Abstract
Mo-nitrogenase catalyzes the reduction of dinitrogen to ammonia at the cofactor (i.e., FeMoco) site of its MoFe protein component. Biosynthesis of FeMoco involves NifEN, a scaffold protein that hosts the maturation of a precursor to a mature FeMoco before it is delivered to the target location in the MoFe protein. Previously, we have shown that the NifEN-bound precursor could be converted, in vitro, to a fully complemented “FeMoco” in the presence of 2 mM dithionite. However, such a conversion was incomplete and Mo was only loosely associated to the NifEN-bound “FeMoco”. Here we report the optimized maturation of NifEN-associated precursor in 20 mM dithionite. Activity analyses show that, upon the optimal conversion of precursor to “FeMoco”, NifEN is capable of activating a FeMoco-deficient form of MoFe protein to the same extent as the isolated FeMoco. Further, EPR and XAS/EXAFS analyses reveal the presence of a tightly organized Mo site in NifEN-bound “FeMoco”, which allows the observation of a FeMoco-like, S = 3/2 EPR signal and the modeling of a NifEN-bound “FeMoco” that adopts a very similar conformation to that of the MoFe protein-associated FeMoco. The sensitivity of FeMoco maturation to dithionite concentration suggests an essential role of redox chemistry in this process, and the optimal potential of dithionite solution could serve as a guideline for future identification of in vivo electron donors for FeMoco maturation.
Keywords: Nitrogenase, NifEN, FeMoco biosynthesis
1. Introduction
Nitrogenase provides the biochemical machinery for ATP-dependent reduction of N2 to NH3. The best-characterized Mo-nitrogenase1 is a binary system comprising the Fe protein and the MoFe protein. The Fe protein is an α2-homodimer containing one [Fe4S4] cluster at the subunit interface and one MgATP binding site in each subunit; whereas the MoFe protein is an α2β2-heterotetramer containing one P-cluster ([Fe8S7]) at each α/β-subunit interface and one FeMoco ([MoFe7S9X-homocitrate], where X=C, N or O) within each α subunit. Nitrogenase catalysis involves repeated association and dissociation between Fe protein and MoFe protein and ATP hydrolysis-driven, inter-protein electron transfer from the [Fe4S4] cluster of the Fe protein to the P-cluster, and then the FeMoco of the MoFe protein, where substrate reduction occurs.
Biosynthesis of FeMoco2 is presumably launched by the production of a Fe/S core structure on NifB (encoded by nifB), which is then transferred to NifEN (encoded by nifE and nifN) for further processing before it is delivered to its target location in MoFe protein. NifEN shares significant homology with MoFe protein in primary sequence and cluster sites. It has been proposed that NifEN contains a “P-cluster site” that houses a P-cluster homolog and a “FeMoco site” that hosts the conversion of FeMoco precursor to a mature cluster.2 Previously, we confirmed the identity of a P-cluster homolog as a [Fe4S4]-type cluster and, further, identified a FeMoco precursor as a Mo/homocitrate-free cluster that closely resembled the Fe/S core of FeMoco.3,4 Subsequently, we showed that, in an ATP- and reductant-dependent process, Fe protein inserted Mo and homocitrate into the precursor, leading to the formation of a “FeMoco” on NifEN that could directly activate the FeMoco-deficient MoFe protein.5,6 While these studies have established the key components for FeMoco assembly, a detailed molecular analysis of this process has remained elusive due to the lack of means to quantitatively obtain a fully matured form of NifEN-bound FeMoco. Our data indicated that conversion of precursor was incomplete in 2 mM dithionite, as MoFe protein reconstituted by such a NifEN-bound “FeMoco” species was only ~30% active compared to that by isolated FeMoco. Additionally, we were unable to unambiguously define the pattern of Mo binding on the basis of our XAS/EXAFS data, as Mo appeared to be only loosely associated to this “FeMoco”.5 Clearly, a reconstitution system that facilitates the complete formation of active FeMoco needs to be developed for the elucidation of the molecular details of FeMoco biosynthesis.
