Abstract
The histone chaperone Vps75 forms a complex with, and stimulates the activity of, the histone acetyltransferase Rtt109. However, Vps75 can also be isolated on its own and might therefore possess Rtt109-independent functions. Analysis of epistatic miniarray profiles showed that VPS75 genetically interacts with factors involved in transcription regulation whereas RTT109 clusters with genes linked to DNA replication/repair. Additional genetic and biochemical experiments revealed a close relationship between Vps75 and RNA polymerase II. Furthermore, Vps75 is recruited to activated genes in an Rtt109-independent manner, and its genome-wide association with genes correlates with transcription rate. Expression microarray analysis identified a number of genes whose normal expression depends on VPS75. Interestingly, histone H2B dynamics at some of these genes are consistent with a role for Vps75 in histone H2A/H2B eviction/deposition during transcription. Indeed, reconstitution of nucleosome disassembly using the ATP-dependent chromatin remodeler Rsc and Vps75 revealed that these proteins can cooperate to remove H2A/H2B dimers from nucleosomes. These results indicate a role for Vps75 in nucleosome dynamics during transcription, and importantly, this function appears to be largely independent of Rtt109.
Eukaryotic DNA is packaged into chromatin, comprised of repeating units of nucleosomes each containing two histone H2A-H2B dimers and one histone H3-H4 tetramer around which 147 bp of DNA are wrapped (29). The compact nature of chromatin severely impinges on processes occurring on DNA, such as replication, repair, and transcription. For transcription, several important mechanisms for relieving this inhibition exist, including covalent modification of histones, incorporation of histone variants, and the action of ATP-dependent chromatin remodelers (34). In recent years, a more drastic method to enable efficient transcriptional initiation and elongation has emerged: nucleosomes can be completely disassembled at promoters and, to a lesser extent, within the coding region of genes (70, 73). Nucleosome eviction occurs at most active genes and is often proportional to the transcription rate (5, 33, 76). In contrast to the case with promoters, loss of chromatin structure in the body of genes is highly transient, and in the wake of RNA polymerase II passage, nucleosomes are rapidly reassembled. This process is required to prevent the transcription machinery from having inappropriate access to cryptic sites of initiation within genes, which can result in spurious transcriptional events (36).
Originally thought to be simple histone binding proteins involved in intracellular histone movement and storage and in replication-associated chromatin assembly, histone chaperones are now known to also be involved in all aspects of transcription-related chromatin dynamics (47). This includes nucleosome disassembly at promoters (1, 2, 68) and within coding regions (57), as well as nucleosome reassembly in the wake of RNA polymerase II (RNAPII) passage to prevent internal transcription initiation (22, 25, 39, 57). Furthermore, histone chaperones are likely to promote enrichment of certain histone variants involved in transcription: for example, Chz1 preferentially binds H2A.Z-H2B over H2A-H2B dimers and may be involved in deposition of this histone complex into nucleosomes flanking nucleosome-free promoters throughout the yeast genome (38). In addition, it has been suggested that mammalian HIRA has a preference for the major histone variant H3.3, which is a key substrate for replication-independent chromatin assembly (65). Recent evidence also implicates histone chaperones in covalent modification of histones. For example, nucleosome assembly protein (NAP)-domain containing factors are able to stimulate the histone acetyltransferase (HAT) activity of the p300 coactivator complex (61). Conversely, TEF1/SET, another NAP family member, is a member of the INHAT complex, which inhibits the HAT activity of p300 and PCAF (60). Finally, histone chaperones appear to function in combination with ATP-dependent chromatin-remodeling factors, for example, to promote nucleosome disassembly (37, 68). The importance of these factors is highlighted by their apparent redundancy: in yeast, many nonessential histone chaperones have multiple, overlapping roles in transcription (for a review, see reference 47).
The histone chaperone Vps75 was originally identified in global genetic screens to identify factors involved in vacuolar sorting (6) and telomere maintenance (3). Sequence comparisons revealed that this gene belonged to the NAP domain family of histone chaperones, and its product was shown to bind histones and assemble nucleosomes in vitro (59). The cellular functions of Vps75 have also begun to be elucidated. Of particular note, this protein forms a complex with, and stimulates the activity of, Rtt109, the HAT that was identified as the elusive modifier of lysine 56 on histone H3 (11, 17, 67). This modification occurs on newly synthesized histone H3 molecules and is required for chromatin assembly during DNA replication and repair (8, 35). In addition, acetylation of histone 3 lysine 56 (H3K56ac) plays important roles in telomeric silencing (40, 75) and transcriptional activation (52, 69, 74). However, despite strongly stimulating H3K56ac in vitro, deletion of VPS75 does not significantly affect bulk H3K56ac in vivo or result in sensitivity to drugs affecting replication, phenotypes that are typical of cells lacking Rtt109 (11, 59). These data suggest that Vps75 may have alternative cellular roles, an idea that was supported by two recent reports describing a catalytic function for the Rtt109/Vps75 complex in acetylation of lysine 9 on histone H3 (4, 14).
In this study, we used an unbiased genetic approach as a basis to more thoroughly investigate the function of the Vps75 histone chaperone. This strategy revealed numerous interactions between VPS75 and genes encoding factors involved in transcription. Genomic and biochemical assays were then used to show that Vps75 mediates transcription-associated histone exchange, a function that appears to be independent of Rtt109-mediated histone acetylation. Mechanistically, Vps75-mediated histone exchange might occur in combination with ATP-dependent chromatin remodeling, since Vps75 and the well-known chromatin remodeler RSC can induce partial disassembly of nucleosomes in vitro. These data highlight a new role for Vps75 in transcription, distinct from its function as a cofactor for the Rtt109 HAT.
MATERIALS AND METHODS
Yeast manipulation.
Genotypes of Saccharomyces cerevisiae strains used in this study are listed in Table 1. Tagging and deletion of genes was done using standard yeast genetic methods (details are available on request). Epistatic miniarray profile (E-MAP) analysis was performed as described previously (55). To test for sensitivity to acetic acid, yeast strains based on BY4741 were used. To analyze intragenic transcription of the GAL::FLO8-HIS3 gene, deletions of VPS75 and RTT109 were made in strain FY2452 (44). 5′:3′ RNA ratios were measured in vps75Δ and rtt109Δ strains based on W303-1A. G1 arrest was achieved by addition of α-factor (1 μg/ml) for 2 h.
TABLE 1.
Saccharoymyces cerevisiae strains used in this study
| Name | Genotype | Source or reference |
|---|---|---|
| W303 1A | MATaleu2-3,112 his3-11,15 ade2-1 ura3-1 trp1-1 can1-1 | |
| JSY1099 | W303 1A VPS75-8His-2TEV-9Myc::URA3 | 59 |
| JSY1082 | W303 1A 8His-2TEV-9Myc-VPS75::URA3 | This study |
| JSY1083 | W303 1A 8His-2TEV-9Myc-VPS75::URA3 rtt109Δ::HIS3 | This study |
| JSY1076 | W303 1A vps75Δ::HIS3 | 59 |
| PHY2193 | MATahis3-Δ200 leu2-3,112 rpb1Δ187::HIS3 ura3-52 (pC6; rpb1-104 and pRS416) | 21 |
| JSY1101 | MATahis3-Δ200 leu2-3,112 rpb1Δ187::HIS3 ura3-52 (pC6; rpb1-104 and pRS416) vps75Δ::TRP1 | This study |
| JSY110 | W303 1A dst1Δ::HIS3 | J. Fellows |
| JSY1086 | W303 1A dst1Δ::HIS3 vps75Δ::TRP | This study |
| JSY1102 | W303 1A rtt109Δ::HIS3 | This study |
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | 71 |
| BY4741 vps75Δ | BY4741 vps75Δ::KANMX4 | 71 |
| BY4741 haa1Δ | BY4741 haa1Δ::KANMX4 | 71 |
| BY4741 vps75Δhaa1Δ | BY4741 haa1Δ::KANMX4 vps75Δ::URA3 | This study |
| FY2452 | MATaura3-52 his3 RPB3-HA1::LEU2 KANMX6-PGAL1-FLO8-HIS3 | 44 |
| FY2445 | MATaura3-52 his3 RPB3-HA1::LEU2 spt2::KANMX6 KANMX6-PGAL1-FLO8-HIS3 SPT6-FLAG | 44 |
| JSY1100 | MATaura3-52 his3 RPB3-HA1::LEU2 KANMX6-PGAL1-FLO8-HIS3 vps75Δ::URA3 | This study |
| JSY1103 | MATaura3-52 his3 RPB3-HA1::LEU2 KANMX6-PGAL1-FLO8-HIS3 rtt109Δ::URA3 | This study |
ChIP.
Chromatin immunoprecipitation (ChIP) analyses were performed as described previously (59). Chromatin was immunoprecipitated with 9E10 (monoclonal antibody against the Myc tag of Vps75), 12CA5 (monoclonal antibody against the hemagglutinin (HA) tag of histones H2B and H3), and polyclonal antibodies specific for the carboxy terminus of histone H3 (a kind gift of Alain Verreault) or for histone H3 acetylated at lysine 56 (Upstate). To measure recruitment of Vps75 to induced genes, the amount of precipitated DNA was divided by the amount in the input sample and corrected to a nontranscribed region (telomeric DNA on chromosome 6) and, in most cases, the equivalent signal from an untagged strain. Primer sequences are available on request.
To determine the genome-wide distribution of Vps75 (ChIP-chip), chromatin immunoprecipitates were prepared and hybridized to Affymetrix S. cerevisiae whole-genome forward tiling arrays (Tiling 1.0F array; P/N 520286) as described previously (45). Two replicates were hybridized to separate whole-genome tiling arrays and normalized to the signals obtained from their own respective supernatant (unbound fraction) controls (also hybridized on separate tiling arrays).
Microarray and RNA analyses.
Cells were grown in yeast extract-peptone-dextrose (YPD) medium to 1 × 107 cells/ml at 30°C. RNA was prepared using a Qiagen RNeasy minikit, labeled with One Cycle target labeling (Affymetrix), and hybridized to oligonucleotide arrays (GeneChip Yeast Genome 2.0 arrays; Affymetrix) using standard techniques. Three independent experiments were performed and an average change in expression calculated.
For determination of 5′:3′ RNA ratios at various genes in mutant and wild-type yeast strains, RNA was extracted as described above and quantitated by reverse transcriptase PCR using the ABsolute QPCR SYBR green reagent (ABgene) with Multiscribe reverse transcriptase (Applied Biosystems) and a Bio-Rad MyIQ iCycler. Primer sequences are available on request.
Nucleosome disassembly assays.
Nucleosome disassembly assays were carried out as described previously (37). His-tagged Vps75 and Nap1 proteins were prepared as described previously (67, 72). The Vps75-His expression vector was a kind gift from P. Kaufman.
Microarray data accession number.
Microarray data (MIAME compliant) are available at http://bioinformatics.picr.man.ac.uk/vice/ExternalReview.vice?k=kIa2QDT0u51tX4IShdNgz8ZB8rPdQErmOq91xD1ROtFjULbx4Q8rgHVoBngxfus4yWcTKjqbgOjm%0D%0A4aujRwrhww%3D%3D. ChIP-chip data were deposited in NCBI's Gene Expression Omnibus (12) and are accessible through GEO Series accession number GSE15607 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE15607).
