Abstract
Elucidating mechanisms that differentiate motor neurons from interneurons is fundamental to understanding CNS development. Here we demonstrate that within the Drosophila NB 7-3/serotonergic lineage, different levels of Zfh-1 are required to specify unique properties of both motor neurons and interneurons. We present evidence that Zfh-1 is induced by Notch signaling and suppressed by the transcription factor Eagle. The antagonistic regulation of zfh-1 by Notch and Eagle results in Zfh-1 being expressed at low levels in the NB 7-3 interneurons and at higher levels in the NB 7-3 motor neurons. Furthermore, we present evidence that the induction of Zfh-1 by Notch occurs independently from canonical Notch signaling. We present a model where the differentiation of cell fates within the NB 7-3 lineage requires both canonical and non-canonical Notch signaling. Our observations on the regulation of Zfh-1 provide a new approach for examining the function of Zfh-1 in motor neurons and larval locomotion.
Keywords: Zfh-1, Notch, Eagle, NB7-3, Serotonin
Introduction
Zfh-1 is a zinc-finger/homeodomain protein (Fortini et al., 1991) conserved across the animal kingdom, that functions as a transcriptional repressor by binding to E boxes (Postigo et al., 1999). In Drosophila, it is expressed throughout the embryonic mesoderm, mesodermal-derived structures, and in the developing CNS including most, if not all, motor neurons (Lai et al., 1991; Layden et al., 2006). Zfh-1 has been shown to be required for gonad, muscle, and cardiac tissue differentiation as well as axonal guidance (Broihier et al., 1998; Garces and Thor, 2006; Lai et al., 1993; Layden et al., 2006; Moore et al., 1998; Postigo et al., 1999; Su et al., 1999). The C. elegans homolog of Zfh-1, ZAG-1, is expressed both in muscle and the nervous system and is required for differentiation and axonal guidance of both motor neurons and interneurons (Clark and Chiu, 2003; Wacker et al., 2003). Vertebrates have two Zfh-1 homologs that are widely expressed and have been shown to affect development in a variety of tissues such as thymus, skeletal, muscle, lens and specific neural crest cells (Bassez et al., 2004; Higashi et al., 1997; Maruhashi et al., 2005; Miyoshi et al., 2006; Muraoka et al., 2000; Postigo et al., 1999; Takagi et al., 1998; Van de Putte et al., 2003; Yoshimoto et al., 2005). The mouse homologs have been shown to bind Smads and can regulate BMP signaling (Postigo, 2003). In this manuscript we investigate the role of Zfh-1 in the specification and differentiation of the Drosophila serotonergic lineage.
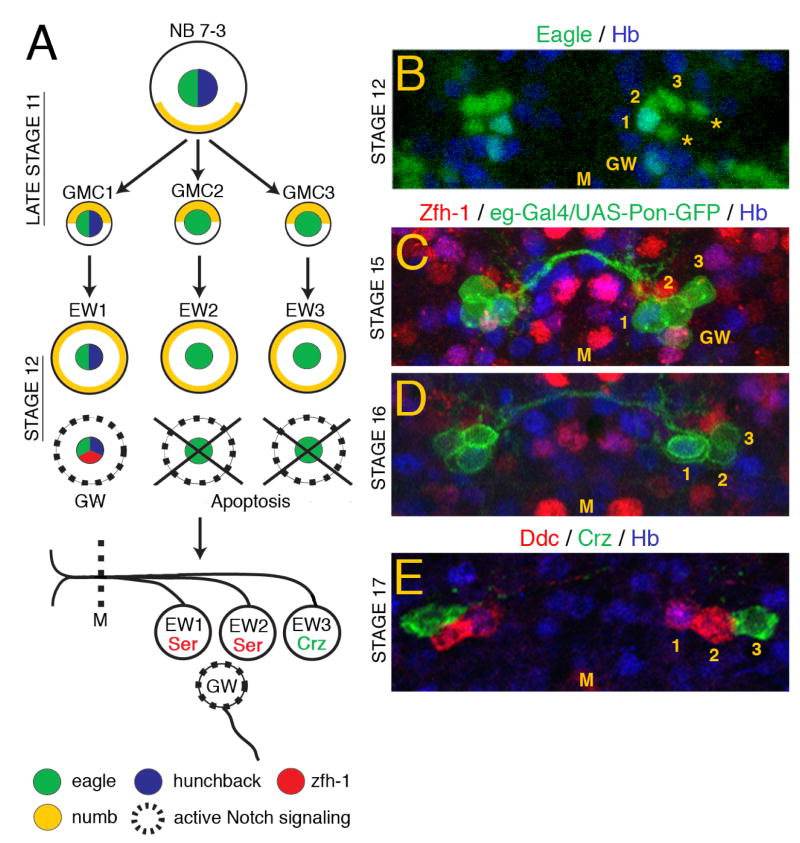
The serotonergic neurons of the Drosophila ventral nerve cord (VNC) arise from neuroblast 7-3 (NB 7-3) (Lundell et al., 1996). Neuroblasts are stem cells that undergo an invariant number of asymmetric cell divisions to produce several ganglion mother cells (GMCs). GMCs divide once to form a pair of neuronal or glial progeny (Hartenstein et al., 1987). There are 30 neuroblasts in each hemisegment of the VNC (Doe, 1992) that give rise to approximately 350 neurons and 30 glial cells by the end of embryogenesis (Schmid et al., 1999). Fig. 1A schematically outlines the divisions of NB 7-3 (Bossing et al., 1996; Dittrich et al., 1997; Higashijima et al., 1996; Isshiki et al., 2001; Karcavich and Doe, 2005; Lundell and Hirsh, 1998; Lundell et al., 2003; Novotny et al., 2002; Schmid et al., 1999). Three GMCs are derived from NB 7-3. GMC-1 produces two cells; GW, a motor neuron, and EW1, the more medial serotonergic neuron. GMC-2 produces EW2, the more lateral serotonergic neuron, and a sister cell that undergoes apoptosis. GMC-3 produces EW3, a neuron that synthesizes the neuropeptide corazonin and a sister cell that undergoes apoptosis. It has also been suggested that GMC-3 may not always undergo division but differentiate directly into EW3 (Karcavich and Doe, 2005). The three EW interneurons project axons across the midline to the contralateral side of the VNC and bifurcate in both the anterior and posterior directions. The GW motor neuron projects an ipsilateral axon that exits the CNS via the intersegmental nerve and innervates muscles 15–17 (Bossing et al., 1996; Dittrich et al., 1997; Higashijima et al., 1996; Schmid et al., 1999). The GW motor neuron can be distinguished from the EW interneurons by its high level of Zfh-1 immunoreactivity (Isshiki et al., 2001).