Here we report the optimization of FeMoco maturation on NifEN at an elevated reductant concentration (i.e., 20 mM dithionite), which not only suggests a key role of redox chemistry in this process, but also permits the first detailed structural analysis of the NifEN-associated FeMoco species. Together, these studies constitute an important step in establishing the mechanism of FeMoco assembly, which is one of the most outstanding questions in the field of metalloprotein biochemistry.
2. Results and Discussion
Three forms of NifEN were used in this study. One, designated ΔnifB NifEN, contains two permanent [Fe4S4] clusters but no FeMoco precursor due to the deletion of the nifB gene. Another, designated NifENPrecursor, contains, in addition to the two [Fe4S4] clusters, a FeMoco precursor. The third, designated NifEN“FeMoco”(20 mM), contains a “FeMoco” matured in 20 mM dithionite. The elevated dithionite concentration allows the optimal conversion of precursor to “FeMoco” (Fig. 1), as NifEN“FeMoco”(20 mM) is capable of activating the FeMoco-deficient MoFe protein to the same extent as the isolated FeMoco (Table 1). Such a dramatic increase is not observed when the NifEN-bound precursor is matured in the absence of homocitrate (data not shown), suggesting that the matured cluster on NifEN is fully complemented with both Mo and homocitrate.
Figure 1.
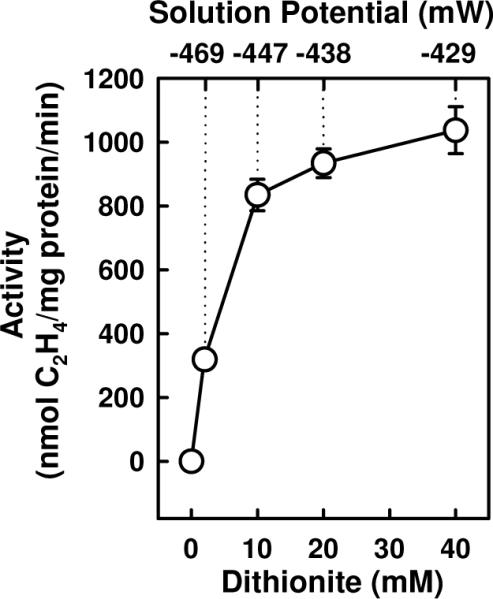
Dithionite-dependency of precursor maturation on NifEN. NifENPrecursor was first converted to NifEN“FeMoco” in varying concentrations of dithionite (0, 2, 10, 20 and 40 mM, respectively) and, subsequently, NifEN“FeMoco” was re-isolated and used as a FeMoco source for the reconstitution of FeMoco-deficient ΔnifB MoFe protein. Shown are the specific C2H2-reducing activities of MoFe protein upon reconstitution with NifEN“FeMoco” prepared at different dithionite concentrations. The calculated
solution potentials9 are indicated.
Table 1.
Reconstitution of FeMoco-deficient MoFe protein by various FeMoco sources.