RESULTS
VPS75 genetically interacts with RNAPII and other factors involved in transcription and chromatin function.
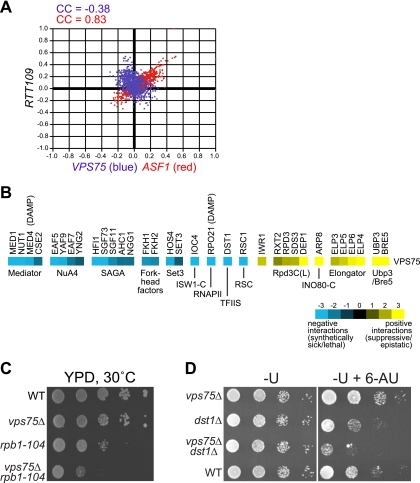
Almost all research on Vps75 to date has focused on the mechanism by which it regulates Rtt109-mediated histone acetylation (14, 18, 19, 67), but little is known about whether it plays other roles in the cell. To gain a more complete understanding of the function of VPS75, we analyzed a high-density E-MAP generated using synthetic genetic array technology (9). Each gene on the E-MAP possesses a genetic interaction profile that describes its interactions with all other genes on the map. The E-MAP profile thus provides a high-resolution phenotype, and functionally related genes often have similar interaction profiles. Interestingly, even though Rtt109 and Vps75 are physically associated, the corresponding mutants gave very different genetic interaction profiles that were in fact weakly anticorrelated (Pearson correlation coefficient [CC] = −0.38) (Fig. 1A). It has previously been shown (9) that deletion of RTT109 gives a profile that is similar to those seen with mutants known to function in DNA repair/replication (e.g., for ASF1, CC = 0.838) (Fig. 1A). In contrast, VPS75 generally interacts with factors linked to transcriptional regulation, as shown by clustering of the most strongly interacting genes by their gene ontology term (Table 2). For example, deletion of VPS75 was associated with significant growth defects when combined with mutations in components of the Mediator complex, the SAGA and NuA4 HATs and SET3 histone deacetylase, the RSC and ISW1B ATP-dependent chromatin remodeling complexes, and forkhead transcription factors, which influence the elongation phase of transcription (Fig. 1B) (41). Strong negative interactions were also observed with a DAmP allele of RPO21 (54), encoding the largest subunit of RNAPII, and with DST1, which encodes the archetypal transcription elongation factor TFIIS. Conversely, the most striking positive interaction (e.g., suppression) was observed with mutants of the Elongator HAT complex, which functions in such diverse processes as transcription and tRNA modification (64). Other transcription-related genes displaying positive genetic interactions with VPS75 included UBP3 and BRE5, which encode factors involved in RNAPII deubiquitination (32), members of the Rpd3C(L) histone deacetylase complex, a component of the INO80 chromatin remodeling complex (ARP8), and IWR1, a factor of unknown function which interacts with RNAPII (31) (Fig. 1B). Collectively, these genetic data suggest that Vps75 directly functions in pathways contributing to transcriptional control.
FIG. 1.
VPS75 interacts genetically with genes involved in transcription. (A) Scatter plot of the CCs of vps75Δ, asf1Δ, and rtt109Δ with all genetic profiles from the chromosome function E-MAP (9). The profile of RTT109 correlates with that of ASF1 (red) but not that of VPS75 (blue). (B) A subset of the genetic interaction profile for VPS75 as determined by E-MAP analysis, showing strongly interacting genes involved in transcription. Blue and yellow represent negative and positive genetic interactions, respectively. The DAmP (decreased abundance by mRNA perturbation) technique was used to generate hypomorphic alleles of essential genes (54). (C) VPS75 interacts genetically with a conditional allele of the gene encoding Rpb1, rpb1-104. Serial dilutions of the indicated strains were grown on YPD at 30°C for 2 days. (D) Deletion of VPS75 enhances the 6-AU sensitivity of a dst1Δ strain. Serial dilutions of the indicated strains were grown on medium lacking uracil (−U) or −U containing 50 μg/ml 6-AU (−U + 6-AU) at 30°C for 2 days.
TABLE 2.
Five most statistically overrepresented gene ontology (GO) terms scored by GOstat analysis of top 100 genes (top 50 positive and top 50 negative) interacting with VPS75 in an epistatic miniarray profile
| GO term | GO category | No. of genesa | P value |
|---|---|---|---|
| Chromosome organization | GO:0051276 | 41 | 1.45e−26 |
| Transcription, DNA dependent | GO:0006351 | 38 | 3.29e−25 |
| RNA biosynthetic process | GO:0032774 | 38 | 3.29e−25 |
| Transcription from RNA polymerase II promoter | GO:0006366 | 30 | 3.32e−25 |
| Transcription | GO:0006350 | 38 | 1.41e−22 |
Number of genes (out of 100) present in gene ontology group.
To verify the E-MAP data and to further investigate the putative function of Vps75 in transcription, we examined two of the interactions in more detail by performing growth assays. In the first experiment, a deletion of VPS75 was constructed in the slow-growing rpb1-104 strain, which expresses a modified Rpb1 protein containing just 11 repeats of the C-terminal domain heptad repeat (YSPTSPS) (43). Even though deletion of VPS75 itself had little or no consequence for growth, it negatively affected the growth of rpb1-104 such that the vps75Δ rpb1-104 double mutant grew more slowly than either single mutant alone (Fig. 1C). We next tested the functional relationship between VPS75 and DST1. Deletion of DST1 (encoding TFIIS) causes sensitivity to 6-azauracil (6-AU), a drug that depletes intracellular nucleotide pools and thereby inhibits transcript elongation (42). Interestingly, even though the vps75Δ single mutant grew normally on 6-AU, a vps75Δ dst1Δ double mutant exhibited significantly enhanced sensitivity to 6-AU compared to that of the dst1Δ single mutant (Fig. 1D). These results confirm and extend the E-MAP data and further indicate that VPS75 influences transcription by RNAPII.
Vps75 is recruited to genes following their activation.
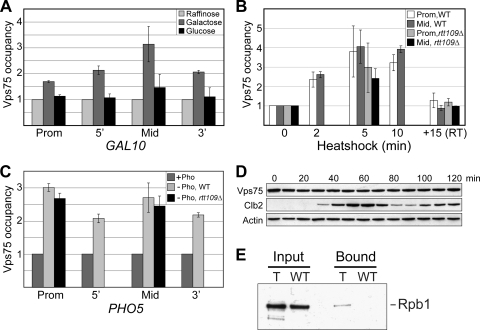
Previous work showed that Vps75 is associated with chromatin at both active and inactive genes and at telomeric DNA (59), potentially pointing to an involvement in multiple chromatin-related processes, including replication, telomere silencing, and transcription. To more specifically examine its putative role in transcription, the presence of Vps75 at GAL10, PHO5, and HSP104 before and after gene induction was determined by ChIP using a strain expressing Myc-tagged Vps75 from its native chromosomal location. Vps75 was detected at each of these genes even under repressive conditions (59) (data not shown). Upon induction of GAL10 by addition of galactose, however, Vps75 density in both the promoter and open reading frame (ORF) increased markedly (Fig. 2A). A similar increase in density was also consistently observed at HSP104 after heat shock (Fig. 2B) and at PHO5 in response to phosphate starvation (Fig. 2C), suggesting that Vps75 recruitment to genes following transcriptional activation is a general phenomenon. Furthermore, the rapid rate of Vps75 recruitment to GAL10 and HSP104 was comparable to that previously reported for RNAPII (57, 58), suggesting that Vps75 is directly involved in the transcription process. This idea was reinforced by the observation that rerepression of GAL10 (by glucose addition) and HSP104 (by reverting to growth at room temperature) caused a loss of Vps75 from these genes to background levels (Fig. 2A and B, respectively).
FIG. 2.
Vps75 is physically associated with sites of active transcription, and its expression remains constant throughout the cell cycle. (A) Vps75 is recruited to GAL10 following galactose induction. Cells expressing a tagged form of Vps75 were grown in medium containing raffinose at 30°C, and GAL10 was induced by the addition of galactose for 1 h. Glucose was then added and the cells grown for a further 20 min to repress GAL10. The occupancy of Vps75 at the promoter (prom), 5′ end (5′), middle (mid), and 3′ end (3′) of GAL10 in raffinose, galactose, or glucose was measured by ChIP followed by quantitative PCR. Occupancy in raffinose was set to 1. Values shown are the averages, with standard errors, of four independent experiments. (B) Vps75 is recruited to HSP104 following heat shock. Wild-type (WT) or rtt109Δ cells expressing a tagged form of Vps75 were grown in YPD at 30°C, switched to 39°C for 10 min to induce HSP104, and then returned to room temperature for 15 min [+15 (RT)] to repress the gene. At the indicated time points, cells were cross-linked and the level of Vps75 at the HSP104 promoter (Prom) or ORF (Mid) was measured by ChIP followed by quantitative PCR. Occupancy prior to heat shock (0 min) was set to 1. Values shown are the averages, with standard errors, of three independent experiments. (C) Vps75 is recruited to PHO5 following induction by phosphate starvation. Wild-type (WT) or rtt109Δ cells expressing a tagged form of Vps75 were grown in YPD, spun down, and resuspended in either YPD (+Pho) or medium lacking phosphate (−Pho). After 2 h of induction, the occupancy of Vps75 at the promoter (Prom), 5′ end (5′), middle (Mid), and 3′ end (3′) of PHO5 was measured as described above. Occupancy in YPD was set to 1. Values shown are the averages, with standard errors, of four independent experiments. (D) Expression of Vps75 does not change during the cell cycle. A strain expressing Vps75-Myc was arrested in G1 with α-factor and then released. Samples were taken at the indicated time points for Western analysis. Clb2 is shown as a cell cycle-regulated G2-M marker, and actin as a loading control. (E) Vps75 can be cross-linked to Rpb1. Cells from a Vps75-Myc (T) or untagged wild-type (WT) strain were cross-linked with formaldehyde and chromatin prepared as for ChIPs. Extracts were immunoprecipitated with 9E11 (anti-Myc) and coprecipitating Rpb1 identified by Western analysis following reversal of cross-links.
The finding that Vps75 is associated with sites of active transcription and can be rapidly recruited to a gene following induction prompted us to examine whether this occurs in the context of a Vps75/Rtt109 complex. Initially, we attempted to analyze the occupancy of TAP-tagged Rtt109 at the induced GAL10 and HSP104 genes by ChIP, but despite repeated attempts, a signal for Rtt109 above the background was never observed (data not shown). This is consistent with a previous study which reported no binding of Rtt109 to the promoter of the activated PHO5 gene (69) and also appears to be compatible with the finding that Rtt109 acetylates free, but not nucleosomal, histones (19, 67). As an alternative approach, RTT109 was therefore deleted in the strain expressing tagged Vps75 to investigate whether Vps75 recruitment requires Rtt109. Interestingly, Vps75 was recruited to HSP104 and PHO5 in the absence of Rtt109 (Fig. 2B and C), suggesting that the function of Vps75 at active genes may be unrelated to histone H3 acetylation and certainly that it does not require concomitant recruitment of Rtt109. There did appear to be slightly less Vps75 at these activated genes in the absence of Rtt109, but this was within the error of the experiment. However, we cannot completely rule out the possibility that recruitment of Vps75 following gene induction may be somewhat affected by Rtt109.