Fig. 1. Developmental stages and molecular markers of the NB 7-3 lineage.
(A) Schematic of NB 7-3 development, see text for details. (B) A stage 12 wild-type VNC abdominal segment showing 5–6 Eg positive progeny of the NB 7-3 lineage, only the GMC1 progeny are immunoreactive for Hb. The asterisks indicate sister cells of EW2 and EW3 that will undergo apoptosis. (C) A stage 15 VNC showing that only the GW motor neuron is immunoreactive for Zfh-1. (D) A stage 16 VNC showing that the GW neuron no longer expresses detectable levels of Eg. (E) A stage 17 VNC showing expression of the terminal markers Ddc and Corazonin. The yellow numbers 1, 2 and 3 indicate the three EW neurons. M indicates the midline.
All the cells of the NB 7-3 lineage express the transcription factor Eagle (Eg). Eg is a zinc finger protein homologous to the steroid receptor family (Higashijima et al., 1996; Rothe et al., 1989). It is expressed in only four neuronal lineages including NB 7-3 (Higashijima et al., 1996) and transiently in the embryonic gonad (Rothe et al., 1989). Mutant alleles of eg show the correct number of NB 7-3 progeny early in development, but there is a dramatic reduction in the number of detectable serotonin cells and the few remaining neurons have abnormal axon projections (Dittrich et al., 1997; Higashijima et al., 1996; Lundell and Hirsh, 1998). This suggests that Eg does not have a role in specification of cell identity in the NB 7-3 lineage but is important in the terminal differentiation of the progeny. The target genes of Eg regulation are unknown.
Previously, we have shown that Notch signaling is responsible for inducing apoptosis in the sister cells of EW2 and EW3 (Lundell et al., 2003). During division of the GMCs, Numb, a membrane-associated inhibitor of Notch signaling (Guo et al., 1996), is asymmetrically partitioned into the EW neurons. Numb inactivates the Notch receptors in these cells and prevents apoptosis (Karcavich and Doe, 2005; Lundell et al., 2003). Therefore, Numb inhibition of Notch signaling in NB 7-3 induces an interneuron cell fate, producing either serotonin or corazonin neurons, whereas activation of Notch signaling in NB 7-3 leads to apoptosis or the GW motor neuron cell fate.
Notch signaling is an evolutionarily conserved pathway that plays a critical role in both the specification and differentiation of cells. Various pathologies including cancer have been associated with aberrant regulation of Notch signaling (reviewed in Harper et al., 2003; Weinmaster and Kopan, 2006). Numerous studies in Drosophila have identified a canonical Notch signaling pathway where Delta and Serrate acting as ligands, lead to the proteolytic release of the Notch intracellular domain (NICD). The NICD then translocates to the nucleus and associates with the transcription factor Suppressor of Hairless (Su(H)) and other proteins to form a complex that activates gene transcription (recent reviews Bray, 2006; Le Borgne et al., 2005; Louvi and Artavanis-Tsakonas, 2006; Roegiers and Jan, 2004; Schweisguth, 2004). More recently, Notch signaling that is independent of Su(H) has been described (Fuwa et al., 2006; Hayward et al., 2005; Hori et al., 2004; Langdon et al., 2006; reviewed in Martinez Arias et al., 2002; Wilkin and Baron, 2005). The molecular mechanism of Su(H)-independent Notch signaling is uncertain and it is possible there may be more than one mechanism. It is unclear how different intracellular Notch-signaling mechanisms would be integrated in specific developmental pathways. This paper supports a model where both Su(H)-dependent and Su(H)-independent Notch signaling are required for the specification and maturation of cells within the serotonergic cell lineage of the Drosophila CNS.
In this manuscript we report that Zfh-1 is necessary for development of both the GW motor neuron and the EW interneurons in the NB7-3 cell lineage and that zfh-1 expression is regulated antagonistically by Notch and Eg. We also observe that both Zfh-1 and Notch signaling influence the direction of axonal projection in the NB 7-3 cell lineage. Finally we use our observations on the regulation of Zfh-1 to specifically reduce Zfh-1 expression in motor neurons and examine its role in larval locomotion.
