Abstract
Recent research has cast doubt on the reliability of bones and teeth for reconstructing phylogenetic relationships among higher primate species and genera. Herein, we investigate whether this problem is confined to hard tissues by examining the utility of higher primate soft-tissue characters for reconstructing phylogenetic relationships at low taxonomic levels. We use cladistic methods to analyze 197 soft-tissue characters for the extant hominoids and then compare the resulting phylogenetic hypotheses with the group's consensus molecular phylogeny, which is widely considered to be accurate. We show that the soft-tissue characters yield robust phylogenetic hypotheses that are compatible with the molecular phylogeny. Given the strength of the evidence for molecular phylogeny, these results indicate that, unlike craniodental hard-tissue characters, soft tissues are reliable for reconstructing phylogenetic relationships among higher primate species and genera. Thus, in higher primates at least, some types of morphological data are more useful than others for phylogeny reconstruction.
The bones, teeth, and soft tissues that make up the primate body have long been assumed to be useful for phylogenetic reconstruction at all relevant taxonomic levels. In recent years, however, researchers have begun to question the reliability of bones and teeth for reconstructing phylogenetic relationships among higher primate species and genera (1–14). This skepticism is based partly on the fact that phylogenetic analyses of fossil primates have thus far yielded conflicting and weakly supported hypotheses of relationships (4, 15, 16), partly on the fact that comparisons between craniodental phylogenies and reliable molecular phylogenies have found that the former disagree with the latter (1, 14), and partly on the fact that we are developing a better understanding of the processes involved in the generation of osteological and dental similarities and differences among primates (5–9, 12, 13, 17, 18). In this article, we describe a study that examined whether the reliability problem is confined to the bones and teeth of higher primates or whether their soft tissues are also unreliable for reconstructing species- and genus-level relationships.
To assess the phylogenetic utility of higher primate soft-tissue morphology, we carried out cladistic analyses of an extensive soft-tissue data set for the extant hominoids and then judged the resulting phylogenetic hypotheses against the group's consensus molecular phylogeny, which is widely considered to be accurate (19). This approach assumes that a match between morphological and molecular phylogenies is evidence that the morphological evidence is reliable, whereas incongruence indicates the converse (1, 14). There are several reasons for assuming that conflicts between the molecular and morphological phylogenetic hypotheses result from limitations of the morphological evidence. First, in phylogenetics, morphology can never be more than a proxy for molecular data, because phylogenetic relationships are genetic relationships. Second, the consensus molecular cladogram for the extant hominoids is supported by several sets of independent data (19). Agreement among multiple independent data sets is the strongest support possible for a phylogenetic hypothesis. Lastly, the methods of molecular phylogenetics have been successfully tested on taxa of known phylogeny, whereas comparable tests of morphological phylogenetic methods have proved unsuccessful (20–22).
Materials and Methods
Soft-tissue characters have been used in a number of phylogenetic analyses of the extant hominoids (23–28), but it was evident that these characters did not exhaust the available information. Therefore, an extensive literature search was conducted on the soft-tissue anatomy of the five extant hominoid genera, Gorilla (mountain and lowland gorillas), Homo (modern humans), Hylobates (gibbons and siamangs), Pan (common chimpanzees and bonobos), and Pongo (Bornean and Sumatran orangutans; ref. 29). The data collected by this method were collated and distilled into a comparative anatomical format. From this database, characters for cladistic analysis were identified by using three criteria: (i) data had to be available for all five genera (this criterion avoided the problem of missing data); (ii) at least two character states had to be present (this criterion excluded invariant characters); and (iii) one of these character states had to be present in two or more species (this last criterion removed characters that were uniquely derived for a given species). Data for all five genera were available for 240 characters, and 197 of these conformed with criteria ii and iii. The character state data for the 197 characters were additively coded, and a taxon-by-character matrix was compiled. Details of the characters and character states are given in the supplemental Appendix (see the supplemental material at www.pnas.org). The matrix is shown in supplemental Table 1 (see the supplemental material at www.pnas.org).