An alternative possibility is that Vps75 in a complex with Rtt109 is more stable than Vps75 alone, since others have shown that Rtt109 is degraded more quickly in the absence of Vps75 (14). To test this idea, we examined whether the expression of Vps75 changes during cell cycle progression. The rationale behind this experiment relates to the observation that the production of Rtt109 is tightly coordinated with the cell cycle so that it peaks just before S phase, ensuring that newly synthesized H3 molecules are maximally acetylated at K56 prior to deposition on replicated DNA (11). Thus, if Vps75 stability is linked to the presence of Rtt109, we might expect that Vps75 levels would increase coordinately with those of Rtt109. Cells were arrested in G1 with α-factor, and samples were then taken at different time points after release. Since an antibody to Rtt109 is currently unavailable, we used the cyclin Clb2 as a marker of cell cycle progression: this protein exhibits a peak of expression during G2-M, approximately 20 min after that of Rtt109 (11). Unlike Clb2 and Rtt109, the level of Vps75 remained stable throughout the cell cycle (Fig. 2D), suggesting that its stability is unaffected by complex formation with Rtt109 and showing that it must exist without Rtt109 much of the time. This is in agreement with the finding that the majority of Vps75 is purified without a partner (59) and further supports the hypothesis that Vps75 possesses functions distinct from its involvement in Rtt109-mediated histone acetylation.
Given its apparent involvement in transcription, we tested whether Vps75 interacts with RNAPII. After immunoprecipitating Vps75, we were unable to detect copurifying RNAPII (Rpb1) either before or after treatments that result in significant changes in overall cellular transcription (for example, galactose addition or heat shock) (data not shown). This suggests that these proteins do not directly interact. A recent study identified new factors involved in transcription by characterizing proteins copurifying with RNAPII after formaldehyde cross-linking (66). To determine if Vps75 could be similarly cross-linked to RNAPII, cells were treated with formaldehyde, and Vps75 was immunoprecipitated after extensive DNA sonication. Rpb1 was detected in the immunoprecipitates from cells expressing tagged Vps75 but not in a control extract from untagged cells (Fig. 2E). This indicates that Vps75 is in close proximity to RNAPII, such that it can be cross-linked to it either directly or via DNA, RNA, or other proteins. These data further support the idea that Vps75 is involved in RNAPII transcription.
Vps75 localizes primarily to sites of active transcription and is associated with higher levels of histone H3 lysine 9 (H3K9)/H3K56 acetylation.
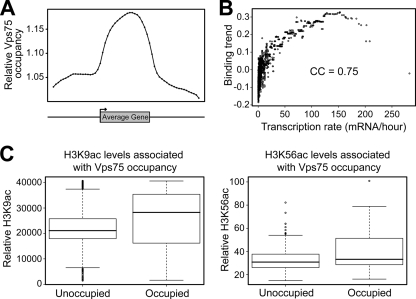
The apparent link between Vps75 and active transcription prompted us to examine its global distribution on chromatin in relation to genes. To achieve this, genome-wide localization studies by ChIP-chip analysis were performed. We found 895 different chromosomal loci enriched for Vps75 (see data set S1 in the supplemental material). Approximately 65% of these loci were in the coding region of protein-coding genes, supporting the idea that Vps75 is targeted to sites of transcription. A composite profile of Vps75 occupancy aligned according to the location of transcriptional start and termination sites at all protein-coding genes containing at least one Vps75 binding site confirmed that it is tightly associated with RNAPII transcription (Fig. 3A). Of the remaining peaks of Vps75 occupancy, we found an unexpectedly high number at tRNA genes (62, compared to 5.7 expected by chance alone) and at snRNA and snoRNA genes (12, compared to 2.4 expected), suggesting that Vps75 might participate in transcription by RNA polymerase III as well (see data set S1 in the supplemental material). In addition, peaks of Vps75 were underrepresented at telomeres (0, compared to 17.1 expected) and transposable elements (2, compared to 29.0 expected) (see data set S1 in the supplemental material).
FIG. 3.
Genome-wide analysis of Vps75 binding sites reveals an association with transcription and histone acetylation. (A) Composite profile of Vps75 occupancy (detected by ChIP) across the average gene. The ends of genes were defined at fixed points according to the positions of transcriptional start and termination sites. The gene and the flanking (upstream and downstream) regions were then subdivided into 20 regions each. For each of the 60 intervals, a mean Vps75 occupancy was calculated and plotted. (B) Vps75 occupancy correlates with the transcription rate genome-wide. The Vps75 binding trend was determined as described previously (44) and plotted against the transcription rate for all yeast genes (20). The Pearson CC is shown. (C) H3K9ac and H3K56ac levels are higher at sites of Vps75 occupancy. Box plots show histone H3K9 (left) and H3K56 (right) acetylation levels at Vps75-occupied and -unoccupied regions, generated by comparing peaks of Vps75 occupancy with published maps of histone acetylation (26, 50). The line in the center of each box represents the median value of the distribution, and the upper and lower ends of the box are the upper (25th) and lower (75th) quartiles, respectively.
Given that Vps75 is enriched at sites of active transcription, we hypothesized that its distribution across the genome might relate more closely to gene activity than to gene function. To test this idea, we analyzed the relationship between its genome-wide occupancy and the transcription rate for all RNAPII-transcribed yeast genes (20). Indeed, we calculated a strong correlation (Pearson CC = 0.75) between the transcription rate and Vps75 occupancy (Fig. 3B), providing further support for a general role in transcription by RNAPII.
Since others have shown that Vps75 facilitates H3K9ac in vivo and H3K56ac in vitro (4, 11, 14, 67), we compared genome-wide maps of these histone modifications (26, 50) to Vps75 occupancy. A Wilcoxon rank sum test was used to assess whether Vps75-occupied and -unoccupied regions showed differences in the levels of H3K9 and H3K56 acetylation. This analysis revealed significantly greater H3K9ac at sites of Vps75 occupancy (Fig. 3C) (P = 2.521 × 10−11). Given that Rtt109/Vps75 acetylates only nonnucleosomal histones (19), this may simply reflect the fact that Vps75 is found at highly transcribed genes, the chromatin of which is often hyperacetylated. However, it is also possible that this overlap exists because Vps75 facilitates acetylation of H3 immediately prior to its deposition onto chromatin. A minor but still statistically significant (P = 0.044) increase in H3K56ac was observed at peaks of Vps75 (Fig. 3C), suggesting a connection between Vps75 and this histone mark in vivo.
Transcriptional defects in the absence of Vps75.
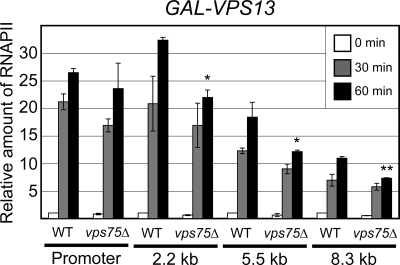
If Vps75 functions in transcription, we might expect that the absence of this factor would affect the expression level of a subset of genes. To test the potential effect of deleting VPS75 on transcription, we first analyzed RNAPII density in a long, galactose-inducible gene, GAL-VPS13 (30). As described previously, RNAPII is rapidly recruited to the promoter and is detected throughout the coding region of this transcription unit as the gene is activated (Fig. 4). In the absence of Vps75, a slight reduction in RNAPII levels was observed at the GAL-VPS13 promoter (Fig. 4). However, the decrease was more pronounced in the coding region of the gene. For example, after 60 min of activation, the density of RNAPII at the promoter in vps75 was approximately 89% of that for the wild type. However, in the coding region (at 2.2, 5.5, and 8.8 kb), it was only 68, 66, and 66%, respectively. This suggests that Vps75 can affect the movement of RNAPII along the chromatin of the GAL-VPS13 gene.
FIG. 4.
Induction of GAL-VPS13 is affected by Vps75. Recruitment of RNAPII to the promoter and through the coding region (2.2 kb, 5.5 kb, and 8.3 kb) of GAL-VPS13 was measured by ChIP assays followed by quantitative PCR in wild-type (WT) and vps75Δ cells after 0, 30, and 60 min of induction. Values shown are the averages, with standard errors, for three independent experiments and are expressed as the amount of immunoprecipitated DNA corrected to input DNA. The value for the wild-type strain at 0 min was set to 1 and other values expressed relative to that. An unpaired t test was used to determine the significance of the difference between mutant and WT cells at each location after 60 min, with one asterisk denoting a P value of <0.05 and two asterisks denoting a P value of <0.01.
The transcription of other inducible, shorter genes, including GAL10, PHO5, and HSP104, appears to be largely unaffected in cells lacking Vps75 (see below; also data not shown), possibly due to the high level of redundancy in the yeast histone chaperone system (47). The observation that VPS75 deletion has a very modest phenotype may also reflect the functional redundancy of these factors. To identify genes which require Vps75 for normal expression, the transcription profiles of wild-type and vps75Δ cells were compared using Affymetrix microarrays. The expression of 57 genes (approximately 1% of the genome) was altered by a factor of 2 or more (data not shown), while 30 genes were changed by at least 2.5-fold in vps75Δ (Table 3). Of these genes, the products of eight are yet to be experimentally verified and the function of most of the remainder is largely unknown. Interestingly, however, many of the differentially expressed genes are predicted to encode membrane proteins, including the most significantly upregulated gene, HSP30, which encodes the integral plasma membrane heat shock protein in yeast.
TABLE 3.