RESULTS
Eagle immunoreactivity decreases more rapidly in the GW motor neuron than in the EW interneurons
Fig. 1A outlines the different stages of NB 7-3 development (Bossing et al., 1996; Dittrich et al., 1997; Higashijima et al., 1996; Isshiki et al., 2001; Karcavich and Doe, 2005; Lundell and Hirsh, 1998; Lundell et al., 2003; Novotny et al., 2002; Schmid et al., 1999). During late stage 11, NB 7-3 delaminates from the neural ectoderm and produces three GMCs. Numb protein is asymmetrically partitioned during division of each GMC, producing three EW progeny that receive Numb and three progeny cells that do not receive Numb, the GW cell and two apoptotic cells. The early NB 7-3 progeny can be detected with either an Eg antibody (Fig. 1B), an eg-Gal4 transgene (egmz360) (Figs. 1C,D) or an eg-lacZ transgene (eg289) (Fig. 3A). Hunchback (Hb) immunoreactivity is detected only in the GMC1 progeny, EW1 and GW (Figs. 1B–E). At stage 12, all six progeny cells can be detected with Eg immunoreactivity (Fig. 1B). The sister cells of EW2 and EW3 rapidly undergo apoptosis such that they are no longer detectable by stage 15 (Fig. 1C). At stage 15, Zfh-1 immunoreactivity is detected only in the GW motor neuron (Fig. 1C). Eg expression begins to decrease after stage 15, and it does so more rapidly in the GW cell such that by stage 16 the GW is no longer detectable with Eg antibody or the eg-Gal4/UAS-PON-GFP transgenes (Fig. 1D). Stable lacZ produced by the eg-lacZ transgene (eg289) can be detected in later stages (Lundell and Hirsh, 1998). By the end of stage 15 the EW cells begin to synthesize terminal products, Dopa decarboxylase (Ddc) and serotonin in EW1 and EW2, and corazonin in EW3. These markers become clearly detectable by stage 17 (Figs. 1E, 2E).
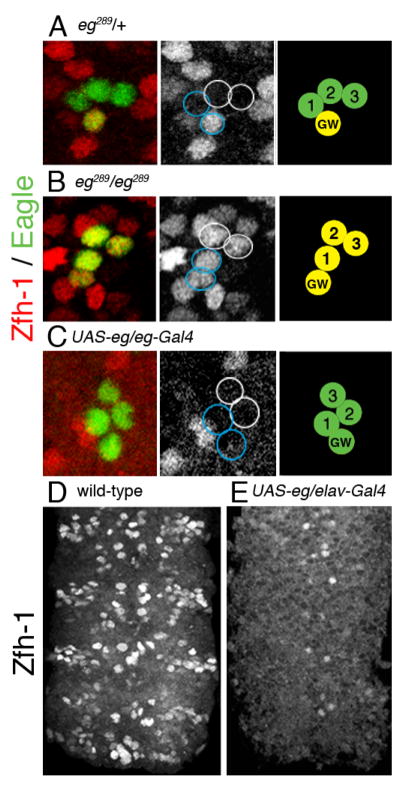
Fig. 3. Eagle suppresses zfh-1 expression.
(A–C) Abdominal hemisegments from stage 15 VNC. All VNC were immunostained for Zfh-1(red). Eg expression (green) was detected with either anti-lacZ antibody for the eg-lacZ (eg289) transgene (A,B) or anti-Eg antibody (C). Black and white panels show the Zfh-1 immunoreactivity separately for the same hemisegment. Blue circles indicate the GMC1 progeny that were determined by Hb immunoreactivity (actual immunoreactivity not shown). In the schematic the numbers 1, 2, and 3 indicate the three EW neurons, green cells are Eg positive and yellow cells are both Eg and Zfh-1 positive. The midline is to the left. (A) A wild-type (eg289/+) VNC showing that only the GW neuron is Zfh-1 positive. (B) Embryos homozygous for eg289 show Zfh-1 immunoreactivity in all four NB 7-3 progeny. (C) Ectopic expression of eg using the eg-Gal4 driver results in a loss of Zfh-1 immunoreactivity in the GW neuron. (D,E) Stage 15 VNC showing that ectopic expression of eg using the elav-Gal4 driver results in a loss of Zfh-1 immunoreactivity throughout the VNC.
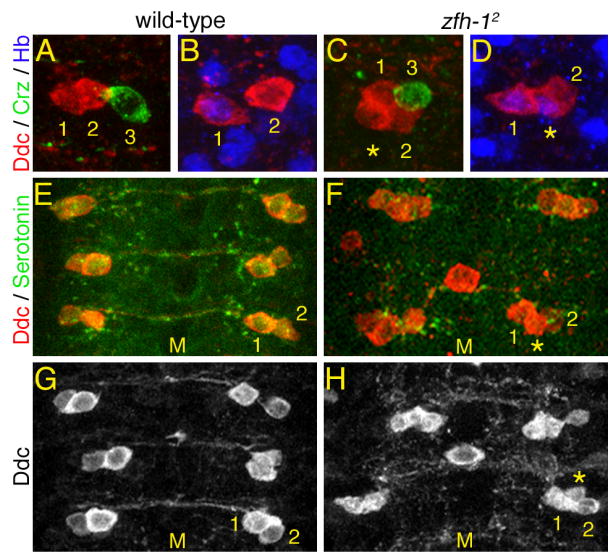
Fig. 2. zfh-12 mutant phenotypes of the NB 7-3 lineage.
