with mouse immunoglobulin G (IgG) agarose and incubated with 1 μl of mouse anti-Flag M2 (Sigma) or 1 μl of mouse α-V5 (Invitrogen) monoclonal antibody on ice for 2 h. After the addition of 50 μl of protein G-Sepharose beads, the lysates were incubated at 4°C overnight. Proteins bound to the beads were then eluted into 1× sodium dodecyl sulfate (SDS) running buffer by heating at 95°C for 5 min. For RNase treatment, 100 μl of RNase V in RNase working buffer (0.5 U) was added prior to addition of the antibody, and the samples were incubated at 37°C for 25 min.
Protein identification by MALDI-TOF/TOF MS/MS analysis.
Pull-down products were separated by one-dimensional SDS-polyacrylamide gel electrophoresis (PAGE). Each protein band was excised, destained, reduced, alkylated, and digested with trypsin. To extract the polypeptides, the gel particles were twice subjected to consecutive 20 mM NH4HCO3 and 5% formic acid (FA) in 50% acetonitrile (ACN) treatments. The supernatants were combined and lyophilized, and dried polypeptides were recovered by adding 10 μl of 0.1% FA, followed by sonication for 1 min. Recovered polypeptides were further purified by using a ZipTip C18 column (Millipore) and eluted in a final volume of 3 μl of ACN. The dissolved polypeptides (0.5 μl) were spotted onto a matrix-assisted laser desorption ionization (MALDI) target plate, followed by an equal volume of 10 mg of α-cyano-4-hydroxycinnamic acid/ml in 50% ACN-0.1% trifluoroacetic acid. Dried samples on target plates were analyzed by using an ABI 4800 Plus MALDI-TOF/TOF analyzer (Applied Biosystems). For mass spectrometric (MS) analyses, 1,000 shots were typically accumulated for each sample. Tandem MS (MS/MS) analyses were performed using nitrogen at a collision energy of 2 kV and a collision gas pressure of ∼3.0 × 10−7 torrs. A stop condition was used so that 2,000 to 10,000 shots were combined for each spectrum, depending on the quality of the data. The MASCOT search engine (version 2.0; Matrix Science) was used to search all of the tandem mass spectra. A database containing all of the available virus and human genomes, as well as the International Protein Index human database (www.ebi.ac.uk/IPI/IPIhelp.html) was used for the search, which was restricted to tryptic peptides. The precursor error tolerance was set to <75 ppm, and the MS/MS fragment error tolerance was set to <0.4 Da.
In vitro binding assay.
His-tagged NF90 was expressed from pET22b(+) plasmid in the Escherichia coli strain BL21(DE3)pLysS. Expression was induced with 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 20°C for 12 h. Bacterial cells were lysed in buffer containing 20 mM Tris-HCl (pH 7.4) and 200 mM NaCl by sonication, and the resulting lysate was cleared by centrifugation at 40,000 × g for 40 min at 4°C. NF90 protein was adsorbed onto an Ni-nitrilotriacetic acid column and eluted with a buffer containing 20 mM Tris-HCl (pH 7.5), 200 mM NaCl, and 500 mM imidazole (Imidazol). The purity of the His-tagged NF90 was checked by SDS-PAGE (see Fig. 2F). Flag-tagged NPs and PA proteins were expressed in HEK 293T cells, as described above. Cell lysates were harvested at 48 h posttransfection and NP-Flag or PA-Flag proteins were pulled down and immobilized using α-Flag M2 agarose beads (Sigma). The beads were washed extensively in lysis buffer. After a washing step, different amounts (0, 1, or 10 μg) of purified His-tagged NF90 protein were added to the beads, and the mixtures were incubated at 4°C overnight. The beads were washed three times and then boiled. Proteins were detected by Western blotting with α-Flag (1:5,000) and α-His antibodies (Novagen) (1:1,000).
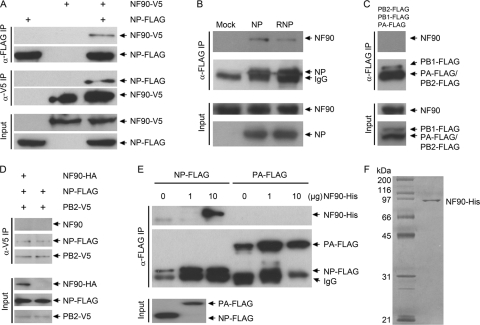
FIG. 2.
Confirmation of physical interaction between NF90 and NP. (A) Reciprocal Co-IP assay of NP-Flag and NF90-V5 in HEK 293T cells. NP-Flag and NF90-V5 expression vectors were cotransfected into HEK 293T cells. Cell lysates were prepared and incubated with α-Flag M2 (Sigma) or α-V5 (Invitrogen) monoclonal antibodies to precipitate NP and NF90, respectively. Proteins coprecipitated with NP and NF90 were analyzed by Western blotting with α-V5 and α-Flag, respectively. (B) Interaction between NP and endogenous NF90 in the presence of other polymerase subunits. NP alone or NP plus polymerase complex subunits (PB1, PB2, and PA) were transfected into HEK 293T cells; cell lysates were prepared 24 h posttransfection and precipitated with α-NP (9). Precipitates were analyzed with α-NF90 (BD) and α-NP antibodies. (C) Coprecipitation of NF90 with polymerase complex subunits (PB1, PB2 and PA). Flag-tagged PB1, PB2, and PA were cotransfected into HEK 293T cells, and cell lysates were prepared 24 h posttransfection and precipitated with α-Flag. (D) Coprecipitation of NP and PB2 in the presence or absence of overexpressed NF90. Flag-tagged NP and V5-tagged PB2 were transiently expressed in HEK 293T cells with or without coexpression of HA-tagged NF90; cell lysates were prepared at 24 h posttransfection and precipitated with α-V5. (E) Coprecipitation of NF90 and NP by individually expressed and purified proteins. His-tagged NF90 was expressed in bacteria, and Flag-tagged NP and PA were expressed in HEK 293T cells. Proteins were purified and used for coprecipitation as described in Materials and Methods. Different amounts of His-tagged NF90 were mixed with bead-immobilized/bead-bound Flag-tagged NP or PA, the mixtures were incubated, the beads were washed and boiled, and the samples were analyzed by Western blotting with α-Flag and α-His antibodies. “Input” indicates 1/10 diluted pre-Co-IP cell lysate samples, similarly analyzed to show the amount of each protein in the lysate. (F) Silver staining of His-tagged NF90 purified from bacterial culture.
Western blotting.
Co-IP products were fractionated by SDS-12% PAGE and then blotted onto nitrocellulose membranes (Bio-Rad). Membranes were incubated with the primary antibodies α-Flag M2 IgG (Sigma), α-V5 IgG (Invitrogen), or α-NP (9, 66) at 1:5,000 dilution; α-NF90 (BD) at a 1:500 dilution; or β-actin (Sigma) at a 1:5,000 dilution. After incubation with horseradish peroxidase-labeled sheep anti-mouse IgG secondary antibody (GE Healthcare), blots were visualized by using ECL Plus Western blotting detection reagents (GE Healthcare).
Knockdown of target gene expression and viral infection.