Genes differentially expressed between vps75Δ and wild-type yeast cells (change of ≥2.5-fold)
| ORF | Gene | Fold change in expressiona
|
Description | |||
|---|---|---|---|---|---|---|
| Expt 1 | Expt 2 | Expt 3 | Avg | |||
| Upregulated | ||||||
| YCR021C | HSP30 | 24.8 | 24.2 | 4.1 | 17.7 | Plasma membrane protein induced by various stresses |
| YDL037C | BSC1 | 21.7 | 13.2 | 2.4 | 12.4 | Transcript shows a high level of stop codon bypass |
| YGR249W | MGA1 | 13.5 | 15.4 | 2.1 | 10.3 | Similar to heat shock transcription factor |
| YDL038C | 13.8 | 13.1 | 3.1 | 10.0 | Putative protein of unknown function | |
| YGR052W | FMP48 | 9.2 | 9.2 | 2.4 | 6.9 | Unknown; localizes to mitochondria |
| YDL039C | PRM7 | 6.5 | 8.6 | 3.0 | 6.0 | Pheromone-regulated membrane protein |
| YPL014W | 6.6 | 7.7 | 3.2 | 5.8 | Putative protein of unknown function | |
| YGR138C | TPO2 | 5.8 | 6.1 | 3.3 | 5.1 | Polyamine transport protein of the major facilitator superfamily |
| YPR157W | 5.2 | 5.3 | 3.6 | 4.7 | Putative protein of unknown function | |
| YDL048C | STP4 | 3.9 | 4.5 | 3.1 | 3.8 | Kruppel-type zinc-finger-domain-containing protein |
| YER150W | SPI1 | 4.5 | 5.7 | 0.8 | 3.7 | GPI-anchored cell wall protein involved in weak acid resistance |
| YOR107W | RGS2 | 3.6 | 4.3 | 3.1 | 3.7 | Regulator of G-protein signaling for Gpa2 |
| YLR297W | 4.0 | 3.3 | 3.0 | 3.4 | Putative protein of unknown function | |
| YOL016C | CMK2 | 4.7 | 4.2 | 1.2 | 3.4 | Calmodulin-dependent protein kinase |
| YMR316W | DIA1 | 4.3 | 4.2 | 1.6 | 3.4 | Unknown, involved in invasive and pseudohyphal growth |
| YMR081C | ISF1 | 3.4 | 4.3 | 1.0 | 2.9 | Serine-rich, hydrophilic protein with similarity to Mbr1p |
| YOL014W | 3.4 | 3.5 | 1.0 | 2.7 | Putative protein of unknown function | |
| YER130C | 3.1 | 2.8 | 2.0 | 2.6 | Putative protein of unknown function | |
| YOR306C | MCH5 | 3.0 | 2.8 | 1.8 | 2.5 | Plasma membrane riboflavin transporter |
| YNL065W | AQR1 | 2.7 | 2.7 | 2.2 | 2.5 | Multidrug transporter of the major facilitator superfamily |
| YER037W | PHM8 | 2.7 | 2.8 | 2.0 | 2.5 | Unknown, possibly involved in phosphate metabolism |
| YOR032C | HMS1 | 3.3 | 2.8 | 1.4 | 2.5 | Basic helix-loop-helix protein |
| Downregulated | ||||||
| YHL048W | COS8 | −1.8 | −1.6 | −4.2 | −2.5 | Nuclear membrane protein, member of the DUP380 subfamily |
| YJR079W | −2.4 | −3.1 | −2.1 | −2.5 | Putative protein of unknown function | |
| YLR327C | TMA10 | −2.7 | −2.6 | −2.2 | −2.5 | Unknown; associates with ribosomes |
| YPR158W | −2.9 | −2.3 | −2.4 | −2.5 | Putative protein of unknown function | |
| YIL037C | PRM2 | −4.0 | −1.6 | −2.5 | −2.7 | Pheromone-regulated protein; regulated by Ste12p |
| YHR177W | −3.0 | −3.2 | −2.2 | −2.8 | Putative protein of unknown function | |
| YNL277W | MET2 | −3.0 | −3.4 | −3.2 | −3.2 | l-homoserine-O-acetyltransferase |
| YJR078W | BNA2 | −4.3 | −3.4 | −2.3 | −3.3 | Putative tryptophan 2,3-dioxygenase |
| YNL246W | VPS75 | −16.8 | −12.7 | −11.6 | −13.7 | NAP domain histone chaperone |
Three independent experiments (1 to 3) were done.
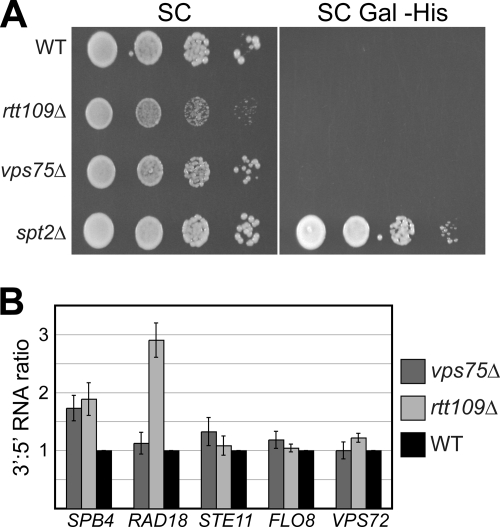
To verify the microarray results, we measured the expression of five of the affected genes by reverse transcription followed by quantitative PCR. This independent analysis confirmed the data obtained in the microarray study (Table 4). To determine whether the expression of these same genes was regulated by Rtt109, we also quantitated RNA isolated from rtt109Δ cells by reverse transcription followed by quantitative PCR. Of the five genes tested, four were unaffected while the expression of BSC1 (significantly upregulated in the absence of Vps75) was repressed in rtt109Δ cells (Table 4). This further highlights the distinct functions of Vps75 and Rtt109.
TABLE 4.
Validation of vps75Δ microarray data with reverse transcription and real-time (quantitative) PCR and comparison with expression in rtt109Δ cells
| Gene | Fold change in expression in:a
|
||
|---|---|---|---|
|
vps75Δ cells
|
rtt109Δ cells (QPCR) | ||
| Microarray | QPCR | ||
| HSP30 | 17.7 | 14.2 | 1.6 |
| BSC1 | 12.4 | 7.4 | 0.2 |
| MGA1 | 10.3 | 12.9 | 1.4 |
| BNA2 | −3.3 | −2.3 | 1.3 |
| MET2 | −3.2 | −2.5 | 1.2 |
Change in expression from that in wild-type cells. QPCR, real-time (quantitative) PCR.
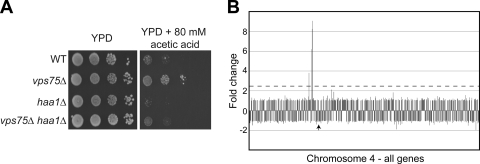
Bioinformatics analysis indicated that the Vps75 regulon is strikingly similar to that of Haa1, a transcriptional activator that controls numerous stress-related membrane proteins (27). However, whereas loss of HAA1 resulted in downregulation of these genes, the opposite was the case in vps75Δ cells. Five of the ten genes most significantly downregulated in haa1Δ cells, namely, YGR138C (TPO2), YPR157W, YLR297W, YER130C, and YER037W (PHM8), were upregulated by a factor of ≥2.5-fold in cells lacking VPS75. Moreover, expression of SPI1, another gene whose expression was increased in vps75Δ cells, was also shown to require Haa1 in a separate study (63). These data could potentially be explained if expression of HAA1 itself was increased in vps75Δ. However, HAA1 expression was largely unaltered (1.38-fold upregulated) in the mutant cells. Many of the genes controlled by Haa1, including those upregulated in response to VPS75 deletion, are required for adaptation to growth in the presence of weak acids (13, 63). Interestingly, the VPS75-regulated HSP30 gene is also induced by sorbic acid, in a manner that is likely to be independent of Haa1 (56). To test the idea that VPS75 is somehow involved in regulating the acid response, we analyzed the growth of cells lacking VPS75 or HAA1 on medium containing acetic acid. As expected, the haa1Δ strain was very sensitive to acetic acid, but in contrast, vps75Δ cells grew better than the wild-type control (Fig. 5A). Interestingly, a vps75Δ haa1Δ double mutant exhibited essentially wild-type growth rates on 80 mM acetic acid (Fig. 5A). These data support the idea that Vps75 and Haa1 have opposing roles in controlling the expression of genes responsible for resistance to acid stress and, more specifically, that Haa1 is required to overcome the negative effect of Vps75 on expression of these genes.
FIG. 5.
Genes affected by deletion of VPS75 belong to an acid-responsive regulon and cluster together. (A) Deletion of VPS75 confers resistance to acetic acid. The indicated strains were grown overnight in YPD, spotted as 10-fold serial dilutions on YPD or YPD containing 100 mM acetic acid, and incubated at 30°C for 1 to 3 days. WT, wild-type. (B) Clustering of genes affected by deletion of VPS75 in chromosome IV. The x axis represents the chromosome on which each gene is shown successively from left to right. The expression ratio of each gene, compared with expression of a wild-type strain, is shown on the y axis. The dotted horizontal line represents a 2.5-fold upregulation. The arrow indicates the centromere.
When analyzing the genomic location of genes affected in vps75Δ cells, we noticed that 9 of the 30 genes (30.0%) changed by ≥2.5-fold were clustered together (Table 5). This phenomenon is most striking on chromosome 4, where three consecutive genes (YDL037C, YDL038C, and YDL039C) and another gene within 14 kb (YDL048C) were upregulated in vps75Δ (Fig. 5B). This observation was not pursued further, but it is tempting to speculate that the modified transcription rates in the affected gene clusters reflect localized changes in chromatin architecture.
TABLE 5.
Clustering of genes affected by deletion of VPS75
| Cluster | ORF | Gene | Fold changea | Chrb | Positionc
|
Typed | |
|---|---|---|---|---|---|---|---|
| Start | End | ||||||
| 1 | YDL039C | PRM7 | 6.0 | 4 | 382330 | 381983 | Consecutive |
| YDL038C | 10.0 | 4 | 384078 | 382327 | Consecutive | ||
| YDL037C | BSC1 | 12.4 | 4 | 385584 | 384598 | Consecutive | |
| 2 | YJR078W | BNA2 | −3.3 | 10 | 578853 | 580214 | Consecutive |
| YJR079W | −2.5 | 10 | 580198 | 581232 | Consecutive | ||
| 3 | YOL016C | CMK2 | 3.4 | 15 | 296121 | 294778 | Clustered |
| YOL014W | 2.7 | 15 | 299694 | 300068 | Clustered | ||
| 4 | YPR157W | 4.7 | 16 | 841262 | 842665 | Consecutive | |
| YPR158W | −2.5 | 16 | 843258 | 844016 | Consecutive | ||
Positive values signify upregulated genes, while negative values signify downregulated genes.
Chromosome.
Nucleotide position on the chromosome.
Genes immediately adjacent were designated “consecutive,” while those within 5 kb were designated “clustered.”
Vps75 functions in replication-independent histone exchange.
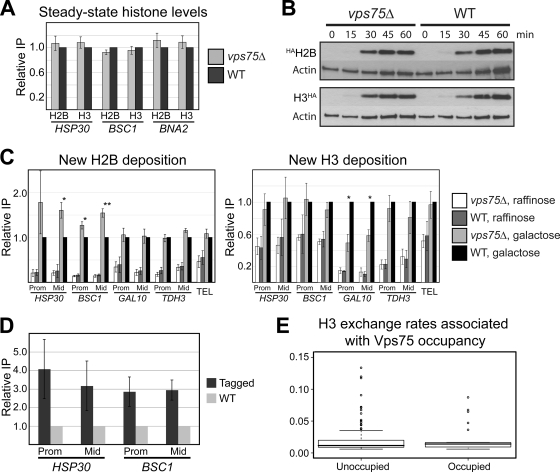
Given its role as a histone chaperone, we hypothesized that transcriptional changes in cells lacking Vps75 might correlate with an aberrant chromatin structure, a notion supported by the observation that some genes affected by VPS75 deletion were clustered together. To test this idea, we first analyzed histone levels at affected loci by ChIP assays using antibodies against endogenous histones. In all cases tested, steady-state levels of H2B and H3 were essentially unchanged at genes with modified transcription (Fig. 6A), indicating that an altered nucleosome density is not the cause of aberrant transcription at these genes in cells lacking Vps75.
FIG. 6.