All images are VNC abdominal segments from stage 17 embryos. The numbers 1, 2 and 3 indicate the EW neurons, the asterisks indicate ectopic Ddc neurons and M indicates the midline. (A,C) Embryos homozygous for zfh-12 show one additional Ddc positive cell per hemisegment. (B,D) The ectopic Ddc cell in the zfh-12 mutant is Hb positive, suggesting the GW has converted to an EW1 cell fate. (E,F) Embryos homozygous for zfh-12 show a severe reduction in serotonin immunoreactivity. (G,H) Embryos homozygous for zfh-12 show defasciculation of the EW axons. The CNS of zfh-12 mutant embryos is extended such that length of three abdominal segments in a wild-type VNC is equal to two abdominal segments in a zfh-12 VNC. Panels F and H also show a midline dopaminergic neuron that is immunoreactive for Ddc.
Zfh-1 is required for the differentiation of both the GW motor neuron and EW interneurons
To investigate the function of Zfh-1 in the NB 7-3 lineage we examined the protein-null allele, zfh-12 (Lai et al., 1993), in stage 17 embryos (Fig. 2. and Table 1). In the wild-type VNC, each hemisegment has two Ddc immunoreactive cells and one corazonin immunoreactive cell (Fig. 2A). In the zfh-12 mutant over 84% of the hemisegments have an extra cell positive for Ddc immunoreactivity (Figs. 2C,D,F, H and Table 1). This additional Ddc cell is also immunoreactive for Hb similar to the EW1 cell (Fig. 2D and Table 1). These results suggest that a loss of Zfh-1 in the GW motor neuron converts it to an EW1 interneuron cell fate and that Zfh-1 is necessary for motor neuron identity in the NB 7-3 lineage. It has previously been suggested that Zfh-1 is not required for specification of cell identity in the CNS (Lai et al., 1993; Layden et al., 2006), clearly this is not the case for the GW motor neuron and there may be other undetected examples.
Table 1. Analysis of molecular marker expression in the NB 7-3 lineage of wild-type and zfh-12 mutant embryos at stage 17.
For each genotype, hemisegments in abdominal segments 1–6 were counted for the number of NB 7-3 cells that expressed the specific antigen. The results are presented as a percentage of total number of hemisegments counted (n) and as an average number of cells per hemisegment
| Number of immunoreactive cells/hemisegment | ||||||||
|---|---|---|---|---|---|---|---|---|
| Antigen | Genotype | 0 | 1 | 2 | 3 | 4 | n | avg. ± s.d. |
| Ddc | wild-type zfh-12/zfh-12 |
0.6% 1.4% |
1.9% --- |
91.9% 14.4% |
5.6% 82.0% |
--- 2.2% |
161 139 |
2.02+0.32 2.80+0.50 |
| hunchback | wild-type zfh-12/zfh-12 |
2.0% 4.0% |
91.2% 15.8% |
6.9% 80.2% |
--- --- |
--- --- |
102 101 |
1.05+0.29 1.76+0.51 |
| serotonin | wild-type zfh-12/zfh-12 |
--- 88.8% |
3.4% 10.5% |
93.9% 0.8% |
2.7% --- |
--- --- |
148 134 |
1.99+0.25 0.12+0.35 |
| corazonin | wild-type zfh-12/zfh-12 |
2.4% 28.9% |
97.6% 68.5% |
--- 2.7% |
--- --- |
--- --- |
166 149 |
0.98+0.15 0.56+0.55 |
Despite the fact that Zfh-1 is not detectable in the EW interneurons with the Zfh-1 antibody, we were surprised to find that the zfh-12 allele also has an effect on terminal differentiation of the EW cells. In the zfh-12 mutant 88% of the detectable Ddc-positive cells, including the ectopic cell, fail to produce serotonin unlike the wild-type EW interneurons (Figs. 2E,F and Table 1). In addition, the number of cells detectable with corazonin antibody decreases 30% (Table 1). The axons of the Ddc-positive neurons, including the ectopic Ddc cell, appear to cross the midline; however, these axon bundles show defasciculation and increased varicosities relative to the wild-type axons (Figs. 2G,H). It has previously been observed that serotonin autoregulates the varicosity density of serotonin neurons (Budnik et al., 1989; Sykes and Condron, 2005). The reduction in serotonin levels induced by the zfh-12 mutation may be responsible for stimulating an increase in varicosity density. In summary, Zfh-1 is required for proper development of both the GW motor neuron and EW interneurons in the NB 7-3 lineage.
Eagle suppresses zfh-1 expression
As demonstrated in Fig. 1, Eg immunoreactivity declines more rapidly in the GW neuron. This suggests that eg expression might need to be suppressed in order to maintain high levels of Zfh-1. To test this possibility, we examined Zfh-1 immunoreactivity in the hypomorphic allele eg289. In the absence of Eg, all four cells in the NB7-3 lineage become Zfh-1 positive, including the EW interneurons (Fig. 3B). In a wild-type stage 15 VNC, the ratio of the number of Zfh-1 immunoreactive cells to the number of Eg immunoreactive cells (Zfh-1/Eg) is 23%, whereas in the eg289 genotype at stage 15 the ratio of Zfh-1/Eg immunoreactivity is 93% (Table 2).
Table 2. Analysis of Eg and Zfh-1 expression in the NB 7-3 lineage at stage 15.