Gene-specific small interfering RNA (siRNA) were designed by using Invitrogen online design tools. NF90-specific siRNAs were identical to those previously reported (48). Nonspecific control oligonucleotides were supplied by Invitrogen. A549 cells or HeLa cells were transfected with siRNA at a concentration of 50 nM by using Lipofectamine RNAiMax transfection reagent (Invitrogen) and incubated for 48 h. Efficiency of NF90 expression knockdown was confirmed by Western blotting. NF90 knockdown cells were washed with 1× PBS and infected at a multiplicity of infection (MOI) of 1 with either the PR8 or the VNM1194 influenza virus strain. Unattached viruses were removed after incubation at 37°C for 30 min, and the culture medium was replenished. Culture supernatant was then collected at fixed intervals, and the virus titer was determined by plaque assay.
Immunofluorescence assay.
HeLa or A549 cells were grown on Lab-Tek chamber slides (Fisher Scientific) and transiently transfected with plasmids expressing NF90 or NP. For indirect immunofluorescence assay, cells were washed with PBS and fixed for 15 min using 4% formaldehyde in PBS, followed by permeabilization with 0.2% Triton X-100 in PBS for 5 min. The cells were then washed with PBS and blocked with 2% bovine serum albumin or 10% normal donkey serum in PBS at 37°C for 30 min, followed by incubation with α-NP (1:1,000), α-V5 (1:100), or α-Flag (1:100) diluted in PBS for 1 h at 37°C. The slides were then washed twice with PBS and incubated with donkey α-mouse conjugates (fluorescein isothiocyanate or rhodamine) for 30 min at room temperature. For costaining procedures, where primary antibodies raised from the same species were used, the primary antibodies were coupled with Zenon mouse IgG labeling reagents (Invitrogen) prior to application onto the cells, and the use of secondary antibodies was omitted. After staining, the cells were washed several times, and the signals examined by using a Nikon Eclipse Microscope. For the virus infection assay, cells were first treated with siRNA targeting NF90 for 24 h and then infected with PR8 or VNM1194 virus, as described above. Cells were fixed and stained for NF90 or viral NP expression at various postinfection time points.
Luciferase reporter assay for polymerase complex activity.
Full-length genomic segments of NP, PA, PB1, and PB2 derived from PR8 or VNM1194 were cloned into pHW2000 (22). NP F412A mutant was generated as described above. HeLa cells were treated with either siRNA specifically targeting NF90 or control siRNA; knockdown of NF90 expression was verified by Western blotting. Components of the RNP complex, comprising NP or NP F412A, plus PA, PB1, and PB2, were cotransfected into siRNA-treated HeLa cells, together with a luciferase reporter plasmid, pHY-Luci, which contains noncoding sequences from the M segment of influenza A virus and the luciferase gene driven by PolI (gifts from R. Webster and E. Hoffmann of St. Jude Children's Research Hospital) (51). At 24 h posttransfection, cell lysates were prepared by using the luciferase assay system (Promega), and luciferase activity was measured by using an E1500 luminometer (Promega). Plasmid phRL-TK (Promega), which expresses Renilla luciferase, was cotransfected as an internal control for data normalization.
Quantitative RT-PCR assays for vRNA and mRNA.
HeLa or A549 cells were treated with NF90-specific siRNA or nonspecific siRNA (Invitrogen) for 48 h. Cells were infected with A/Vietnam/1194/04 H5N1 virus at an MOI of 5. To study the effect of NF90 knockdown on viral genomic RNA (vRNA) and viral mRNA in a single replication cycle, total RNA was extracted at 4 and 8 h postinfection by using an RNeasy kit (Qiagen). A 3-μg portion of total RNA was reverse transcribed by using SuperScript II reverse transcriptase (Invitrogen). For the detection of vRNA, 500 ng of vRNA specific primer complementary to the 3′ end of vRNA was used in the reverse transcription (RT) reaction (23), whereas for mRNA detection, 50 pmol of oligo(dT) primer was used instead. For quantification of NP and NS1 vRNA and mRNA, a SYBR green-based real-time PCR method (Roche) was used. Primers were designed by using the Primer3 website. 1194-NP-F1 (5′-GGTAGGGACAATGGTGATGG-3′) and 1194-NP-R1 (5′-TTCTGCTCTCTCGCACTTGA-3′) were used for NP, and 1194-NS1-F1 (5′-GACCGGTTGGAAACCCTAAT-3′) and 1194-NS1-R1 (5′-TGATTTGGAGGGAGTGGAAG-3′) were used for NS1. All real-time PCR experiments were performed by using the LightCycler system (Roche). β-Actin mRNA was quantified, and the information was used to normalize the total RNA concentration between different samples, as described previously (25). A reaction mix of 20 μl was composed of 10 pmol of each gene-specific primer, 3 mM MgCl2, 2 μl of SYBR green master mix, and 2 μl of 10-fold-diluted cDNA. The amplification program was as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 10 s, 61°C for 5 s, and 72°C for 15 s. The specificity of the assay was confirmed by melting-curve analysis at the end of the amplification program (65 to 95°C, 0.1°C/s). Amplified products were further analyzed by agarose gel electrophoresis and sequencing.
RESULTS
Identification of NF90 as a host factor associating with H5N1 virus NP.
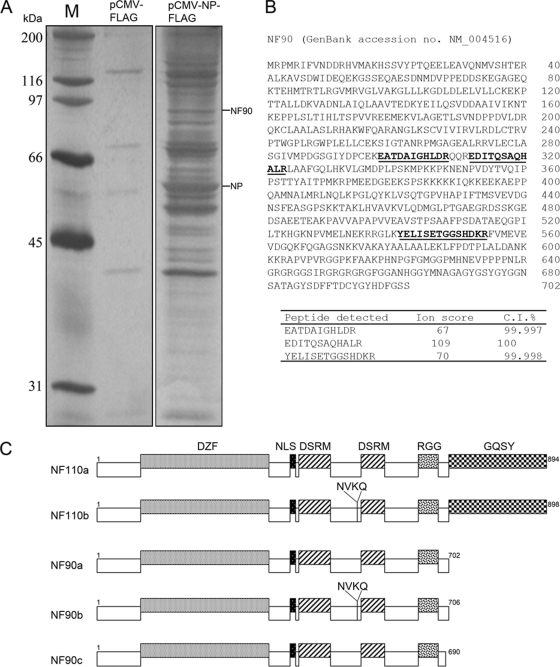
Several cellular factors associated with the influenza virus replication complex have recently been identified by using the yeast-two-hybrid system or Co-IP combined with proteomics procedures (41). However, these studies mainly used human influenza virus genes to identify host interacting factors (28, 36). It is not known whether current H5N1 viruses, which are still avian type and have limited transmissibility in humans, interact with these cellular factors in the same way that human influenza viruses do. In the present study, NP from an H5N1 strain, VNM1194, was used to identify cellular factors in a Co-IP experiment. The viral NP gene was tagged with a Flag epitope; a polymerase complex activity assay was performed to verify that the tagged epitope did not affect NP function (data not shown). Flag-tagged NP was then transiently expressed in HEK 293T cells, and NP-bound proteins were immunoprecipitated with α-Flag antibody. One protein band with an apparent molecular mass of 90 kDa was repeatedly identified in the Co-IP assay compared to the lysate from control HEK 293T cells transfected with vector alone (Fig. 1A). MS analysis identified this protein as nuclear factor 90 (Fig. 1B) (53). NF90, also known as NFAR, MBP4, or DRBP76 (45, 46, 48), belongs to the family of dsRNA-binding proteins (58). Five isoforms, which differ in the C-terminal region, have been reported (Fig. 1C) (50). NF90a, designated NF90 in the present study, is the most common isoform; its interaction with NP and role in virus replication are further investigated in the experiments described below. Other host proteins identified in the Co-IP assay will be described elsewhere.