Changes to chromatin in cells lacking Vps75. (A) Steady-state histone levels are not affected in cells lacking Vps75. The relative levels of histones H2B and H3 in the wild-type (WT) or vps75Δ strain grown in YPD were determined by ChIP assays followed by quantitative PCR. Values shown are the averages with standard errors of four independent experiments and are expressed as the amounts of immunoprecipitated DNA corrected to that of input DNA. The value for the wild-type strain was set to 1 and other values expressed relative to that. (B) Deletion of VPS75 does not significantly affect the production of galactose-induced HAH2B or H3HA proteins. Wild-type (WT) or vps75Δ cells were arrested in G1 phase with α-factor for 2 h, and then galactose was added to the medium. Samples were taken at the indicated time points and cross-linked with formaldehyde. After preparation of chromatin and reversal of cross-links, proteins were fractionated on 12% polyacrylamide gels and the presence of HAH2B or H3HA proteins detected by Western blotting with an antibody directed against the HA tag (12CA5). Actin is shown as a loading control. (C) Vps75 regulates replication-independent incorporation of new histones H2B and H3. Wild-type (WT) or vps75Δ strains containing HA-tagged, galactose-inducible forms of H2B and H3, respectively, were grown in raffinose and arrested in G1 with α-factor. Cells were then treated with formaldehyde before (raffinose) or after (galactose) a 1-h induction with galactose. The relative level of HAH2B (left graph) or H3HA (right graph) was determined by ChIP assays followed by quantitative PCR. Values shown are the averages, with standard errors, of four independent experiments and are expressed as the amounts of immunoprecipitated DNA corrected to input DNA. The value for the wild-type strain in galactose was set to 1 and other values expressed relative to that. Prom, promoter; Mid, middle; TEL, telomeric DNA on chromosome 6. A one-sample t test was used to determine statistical significance, with a single asterisk denoting a P value of <0.05 and two asterisks denoting a P value of <0.01. (D) Vps75 is physically associated with the HSP30 and BSC1 genes. Wild-type (WT) yeast or a strain expressing Myc-tagged Vps75 (Tagged) were cross-linked and subjected to ChIP using an antibody against the Myc epitope. Real-time PCR was used to detect coprecipitated DNA from the promoter (Prom) and middle (Mid) of the HSP30 and BSC1 genes. Values shown are the averages, with standard errors, of three independent experiments and are expressed as amounts of immunoprecipitated DNA corrected to that of input DNA. The value for the untagged strain was set to 1 and other values expressed relative to that. (E) Histone H3 exchange rates are not significantly different at sites of Vps75 occupancy on chromatin. Peaks of Vps75 occupancy were compared to a published map of histone H3 exchange (26). A box plot comparing exchange rates at Vps75-occupied and unoccupied regions is shown. The line in the center of each box represents the median value of the distribution, and the upper and lower ends of the box are the upper (25th) and lower (75th) quartiles, respectively.
Besides being deposited during replication, histones are also turned over in a replication-independent manner that appears to be at least partly proportional to the transcription rate (10, 52). We therefore speculated that Vps75 might affect histone turnover, rather than density, at certain genes. To test this idea, we performed histone exchange assays using strains containing two sources of histones H2B and H3: the untagged, endogenous protein and a HA-tagged, galactose-inducible form (HAH2B and H3HA) (23). In order to restrict the analysis to replication-independent exchange of histones, cells were arrested in G1 with α-factor and induced with galactose, and the amounts of the relevant histone protein at specific loci were determined by ChIP analysis. Given that Vps75 affects the density of RNAPII in at least one galactose-inducible gene (Fig. 4), an important preliminary control experiment was to determine whether the production of HAH2B and H3HA protein was affected in cells lacking this factor. Western blotting of cell extracts after galactose treatment revealed that there was no observable difference in HAH2B and H3HA production between wild-type and vps75Δ cells (Fig. 6B).
As reported by others previously (23), the induced, HA-tagged version of H2B is rapidly incorporated into the coding region and promoter of genes in a manner that is largely independent of gene activity (Fig. 6C, left graph). Interestingly, at two genes upregulated in response to loss of VPS75, HSP30 and BSC1, a small but significant and highly reproducible increase in H2B incorporation was observed in the vps75Δ strain. In contrast, the much lower histone exchange in nontranscribed, telomeric DNA within chromosome VI (TEL) was unaffected by loss of VPS75. Moreover, H2B dynamics were also largely unaffected by VPS75 at the induced GAL10 gene and at TDH3, a constitutive, highly expressed gene. Note, however, that highly expressed genes typically have a low nucleosome occupancy, which might affect our ability to detect small changes in this assay. These results indicate that Vps75 affects histone dynamics so that in its absence, new H2B incorporation is increased at certain genes.
Interestingly, the profile of new H3HA incorporation was significantly different from that for HAH2B at the same loci (Fig. 6C, right graph). New H3 incorporation at HSP30, BSC1, TDH3 and TEL was essentially the same in wild-type and vps75Δ cells. In contrast, deletion of VPS75 caused a significant reduction in the incorporation of new H3 molecules at the GAL10 gene following galactose induction. This indicates that Vps75 can positively influence H3 turnover in some situations.
Taken together, these data indicate that Vps75 affects the transcription-dependent dynamics of both histones H2B and H3. The observation that Vps75 is physically associated with each of the genes exhibiting changes in new histone incorporation (Fig. 2A and 6D) and the fact that the changes at HSP30 and BSC1 correlate with altered transcription levels suggest that this regulation could be direct.
Given the putative involvement of Vps75 in regulating the incorporation of new H3 molecules into chromatin at the active GAL10 gene, we compared our Vps75 chromatin occupancy map to a published map of H3 exchange (26). The rate of H3 exchange was slightly higher at sites of Vps75 occupancy (Fig. 6E), but this difference was not statistically significant as determined by a Wilcoxon rank sum test (P = 0.5708). This result, combined with our observation that new H3 incorporation is not affected at most genes tested in mutant cells, suggests that Vps75 does not play a significant role in regulating H3 exchange. However, we cannot rule out that in certain situations where the requirement for histone exchange factors is great, for example, when a gene is rapidly induced to a high level of expression (e.g., GAL10), Vps75 might have an auxiliary function in the assembly/disassembly of H3/H4.
Reassembly of chromatin in the wake of transcription is required to prevent initiation from cryptic promoters within the coding regions of certain genes, with the best characterized of these being FLO8 (25). Mutations in numerous factors involved in transcription elongation, including histone chaperones such as Spt6 and FACT, result in increased use of these promoter-like sequences, presumably by decreasing nucleosome reassembly in the wake of transcription and thereby allowing inappropriate access of transcription factors to chromatin (7, 22, 25, 39, 57). Given its role in regulating transcription-associated histone exchange, we speculated that VPS75 might affect the level of cryptic initiation. VPS75 was therefore disrupted in a strain expressing a galactose-inducible version of FLO8 in which the 3′ coding region has been replaced with the HIS3 coding region, so that prototrophic growth on galactose medium lacking histidine is possible only if the cryptic promoter is active (44). According to this assay, VPS75 was not required for normal chromatin reassembly within the FLO8 coding region (Fig. 7A). Further, quantitation of mRNA from the 3′ and 5′ ends of several other genes that can initiate transcription from cryptic promoters revealed that a loss of Vps75 did not significantly increase the 3′:5′ RNA ratio at these loci either, although a weak but reproducible effect was seen at SPB4 (Fig. 7B). These two experimental strategies were also used to assess the role of Rtt109 in transcription-coupled chromatin reassembly. The absence of this factor did not activate the cryptic promoter in FLO8 but significantly increased the 3′:5′ RNA ratio at RAD18 and to a lesser extent SPB4 (Fig. 7). These data suggest that Vps75 and Rtt109 are unlikely to have a dramatic and general role in chromatin reassembly in the wake of transcription, although they may function redundantly and/or at specific loci.
FIG. 7.
Effect of deletion of VPS75 and RTT109 on intragenic transcription. (A) Vps75 and Rtt109 do not regulate intragenic transcription from the cryptic promoter of the GAL1::FLO8-HIS3 reporter gene. The indicated wild-type (WT) and deletion strains containing the GAL1::FLO8-HIS3 reporter gene were grown overnight in synthetic complete (SC) medium and spotted as 10-fold serial dilutions on SC or SC lacking histidine and containing galactose as the carbon source (SC Gal −His) and incubated at 30°C for 1 to 3 days. (B) Ratio of 3′ to 5′ RNA levels expressed from the indicated genes in wild-type (WT), vps75Δ, or rtt109Δ cells. RNA levels were normalized to an internal 18S control and represent the averages for three independent experiments, with standard errors shown. The value for the WT strain was set to 1 and other values expressed relative to that.
Vps75 regulates activation-associated H3K56ac enrichment at GAL10.
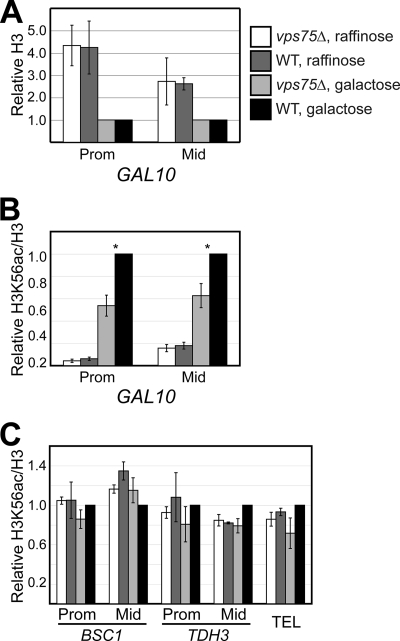
It was recently shown that histone eviction and transcriptional activation at PHO5 is slowed in yeast expressing a form of histone H3 that cannot be acetylated on lysine 56 (H3K56R), suggesting that this modification can affect chromatin disassembly (69). Furthermore, two recent studies indicate that H3K56ac can aid histone deposition during replication and repair (8, 35), while global analyses of histone turnover in yeast cells lacking H3K56ac also indicate a role for this mark in stimulating histone replacement (26). While Vps75 does not appear to play an important role in stimulating H3K56ac in vivo, we could not rule out the possibility that defects in histone exchange (i.e., new histone incorporation) in cells lacking Vps75 were related to localized changes in acetylation of H3K56. To test this idea, we used ChIP with antibodies recognizing total H3 (a C-terminal epitope) or H3 acetylated at K56 to investigate enrichment of this mark at GAL10 following activation. As reported previously (30), H3 levels were significantly reduced in both the promoter and coding region of GAL10 following addition of galactose (Fig. 8A). The extent of chromatin disassembly was essentially the same in wild-type and vps75Δ cells. Interestingly, H3K56ac levels at the GAL10 promoter increased significantly (>15-fold) following induction (Fig. 8B). This parallels the findings at the PHO5 promoter and suggests that H3K56ac may be generally involved in chromatin assembly/disassembly during transcription. These experiments also revealed a similar although less dramatic (>5-fold) increase in H3K56ac within the GAL10 ORF following galactose induction (Fig. 8B), indicating that this mark might play a role in histone dynamics not only during transcriptional initiation but also during elongation. Importantly, when the same ChIP assays were carried out with the vps75Δ strain, we observed a significant decrease in H3K56ac enrichment at both the promoter and ORF of GAL10 compared to results for wild-type cells (Fig. 8B). These experiments indicate that Vps75 affects transcription-associated accumulation of acetylated K56 at the highly transcribed GAL10 gene.