For each genotype, hemisegments in abdominal segments 1–6 were counted for the number of NB 7-3 cells that expressed either the Eg or Zfh-1 antigen. The results are presented as an average number of cells per hemisegment. The two averages for each genotype are also presented as a ratio of Zfh-1 immunoreactive cells to Eg immunoreactive cells
| Genotype | Antigen | n | average number of cells ± s.d. | Zfh-1/Eg |
|---|---|---|---|---|
| eg289/+ (wild-type) | Eg-lacZ Zfh-1 |
113 | 3.81+0.48 0.89+0.43 |
23.4% |
| eg289/eg289 | Eg-lacZ Zfh-1 |
68 | 4.03+0.60 3.74+0.77 |
92.8% |
| eg-gal4/UAS-eg | Eg Zfh-1 |
86 | 4.10+0.51 0.21+0.41 |
5.1% |
| eg-Gal4/UAS-Notch/UAS-p35 | Eg-GFP Zfh-1 |
69 | 4.20+0.83 4.00+0.87 |
95.2% |
| eg-Gal4/UAS-p35 | Eg-GFP Zfh-1 |
87 | 4.43+0.82 1.97+0.75 |
44.5% |
| DlRevF10SerRX82/DlRevF10SerRX82 | Eg Zfh-1 |
74 | 5.12+0.60 2.00+0.60 |
39.1% |
| eg-gal4/UAS-Numb | Eg Zfh-1 |
85 | 5.00+0.73 1.93+0.75 |
38.6% |
| Su(H)1/Su(H)1 | Eg-lacZ Zfh-1 |
59 | 5.29+0.64 2.20+0.69 |
41.7% |
| Su(H)1/Su(H)1:eg289/eg289 | Eg-lacZ Zfh-1 |
26 | 5.42+0.70 5.04+0.72 |
92.9% |
When a UAS-eg allele is over-expressed in the lineage using the eg-Gal4 driver, 95% of the GW neurons are no longer detectable with Zfh-1 immunoreactivity (Fig. 3C, Table 2). Occasionally this results in two EW1-type neurons that are both Ddc and Hb positive (data not shown), which is identical to the zfh-12 mutant phenotype (Fig. 2D). The suppression effect that Eg has on the expression of zfh-1 can be dramatically demonstrated if eg is mis-expressed throughout the CNS using the pan-neural driver elav-Gal4. Zfh-1 immunoreactivity is diminished in nearly all cells except for a few near the midline (Figs. 3D,E).
Together, these results demonstrate that Eg is both necessary and sufficient to suppress Zfh-1 expression in the NB 7-3 lineage. Presumably the preferential loss of Eg in the GW cell in a wild-type embryo (Fig. 1C) allows for the continued expression of zfh-1, whereas the high expression of Eg in the EW neurons reduces zfh-1 expression to very low levels.
Notch induces Zfh-1 expression in the NB 7-3 lineage
Since the GW neuron does not inherit Numb and has active Notch signaling, we asked whether Notch signaling could be responsible for inducing expression of Zfh-1 in the GW motor neuron. First, we investigated whether Zfh-1 immunoreactivity is detectable in the sister cells of EW2 and EW3 prior to apoptosis, since these cells also do not receive Numb. Fig. 4A shows a stage 12 embryo where all six Eg positive cells of the lineage are detected, three of the cells are immunoreactive for Zfh-1. By stage 15 the sisters of EW2 and EW3 have undergone apoptosis and only the GW shows Zfh-1 immunoreactivity (Fig. 4B). Thus, all the cells in the NB 7-3 lineage that have active Notch signaling are positive for Zfh-1 immunoreactivity.
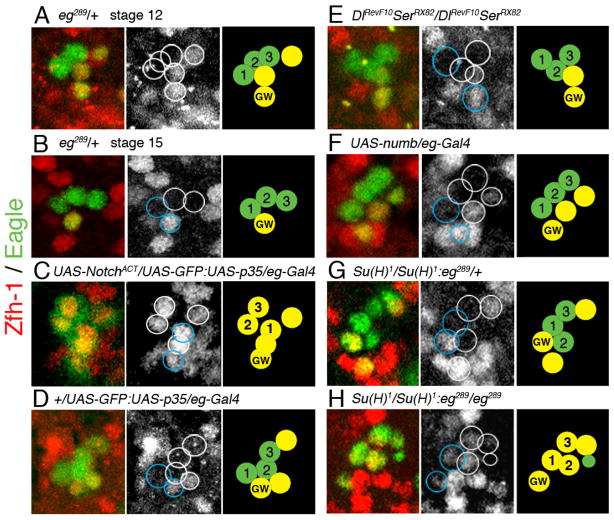
Fig. 4. Notch regulation of zfh-1 expression.
All images are abdominal hemisegments from stage 15 VNC, except for panel A which is stage 12. All VNC were immunostained for Zfh-1(red). Eg expression (green) was detected with either anti-lacZ antibody for the eg-lacZ (eg289) transgene (A,B,G,H), GFP fluorescence for the eg-gal4/UAS-GFP transgenes (C,D), or anti-Eg antibody (E,F). The black and white panels and the schematic are as described in the legend for Fig. 3. Panel B is repeated from Fig. 3. (A) A wild-type (eg289/+) stage 12 VNC showing all six NB 7-3 progeny. The GW neuron and the cells undergoing apoptosis are Zfh-1 positive. (B) A wild-type (eg289/+) stage 15 VNC showing that after apoptosis only the GW neuron is Zfh-1 positive. (C) Ectopic Notch expression induces Zfh-1 immunoreactivity in all NB 7-3 progeny. (D) Ectopic p35 rescues cells from apoptosis but does not alter Zfh-1 immunoreactivity. (E–G) Three genetic conditions that reduce canonical Notch signaling all show rescue of apoptotic cells but Zfh-1 immunoreactivity is identical to wild-type. (H) Embryos homozygous for Su(H)1 and eg289 show both the rescue of apoptotic cells and expansion of Zfh-1 immunoreactivity into the EW neurons. The number of cells that can be rescued from apoptosis is variable: (C,D,F,G) show the rescue of both cells, (E) shows the rescue of only one cell and (H) shows the rescue of one cell and the other undergoing apoptosis.