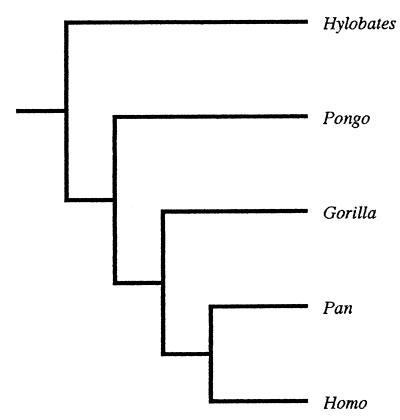
The data matrix was used to perform two tests of the hypothesis that soft-tissue characters are reliable for reconstructing the phylogenetic relationships of the hominoids. The first was based on parsimony analysis, which identifies the cladogram(s) requiring the smallest number of ad hoc hypotheses of character state change to account for the distribution of character states among the taxa. The matrix was subjected to parsimony analysis with PAUP 3.0s (30), and the shortest cladogram was compared with the consensus molecular cladogram for the extant hominoids (Fig. 1). Because parsimony analysis provides no means of discriminating between “true” and “false” clades, we reasoned that the hypothesis would be supported if the analysis favored a fully resolved cladogram that matched the molecular cladogram or a partially resolved cladogram that comprised only molecular clades. We also reasoned that the hypothesis would be supported if the analysis produced several equally parsimonious cladograms whose strict consensus comprised only clades that were compatible with the molecular cladogram.
Figure 1.
Hominoid molecular relationships.
The second test of the hypothesis was based on the phylogenetic bootstrap, which assesses the confidence interval associated with a clade (31, 32). In PAUP 3.0s, 10,000 matrices were derived from each matrix by sampling with a replacement. The new matrices were subjected to parsimony analysis, and a consensus of the most parsimonious cladograms was computed with a confidence region of 70% (33). Thereafter, the clades of the consensus cladogram were compared with the molecular cladogram (Fig. 1). In this test, it was reasoned that, for the hypothesis to be supported, the best supported clades should not be false clades, because it is commonly assumed in primate phylogenetics that the better the bootstrap support for a clade, the more likely the clade is to be true (15, 16).
In both the parsimony and bootstrap analyses, the characters were given equal weights and treated as unordered. No a priori judgements were made as to the primitive or derived condition of characters. The cladograms were obtained with the branch and bound search routine of PAUP 3.0s.
Results
The hypothesis that hominoid soft-tissue characters are reliable for phylogenetic reconstruction was supported by the parsimony analysis of the soft-tissue data set. The analysis yielded a single most parsimonious cladogram whose branching pattern matched that of the hominoid consensus molecular cladogram. When rooted on Hylobates, the cladogram suggested that Pongo is the sister taxon of a clade comprising Homo and the African apes and that Gorilla is the sister taxon of a (Homo and Pan) clade. After the exclusion of 45 uninformative characters, the cladogram had a length of 288, a consistency index of 0.65, and a retention index of 0.34. The next most parsimonious cladogram, in which the positions of Gorilla and Pongo were reversed, had a length of 296. The conventional hard-tissue cladogram, which links Gorilla and Pan to the exclusion of Homo and locates Pongo as the sister taxon of Gorilla, Pan, and Homo, had a length of 301.
The bootstrap analysis also upheld the hypothesis that hominoid soft-tissue characters are reliable for phylogenetic reconstruction. The (Homo and Pan) clade and the (Gorilla, Pan, and Homo) clade were supported by 91% and 84% of the bootstrap samples, respectively. Alternative groupings received little support. Of the bootstrap samples, 13% supported a (Homo, Pongo, and Pan) clade, 4% supported a (Gorilla and Homo) clade, 3% supported a (Pan and Pongo) clade, and 2% supported a (Gorilla and Pongo) clade. Of the bootstrap cladograms, 1% supported a (Gorilla, Homo, and Pongo) clade, a (Gorilla, Pongo, and Pan) clade, and a (Homo and Pongo) clade. A clade comprising Gorilla and Pan was also supported by just 1% of the bootstrap samples.
Discussion
The results of the parsimony and bootstrap tests strongly support the hypothesis that soft-tissue characters can be relied on to reconstruct the phylogenetic relationships of the extant hominoids. The parsimony analysis unambiguously favored a cladogram with the same topology as the molecular cladogram, and the bootstrap analysis returned high levels of support for clades with the same membership as the clades of the molecular cladogram. It is noteworthy that the two main alternative hypotheses of relationship that have been posited for the extant hominoids received extremely low levels of support in the bootstrap test. Neither the (Gorilla and Pan) clade that until recently was favored by most morphologists (24–26) nor the (Homo and Pongo) clade that has been supported by Schwartz (34, 35) appeared in more than 1% of the bootstrap cladograms. In view of the hominoid soft-tissue characters' strong support for the group's true phylogeny, we infer from the results of the tests that soft-tissue characters, unlike the hard-tissue characters, can be relied on to reconstruct the phylogenetic relationships of higher primates, even at low taxonomic levels.