FIG. 1.
Identification of host NF90 association with influenza viral NP by MS. (A) HEK 293T cells were transfected with expression vectors for A/Vietnam/1194/04 (H5N1) NP-Flag and cultured for 48 h. Cell lysate was precipitated with α-Flag M2 (Sigma) monoclonal antibody, and precipitates were separated by SDS-PAGE and visualized by silver staining. Each of the separated protein bands was individually excised and analyzed by MS. (B) All resulting MS and MS/MS spectra were searched against human and virus databases. Three peptides detected from a band at the 90-kDa position indicated the identity of NF90 protein; the peptide sequences identified covered 5.3% of the NF90 protein sequence, and the MS/MS spectra matched NF90 with a >99% confidence interval. (C) Illustration of different NF90 isoforms.
NF90 interacts with the RNP complex through NP.
To confirm a direct physical interaction between NF90 and the NP of A/VNM/1194/04 H5N1 virus, Flag-tagged NP and V5-tagged NF90 expression vectors were cotransfected into HEK 293T cells, and Co-IP of cell lysates with either α-Flag or α-V5 antibodies was performed. Immunoprecipitates were then analyzed reciprocally by Western blotting, using α-V5 antibodies to probe α-Flag immunoprecipitates and α-Flag antibodies to detect α-V5 immunoprecipitates, to confirm coprecipitation of NF90 with NP. As shown in Fig. 2A, immunoprecipitation with antibodies targeting either NP or NF90 results in pull-down of both proteins, confirming their physical interaction in cells. Coprecipitation of NF90 with NP from PR8 H1N1 human influenza virus was also observed (data not shown). A monoclonal antibody recognizing NP also coprecipitated NF90 and NP from cell lysates (Fig. 2B). To test the interaction of NF90 with NP in the context of the RNP complex, NF90 was coexpressed either together with NP and other polymerase subunits (Fig. 2B), or with PB1, PB2, and PA alone (Fig. 2C). NF90 could only be coprecipitated with the viral replication complex when NP was present, which suggests that NF90 interacts with the RNP complex through NP.
Interaction between PB2 and NP within the RNP complex has been described previously (49); we examined whether overexpressing NF90 in cells may interfere with NP and PB2 interaction. To this end, HA-tagged NF90 and V5-tagged PB2 were coexpressed along with Flag-tagged NP in HEK 293T cells, and cell lysates were pulled down with α-V5 antibody and then analyzed by Western blotting with either α-V5 or α-Flag antibodies. As demonstrated in Fig. 2D, PB2 was able to pull down NP in the presence or absence of overexpressed NF90, but NF90 was not simultaneously pulled down with the NP-PB2 complex, suggesting that the interaction between NF90 and NP may be independent of the interaction between NP and PB2. It remains to be investigated how NF90 might engage with NP in the context of the RNP complex. To further verify whether the interaction between NF90 and NP is direct, rather than through the mediation of other host proteins, we examined the NF90-NP interaction using individually expressed and purified proteins. His-tagged NF90 was expressed in bacteria and purified as described above (Fig. 2F). Because NP cannot be expressed in functional form in bacterial systems, Flag-tagged NP was expressed in HEK 293T cells and purified by immunoprecipitation with α-Flag antibody. Purified Flag-tagged NP and His-tagged NF90 were examined for interaction in an in vitro binding assay. As shown in Fig. 2E, Flag-tagged NP was able to pull down bacterially expressed His-tagged NF90. In contrast, Flag-tagged PA did not pull down NF90, which is consistent with the coprecipitation results obtained in transient-transfection assays (Fig. 2B and C). These results provide evidence supporting a direct interaction between NF90 and NP and suggest that the interaction is independent of the interaction between NP and PB2.
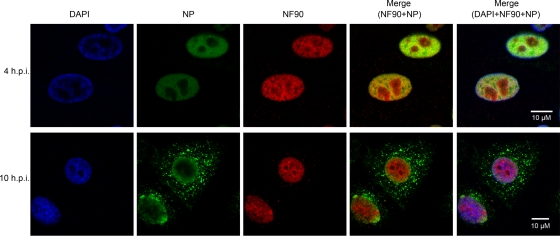
Because NP shuttles between the nucleus and cytoplasm during different stages of virus replication, we examined the intracellular colocalization of NF90 and NP proteins during virus infection. A549 cells were infected with A/Vietnam/1194/04 and cellular localization of NP and NF90 examined by indirect immunofluorescence assay at early and late time points. Both NP and NF90 proteins localized in the nucleus at 4 h postinfection, whereas NP was predominantly located in the cytoplasm at 10 h postinfection (Fig. 3). Confocal imaging revealed areas of colocalization of NP and NF90 within the nucleus in the early phase of infection (Fig. 3, merge images). A similar pattern of NF90 and NP cellular localization was seen in HeLa cells (data not shown).
FIG. 3.
Intracellular localization of NP and NF90 during influenza A virus infection. A549 cells were infected with VNM1194 at an MOI of 5. Infected cells were processed for microscopy at 4 and 10 h postinfection (h.p.i.). After fixation, the cells were stained with anti-NP and anti-NF90 monoclonal antibodies coupled with Zenon mouse IgG Alexa 488 and Alexa 594, respectively (Invitrogen). DAPI (4′,6′-diamidino-2-phenylindole) was used to visualize the nuclei of the cells. Imaging of cells was performed by using an LSM 510 Meta confocal laser scanning microscope (Carl Zeiss).
Taken together, these data confirm the MS result that the host nuclear protein NF90 interacts with influenza virus RNP complex by directly binding to NP and furthermore, suggest that the interaction between NF90 and NP occurs during the early phase of virus replication.
NP-NF90 interaction requires a functional NP C-terminal but is independent of NP RNA binding.