| Activities (nmol/mg protein/min) |
||||
|---|---|---|---|---|
| FeMoco Source | C2H2 formation under C2H2/Ar | H2 formation under Ar | NH3 formation under N2 | H2 formation under N2 |
| NifENPrecursor | 0 | 0 | 0 | 0 |
| Isolated FeMoco | 917 ± 105 | 1269 ± 154 | 305 ± 75 | 292 ± 20 |
| NifEN“FeMoco”(20mM) | 1065 ± 88 | 1100 ± 19 | 334 ± 40 | 323 ± 23 |
| NifEN“FeMoco”[Ti(III) citrate] | 680 ± 28 | 890 ± 35 | 272 ± 32 | 225 ± 9 |
| NifEN“FeMoco”(Fld_1) | 538 ± 20 | 520 ± 21 | 210 ± 17 | 120 ± 5 |
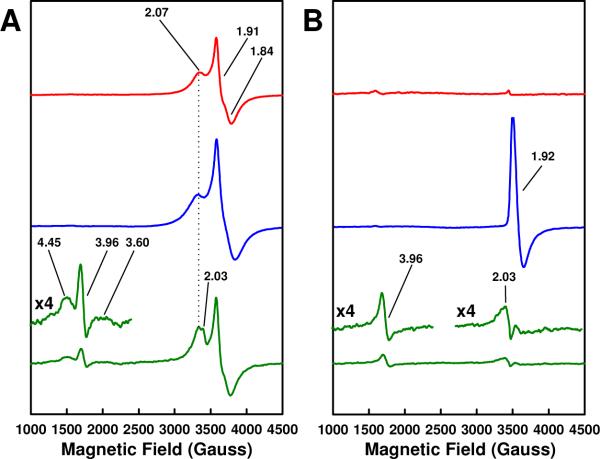
Consistent with the significant improvement in its reconstitution capacity, NifEN“FeMoco”(20 mM) displays EPR features (Fig. 2; also see Figs. S1 and S2) that have not been directly observed following the maturation of precursor in 2 mM dithionite (referred to as NifEN“FeMoco”(2 mM) in the following text).5 In the dithionite-reduced state, NifEN“FeMoco”(20 mM) exhibits an S = 3/2 EPR signal with g values of 4.45, 3.96 and 3.60 at 6 K, which is absent from the spectra of both ΔnifB NifEN and NifENPrecursor (Fig. 2A). In addition, a new g = 2.03 feature appears in the S = 1/2 signal of NifEN“FeMoco”(20 mM) (Fig. 2A). Clearly, the additional features in both S = 3/2 and S = 1/2 regions of NifEN“FeMoco”(20 mM) spectrum are intimately associated with the attachment of Mo and homocitrate to the NifEN-bound precursor. Further, the appearance of a FeMoco-like, S = 3/2 signal signifies the presence of a “FeMoco” species in NifEN that is very similar to the native FeMoco in MoFe protein.
Figure 2.
EPR properties of ΔnifB NifEN (red), NifENPrecursor (blue) and NifEN“FeMoco”(20 mM) (green) in dithionite-reduced (A) and IDS-oxidized (B) states. The dithionite-reduced samples were measured at 6 K, whereas the IDS-oxidized samples were measured at 15 K. The features of NifEN“FeMoco”(20 mM) are enlarged, and the g values are indicated.
In the indigodisulfonate (IDS)-oxidized state, NifENPrecursor shows a distinct g = 1.92 signal at 15 K, which is absent from the spectrum of the precursor-free ΔnifB NifEN (Fig. 2B). This precursor-specific signal disappears in the case of NifEN“FeMoco”(20 mM), suggesting a change in the electronic properties of the cluster upon insertion of Mo and homocitrate (Fig. 2B). Interestingly, while the broad g = 4.45 and 3.60 features of NifEN“FeMoco”(20 mM) disappear upon oxidation, the g = 3.96 and 2.03 features remain intact (Fig. 2B). Thus, the Mo/homocitrate-associated features of NifEN“FeMoco”(20 mM) do not always behave in sync, particularly those in the S = 3/2 region (i.e., features at g = 4.45, 3.96 and 3.60). This observation suggests that, unlike the S = 3/2 signal of MoFe protein-associated FeMoco, the S = 3/2 signal of NifEN-bound “FeMoco” may be a mixture of S = 3/2 species, likely reflecting its interactions with a cofactor site that is homologous to yet different than that in MoFe protein.2 An indirect support for this argument comes from the EPR properties of FeVco, another FeMoco homolog that exhibits an S = 3/2 signal with mixed composition.7
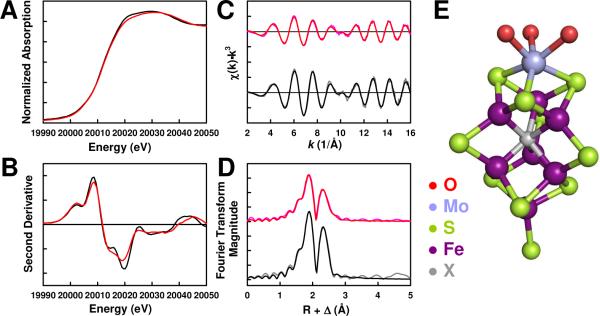
XAS/EXAFS spectroscopy of NifEN“FeMoco”(20 mM) provides further structural proof that the “FeMoco” on NifEN closely resembles the native FeMoco in MoFe protein (designated MoFe proteinFeMoco) while having some unique features of its own. The Mo K-edge XAS spectra of NifEN“FeMoco”(20 mM) and MoFe proteinFeMoco show no difference in energy in the rising edge (Fig. 3A) and strong similarity in shape in the second derivative (Fig. 3B), indicating that the effective charge and electronic environment of Mo in NifEN“FeMoco”(20 mM) are very close to those in MoFe proteinFeMoco. In contrast, NifEN“FeMoco”(2 mM) shows a ~0.6 eV positive shift of the rising edge from that of MoFe proteinFeMoco, implying a lesser degree of resemblance of this NifEN-bound “FeMoco” to the native cofactor in MoFe protein.5
Figure 3.