FIG. 8.
Vps75 regulates transcription-dependent accumulation of histone H3 acetylated at lysine 56 (H3K56ac) at GAL10. (A) Loss of histone H3 at GAL10 following induction. Wild-type (WT) or vps75Δ cells were grown in raffinose and arrested in G1 phase with α-factor. Cells were then treated with formaldehyde before (raffinose) or after (galactose) a 1-h induction with galactose. The relative levels of histone H3 at the induced GAL10 gene were then determined by ChIP assays followed by quantitative PCR. Values shown are the averages, with standard errors, of four independent experiments and are expressed as the amounts of immunoprecipitated DNA corrected to input DNA. Values in galactose were set to 1 and other values expressed relative to that. Prom, promoter; Mid, middle of ORF. (B) Defects in H3K56ac accumulation at induced GAL10 in cells lacking Vps75. Wild-type or vps75Δ cells were grown and treated with formaldehyde as described for panel A. The relative levels of H3K56ac at the induced GAL10 gene in were then determined by ChIP assays followed by quantitative PCR. Values shown are the averages, with standard errors, of four independent experiments and are expressed as the amounts of immunoprecipitated DNA corrected to that of input DNA and normalized to total histone H3 levels. The value for the wild-type strain in galactose was set to 1 and other values expressed relative to that. Prom, promoter; Mid, middle of ORF. A one-sample t test was used to determine statistical significance, with an asterisk denoting a P value of <0.05. The key is as shown in panel A. (C) Normal H3K56ac at genes exhibiting altered H2B exchange. H3K56ac levels in wild-type and vps75Δ cells were measured as described for panel B. The value for the wild-type strain in galactose was set to 1 and other values expressed relative to that. Prom, promoter; Mid, middle of ORF; TEL, telomeric DNA on chromosome 6. The key is as shown for panel A.
We next tested whether loss of Vps75 might also cause changes in H3K56ac levels at HSP30 and BSC1, where changes in new H2B incorporation were observed (see Fig. 6C). However, vps75Δ and wild-type cells contained the same low level of acetylated K56 at these loci, and this did not change significantly following galactose induction (Fig. 8C). Therefore, the changes in H2B exchange at these genes caused by loss of VPS75 are probably unrelated to K56 acetylation.
Vps75 and RSC can mediate removal of H2A/H2B dimers from nucleosomes.
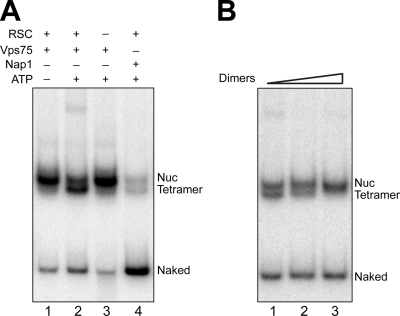
The SWI/SNF-related chromatin remodeling complex RSC can disassemble nucleosomes in vitro by a stepwise transfer of the octamer to a NAP domain histone chaperone, Nap1 (37). It has been proposed that this ability of a chromatin remodeler and a histone chaperone to remove nucleosomes might facilitate transcriptional activation (37). We previously showed that Vps75 can assemble nucleosomes in vitro (59), but given the pleiotropic effect of vps75Δ on nucleosome dynamics described above, we investigated another biochemical mechanism that might underlie the role of Vps75 in transcription and its apparent affect on histone/nucleosome equilibria, namely, its ability to act as a histone acceptor during chromatin remodeler-mediated nucleosome disassembly. As reported previously (37), the combined action of Nap1 and RSC resulted in a complete release of histone proteins from the nucleosome, generating naked DNA (Fig. 9A, lane 4). When incubated with nucleosomal DNA and ATP, neither Vps75 nor RSC alone could remove histone proteins from nucleosomes (Fig. 9A, lanes 1 and 3). However, when both factors were added to the reaction mixture, a band representing a nucleosome devoid of H2A/H2B dimers rapidly appeared (Fig. 9A, lane 2). The identity of this species was verified by the addition of excess dimer, which restored the complete nucleosome (Fig. 9B). These data provide support for the idea that histone chaperones act in combination with ATP-dependent chromatin remodeling complexes to achieve transcription-associated nucleosome disassembly (37) and also provide an important biochemical basis on which to understand the diverse effects on histone dynamics observed in cells lacking VPS75.
FIG. 9.
Vps75 promotes loss of H2A/H2B dimers from mononucleosomes in the presence of RSC and ATP. (A) Nucleosomes were treated for 2 h at 30°C with the indicated proteins and then fractionated by gel electrophoresis. An autoradiograph of the gel is shown. Bands corresponding to nucleosomes (Nuc), nucleosomes lacking H2A/H2B dimers (Tetramer), and free DNA (Naked) are indicated. (B) Reversal of histone depletion by RSC and Vps75. After a 2-h incubation (as in lane 2 of panel A), increasing amounts of yeast H2A/H2B dimer were added and the reaction mixtures incubated for an additional 2 h at 30°C. Gel electrophoresis and autoradiography was performed as described for panel A. Bands corresponding to nucleosomes (Nuc), nucleosomes lacking H2A/H2B dimers (Tetramer), and free DNA (Naked) are indicated.
DISCUSSION
An Rtt109-independent role for Vps75 in transcription.
Previous studies have shown that Vps75 is a histone chaperone that acts as a potent stimulator of the Rtt109 HAT, which acetylates histone H3 primarily on lysines 9 and 56 (4, 14, 67). However, elucidating the precise function of Vps75 in vivo has been difficult. The absence of Vps75 does not affect bulk H3K56ac levels (59), suggesting that its ability to enhance Rtt109-catalyzed modification of this residue may be an in vitro artifact. This idea is supported by the finding that Asf1, another histone chaperone, is absolutely required for Rtt109-mediated H3K56ac in vivo (51). In addition, while clearly required for efficient H3K9 acetylation, it is unlikely that this is the only role of Vps75 given the pleiotropic phenotypes of vps75Δ cells, which include defects in telomere maintenance (3), intracellular protein sorting (6), and nonhomologous end joining (24).
In this study, we present evidence to support the idea that Vps75 functions in transcription independently of its binding partner, Rtt109. First, epistasis measurements show that it genetically interacts with many genes involved in transcription, whereas Rtt109 (and as expected Asf1) is found in a network that consists primarily of factors involved in DNA repair/replication. Second, Vps75 is recruited to genes following their induction in the absence of Rtt109, and its genome-wide chromatin association profile correlates with transcription activity. Third, the absence of this factor deregulates transcription at a number of genes, none of which (tested thus far) are affected by deletion of RTT109. Finally, Vps75 affects replication-independent nucleosome dynamics at certain genes, providing a mechanism by which it might regulate transcription. This last function is also unlikely to involve Rtt109, since the absence of Vps75 promotes new H2B incorporation at certain loci (this study) whereas deletion of RTT109 causes an overall decrease in histone exchange (26; our unpublished data). Other indirect evidence supports the idea that Vps75 is not simply a cofactor for Rtt109-catalyzed histone acetylation but has alternative cellular functions: while expression of Rtt109 is tightly regulated by the cell cycle (11), Vps75 is expressed ubiquitously (this study), meaning that it will frequently exist separately from Rtt109. This is supported by our earlier finding that purification of Vps75 yields a significant fraction of protein not complexed with Rtt109 (59).
We propose that during S phase, Vps75 exists primarily within a Rtt109/Vps75 heterodimer. At this time, the primary role of Vps75 is to enable efficient H3K9 acetylation by Rtt109, a modification that may be important for transcription of S-phase-specific genes (14). It is important to note that Vps75 facilitates histone acetylation not only by presenting the substrate to Rtt109 but also by stabilizing this enzyme (14). In addition, the possibility that the Rtt109/Vps75 HAT complex also enhances Rtt109-catalyzed H3K56 acetylation at some loci during S phase to regulate transcription and/or promote genome stability cannot be ruled out. However, during the remainder of the cell cycle, when Rtt109 levels are dramatically reduced, Vps75 functions independently in the regulation of replication-independent chromatin assembly/disassembly and possibly other, as yet undiscovered processes. Thus, future studies of Vps75 should not be limited to analyses of Rtt109-catalyzed histone acetylation.
How does Vps75 regulate transcription?
Our results suggest that Vps75 functions in transcription by modifying chromatin structure. Previous biochemical data indicated that Vps75 can assemble nucleosomes (59), and here we show that it is also a potent acceptor of histone H2A/H2B dimers during RSC-mediated nucleosome remodeling. Thus, Vps75 can act as both a nucleosome assembly and disassembly factor in vitro. Our analyses of histone dynamics in yeast cells lacking Vps75 suggest that this factor also mediates equivalent changes to chromatin in vivo. At genes upregulated in response to deletion of VPS75 (BSC1 and HSP30), new histone H2B incorporation is more rapid, suggesting that in wild-type cells Vps75 promotes nucleosome stability or acts as a histone eviction factor, thereby slowing new H2B deposition. It could be argued that this observation simply reflects higher levels of transcription at these genes, which may result in increased displacement of H2A/H2B dimers (28). However, the role of Vps75 as a histone chaperone, its physical presence at these genes, and the finding that H3 turnover is not affected at BSC1 or HSP30 all suggest that reduced incorporation of new H2B molecules is a cause, and not simply an indirect consequence, of changes in transcription. The idea that Vps75 inhibits transcription by suppressing histone exchange is supported by a recent study, published while the manuscript of this article was undergoing review, reporting that the absence of Vps75 increased H3 turnover at certain “hot” nucleosomes (26).
Interestingly, we also identified a locus (activated GAL10) where loss of Vps75 reduced the rate of new H3 incorporation, suggesting that in some circumstances it facilitates H3 turnover. On the surface, this observation seems difficult to reconcile with Vps75's putative role as a negative regulator of H2B and H3 exchange (discussed above). However, accumulating evidence from in vitro assays and in vivo experiments indicates that other histone chaperones, such as Asf1, can mediate replication-independent nucleosome assembly (15, 57) and disassembly (1, 53, 57) and moreover that these seemingly opposing functions can occur even on the same gene. We propose that Vps75 can also promote both histone deposition and eviction depending on spatial context and in all likelihood associated factors. For example, Vps75 may act coordinately with chromatin remodelers at certain genes (i.e., HSP30 and BSC1) to promote H2A/H2B eviction and inhibit new H2A/H2B incorporation, a hypothesis supported by our observation that RSC and Vps75 can catalyze dimer removal from nucleosomes in vitro. At other loci, Vps75 could enhance deposition of new H3/H4, either alone or in concert with other remodeling factors.