Previously we reported that expression of a constitutively active Notch allele, UAS-NotchACT (Doherty et al., 1996), within the NB 7-3 lineage, induces apoptosis of EW2 and EW3 and produces a second cell immunoreactive for Zfh-1 (Lundell et al., 2003). This result suggests that in the presence of ectopic Notch the EW1 cell converts to a GW cell fate. Here we show that if we express both UAS-NotchACT and UAS-p35 (an apoptosis inhibitor) with an eg-Gal4 driver, all detectable cells of the lineage are immunoreactive for Zfh-1 (Fig. 4C). The ratio of Zfh-1/Eg immunoreactivity is 95% (Table 2). Even the EW neurons that normally lack Zfh-1 are positive for ZFh-1 immunoreactivity when Notch is ectopically expressed. The eg-Gal4/UAS-p35 control shows that the rescued apoptotic cells are Zfh-1 positive, but that the EW neurons remain Zfh-1 negative (Fig. 4D). These results demonstrate that in the NB 7-3 lineage Notch signaling is sufficient to induce zfh-1 expression and shows a positive correlation between cells that have active Notch receptors and Zfh-1 expression.
Canonical Notch signaling in the NB 7-3 lineage is necessary for apoptosis but not for Zfh-1 expression
To demonstrate that Notch is required for zfh-1 expression we tested three conditions where canonical Notch signaling is inhibited: a Dl/Ser double mutant null allele (Fig. 4E), the ectopic expression of numb (Fig. 4F) and a Su(H) null allele (Fig. 4G). We found that for all three genotypes, 74–90% of the hemisegments at stage 15 show rescue of at least one of the apoptotic sisters of EW2 and EW3. Zfh-1 immunoreactivity is still detected in the GW neuron and is also detected in the rescued apoptotic cells. The ratio of Zfh-1/Eg immunoreactivity is approximately 40% (Table 2). Although there is variability in the number of cells rescued from apoptosis when Notch signaling is inhibited, these rescued cells are always positive for Zfh-1 immunoreactivity. A Su(H)1:eg289 double mutant not only shows rescue of the apoptotic cells, but also shows that the expanded Zfh-1 immunoreactivity detected in the EW neurons due to eg289, also occurs in the absence of Su(H) (Fig. 4H, Table 2). Thus, while Notch-induced apoptosis in the NB 7-3 lineage is clearly dependent on the Notch/Delta/Numb/Su(H) canonical signaling pathway, disruptions in canonical Notch signaling do not effect the expression of zfh-1.
Mis-expression of Zfh-1 and Notch prevent axonal midline crossing of the EW neurons
We next asked whether Notch signaling or Zfh-1 expression is important for the distinction between the direction of GW and EW axonal projection. The EW interneurons send axons across the midline to the contralateral side, whereas the GW motor neuron extends an axon on the ipsilateral side (Bossing et al., 1996; Dittrich et al., 1997; Higashijima et al., 1996; Schmid et al., 1999). To visualize the axons, we used eg-Gal4 to drive a membrane bound GFP (UAS-Pon-GFP or UAS-mCD8-GFP). Fig. 5A shows the wild-type axonal projection at stage 15 of both NB 7-3 and another Eg immunoreactive lineage, NB 3-3. The three EW axons cross the midline through the posterior commisure (closed arrow), the single GW axon remains ipsilateral (arrowheads) and the axons of the NB 3-3 lineage cross the midline through the anterior commisure (open arrow). We focused our study on the midline crossing of the EW neurons since the single axon of the GW neuron is more difficult to detect.
Fig. 5. Zfh-1 and Notch regulate axonal projections of NB 7-3 progeny.
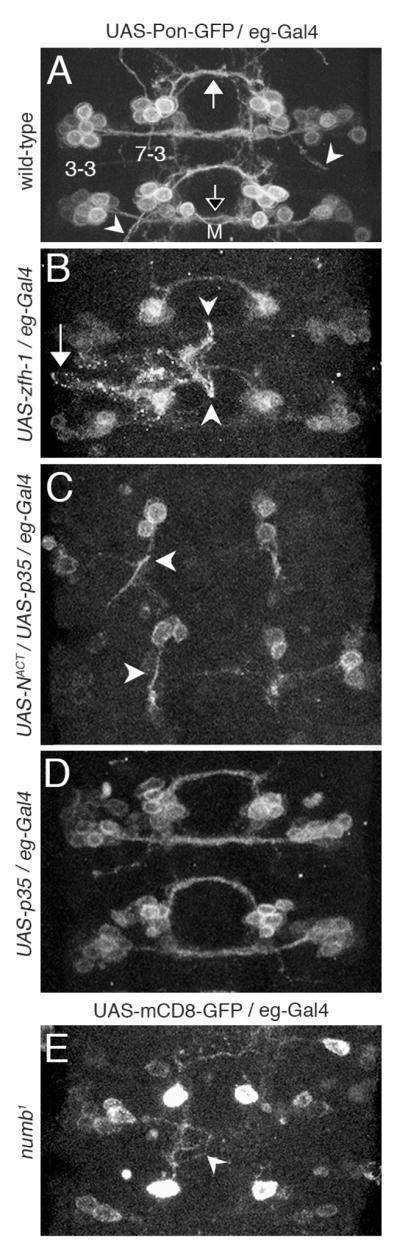
All images are VNC abdominal segments from stage 15 embryos. To visualize axons eg-Gal4 was used to drive expression of a membrane bound GFP marker (either UAS-Pon-GFP or UAS-mCD8-GFP). M indicates the midline. (A) A wild-type VNC showing the EW axonal bundle of the NB 7-3 lineage crossing the midline through the posterior commissure (closed arrow), the NB 3-3 axonal bundle crossing the midline through the anterior commissure (open arrow) and the GW ipsilateral axons (arrowheads). (B) The ectopic expression of zfh-1 shows aberrant EW axon projections (arrow and arrowheads) but all hemisegments maintain some EW midline crossing. (C) The ectopic expression of NotchACT blocks all midline crossing of the EW neurons and produces a thick ipsilateral projection (arrowheads). (D) The ectopic expression of p35 rescues cells from apoptosis but has no effect on EW midline crossing. (E) The numb1 mutation blocks midline crossing of EW1.