Mo K-edge x-ray absorption spectra (A) and smoothed second derivatives (B) of NifEN“FeMoco”(20 mM) (red) and MoFe proteinFeMoco (black); Mo K-edge EXAFS (C) and Fourier transforms (D) of data (pink) and fits (red) for NifEN“FeMoco”(20 mM), and data (gray) and fits (black) for MoFe proteinFeMoco; and structural model of NifEN-bound “FeMoco” (E). The structural model was adapted from the crystallographic coordinates of the MoFe protein14 but modified for distances <3 Å from Mo on the basis of the EXAFS fits. S is shown as the ligand at the Fe-end of the “FeMoco”, as the Cys residue that ligates FeMoco in the MoFe protein is conserved in the NifE primary sequence.2 The ligand at the Mo-end of “FeMoco” is not given, but it is likely an Asn residue instead of the His ligand in the MoFe protein.2
As is observed from their XAS spectra, there is a considerable degree of similarity between the EXAFS data of NifEN“FeMoco”(20 mM) and MoFe proteinFeMoco in both frequency and beat pattern. The key beat region at k = 10 Å-1 is conserved between the two species (Fig. 3C), suggesting, yet again, that the Mo in NifEN“FeMoco”(20 mM) is inserted in a coordination environment similar to that in MoFe proteinFeMoco. The EXAFS intensity of NifEN“FeMoco”(20 mM) is improved compared to that of NifEN“FeMoco”(2 mM);5 however, it is still diminished compared to that of MoFe proteinFeMoco (Fig. 3C). Additionally, the backscattering peak at ~5 Å, which corresponds to the distant, cross-cluster Mo-Fe backscattering in MoFe proteinFeMoco, is absent from the Fourier transform of NifEN“FeMoco”(20 mM) (Fig. 3D), suggesting that the Mo environment in NifEN“FeMoco”(20 mM) is either less ordered or less symmetrical than that in MoFe proteinFeMoco. The high reconstitution activity of NifEN“FeMoco”(20 mM) apparently points to the presence of a well-ordered Mo environment in NifEN-associated “FeMoco”, one that has the Mo sufficiently secured in the “FeMoco” and therefore not easily lost during the transfer of “FeMoco” to the FeMoco-deficient MoFe protein. Moreover, despite their similarity, the “FeMoco” site in NifEN is different from that in MoFe protein, particularly at the Mo end.2 As such, it is very likely that Mo in NifEN-bound “FeMoco” has a somewhat different coordination than that in the MoFe protein-bound FeMoco.