Another way that Vps75 could modify the chromatin structure to regulate transcription was revealed by our microarray data. Genes affected by deletion of VPS75 tend to group together within the genome, implying that the transcription defects are due to localized changes in higher-order chromatin architecture. In this model, upregulation of a gene cluster would relate to a looser local chromatin structure, whereas downregulation would result from a more compact assembly. It is important to note that this idea is compatible with the observed defects in new histone incorporation in vps75Δ, since altered histone exchange rates may be a manifestation of changes in higher-order structure and vice versa. Interestingly, loss of Nap1 also affects clusters of genes, suggesting that proteins in the NAP domain family of histone chaperones may have a general role in maintaining higher-order chromatin structure in yeast (46). Acetylation of H4 lysine 16 is known to decompact chromatin (62), and Rtt109/Vps75 can weakly acetylate H4 tails in vitro (4), suggesting a mechanism by which this factor could also regulate the transcription of multiple consecutive genes. Future work should be aimed at investigating this possibility by genome-wide analyses of nucleosome density and positioning and histone acetylation patterns in cells lacking Vps75.
One possibility that remains to be investigated is whether the role of Vps75 in promoting new H3 incorporation at GAL10 reflects its ability to stimulate Rtt109-catalyzed H3K56 acetylation. This is particularly pertinent since this modification was recently shown to be required for efficient promoter chromatin disassembly, and subsequent activation, of PHO5 (69). In this study, we showed that Vps75 is required for normal enrichment of H3K56ac at the promoter and ORF of GAL10 following galactose induction. It is tempting to speculate that Vps75-mediated deposition of H3K56ac is a direct process whereby Rtt109/Vps75 (possibly in a complex with Asf1) acetylates free H3 molecules, which are subsequently incorporated into chromatin. Such a model is particularly attractive because H3K56ac is known to facilitate nucleosome assembly on plasmids and replicating DNA (35) and Rtt109 was recently shown to stimulate the histone deposition activity of Vps75, albeit in a manner that appears to be independent of acetylation (4). However, given that Vps75 does not affect bulk H3K56ac levels in cells (14, 59) and that its ability to regulate H2B exchange appears to be unrelated to this modification (this study), it is equally plausible that reduced incorporation of H3K56ac at GAL10 simply reflects the function of Vps75 as a dedicated histone exchange factor (discussed above). In other words, Vps75 may be required for disassembly of nucleosomes at GAL10, so that its absence would only indirectly reduce incorporation of new H3 acetylated at K56. It is important to note that these two models are not mutually exclusive and that Vps75 might regulate new H3 incorporation by stimulating histone acetylation at some genes and at others by promoting acetylation-independent histone exchange.
Another interesting observation to emerge from this study is the apparent ability of Vps75 to regulate the replication-independent dynamics of both H2B and H3. In general, histone chaperones are thought to target specific subnucleosomal components, with distinct activities being responsible for H2A/H2B and H3/H4 exchange (57). However, Vps75 can bind both H2A/H2B and H3/H4 in vitro (49, 59), and there is considerable evidence that Nap1, its closest homologue in yeast, can also act as a chaperone for all core histones and histone H1 (for a review, see reference 48). The idea that histone chaperones facilitate the transfer of both subnucleosomal histone complexes is also supported by the finding that most of these factors, including Vps75 (59), can assemble nucleosomes in vitro, a process that is likely to occur in two steps with deposition of H3/H4 (to form the tetrasome) preceding addition of H2A/H2B dimers (16). Clearly, there is still much to learn regarding the means by which histone chaperones contribute to chromatin assembly and disassembly. Having taken the first important step in identifying the majority of these factors in eukaryotes, it will now be important to elucidate these mechanistic details.
Supplementary Material
Acknowledgments
This work was supported by a generous in-house grant (to J.Q.S.) and by an EMBO long-term fellowship (to L.A.S.). M.T.O.-H. is currently funded by a Marie Curie Incoming International fellowship.
We thank the Cancer Research UK GeneChip Microarray Facility, Alain Verreault for the generous gift of anti-H3 antibody, Amine Nourani for yeast strains, and Paul Kaufman for the His-Vps75 expression plasmid. Members of the Svejstrup laboratory are thanked for helpful comments on the manuscript.
Footnotes
Published ahead of print on 26 May 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adkins, M. W., S. R. Howar, and J. K. Tyler. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14657-666. [DOI] [PubMed] [Google Scholar]
- 2.Adkins, M. W., S. K. Williams, J. Linger, and J. K. Tyler. 2007. Chromatin disassembly from the PHO5 promoter is essential for the recruitment of the general transcription machinery and coactivators. Mol. Cell. Biol. 276372-6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Askree, S. H., T. Yehuda, S. Smolikov, R. Gurevich, J. Hawk, C. Coker, A. Krauskopf, M. Kupiec, and M. J. McEachern. 2004. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc. Natl. Acad. Sci. USA 1018658-8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berndsen, C. E., T. Tsubota, S. E. Lindner, S. Lee, J. M. Holton, P. D. Kaufman, J. L. Keck, and J. M. Denu. 2008. Molecular functions of the histone acetyltransferase chaperone complex Rtt109-Vps75. Nat. Struct. Mol. Biol. 15948-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein, B. E., C. L. Liu, E. L. Humphrey, E. O. Perlstein, and S. L. Schreiber. 20 August 2004, posting date. Global nucleosome occupancy in yeast. Genome Biology 5R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonangelino, C. J., E. M. Chavez, and J. S. Bonifacino. 2002. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol. Biol. Cell 132486-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrozza, M. J., B. Li, L. Florens, T. Suganuma, S. K. Swanson, K. K. Lee, W. J. Shia, S. Anderson, J. Yates, M. P. Washburn, and J. L. Workman. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123581-592. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C. C., J. J. Carson, J. Feser, B. Tamburini, S. Zabaronick, J. Linger, and J. K. Tyler. 2008. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell 134231-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins, S. R., K. M. Miller, N. L. Maas, A. Roguev, J. Fillingham, C. S. Chu, M. Schuldiner, M. Gebbia, J. Recht, M. Shales, H. Ding, H. Xu, J. Han, K. Ingvarsdottir, B. Cheng, B. Andrews, C. Boone, S. L. Berger, P. Hieter, Z. Zhang, G. W. Brown, C. J. Ingles, A. Emili, C. D. Allis, D. P. Toczyski, J. S. Weissman, J. F. Greenblatt, and N. J. Krogan. 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446806-810. [DOI] [PubMed] [Google Scholar]
- 10.Dion, M. F., T. Kaplan, M. Kim, S. Buratowski, N. Friedman, and O. J. Rando. 2007. Dynamics of replication-independent histone turnover in budding yeast. Science 3151405-1408. [DOI] [PubMed] [Google Scholar]
- 11.Driscoll, R., A. Hudson, and S. P. Jackson. 2007. Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315649-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edgar, R., M. Domrachev, and A. E. Lash. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandes, A. R., N. P. Mira, R. C. Vargas, I. Canelhas, and I. Sa-Correia. 2005. Saccharomyces cerevisiae adaptation to weak acids involves the transcription factor Haa1p and Haa1p-regulated genes. Biochem. Biophys. Res. Commun. 33795-103. [DOI] [PubMed] [Google Scholar]
- 14.Fillingham, J., J. Recht, A. C. Silva, B. Suter, A. Emili, I. Stagljar, N. J. Krogan, C. D. Allis, M. C. Keogh, and J. F. Greenblatt. 2008. Chaperone control of the activity and specificity of the histone H3 acetyltransferase Rtt109. Mol. Cell. Biol. 284342-4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green, E. M., A. J. Antczak, A. O. Bailey, A. A. Franco, K. J. Wu, J. R. Yates III, and P. D. Kaufman. 2005. Replication-independent histone deposition by the HIR complex and Asf1. Curr. Biol. 152044-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gruss, C., J. Wu, T. Koller, and J. M. Sogo. 1993. Disruption of the nucleosomes at the replication fork. EMBO J. 124533-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han, J., H. Zhou, B. Horazdovsky, K. Zhang, R. M. Xu, and Z. Zhang. 2007. Rtt109 acetylates histone H3 lysine 56 and functions in DNA replication. Science 315653-655. [DOI] [PubMed] [Google Scholar]
- 18.Han, J., H. Zhou, Z. Li, R. M. Xu, and Z. Zhang. 2007. Acetylation of lysine 56 of histone H3 catalyzed by RTT109 and regulated by ASF1 is required for replisome integrity. J. Biol. Chem. 28228587-28596. [DOI] [PubMed] [Google Scholar]
- 19.Han, J., H. Zhou, Z. Li, R. M. Xu, and Z. Zhang. 2007. The Rtt109-Vps75 histone acetyltransferase complex acetylates non-nucleosomal histone H3. J. Biol. Chem. 28214158-14164. [DOI] [PubMed] [Google Scholar]
- 20.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95717-728. [DOI] [PubMed] [Google Scholar]
- 21.Howard, S. C., Y. V. Budovskaya, Y. W. Chang, and P. K. Herman. 2002. The C-terminal domain of the largest subunit of RNA polymerase II is required for stationary phase entry and functionally interacts with the Ras/PKA signaling pathway. J. Biol. Chem. 27719488-19497. [DOI] [PubMed] [Google Scholar]
- 22.Imbeault, D., L. Gamar, A. Rufiange, E. Paquet, and A. Nourani. 2008. The Rtt106 histone chaperone is functionally linked to transcription elongation and is involved in the regulation of spurious transcription from cryptic promoters in yeast. J. Biol. Chem. 28327350-27354. [DOI] [PubMed] [Google Scholar]
- 23.Jamai, A., R. M. Imoberdorf, and M. Strubin. 2007. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol. Cell 25345-355. [DOI] [PubMed] [Google Scholar]
- 24.Jessulat, M., M. Alamgir, H. Salsali, J. Greenblatt, J. Xu, and A. Golshani. 2008. Interacting proteins Rtt109 and Vps75 affect the efficiency of non-homologous end-joining in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 469157-164. [DOI] [PubMed] [Google Scholar]
- 25.Kaplan, C. D., L. Laprade, and F. Winston. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 3011096-1099. [DOI] [PubMed] [Google Scholar]
- 26.Kaplan, T., C. L. Liu, J. A. Erkmann, J. Holik, M. Grunstein, P. D. Kaufman, N. Friedman, and O. J. Rando. 2008. Cell cycle- and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genet. 4e1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keller, G., E. Ray, P. O. Brown, and D. R. Winge. 2001. Haa1, a protein homologous to the copper-regulated transcription factor AceI, is a novel transcriptional activator. J. Biol. Chem. 27638697-38702. [DOI] [PubMed] [Google Scholar]