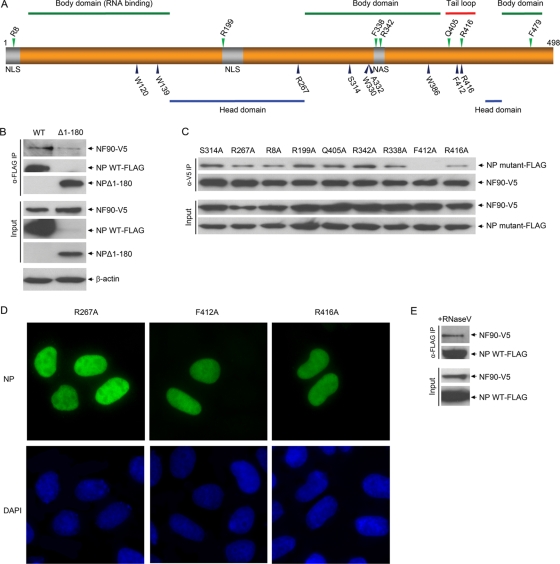
Viral NP interacts with the polymerase complex during viral genome replication and transcription in the nucleus of infected cells. NP binds to newly synthesized vRNA to form the viral RNP complex and interacts with cellular factors as the viral RNP complex is exported from nucleus to cytoplasm for new virion assembly. Several functional elements associated with RNA binding, nuclear localization and viral polymerase interaction have been mapped on the NP molecule based on crystal structure and mutagenesis studies (Fig. 4A) (13, 40, 64). To further characterize the interaction between NF90 and NP, we made a truncated version of NP by deleting the body domain-containing first 180 amino acids from the N-terminal end and then examined its interaction with NF90 in cells. Residues 1 to 180 represent the major part of the RNA binding region, and although NP structural studies have shown that additional residues, such as residues 181 to 186 and R221, and to a lesser extent, R382 and R384, are also involved in RNA binding (64), deletion of residues 1 to 180 removes the majority of the RNA domain and therefore is expected to greatly reduce RNA binding ability. Co-IP assay indicated that deletion of the 180 N-terminal amino acids did not abolish the interaction between mutated NP and NF90 (Fig. 4B), suggesting that the N-terminal NP body domain is not the major region involved in NF90 interaction. As we observed Δ1-180 NP to be less stable than wild-type (WT) NP, we cannot exclude the possibility that this deletion may partially affect NP conformation, resulting in reduced affinity of NF90 binding, since the amount of NF90 pulled down by the Δ1-180 version of NP is about one-third of that pulled down by WT NP. Influenza viral polymerase subunits bind to at least three regions of the NP and the C-terminal domain has been previously reported to mediate the interaction between PB2 and NP molecules (5). However, we were unable to express the C-terminal truncated version (Δ346-498) of NP in sufficient amounts, presumably due to its instability in cells, and the interaction between NF90 and C-terminal truncated NP could not be investigated. In order to verify whether NP interacts with NF90 through the C-terminal domain, several single amino acid mutants of NP were generated by site-directed mutagenesis, and their interactions with NF90 were examined in coexpression/Co-IP assays. Of the nine single-amino-acid mutants tested, only NP F412A was found to abolish interaction with NF90; no apparent change in NF90 interaction pattern was observed for other mutant NPs (Fig. 4C). Intracellular immunofluorescence staining showed that the mutant NPs were still localized in the nucleus (Fig. 4D), demonstrating that the F412A mutation's abolishment of NP-NF90 interaction was not due to a change in cellular localization. Residue F412 has previously been demonstrated to be essential for RNA binding (13). However, another NP mutant, R267A, previously reported to abolish RNA-binding activity, retained interaction with NF90. To further confirm the notion that interaction between NP and NF90 is independent of RNA binding, we pretreated cell lysates with RNase V, since NP does not protect encapsidated RNA from RNase digestion (3). NF90 was still coprecipitated with NP despite treatment with 100 μl of RNase V prior to antibody precipitation (Fig. 4E). F412 is an interior residue, and its role in maintaining the functional structure of NP for interaction with NF90 remains to be investigated. An X-ray crystallographic study of the structure of NP oligomers found that F412 and R416 residues are critical for NP intermolecule interaction (64), indicating that NF90 may interact with oligomeric rather than monomeric NP. Therefore, the interaction between NP and NF90 is dependent on the functionality of the NP C-terminal region but occurs independently of NP RNA binding.
FIG. 4.
Identification of NP domains required for NF90 interaction. (A) Illustration of NP, including known functional domains identified in the crystal structure and critical residues described previously (13, 64). The upper numbers represent residues that are critical for protein interaction, while the lower numbers represent NP residues which are important for RNA binding (13). (B) NF90 interacts with N-terminal truncated NP (Δ1-180). Flag-tagged N-terminal truncated NP (Δ1-180) was coexpressed with V5-tagged NF90 in HEK 293T cells and Co-IP with α-Flag, followed by Western blotting with α-V5 monoclonal antibody. (C) F412A single amino acid mutant abolishes NP interaction with NF90. Nine single amino acid mutants—R8A, R199A, R267A, S314A, R338A, R342A, Q405A, F412A, and R416A—were made by site mutagenesis of Flag-tagged NP and used to examine NP interaction with V5-tagged NF90 in Co-IP assays. (D) Cellular localization of R267A, F412A, and R416A mutated NPs. Mutated NP-Flag expression vectors were transfected into HeLa cells. At 24 h posttransfection, the cells were fixed and stained with anti-NP monoclonal antibody, followed by detection with secondary anti-mouse IgG FITC antibodies. DAPI was used to stain the nuclei of the cells. (E) NP-NF90 interaction is independent of RNA binding. Flag-tagged NP and V5-tagged NF90 expression vectors were coexpressed in HEK 293T cells, and cell lysates were pretreated with RNase V prior to coprecipitation with α-Flag antibody.
NF90 negatively regulates influenza virus replication.
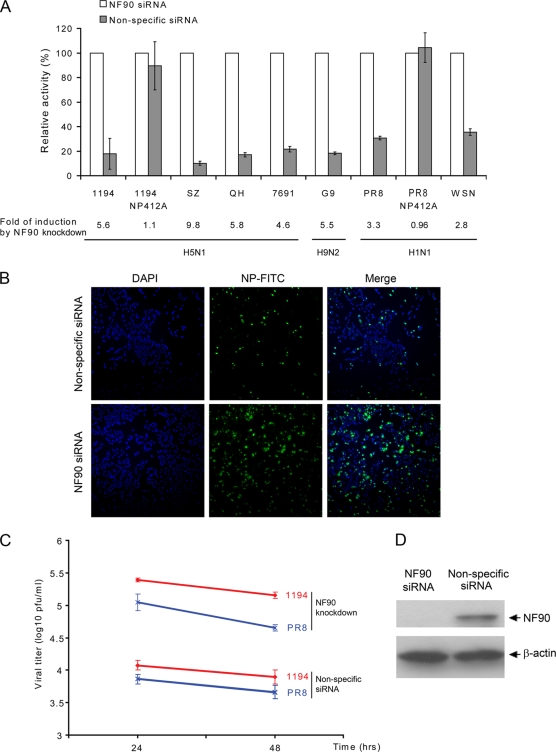
Several host factors interact with the viral NP complex during influenza virus transcription and replication (11, 18, 42). To understand the biological significance of the NP-NF90 interaction, we examined the effect of NF90 on influenza virus polymerase complex activity and replication efficiency. Because NF90 is ubiquitously expressed in many cell lines, we tested the effect of NF90 on influenza virus replication complex activity in A549 and HeLa cells in which NF90 expression was targeted by siRNA knockdown. Western blot confirmed that the expression of NF90 protein was significantly reduced in siRNA-treated cells but not in cells treated with nonspecific siRNA or siRNA targeting β-actin (Fig. 5D and data not shown). HeLa cells treated with either siRNA targeting NF90 or control siRNA were cotransfected with RNP complex genes (NP, PB1, PB2, and PA) derived from either avian H5N1 (1194, SZ, QH, and 769.1) or H9N2 (G9) strains or human H1N1 (PR8 and WSN) strains of influenza virus (virus abbreviations are given in detail in the legend to Fig. 5A), along with a reporter plasmid containing noncoding sequences from the influenza virus M segment and the luciferase gene driven by the human RNA polymerase I promoter (21, 51). For all of the strains tested, RNP activity was found to be significantly higher (3- to 10-fold) in NF90 expression knockdown cells than in cells treated with nonspecific siRNA (Fig. 5A) or left untreated (data not shown), showing that NF90 inhibits RNP activity. The F412A mutant NP described in the preceding section had lost the ability to interact with NF90, and we attempted to use this mutant to examine the inhibitory effect of NF90 on RNP polymerase activity. When F412A mutant NP was tested in the polymerase assay, the polymerase activity of RNP containing F412A mutant NP derived from either PR8 or VNM1194 strains was not affected by NF90 (Fig. 5A). However, because the RNP complexes containing F412A had only ca. 10% of the activity of those containing WT NP (data not shown), these attenuated RNP complexes may not be a good model for examining the effects of NF90 on polymerase activity. The effect of NF90 on the replication of influenza viruses was also investigated. HeLa and A549 cells treated with siRNA targeting NF90 or control siRNAs, or without siRNA treatment, were infected with either avian influenza H5N1 virus (VNM1194) or human influenza H1N1 virus (PR8), and virus replication was examined. Cell culture supernatants were collected at 24 and 48 h postinfection, and virus titers were determined by plaque assay. Reflecting the RNP activity assay result (Fig. 5A), virus titers of both H5N1 and H1N1 strains were significantly higher (10- to 20-fold) in NF90 knockdown cell cultures than in those treated with nonspecific siRNA or for mock-treated cells (Fig. 5C). Immunostaining for NP expression at 6 h postinfection in NF90 knockdown and control A549 cells infected with PR8 or H5N1 influenza virus also confirmed the inhibitory effect of NF90 on virus replication (Fig. 5B and data not shown). In light of these findings, we have found that NF90 exhibits an inhibitory effect on influenza virus replication, possibly interfering with viral RNP activity through direct interaction with NP.