Indeed, in contrast to Mo in MoFe proteinFeMoco, which has three equidistant Fe backscatterers at 2.69 Å, Mo in NifEN“FeMoco”(20 mM) can be best modeled by placing one Fe atom at 2.90 Å and another two at 2.71 Å (Table 2), suggesting a slightly “off-center” coordination of Mo in NifEN“FeMoco”(20 mM). Interestingly, NifEN“FeMoco”(2 mM) does not have the Fe backscatterer at 2.90 Å.5 This observation suggests a much stronger association of Mo and a more tightly organized Mo site in NifEN“FeMoco”(20 mM). Further, compared to Mo in NifEN“FeMoco”(2 mM), which has three O atoms at a consistently shorter average distance,5 Mo in NifEN“FeMoco”(20 mM) has one O at 2.12 Å and the other two at 2.24 Å, two of which are likely contributed by homocitrate. All of these Mo-O distances are longer than those in NifEN“FeMoco”(2 mM) and resemble the Mo-O coordination in MoFe proteinFeMoco more closely (Table 2). Finally, three S atoms can be modeled at an average distance of 2.37 Å to Mo in NifEN“FeMoco”(20 mM), which is very similar to the three Mo-S distances in MoFe proteinFeMoco (Table 2). Taken together, these results suggest a similar yet asymmetrical coordination of Mo in NifEN“FeMoco”(20 mM) as compared to that in MoFe proteinFeMoco. More importantly, NifEN“FeMoco”(20 mM) has a much improved Mo coordination than NifEN“FeMoco”(2 mM), allowing the proposal of a model for the NifEN-bound “FeMoco” (Fig. 3E).
Table 2.
Final EXAFS fit results over a k range of 2-16 Å1. The variables are coordination number, N; interatomic distance, R; mean-square thermal and static deviation, σ2; and the shift in the threshold energy from 20025 eV, ΔE0. The estimated uncertainties in R, σ2 and N are ± 0.02 Å, ± 0.0001 Å2, and ± 20%, respectively. The goodness of fit, F, is defined as F = [Σk6 (χexptl. - ˇcalcd.)2/Σk6 χexptl.)2]0.5.
| MoFe proteinFeMoco |
NifEN“FeMoco”(20 mM) |
|||||
|---|---|---|---|---|---|---|
| N | R(Å) | σ2;(Å2) | N | R(Å) | σ2;(Å2) | |
| Scatterer | ||||||
| Mo-O short | - | - | - | 1 | 2.12 | 0.0055 |
| Mo-O long | 3 | 2.20 | 0.0053 | 2 | 2.24 | 0.0012 |
| Mo-S | 3 | 2.36 | 0.0022 | 3 | 2.37 | 0.0059 |
| Mo-Fe short | 3 | 2.69 | 0.0033 | 2 | 2.71 | 0.0029 |
| Mo-Fe long | - | - | - | 1 | 2.90 | 0.0058 |
| ΔE0 (eV) | -11.0 | -8.9 | ||||
| Weighted F | 0.163 | 0.150 | ||||
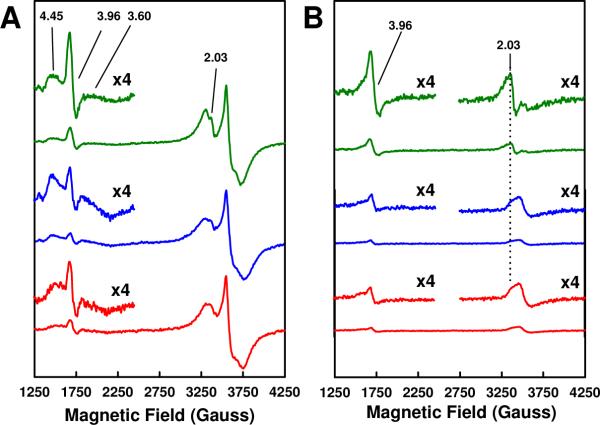
Combined outcome of these studies suggests that an increase in dithionite concentration results in a much tighter association of Mo to the NifEN-bound “FeMoco”, allowing the observation of a FeMoco-like, S = 3/2 EPR signal and the modeling of a NifEN-bound “FeMoco” that adopts a very similar conformation to that of the MoFe protein-associated FeMoco. Such a reductant-dependency of FeMoco maturation is further exemplified by the ability of other non-physiological reductants (e.g.,Ti(III) citrate) and some physiological electron donors (e.g., flavodoxin 1 of Azotobacter vinelandii) to perform in the capacity of dithionite in FeMoco maturation (Fig. 4; also see Figs. S3 and S4). NifEN“FeMoco” matured in the presence of 1.5 mM Ti(III) citrate (designated NifEN“FeMoco”[Ti(III) citrate]) or 40 μM reduced flavodoxin 1 (designated NifEN“FeMoco”(Fld_1)) displays the same characteristic EPR signals as those of NifEN“FeMoco”(20 mM) in dithionite-reduced (Fig. 4A) or IDS-oxidized (Fig. 4B) state. However, compared to NifEN“FeMoco”(20 mM), both NifEN“FeMoco”[Ti(III) citrate] and NifEN“FeMoco”(Fld_1) display EPR signals with reduced magnitudes and exhibit lower activities in the