Layden et. al. (2006) previously concluded that when Zfh-1 is ectopically expressed in the EW neurons using UAS-zfh-1 and the eg-Gal4 driver, many of the EW neurons abnormally extend axons laterally out of the CNS. Using the same genetic background we find a similar result that 25% of the hemisegments (n=130) show EW neurons with aberrant projections and varicosities (Fig. 5B arrow and arrowheads). Some of these axons project laterally as previously reported (arrow), but we additionally see that some axons project to the midline but are inhibited from crossing (arrowheads). Apparently ectopic Zfh-1 either represses some factor required for midline crossing or triggers a repulsive cue that prohibits midline crossing.
We obtain a more dramatic result on the projection of EW axons when NotchACT and p35 are coexpressed using the eg-Gal4 driver. In this genetic background midline crossing of EW axons is completely abolished (n=202). Instead, the NB 7-3 cluster makes a single thick ipsilateral projection (Fig. 5C). A control showing ectopic expression of p35 alone shows rescue of the apoptotic cells but does not alter midline crossing of the EW neurons (Fig. 5D). In a numb1 mutant, Notch repression is lost in the EW neurons and only the EW1 cell survives Notch-induced apoptosis (Lundell et al., 2003). In 91% of the numb1 hemisegments (n = 69) the EW1 axons fail to cross the midline. Often the axons seem to wander aimlessly (Fig. 5E arrowhead). These results suggest that Notch signaling must be repressed by Numb for midline crossing of the EW neurons and that active Notch signaling in the GW neurons may direct ipsilateral projection of the axon. In summary, both Notch and Zfh-1 can influence axon projections in the NB 7-3 lineage.
Ectopic expression of Eagle in motor neurons decreases OK6 gene expression and larval locomotion
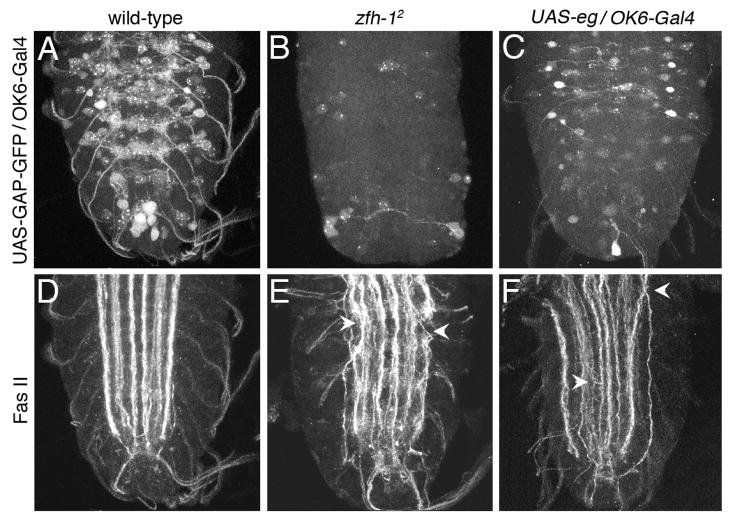
Although Zfh-1 has been shown to be expressed in most motor neurons its role in locomotor activity has not be tested directly because all zfh-1 mutant alleles, including zfh-12, are embryonic lethal. Having established that ectopic expression of Eg can suppress Zfh-1 expression (Fig. 3), we asked whether we could use Eg suppression to reduce Zfh-1 levels specifically in motor neurons, thus avoiding mesodermal defects and maintaining viability. To test this we mis-expressed Eg with the OK6-Gal4 enhancer trap that has been shown to express in most identifiable motor neurons (Aberle et al., 2002; Sanyal et al., 2003). When the OK6-Gal4 is used to drive expression of UAS-GAP-GFP a large number of cell bodies and axons are detected in a wild-type stage 17 embryonic VNC (Fig. 6A). In the zfh-12 mutant this same GFP fluorescence is dramatically reduced (Fig. 6B), suggesting that the loss of Zfh-1 substantially alters development of these cells so that they are no longer capable of expressing the motor neuron marker OK6. This could be due either to an intrinsic alteration of cell fate or due to alterations in axonal projections that prevent the cells from responding to extrinsic signals responsible for OK6 induction. When UAS-eg is mis-expressed in this same set of cells with the OK6-Gal4 driver the level of fluorescence is intermediate between wild-type and the zfh-12 mutant (Fig. 6C). This effect of ectopic Eg is presumably due to its ability to suppress zfh-1(Fig. 3).
Fig. 6. Zfh-1 is required for motor neuron development and axon fasciculation.
All images show a VNC from stage 17 embryos. (A–C) Motor neuron cell bodies and axons are detected using OK6-Gal4 to drive expression of UAS-GAP-GFP. Both the zfh-12 mutation and ectopic expression of eg reduces the number of cells detected with OK6-GFP. (D–F) Lateral projecting motor neurons and longitudinal axon bundles are detected with FasII immunoreactivity. Both the zfh-12 mutation and the ectopic expression of eg result in disorganized lateral motor neurons and defasciculation of the longitudinal bundles. Arrowheads indicate aberrant crossing between adjacent longitudinal bundles.