FIG. 5.
NF90 inhibits polymerase complex activity and replication of influenza viruses. (A) Polymerase complex components derived from four H5N1 (1194, SZ, QH, and 7691), one H9N2 (G9), and two H1N1 (PR8 and WSN) viruses, together with their NP or NP F412A mutants (1194 and PR8 only) were cotransfected alongside a reporter plasmid that contains noncoding sequence from the M segment of influenza A virus and the luciferase gene driven by the PolI promoter into HeLa cells pretreated with either siRNA specifically targeting NF90 or nonspecific siRNA. The luciferase activity in triplicate cultures was estimated at 24 h posttransfection, and the relative activities from cells with or without NF90 knockdown were compared. Plasmid phRL-TK (Promega), which expresses Renilla luciferase, was cotransfected as an internal control for data normalization. (B) Immunostaining of NP expression in PR8 infected A549 cells with or without NF90 knockdown. A549 cells were treated with siRNA specifically targeting NF90 48 h prior to infection, and cells were later immunostained for NP expression at 6 h postinfection, as described above. (C) NF90 inhibits influenza virus replication. Replication of VNM1194 and PR8 strains were estimated in HeLa cells with or without siRNA-mediated NF90 knockdown. Virus titers in infected cell culture supernatants were measured 24 and 48 h postinfection by plaque assay. (D) Western blot to confirm knockdown of NF90 expression in HeLa cells. Abbreviations: 1194, A/VNM/1194/04; QH, A/BH goose/QH15/05; SZ, A/SZ/406H/06; 7691, A/Ck/HK/769.1/02; G9, A/Ck/HK/G9/97; PR8, A/PR/8/34; WSN, A/WSN/33.
NF90 inhibits both viral genome replication and mRNA transcription.
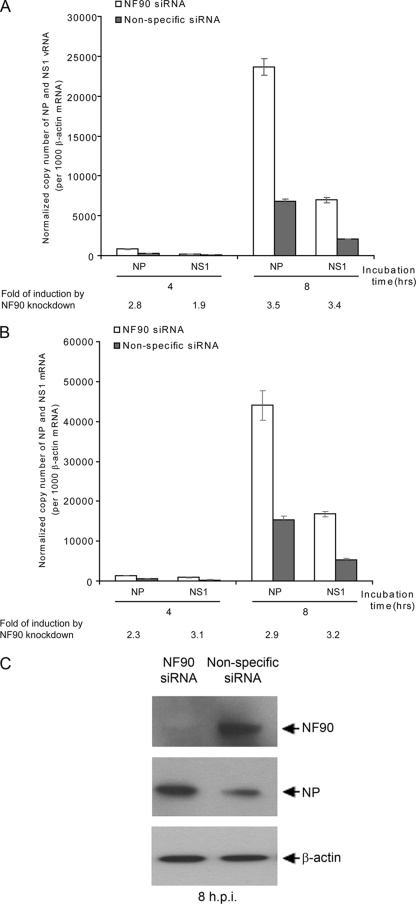
To further understand the mechanism by which NF90 inhibits influenza virus replication, we examined viral genome replication (vRNA) and transcription (mRNA) of NP and NS1 genes in H5N1 virus-infected A549 cells, with or without NF90 knockdown, by using quantitative RT-PCR. As shown in Fig. 6, both vRNA and mRNA levels were increased (two- to threefold) in A549 cells with NF90 knockdown at 4 and 8 h postinfection (Fig. 6A and B). Western blot analysis for NP expression in virus-infected NF90 knockdown A549 cells also showed increased expression of viral protein (∼4-fold) at 8 h postinfection (Fig. 6C). Experiments with HeLa cells gave similar results (data not shown). These results further confirm that the negative effect of NF90 on virus replication involves inhibition of virus genome replication and transcription during the early phase of infection.
FIG. 6.
NF90 inhibits viral genome replication and gene expression. A549 cells were treated with NF90 specific or nonspecific siRNAs for 24 h, and cells were then infected with H5N1 virus (A/Vietnam/1194/04) at an MOI of 1, as described in Materials and Methods. NP and NS1 vRNA (A) and mRNA (B) levels in infected cells were estimated by quantitative RT-PCR at 4 and 8 h postinfection. (C) The expression of NP in virus-infected NF90 knockdown and control cells was compared by Western blot at 8 h postinfection.
DISCUSSION
Sporadic but continuous animal to human transmissions of avian H5N1 virus occurring in recent years have raised concern that another influenza pandemic may be in the making (1, 61). Experiments with a forced reassortant virus containing HA and NA genes from a human virus and internal genes from a 1997 avian H5N1 influenza virus strain found that the resultant virus exhibited poor transmission in ferrets, compared to H3N2 human influenza virus (34). This suggests that a lack of adaptation of internal viral elements, in addition to the constraints of receptor specificity, may impede transmission of avian H5N1 virus, including current strains, in mammalian hosts, including humans. Host factors are known to inhibit virus replication and may also serve as a barrier to limit adaptation of viruses from animals to humans (40, 63). The influenza virus polymerase complex interacts with host replication machinery to replicate and transcribe the viral genome and mRNA in the nucleus of infected cells. It has been suggested that viral polymerase complex plays a critical role in adaptation to a new host by circumventing host restriction barriers (17, 40). Host factors exhibiting anti-influenza effects have been extensively investigated for their interactions with the NS1 protein (30, 33, 38, 43, 44). However, host factors directly targeting the viral replication mechanism to limit virus replication are less well described. Several host factors that associate with viral RNP have been identified using either the yeast-two-hybrid system or MS analyses of host interacting proteins pulled down in Co-IP experiments (12, 15, 24, 28, 36, 39, 59). The majority of identified host factors are associated with viral protein/complex import and export processes (11, 18, 20). The influenza virus replication complex is comprised of NP and three polymerase subunits: PA, PB1, and PB2. Studies based on phylogenetic analyses of influenza viruses from different hosts found that NP has formed host specific lineages, and it is suggested to be a determinant for influenza virus host range (4, 54, 56, 62). Therefore, understanding the host factors that interact with NP may shed light on the mechanism of host range restriction and host adaptation of influenza viruses. Previous studies have mainly used human influenza virus to screen for host interacting factors; there is a possibility that some host factors interact differently with avian influenza viruses, including current H5N1 virus, and have not been identified. It is thus important to screen for host interacting factors using bona fide avian influenza H5N1 viral components.