reconstitution assays (Table 1). The inability of Ti(III) citrate to completely mature the NifEN-bound precursor may originate from the harmful effect of this strong reductant on the integrity of NifEN and its associated clusters at higher concentrations, which is reflected by a sharp decrease in the reconstitution activity of NifEN“FeMoco”[Ti(III) citrate] when it is matured in higher than 1.5 mM Ti(III) citrate (data not shown); whereas the sub-optimal capacity of flavodoxin 1 in this process can be interpreted by the “incorrect” redox potential of this electron donor (-330 mV8), which renders it less efficient in FeMoco maturation. The latter observation is particularly interesting in light of the optimal solution potential of dithionite for FeMoco maturation (ca -440 mV, calculated for 20 mM dithionite9). More importantly, it suggests an essential role of redox chemistry in the in vivo process of FeMoco assembly. A solution potential of ca -440 mV may produce a “correct” redox state of Mo before it can be mobilized by Fe protein, generate a certain redox state of the Fe protein that is favorable for Mo mobilization, or render the NifEN-associated precursor in an optimal redox state for Mo attachment, all of which could account for the improved association of Mo to NifEN-bound “FeMoco”. Further investigation along this line is in progress, and the optimal solution potential of dithionite will serve as a guideline for identifying in vivo electron donors for FeMoco maturation in the future.
Figure 4.
EPR properties of NifEN“FeMoco”(20 mM) (green), NifEN“FeMoco”[Ti(III) citrate] (blue) and NifEN“FeMoco”(Fld_1) (red) in dithionite-reduced (A) and IDS-oxidized (B) states. The dithionite-reduced samples were measured at 6 K, whereas the IDS-oxidized samples were measured at 15 K. The g values are indicated.
3. Experimental Procedures
FeMoco Maturation
The conversion of NifEN-bound precursor to “FeMoco” was performed in a 50 ml maturation assay containing 25 mM Tris-HCl (pH 8.0), 100 mg NifEN containing precursor (designated NifENPrecursor, which was isolated from Azotobacter vinelandii strain DJ 10413), 120 mg Fe protein, 0.4 mM homocitrate, 0.4 mM Na2MoO4, 2.4 mM ATP, 4.8 mM MgCl2, 30 mM creatine phosphate, 24 units/ml creatine phosphokinase, and varying concentrations of dithionite (Na2S2O4) (0, 2, 10, 20 and 40 mM, respectively) or Ti(III) citrate (0, 0.5, 1.5, 3 and 7.5 mM, respectively). In the case of A. vinelandii flavodoxin 1, 40 mg (or 40 M) reduced protein was added to the assay as a reductant to achieve a flavodoxin 1:NifEN molar ratio of 4:1. Reduced flavodoxin 1 of A. vinelandii was prepared by purifying the protein as described elsewere,8 and removing excess dithionite by a single passage over a Sephadex G-50 column. In all cases, the maturation mixture was stirred for 1 hr at 30°C and then NifEN was re-isolated (categorically designated NifEN“FeMoco”) and used for reconstitution of FeMoco-deficient MoFe protein (see below). NifEN re-isolated following maturation in 20 mM Na2S2O4 (designated NifEN“FeMoco”(20 mM)) was examined further by EPR and XAS analyses (see below); whereas NifEN re-isolated following maturation in 1.5 mM Ti(III) citrate (designated NifEN“FeMoco”[Ti(III) citrate]) or 40 μM reduced A. vinelandii flavodoxin 1 (designated NifEN“FeMoco”(Fld_1)) were analyzed further by EPR (see below). The purity of Na2S2O4 (Sigma) is 85%, and the dithionite concentrations described above reflect the actual Na2S2O4 contents based on this purity.