Why do higher primate hard-tissue and soft-tissue characters differ in their phylogenetic utility? Experiments in which rhombomere quail-to-chick grafts have been used to investigate the influence of hindbrain segmentation on craniofacial patterning (36) may provide part of the answer to this question. These experiments demonstrate that each rhombomeric population remains coherent throughout ontogeny, with rhombomere-specific matching of muscle connective tissue and their attachment sites for all branchial and tongue muscles. If this system operates elsewhere in the body, it would help explain how muscle gross morphology can be conserved despite phylogeny-obscuring changes in the shapes of skeletal elements.
Another part of the answer may be that the type of soft-tissue characters we have used are not as prone to homoiology (37) as skeletal characters. Homoiologies result from phenotypic similarities in the way that different genotypes interact with the environment (e.g., remodeling in response to mechanical loading), and it has been claimed that, because bone is a dynamic tissue, many osseous morphologies may be homoiologous (5–9). We suspect that if homoiology has played a role in the generation of the character states in our soft-tissue data set, it has been only a very minor one. Although the mass of a muscle may be affected by activity or the lack of it, the manner in which it originates or the number of component bellies is unlikely to be. Likewise, mechanical loading is unlikely to affect the branching pattern of an artery or the number of digits supplied by a given nerve.
Although homoiology may account for some of the difference in phylogenetic utility between the hard and soft tissues, it cannot be the whole explanation. Dental enamel is not prone to homoiology, because it does not remodel, and yet Hartman (1) found that molar morphology is unreliable for reconstructing the phylogenetic relationships of the extant hominoids. Thus, other factors must also be involved in reducing the phylogenetic utility of teeth relative to that of soft tissues. Function has been posited as a cause of phylogeny-obscuring change in tooth morphology by several authors (1, 38), but recent work on the dentition of the Lake Lagoda seal suggests that developmental constraints may also be a reason why tooth morphology is prone to homoplasy and is therefore a poor guide to low-level phylogenetic relationships (39).
This study has shown that, for the extant hominoids and, by extension, other higher primates, the classic molecules versus morphology conflict (40) does not hold. Rather, the contrast is between molecules and soft-tissue morphology on the one hand and hard-tissue morphology on the other. The next phase of this research will investigate whether the phylogenetic signal varies among regions, systems, and tissues within the soft tissue data set.
Supplementary Material
Acknowledgments
This study benefited from the advice of M. C. Dean, P. O'Higgins, D. R. Pilbeam, J. Shaw-Dunn, and M. Gunther. S.G. was funded by a studentship from the Anatomical Society of Great Britain and Ireland. M.C. is funded by a Wellcome Trust Bioarchaeology Fellowship. B.W. was funded by The Leverhulme Trust and is now funded by The Henry Luce Foundation.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 10684.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.190252697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.190252697
References
- 1.Hartman S E. J Hum Evol. 1988;17:489–502. [Google Scholar]
- 2.Trinkaus E. Am J Phys Anthropol. 1990;83:1–11. doi: 10.1002/ajpa.1330830102. [DOI] [PubMed] [Google Scholar]
- 3.Trinkaus E. In: Continuity or Replacement: Controversies in Homo sapiens Evolution. Bräuer G, Smith F H, editors. Rotterdam: Balkema; 1992. pp. 1–7. [Google Scholar]
- 4.Harrison T. In: Species, Species Concepts and Primate Evolution. Kimbel W H, Martin L B, editors. New York: Plenum; 1993. pp. 345–371. [Google Scholar]
- 5.Lieberman D E. Curr Anthropol. 1995;36:159–197. [Google Scholar]
- 6.Lieberman D E. Annu Rev Anthropol. 1997;26:185–210. [Google Scholar]
- 7.Lieberman D E. Evol Anthropol. 1999;7:142–151. [Google Scholar]