- 28.Kireeva, M. L., W. Walter, V. Tchernajenko, V. Bondarenko, M. Kashlev, and V. M. Studitsky. 2002. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol. Cell 9541-552. [DOI] [PubMed] [Google Scholar]
- 29.Kornberg, R. D. 1974. Chromatin structure: a repeating unit of histones and DNA. Science 184868-871. [DOI] [PubMed] [Google Scholar]
- 30.Kristjuhan, A., and J. Q. Svejstrup. 2004. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 234243-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krogan, N. J., G. Cagney, H. Yu, G. Zhong, X. Guo, A. Ignatchenko, J. Li, S. Pu, N. Datta, A. P. Tikuisis, T. Punna, J. M. Peregrin-Alvarez, M. Shales, X. Zhang, M. Davey, M. D. Robinson, A. Paccanaro, J. E. Bray, A. Sheung, B. Beattie, D. P. Richards, V. Canadien, A. Lalev, F. Mena, P. Wong, A. Starostine, M. M. Canete, J. Vlasblom, S. Wu, C. Orsi, S. R. Collins, S. Chandran, R. Haw, J. J. Rilstone, K. Gandi, N. J. Thompson, G. Musso, P. St Onge, S. Ghanny, M. H. Lam, G. Butland, A. M. Altaf-Ul, S. Kanaya, A. Shilatifard, E. O'Shea, J. S. Weissman, C. J. Ingles, T. R. Hughes, J. Parkinson, M. Gerstein, S. J. Wodak, A. Emili, and J. F. Greenblatt. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440637-643. [DOI] [PubMed] [Google Scholar]
- 32.Kvint, K., J. P. Uhler, M. J. Taschner, S. Sigurdsson, H. Erdjument-Bromage, P. Tempst, and J. Q. Svejstrup. 2008. Reversal of RNA polymerase II ubiquitylation by the ubiquitin protease Ubp3. Mol. Cell 30498-506. [DOI] [PubMed] [Google Scholar]
- 33.Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl, and J. D. Lieb. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36900-905. [DOI] [PubMed] [Google Scholar]
- 34.Li, B., M. Carey, and J. L. Workman. 2007. The role of chromatin during transcription. Cell 128707-719. [DOI] [PubMed] [Google Scholar]
- 35.Li, Q., H. Zhou, H. Wurtele, B. Davies, B. Horazdovsky, A. Verreault, and Z. Zhang. 2008. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 134244-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lieb, J. D., and N. D. Clarke. 2005. Control of transcription through intragenic patterns of nucleosome composition. Cell 1231187-1190. [DOI] [PubMed] [Google Scholar]
- 37.Lorch, Y., B. Maier-Davis, and R. D. Kornberg. 2006. Chromatin remodeling by nucleosome disassembly in vitro. Proc. Natl. Acad. Sci. USA 1033090-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luk, E., N. D. Vu, K. Patteson, G. Mizuguchi, W. H. Wu, A. Ranjan, J. Backus, S. Sen, M. Lewis, Y. Bai, and C. Wu. 2007. Chz1, a nuclear chaperone for histone H2AZ. Mol. Cell 25357-368. [DOI] [PubMed] [Google Scholar]
- 39.Mason, P. B., and K. Struhl. 2003. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell. Biol. 238323-8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, A., B. Yang, T. Foster, and A. L. Kirchmaier. 2008. Proliferating cell nuclear antigen and ASF1 modulate silent chromatin in Saccharomyces cerevisiae via lysine 56 on histone H3. Genetics 179793-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morillon, A., J. O'Sullivan, A. Azad, N. Proudfoot, and J. Mellor. 2003. Regulation of elongating RNA polymerase II by forkhead transcription factors in yeast. Science 300492-495. [DOI] [PubMed] [Google Scholar]
- 42.Nakanishi, T., M. Shimoaraiso, T. Kubo, and S. Natori. 1995. Structure-function relationship of yeast S-II in terms of stimulation of RNA polymerase II, arrest relief, and suppression of 6-azauracil sensitivity. J. Biol. Chem. 2708991-8995. [DOI] [PubMed] [Google Scholar]
- 43.Nonet, M., D. Sweetser, and R. A. Young. 1987. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell 50909-915. [DOI] [PubMed] [Google Scholar]
- 44.Nourani, A., F. Robert, and F. Winston. 2006. Evidence that Spt2/Sin1, an HMG-like factor, plays roles in transcription elongation, chromatin structure, and genome stability in Saccharomyces cerevisiae. Mol. Cell. Biol. 261496-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ocampo-Hafalla, M. T., Y. Katou, K. Shirahige, and F. Uhlmann. 2007. Displacement and re-accumulation of centromeric cohesin during transient pre-anaphase centromere splitting. Chromosoma 116531-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohkuni, K., K. Shirahige, and A. Kikuchi. 2003. Genome-wide expression analysis of NAP1 in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 3065-9. [DOI] [PubMed] [Google Scholar]
- 47.Park, Y. J., and K. Luger. 2008. Histone chaperones in nucleosome eviction and histone exchange. Curr. Opin. Struct. Biol. 18282-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park, Y. J., and K. Luger. 2006. Structure and function of nucleosome assembly proteins. Biochem. Cell Biol. 84549-558. [DOI] [PubMed] [Google Scholar]
- 49.Park, Y. J., K. B. Sudhoff, A. J. Andrews, L. A. Stargell, and K. Luger. 2008. Histone chaperone specificity in Rtt109 activation. Nat. Struct. Mol. Biol. 15957-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pokholok, D. K., C. T. Harbison, S. Levine, M. Cole, N. M. Hannett, T. I. Lee, G. W. Bell, K. Walker, P. A. Rolfe, E. Herbolsheimer, J. Zeitlinger, F. Lewitter, D. K. Gifford, and R. A. Young. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122517-527. [DOI] [PubMed] [Google Scholar]
- 51.Recht, J., T. Tsubota, J. C. Tanny, R. L. Diaz, J. M. Berger, X. Zhang, B. A. Garcia, J. Shabanowitz, A. L. Burlingame, D. F. Hunt, P. D. Kaufman, and C. D. Allis. 2006. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc. Natl. Acad. Sci. USA 1036988-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rufiange, A., P. E. Jacques, W. Bhat, F. Robert, and A. Nourani. 2007. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell 27393-405. [DOI] [PubMed] [Google Scholar]
- 53.Schermer, U. J., P. Korber, and W. Horz. 2005. Histones are incorporated in trans during reassembly of the yeast PHO5 promoter. Mol. Cell 19279-285. [DOI] [PubMed] [Google Scholar]
- 54.Schuldiner, M., S. R. Collins, N. J. Thompson, V. Denic, A. Bhamidipati, T. Punna, J. Ihmels, B. Andrews, C. Boone, J. F. Greenblatt, J. S. Weissman, and N. J. Krogan. 2005. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 123507-519. [DOI] [PubMed] [Google Scholar]
- 55.Schuldiner, M., S. R. Collins, J. S. Weissman, and N. J. Krogan. 2006. Quantitative genetic analysis in Saccharomyces cerevisiae using epistatic miniarray profiles (E-MAPs) and its application to chromatin functions. Methods 40344-352. [DOI] [PubMed] [Google Scholar]
- 56.Schuller, C., Y. M. Mamnun, M. Mollapour, G. Krapf, M. Schuster, B. E. Bauer, P. W. Piper, and K. Kuchler. 2004. Global phenotypic analysis and transcriptional profiling defines the weak acid stress response regulon in Saccharomyces cerevisiae. Mol. Biol. Cell 15706-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwabish, M. A., and K. Struhl. 2006. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol. Cell 22415-422. [DOI] [PubMed] [Google Scholar]
- 58.Schwabish, M. A., and K. Struhl. 2007. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol. Cell. Biol. 276987-6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Selth, L., and J. Q. Svejstrup. 2007. Vps75, a new yeast member of the NAP histone chaperone family. J. Biol. Chem. 28212358-12362. [DOI] [PubMed] [Google Scholar]
- 60.Seo, S. B., P. McNamara, S. Heo, A. Turner, W. S. Lane, and D. Chakravarti. 2001. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell 104119-130. [DOI] [PubMed] [Google Scholar]
- 61.Shikama, N., H. M. Chan, M. Krstic-Demonacos, L. Smith, C. W. Lee, W. Cairns, and N. B. La Thangue. 2000. Functional interaction between nucleosome assembly proteins and p300/CREB-binding protein family coactivators. Mol. Cell. Biol. 208933-8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shogren-Knaak, M., H. Ishii, J. M. Sun, M. J. Pazin, J. R. Davie, and C. L. Peterson. 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311844-847. [DOI] [PubMed] [Google Scholar]
- 63.Simoes, T., N. P. Mira, A. R. Fernandes, and I. Sa-Correia. 2006. The SPI1 gene, encoding a glycosylphosphatidylinositol-anchored cell wall protein, plays a prominent role in the development of yeast resistance to lipophilic weak-acid food preservatives. Appl. Environ. Microbiol. 727168-7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Svejstrup, J. Q. 2007. Elongator complex: how many roles does it play? Curr. Opin. Cell Biol. 19331-336. [DOI] [PubMed] [Google Scholar]
- 65.Tagami, H., D. Ray-Gallet, G. Almouzni, and Y. Nakatani. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 11651-61. [DOI] [PubMed] [Google Scholar]
- 66.Tardiff, D. F., K. C. Abruzzi, and M. Rosbash. 2007. Protein characterization of Saccharomyces cerevisiae RNA polymerase II after in vivo cross-linking. Proc. Natl. Acad. Sci. USA 10419948-19953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsubota, T., C. E. Berndsen, J. A. Erkmann, C. L. Smith, L. Yang, M. A. Freitas, J. M. Denu, and P. D. Kaufman. 2007. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol. Cell 25703-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Walfridsson, J., O. Khorosjutina, P. Matikainen, C. M. Gustafsson, and K. Ekwall. 2007. A genome-wide role for CHD remodelling factors and Nap1 in nucleosome disassembly. EMBO J. 262868-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Williams, S. K., D. Truong, and J. K. Tyler. 2008. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc. Natl. Acad. Sci. USA 1059000-9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Williams, S. K., and J. K. Tyler. 2007. Transcriptional regulation by chromatin disassembly and reassembly. Curr. Opin. Genet. Dev. 1788-93. [DOI] [PubMed] [Google Scholar]
- 71.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, N. Liebundguth, D. J. Lockhart, A. Lucau-Danila, M. Lussier, N. M'Rabet, P. Menard, M. Mittmann, C. Pai, C. Rebischung, J. L. Revuelta, L. Riles, C. J. Roberts, P. Ross-MacDonald, B. Scherens, M. Snyder, S. Sookhai-Mahadeo, R. K. Storms, S. Veronneau, M. Voet, G. Volckaert, T. R. Ward, R. Wysocki, G. S. Yen, K. Yu, K. Zimmermann, P. Philippsen, M. Johnston, and R. W. Davis. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285901-906. [DOI] [PubMed] [Google Scholar]
- 72.Wittmeyer, J., A. Saha, and B. Cairns. 2004. DNA translocation and nucleosome remodeling assays by the RSC chromatin remodeling complex. Methods Enzymol. 377322-343. [DOI] [PubMed] [Google Scholar]
- 73.Workman, J. L. 2006. Nucleosome displacement in transcription. Genes Dev. 202009-2017. [DOI] [PubMed] [Google Scholar]
- 74.Xu, F., K. Zhang, and M. Grunstein. 2005. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 121375-385. [DOI] [PubMed] [Google Scholar]
- 75.Xu, F., Q. Zhang, K. Zhang, W. Xie, and M. Grunstein. 2007. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol. Cell 27890-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yuan, G. C., Y. J. Liu, M. F. Dion, M. D. Slack, L. F. Wu, S. J. Altschuler, and O. J. Rando. 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309626-630. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.