In the present study, a host nuclear factor, NF90, was found to interact with NP of the H5N1 influenza virus strain VNM1194. NF90 was first identified to bind to the antigen response element (ARRE-2/NFAT) of the interleukin-2 promoter and activate transcription (29). It belongs to a superfamily of dsRNA-binding proteins that bind to double-stranded or highly structured single-stranded RNAs and have been implicated in cellular defense systems against foreign RNA or viruses (52, 58). NF90 has been shown to regulate gene expression by transcriptional control of mRNA processing and stability (58) and to interact with another dsRNA-binding protein, the protein kinase PKR, through RNA-dependent and RNA-independent mechanisms to become phosphorylated (45, 46). We have shown that NF90 interacts with NP and that the interaction requires functionality of the C-terminal region of NP and is independent of NP RNA binding. NF90 has been identified as exhibiting antiviral activity against HCV, human immunodeficiency virus and influenza virus, but mechanistic details remain elusive (2, 26, 27, 48). Immunostaining of NF90 and NP in virus-infected cells showed that these two proteins only colocalize in the nucleus during the early phase of virus infection, suggesting that the interaction between NF90 and NP occurs during virus replication in the nucleus. Furthermore, we found that H5N1 and H1N1 viruses replicate more efficiently in NF90 knockdown cells than in cells not treated with siRNA targeting NF90 (Fig. 5C). Therefore, NF90 may act as a negative regulator during the early stage of influenza virus replication. This notion was supported by evidence that both viral genome RNA (i.e., vRNA) and mRNA levels were increased after siRNA-mediated NF90 knockdown in A549 cells.
NP plays a central role in influenza virus replication and transcription (32). An X-ray crystallographic structure study of oligomeric NP has shown that the C-terminal tail of NP mediates interactions between NP molecules, while the N-terminal portion and central parts bind to viral RNA (64). Single-residue mutations in the C-terminal tail of NP lead to a complete loss of NP oligomerization (64). We found that the interactions between NF90 and F412A NP mutants were diminished in Co-IP assays (Fig. 2C). Because both F412 and R416 residues are critical for NP subunit interactions, it is possible that NF90 interacts with oligomers of NP rather than monomeric NP.
We further demonstrated that NF90 can interact with NP in both the presence and the absence of other viral polymerase complex subunits. Therefore, NF90 may interfere with viral genome replication and transcription processes through interaction with both non-RNP associated NP molecules and with NP in the context of the RNP complex. In NF90 knockdown HeLa cells, influenza virus polymerase complexes display higher activity than in cells without NF90 knockdown (Fig. 5A), suggesting that the inhibitory effect of NF90 may be due to interference with the function of viral polymerase complexes. It is possible that NF90 binds to the C-terminal of NP, interfering with NP-PB2 interactions, which also occur via the body domain of NP (40, 64). However, overexpression of NF90 in cells does not inhibit interaction between NP and PB2 (Fig. 2D), which may imply that NP interacts with NF90 independent of its interaction with PB2. Nevertheless, it remains possible that engagement of NF90 with NP may alter NP function in the RNP complex and affect the efficiency of viral polymerase complexes. The precise mechanism by which NF90 interaction with NP inhibits influenza virus replication requires further investigation. As mentioned above, NF90 has been identified as a substrate of phosphorylation by PKR (45, 46); it will be important to investigate whether the inhibition of virus replication resulting from the interaction of NF90 with NP is mediated through PKR-associated cellular signaling pathways.
Differential inhibitory activities against PB2 polymerases from avian and human type influenza viruses have been observed in human cells during the early phase of viral replication (37). We have provided evidence showing that NF90 interacts with NP in the cell nucleus and inhibits influenza virus genome replication and transcription during the early stage of virus infection. NF90 was observed to inhibit both avian and human influenza viruses; however, a comprehensive comparison is necessary to understand whether there are any differences in the interactions between NF90 and NP from avian and human influenza viruses.
Acknowledgments
This study was supported by the Research Grants Council of the Hong Kong Special Administrative Region (SAR), China (HKU 74888/05M and 7500/06M), the Area of Excellence Scheme of the University Grants Committee (grant AoE/M-12/06), the National Institutes of Health (NIAID contract HHSN2662007 00005C), the Research Fund for the Control of Infectious Diseases of the Health, Welfare and Food Bureau of the Hong Kong SAR, and the Li Ka Shing Foundation.
Footnotes
Published ahead of print on 3 June 2009.
REFERENCES
- 1.Abdel-Ghafar, A. N., T. Chotpitayasunondh, Z. Gao, F. G. Hayden, D. H. Nguyen, M. D. de Jong, A. Naghdaliyev, J. S. Peiris, N. Shindo, S. Soeroso, and T. M. Uyeki. 2008. Update on avian influenza A (H5N1) virus infection in humans. N. Engl. J. Med. 358261-273. [DOI] [PubMed] [Google Scholar]
- 2.Agbottah, E. T., C. Traviss, J. McArdle, S. Karki, G. C. St Laurent III, and A. Kumar. 2007. Nuclear factor 90 (NF90) targeted to TAR RNA inhibits transcriptional activation of HIV-1. Retrovirology 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baudin, F., C. Bach, S. Cusack, and R. W. Ruigrok. 1994. Structure of influenza virus RNP. I. Influenza virus nucleoprotein melts secondary structure in panhandle RNA and exposes the bases to the solvent. EMBO J. 133158-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bean, W. J. 1984. Correlation of influenza A virus nucleoprotein genes with host species. Virology 133438-442. [DOI] [PubMed] [Google Scholar]
- 5.Biswas, S. K., P. L. Boutz, and D. P. Nayak. 1998. Influenza virus nucleoprotein interacts with influenza virus polymerase proteins. J. Virol. 725493-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, E. G., H. Liu, L. C. Kit, S. Baird, and M. Nesrallah. 2001. Pattern of mutation in the genome of influenza A virus on adaptation to increased virulence in the mouse lung: identification of functional themes. Proc. Natl. Acad. Sci. USA 986883-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butt, K. M., G. J. Smith, H. Chen, L. J. Zhang, Y. H. Leung, K. M. Xu, W. Lim, R. G. Webster, K. Y. Yuen, J. S. Peiris, and Y. Guan. 2005. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J. Clin. Microbiol. 435760-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, L. M., C. T. Davis, H. Zhou, N. J. Cox, and R. O. Donis. 2008. Genetic compatibility and virulence of reassortants derived from contemporary avian H5N1 and human H3N2 influenza A viruses. PLoS Pathog. 4e1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, Y., K. Qin, W. L. Wu, G. Li, J. Zhang, H. Du, M. H. Ng, J. W. Shih, J. S. Peiris, Y. Guan, H. Chen, and N. Xia. 2009. Broad cross-protection against H5N1 avian influenza virus infection by means of monoclonal antibodies that map to conserved viral epitopes. J. Infect. Dis. 19949-58. [DOI] [PubMed] [Google Scholar]