Reconstitution Analysis
The reconstitution of MoFe protein was performed in a 0.8 ml assay containing 25 mM Tris-HCl (pH 8.0), 20 mM Na2S2O4 and 0.5 mg FeMoco-deficient ΔnifB MoFe protein (isolated from A. vinelandii strain DJ114310). FeMoco insertion was initiated with the addition of 2 mg NifEN“FeMoco” to the assay, and the reaction mixture was then incubated for 30 min at 30°C before being terminated and examined for enzymatic activities as described earlier.11
EPR Spectroscopy
All electron paramagnetic resonance spectroscopy (EPR) samples were prepared in a Vacuum Atmospheres dry box (Hawthorne, CA, USA) at an oxygen level of less than 4 ppm. All samples contained 15 mg/ml protein, 10% glycerol, 2 mM Na2S2O4 and 25 mM Tris-HCl (pH 8.0). The oxidized samples were prepared by incubating proteins with excess IDS for 30 min and subsequently removing IDS by an anion-exchange column. Spectra were collected in perpendicular mode using a Bruker ESP 300 Ez spectrophotometer (Bruker, Billerica, MA, USA) interfaced with an Oxford Instruments ESR-9002 liquid helium continuous-flow cryostat (Oxford Instruments, Oxford, UK). All spectra were recorded using a gain of 5 × 104, a modulation frequency of 100 kHz, a modulation amplitude of 5 G, and a microwave frequency of 9.62 GHz.
XAS Data Acquisition
X-ray absorption spectroscopy (XAS) data were measured at the 20-pole wiggler BL7-3 biological XAS station at Stanford Synchrotron Radiation Lightsource (SSRL) with storage ring parameters 3 GeV and 80-100 mA. A pre-monochromator flat bent Rh-coated mirror provided rejection of higher harmonics and vertical collimation, and a Si(220) double-crystal monochromator was used for energy selection. Samples were stored in LN2 prior to data collection and held at a constant temperature of 10 K during data collection via an Oxford Instruments CF1208 liquid-helium continuous-flow cryostat. A Canberra 30-element solid-state Ge detector array was used to record Mo Kα fluorescence data. By using Soller slits and a Zr filter secured between the sample cryostat and the detector window, signal intensity from inelastic scattering and Mo Kβ fluorescence was substantially diminished. Internal energy calibration was performed by simultaneous measurement of the absorption of a Mo foil placed between two ionization chambers filled with Ar located after the sample. The first inflection point of the foil XAS edge was assigned to 20003.9 eV. No signs of photoreduction of the metal sites, as observed by shifts in edge energy with time, were noted. A total of 38 and 12 scans were measured for NifEN“FeMoco”(20 mM) and MoFe proteinFeMoco, respectively.
XAS Data Analysis
After inspection of raw data and averaging, the average data files for each sample were normalized using the program PYSPLINE12 by fitting a second-order polynomial to the pre-edge region and subtracting from the entire data range with control points, followed by fitting a four-region spline function of orders 2, 3, and 3 over the post-edge region. The data were normalized to an edge jump of 1.0 at 20025 eV. Though some data sets extended further, the data range selected for EXAFS fits was limited to the shortest k-range (16 Å-1) to perform internally consistent fits. By means of the least squares fitting program OPT, a component of the EXAFSPAK suite of software,13 EXAFS data of k = 2-16 Å-1 were fit using initial ab initio theoretical phase and amplitude functions calculated from FEFF 7.014 based on the 1M1N15 crystallographic starting model. Atomic coordinates from the crystal model were adjusted as necessary as fits were further refined. During fit optimization, the interatomic distance between the absorbing and backscattering atom (R) and the mean-square thermal and static deviation in R (σ2) were varied for all components. The threshold energy (ΔE0) was allowed to vary for each fit but constrained to the same value for all components. The amplitude reduction factor (S02) was maintained at a value of 1.0 throughout analysis. Coordination numbers (N) were methodically adjusted from crystallographic values to provide the best chemically viable agreement to the EXAFS data and their Fourier transform. Inclusion or exclusion of various scattering paths was systematically tested to fully explore the atomic environment at Mo.