- 8.Lieberman D E. In: Development, Growth and Evolution. O'Higgins P, Cohn M J, editors. London: Linn. Soc.; 2000. pp. 85–122. [Google Scholar]
- 9.Lieberman D E, Pilbeam D R, Wood B A. J Hum Evol. 1996;17:503–511. [Google Scholar]
- 10.Pilbeam D R. Mol Phyl Evol. 1996;5:155–168. doi: 10.1006/mpev.1996.0010. [DOI] [PubMed] [Google Scholar]
- 11.Jablonski N. Curr Biol. 1999;9:122. doi: 10.1016/s0960-9822(99)80077-3. [DOI] [PubMed] [Google Scholar]
- 12.Lovejoy C O, Cohn M J, White T D. Proc Natl Acad Sci USA. 1999;96:13247–13252. doi: 10.1073/pnas.96.23.13247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCollum M. Science. 1999;284:301–305. doi: 10.1126/science.284.5412.301. [DOI] [PubMed] [Google Scholar]
- 14.Collard M, Wood B A. Proc Natl Acad Sci USA. 2000;97:5003–5006. doi: 10.1073/pnas.97.9.5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corruccini R S. In: Integrative Approaches to the Past: Paleoanthropological Advances in Honor of F. Clark Howell. Corruccini R S, Ciochon R L, editors. Englewood Cliffs, NJ: Prentice Hall; 1994. pp. 167–183. [Google Scholar]
- 16.Wood B A, Collard M. Science. 1999;284:65–71. doi: 10.1126/science.284.5411.65. [DOI] [PubMed] [Google Scholar]
- 17.Wood B A, Abbott S A, Uytterschaut H. J Anat. 1988;156:107–139. [PMC free article] [PubMed] [Google Scholar]
- 18.Wood B A. In: Paleoclimate and Evolution with Emphasis on Human Origins. Vrba E S, Denton G H, Partridge T C, Burckle L H, editors. New Haven, CT: Yale Univ. Press; 1995. pp. 438–448. [Google Scholar]
- 19.Ruvolo M. Mol Biol Evol. 1997;14:248–265. doi: 10.1093/oxfordjournals.molbev.a025761. [DOI] [PubMed] [Google Scholar]
- 20.Fitch W M, Atchley W R. In: Molecules and Morphology in Evolution: Conflict or Compromise? Patterson C, editor. Cambridge, U.K.: Cambridge Univ. Press; 1987. pp. 203–216. [Google Scholar]
- 21.Atchley W R, Fitch W M. Science. 1991;254:554–558. doi: 10.1126/science.1948030. [DOI] [PubMed] [Google Scholar]
- 22.Hillis D M, Bull J J, White M E, Badgett M R, Molineux I J. Science. 1992;255:589–592. doi: 10.1126/science.1736360. [DOI] [PubMed] [Google Scholar]
- 23.Groves C P. In: Comparative Primate Biology: Systematics, Evolution and Anatomy. Swindler D R, Erwin J, editors. Vol. 1. New York: Liss; 1986. pp. 187–217. [Google Scholar]
- 24.Andrews P. In: Molecules and Morphology in Evolution: Conflict or Compromise? Patterson C, editor. Cambridge, U.K.: Cambridge Univ. Press; 1987. pp. 23–53. [Google Scholar]
- 25.Andrews P, Martin L B. J Hum Evol. 1987;16:101–118. [Google Scholar]
- 26.Andrews P. Nature (London) 1992;360:641–648. doi: 10.1038/360641a0. [DOI] [PubMed] [Google Scholar]
- 27.Shoshani J, Groves C P, Simons E L, Gunnell G F. Mol Phyl Evol. 1996;5:102–154. doi: 10.1006/mpev.1996.0009. [DOI] [PubMed] [Google Scholar]
- 28.Barriel V. Folia Primatol. 1997;68:50–56. doi: 10.1159/000157232. [DOI] [PubMed] [Google Scholar]
- 29.Gibbs S. Ph.D. thesis. Liverpool, U.K.: Univ. of Liverpool; 1999. [Google Scholar]
- 30.Swofford D L. paup, Phylogenetic Analysis Using Parsimony. Champaign, IL: Illinois Nat. Hist. Surv.; 1991. , Version 3.0s. [Google Scholar]
- 31.Felsenstein J. Evolution (Lawrence, Kans) 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 32.Sanderson M J. Syst Biol. 1995;44:299–320. [Google Scholar]
- 33.Hillis D M, Bull J J. Syst Biol. 1993;42:182–192. [Google Scholar]
- 34.Schwartz J H. Nature (London) 1984;308:501–506. doi: 10.1038/308501a0. [DOI] [PubMed] [Google Scholar]
- 35.Schwartz J H. The Red Ape: Orangutans and Human Origins. Boston: Houghton Mifflin; 1987. [Google Scholar]
- 36.Köntges G, Lumsden A. Development (Cambridge, UK) 1996;122:3229–3242. doi: 10.1242/dev.122.10.3229. [DOI] [PubMed] [Google Scholar]
- 37.Reidl R J. Order in Living Organisms. New York: Wiley; 1978. [Google Scholar]
- 38.Hunter J P, Jernvall J. Proc Natl Acad Sci USA. 1995;92:10718–10722. doi: 10.1073/pnas.92.23.10718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jernvall J. Proc Natl Acad Sci USA. 2000;97:2641–2645. doi: 10.1073/pnas.050586297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patterson C, editor. Molecules and Morphology in Evolution: Conflict or Compromise? Cambridge, U.K.: Cambridge Univ. Press; 1987. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.