- 10.Claas, E. C., A. D. Osterhaus, R. van Beek, J. C. De Jong, G. F. Rimmelzwaan, D. A. Senne, S. Krauss, K. F. Shortridge, and R. G. Webster. 1998. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet 351472-477. [DOI] [PubMed] [Google Scholar]
- 11.Deng, T., O. G. Engelhardt, B. Thomas, A. V. Akoulitchev, G. G. Brownlee, and E. Fodor. 2006. Role of ran binding protein 5 in nuclear import and assembly of the influenza virus RNA polymerase complex. J. Virol. 8011911-11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Digard, P., D. Elton, K. Bishop, E. Medcalf, A. Weeds, and B. Pope. 1999. Modulation of nuclear localization of the influenza virus nucleoprotein through interaction with actin filaments. J. Virol. 732222-2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elton, D., L. Medcalf, K. Bishop, D. Harrison, and P. Digard. 1999. Identification of amino acid residues of influenza virus nucleoprotein essential for RNA binding. J. Virol. 737357-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelhardt, O. G., and E. Fodor. 2006. Functional association between viral and cellular transcription during influenza virus infection. Rev. Med. Virol. 16329-345. [DOI] [PubMed] [Google Scholar]
- 15.Engelhardt, O. G., M. Smith, and E. Fodor. 2005. Association of the influenza A virus RNA-dependent RNA polymerase with cellular RNA polymerase II. J. Virol. 795812-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fouchier, R. A., P. M. Schneeberger, F. W. Rozendaal, J. M. Broekman, S. A. Kemink, V. Munster, T. Kuiken, G. F. Rimmelzwaan, M. Schutten, G. J. Van Doornum, G. Koch, A. Bosman, M. Koopmans, and A. D. Osterhaus. 2004. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc. Natl. Acad. Sci. USA 1011356-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gabriel, G., B. Dauber, T. Wolff, O. Planz, H. D. Klenk, and J. Stech. 2005. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc. Natl. Acad. Sci. USA 10218590-18595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gabriel, G., A. Herwig, and H. D. Klenk. 2008. Interaction of polymerase subunit PB2 and NP with importin α1 is a determinant of host range of influenza A virus. PLoS Pathog. 4e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ginsberg, M., J. Hopkins, A. Maroufi, et al. 2009. Swine-origin influenza A (H1N1) virus infections in a school—New York City, April 2009. MMWR Morb. Mortal. Wkly. Rep. 58470-472. [PubMed] [Google Scholar]
- 20.Hirayama, E., H. Atagi, A. Hiraki, and J. Kim. 2004. Heat shock protein 70 is related to thermal inhibition of nuclear export of the influenza virus ribonucleoprotein complex. J. Virol. 781263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann, E., G. Neumann, G. Hobom, R. G. Webster, and Y. Kawaoka. 2000. “Ambisense” approach for the generation of influenza A virus: vRNA and mRNA synthesis from one template. Virology 267310-317. [DOI] [PubMed] [Google Scholar]
- 22.Hoffmann, E., G. Neumann, Y. Kawaoka, G. Hobom, and R. G. Webster. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. USA 976108-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffmann, E., J. Stech, Y. Guan, R. G. Webster, and D. R. Perez. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 1462275-2289. [DOI] [PubMed] [Google Scholar]
- 24.Huarte, M., J. J. Sanz-Ezquerro, F. Roncal, J. Ortin, and A. Nieto. 2001. PA subunit from influenza virus polymerase complex interacts with a cellular protein with homology to a family of transcriptional activators. J. Virol. 758597-8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hui, K. P., S. M. Lee, C. Y. Cheung, I. H. Ng, L. L. Poon, Y. Guan, N. Y. Ip, A. S. Lau, and J. S. Peiris. 2009. Induction of proinflammatory cytokines in primary human macrophages by influenza A virus (H5N1) is selectively regulated by IFN regulatory factor 3 and p38 MAPK. J. Immunol. 1821088-1098. [DOI] [PubMed] [Google Scholar]
- 26.Isken, O., M. Baroth, C. W. Grassmann, S. Weinlich, D. H. Ostareck, A. Ostareck-Lederer, and S. E. Behrens. 2007. Nuclear factors are involved in hepatitis C virus RNA replication. RNA 131675-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isken, O., C. W. Grassmann, R. T. Sarisky, M. Kann, S. Zhang, F. Grosse, P. N. Kao, and S. E. Behrens. 2003. Members of the NF90/NFAR protein group are involved in the life cycle of a positive-strand RNA virus. EMBO J. 225655-5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jorba, N., S. Juarez, E. Torreira, P. Gastaminza, N. Zamarreno, J. P. Albar, and J. Ortin. 2008. Analysis of the interaction of influenza virus polymerase complex with human cell factors. Proteomics 82077-2088. [DOI] [PubMed] [Google Scholar]
- 29.Kao, P. N., L. Chen, G. Brock, J. Ng, J. Kenny, A. J. Smith, and B. Corthesy. 1994. Cloning and expression of cyclosporin A- and FK506-sensitive nuclear factor of activated T cells: NF45 and NF90. J. Biol. Chem. 26920691-20699. [PubMed] [Google Scholar]
- 30.Krug, R. M., W. Yuan, D. L. Noah, and A. G. Latham. 2003. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology 309181-189. [DOI] [PubMed] [Google Scholar]
- 31.Labadie, K., E. Dos Santos Afonso, M. A. Rameix-Welti, S. van der Werf, and N. Naffakh. 2007. Host-range determinants on the PB2 protein of influenza A viruses control the interaction between the viral polymerase and nucleoprotein in human cells. Virology 362271-282. [DOI] [PubMed] [Google Scholar]
- 32.Lamb, R. A., and R. M. Krug. 2001. Orthomyxoviridae: the viruses and their replication, p. 1487-1531. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 33.Li, S., J. Y. Min, R. M. Krug, and G. C. Sen. 2006. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology 34913-21. [DOI] [PubMed] [Google Scholar]
- 34.Maines, T. R., L. M. Chen, Y. Matsuoka, H. Chen, T. Rowe, J. Ortin, A. Falcon, T. H. Nguyen, Q. Mai le, E. R. Sedyaningsih, S. Harun, T. M. Tumpey, R. O. Donis, N. J. Cox, K. Subbarao, and J. M. Katz. 2006. Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc. Natl. Acad. Sci. USA 10312121-12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matrosovich, M. N., S. Krauss, and R. G. Webster. 2001. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology 281156-162. [DOI] [PubMed] [Google Scholar]
- 36.Mayer, D., K. Molawi, L. Martinez-Sobrido, A. Ghanem, S. Thomas, S. Baginsky, J. Grossmann, A. Garcia-Sastre, and M. Schwemmle. 2007. Identification of cellular interaction partners of the influenza virus ribonucleoprotein complex and polymerase complex using proteomic-based approaches. J. Proteome Res. 6672-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehle, A., and J. A. Doudna. 2008. An inhibitory activity in human cells restricts the function of an avian-like influenza virus polymerase. Cell Host Microbe 4111-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Min, J. Y., and R. M. Krug. 2006. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: inhibiting the 2′-5′ oligo(A) synthetase/RNase L pathway. Proc. Natl. Acad. Sci. USA 1037100-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Momose, F., T. Naito, K. Yano, S. Sugimoto, Y. Morikawa, and K. Nagata. 2002. Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J. Biol. Chem. 27745306-45314. [DOI] [PubMed] [Google Scholar]