Supplementary Material
Acknowledgment
This work was supported by NIH Grants GM 67626 (M.W.R.) and RR 01209 (K.O.H.). SSRL operations are funded by the DOE BES, and the SSRL Structural Molecular Biology Program by NIH NCRR BTP and DOE BER.
Footnotes
Supporting Information Available: Four figures of temperature-dependent EPR spectra. This material is available free of charge via the Internet at http://pubs.acs.org.
BRIEFS: A FeMoco with a slightly asymmetrically ligated Mo can be formed on NifEN in the presence of 20 mM dithionite.
REFERENCES
- (1).Burgess BK, Lowe DJ. Chem. Rev. 1996;96:2983–3012. doi: 10.1021/cr950055x. [DOI] [PubMed] [Google Scholar]
- (2).Hu Y, Fay AW, Lee CC, Yoshizawa J, Ribbe MW. Biochemistry. 2008;47:3973–3981. doi: 10.1021/bi7025003. [DOI] [PubMed] [Google Scholar]
- (3).Hu Y, Fay AW, Ribbe MW. Proc. Natl. Acad. Sci. USA. 2005;102:3236–3241. doi: 10.1073/pnas.0409201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Corbett MC, Hu Y, Fay AW, Ribbe MW, Hedman B, Hodgson KO. Proc. Natl. Acad. Sci. USA. 2006;103:1238–1243. doi: 10.1073/pnas.0507853103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Hu Y, Corbett MC, Fay AW, Webber JA, Hodgson KO, Hedman B, Ribbe MW. Proc. Natl. Acad. Sci. USA. 2006;103:17119–17124. doi: 10.1073/pnas.0602647103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Hu Y, Corbett MC, Fay AW, Webber JA, Hodgson KO, Hedman B, Ribbe MW. Proc. Natl. Acad. Sci. USA. 2006;103:17125–17130. doi: 10.1073/pnas.0602651103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Eady RR. Chem. Rev. 1996;96:3013–3030. doi: 10.1021/cr950057h. [DOI] [PubMed] [Google Scholar]
- (8).Gangeswaran R, Eady RR. Biochem. J. 1996;317:103–108. doi: 10.1042/bj3170103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Mayhew SG. Eur. J. Biochem. 1978;85:535–547. doi: 10.1111/j.1432-1033.1978.tb12269.x. [DOI] [PubMed] [Google Scholar]
- (10).Schmid B, Ribbe MW, Einsle O, Yoshida M, Thomas LM, Dean DR, Rees DC, Burgess BK. Science. 2002;296:352–356. doi: 10.1126/science.1070010. [DOI] [PubMed] [Google Scholar]
- (11).Ribbe MW, Burgess BK. Proc. Natl. Acad. Sci. USA. 2001;98:5521–5525. doi: 10.1073/pnas.101119498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Tenderholt A. PYSPLINE. Stanford Synchrotron Radiation Laboratory; Stanford, CA: 2006. [Google Scholar]
- (13).George GN. EXAFSPAK. Stanford Synchrotron Radiation Laboratory; Stanford, CA: 1990. [Google Scholar]
- (14).Rehr JJ, Albers RC. Rev. Mod. Phys. 2000;72:621–654. [Google Scholar]
- (15).Einsle O, Tezcan FA, Andrade SLA, Schmid B, Yoshida M, Howard JB, Rees DC. Science. 2002;297:1696–1700. doi: 10.1126/science.1073877. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.