- 40.Naffakh, N., A. Tomoiu, M. A. Rameix-Welti, and S. van der Werf. 2008. Host restriction of avian influenza viruses at the level of the ribonucleoproteins. Annu. Rev. Microbiol. 62403-424. [DOI] [PubMed] [Google Scholar]
- 41.Nagata, K., A. Kawaguchi, and T. Naito. 2008. Host factors for replication and transcription of the influenza virus genome. Rev. Med. Virol. 18247-260. [DOI] [PubMed] [Google Scholar]
- 42.Naito, T., Y. Kiyasu, K. Sugiyama, A. Kimura, R. Nakano, A. Matsukage, and K. Nagata. 2007. An influenza virus replicon system in yeast identified Tat-SF1 as a stimulatory host factor for viral RNA synthesis. Proc. Natl. Acad. Sci. USA 10418235-18240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nemeroff, M. E., S. M. Barabino, Y. Li, W. Keller, and R. M. Krug. 1998. Influenza virus NS1 protein interacts with the cellular 30-kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol. Cell 1991-1000. [DOI] [PubMed] [Google Scholar]
- 44.Opitz, B., A. Rejaibi, B. Dauber, J. Eckhard, M. Vinzing, B. Schmeck, S. Hippenstiel, N. Suttorp, and T. Wolff. 2007. IFNbeta induction by influenza A virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell Microbiol. 9930-938. [DOI] [PubMed] [Google Scholar]
- 45.Parker, L. M., I. Fierro-Monti, and M. B. Mathews. 2001. Nuclear factor 90 is a substrate and regulator of the eukaryotic initiation factor 2 kinase double-stranded RNA-activated protein kinase. J. Biol. Chem. 27632522-32530. [DOI] [PubMed] [Google Scholar]
- 46.Patel, R. C., D. J. Vestal, Z. Xu, S. Bandyopadhyay, W. Guo, S. M. Erme, B. R. Williams, and G. C. Sen. 1999. DRBP76, a double-stranded RNA-binding nuclear protein, is phosphorylated by the interferon-induced protein kinase, PKR. J. Biol. Chem. 27420432-20437. [DOI] [PubMed] [Google Scholar]
- 47.Peiris, M., K. Y. Yuen, C. W. Leung, K. H. Chan, P. L. Ip, R. W. Lai, W. K. Orr, and K. F. Shortridge. 1999. Human infection with influenza H9N2. Lancet 354916-917. [DOI] [PubMed] [Google Scholar]
- 48.Pfeifer, I., R. Elsby, M. Fernandez, P. A. Faria, D. R. Nussenzveig, I. S. Lossos, B. M. Fontoura, W. D. Martin, and G. N. Barber. 2008. NFAR-1 and -2 modulate translation and are required for efficient host defense. Proc. Natl. Acad. Sci. USA 1054173-4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rameix-Welti, M. A., A. Tomoiu, E. Dos Santos Afonso, S. van der Werf, and N. Naffakh. 2009. Avian Influenza A virus polymerase association with nucleoprotein, but not polymerase assembly, is impaired in human cells during the course of infection. J. Virol. 831320-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reichman, T. W., A. M. Parrott, I. Fierro-Monti, D. J. Caron, P. N. Kao, C. G. Lee, H. Li, and M. B. Mathews. 2003. Selective regulation of gene expression by nuclear factor 110, a member of the NF90 family of double-stranded RNA-binding proteins. J. Mol. Biol. 33285-98. [DOI] [PubMed] [Google Scholar]
- 51.Salomon, R., J. Franks, E. A. Govorkova, N. A. Ilyushina, H. L. Yen, D. J. Hulse-Post, J. Humberd, M. Trichet, J. E. Rehg, R. J. Webby, R. G. Webster, and E. Hoffmann. 2006. The polymerase complex genes contribute to the high virulence of the human H5N1 influenza virus isolate A/Vietnam/1203/04. J. Exp. Med. 203689-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saunders, L. R., and G. N. Barber. 2003. The dsRNA binding protein family: critical roles, diverse cellular functions. FASEB J. 17961-983. [DOI] [PubMed] [Google Scholar]
- 53.Saunders, L. R., D. J. Perkins, S. Balachandran, R. Michaels, R. Ford, A. Mayeda, and G. N. Barber. 2001. Characterization of two evolutionarily conserved, alternatively spliced nuclear phosphoproteins, NFAR-1 and -2, that function in mRNA processing and interact with the double-stranded RNA-dependent protein kinase, PKR. J. Biol. Chem. 27632300-32312. [DOI] [PubMed] [Google Scholar]
- 54.Scholtissek, C., H. Burger, O. Kistner, and K. F. Shortridge. 1985. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology 147287-294. [DOI] [PubMed] [Google Scholar]
- 55.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69531-569. [DOI] [PubMed] [Google Scholar]
- 56.Snyder, M. H., A. J. Buckler-White, W. T. London, E. L. Tierney, and B. R. Murphy. 1987. The avian influenza virus nucleoprotein gene and a specific constellation of avian and human virus polymerase genes each specify attenuation of avian-human influenza A/Pintail/79 reassortant viruses for monkeys. J. Virol. 612857-2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taubenberger, J. K., J. V. Hultin, and D. M. Morens. 2007. Discovery and characterization of the 1918 pandemic influenza virus in historical context. Antivir. Ther. 12581-591. [PMC free article] [PubMed] [Google Scholar]
- 58.Tian, B., P. C. Bevilacqua, A. Diegelman-Parente, and M. B. Mathews. 2004. The double-stranded-RNA-binding motif: interference and much more. Nat. Rev. Mol. Cell. Biol. 51013-1023. [DOI] [PubMed] [Google Scholar]
- 59.Wang, P., P. Palese, and R. E. O'Neill. 1997. The NPI-1/NPI-3 (karyopherin alpha) binding site on the influenza a virus nucleoprotein NP is a nonconventional nuclear localization signal. J. Virol. 711850-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Watanabe, T., S. Watanabe, K. Shinya, J. Hyun Kim, M. Hatta, and Y. Kawaoka. 2009. Viral RNA polymerase complex promotes optimal growth of 1918 virus in the lower respiratory tract of ferrets. Proc. Natl. Acad. Sci. USA 106588-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Webby, R. J., and R. G. Webster. 2003. Are we ready for pandemic influenza? Science 3021519-1522. [DOI] [PubMed] [Google Scholar]
- 62.Webster, R. G., W. J. Bean, O. T. Gorman, T. M. Chambers, and Y. Kawaoka. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56152-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wolf, D., and S. P. Goff. 2008. Host restriction factors blocking retroviral replication. Annu. Rev. Genet. 42143-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ye, Q., R. M. Krug, and Y. J. Tao. 2006. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature 4441078-1082. [DOI] [PubMed] [Google Scholar]
- 65.Yuen, K. Y., P. K. Chan, M. Peiris, D. N. Tsang, T. L. Que, K. F. Shortridge, P. T. Cheung, W. K. To, E. T. Ho, R. Sung, and A. F. Cheng. 1998. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet 351467-471. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, Z., J. Zhang, K. Huang, K. S. Li, K. Y. Yuen, Y. Guan, H. Chen, and W. F. Ng. 2009. Systemic infection of avian influenza A virus H5N1 subtype in human. Hum. Pathol. 40735-739. [DOI] [PMC free article] [PubMed] [Google Scholar]