The similarity of zfh-12 and UASeg/OK6-gal4 phenotype is also observed with FasII immunoreactivity, which detects three interneuronal fasicles in the longitudinal connectives and lateral projecting motor neuron axons (Fig. 6D). In both the zfh-12 mutant (Fig. 6E) and the UAS-eg/OK6-Gal4 line (Fig. 6F) the lateral projecting motor neuron axon bundles are disorganized and not always detectable. The interneuronal longitudinal axonal bundles are also disorganized and show defasciculation. Some axons leave their appropriate tract and cross to a neighboring tract (Fig. 6E,F arrowheads). The defasciculation of the longitudinal bundles is similar to that what we previously observed for the EW neurons (Fig. 2H). Thus the loss of Zfh-1 and ectopic expression of Eg with OK6-Gal4 have similar detrimental effects on motor neuron development and fasciculation throughout the VNC.
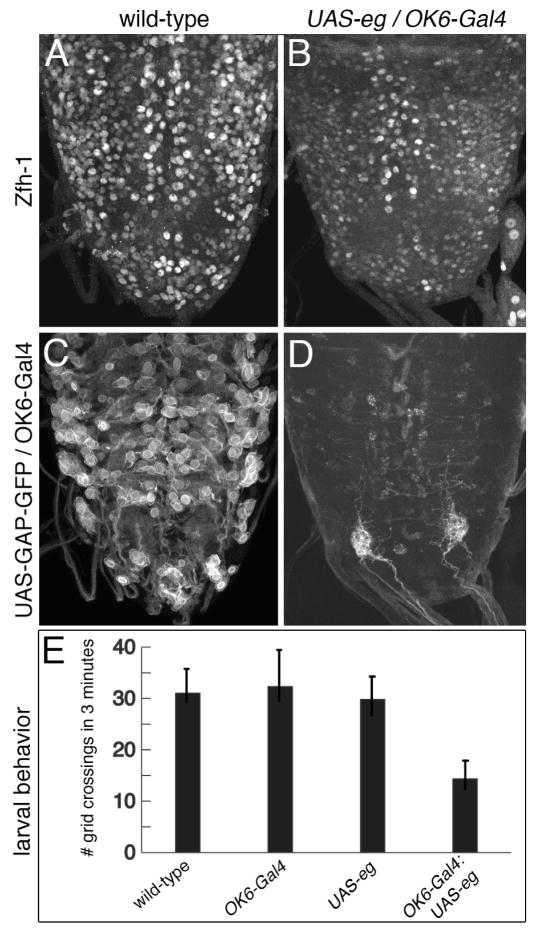
Whereas the zfh-12 mutation is an embryonic lethal the ectopic expression of eg with OK6-Gal4 produces viable larvae (77% survival). Compared to wild-type the UAS-eg/OK6-Gal4 larval VNC shows a significant reduction in both Zfh-1 immunoreactivity (Figs. 7A,B) and OK6-Gal4/UAS-GAP-GFP fluorescence (Figs. 7C,D). When these same larvae are tested in a standard locomotor crawling assay, the activity of the UAS-eg/OK6-Gal4 larvae is reduced by 50% compared to the control larvae (Fig. 7E). The ectopic expression of eg appears to immobilize the posterior end of the larvae. Whereas wild-type larvae will roam over the entire surface of the Petri plate, the locomotor activity in the ectopic Eg larvae is restricted to about 25% of the test plate area. This loss in locomotor activity is consistent with a disruption of the motor system, although the exact nature of the motor neuron defect induced by ectopic eg expression will require further investigation. Using Eg to suppress expression of Zfh-1 in specific subsets of cells will allow further examination of the role of Zfh-1 in motor neuron development without the contributing mesodermal defects that exist in zfh-1 mutant embryos.
Fig. 7. Reduced levels of Zfh-1 alter larval locomotor behavior.
(A,C) A third instar larval VNC showing wild-type Zfh-1 immunoreactivity and OK6-Gal4/UAS-GAP-GFP fluorescence. (B,D) A third instar larval VNC showing that ectopic expression of eg reduces both Zfh-1 immunoreactivity and OK6-GFP. (E) Ectopic expression of eg in motor neurons that express OK6 reduces third instar larval locomotor activity by 50% relative to three control lines with a significant difference in the mean activity of P<0.0001. At least 20 larvae were tested for each line.
Discussion
Eagle/Notch regulation of zfh-1 expression in NB 7-3 lineage
Our data shows that in the NB 7-3 lineage, zfh-1 is induced by Notch and repressed by Eg. Zfh-1 is expressed in all cells where Notch signaling is active (GW and apoptotic cells, Fig. 4A) and ectopic expression of Notch can induce ectopic Zfh-1 in the EW neurons (Fig. 4C). In addition we have previously shown that a loss of Notch signaling with a sanpodo mutant can lead to a conversion of the GW neuron to an EW1 neuron (Lundell et al., 2003) similar to the Zfh-1 mutation presented in Fig. 2. However, we have also shown that three different mutations (DlRevF10SerRX82, UAS-numb and Su(H)1) in the canonical Notch signaling pathway do not reduce Zfh-1 expression (Figs. 4E–G), suggesting Zfh-1 expression is independent of canonical Notch signaling. We propose that an alternative non-canonical Notch pathway can induce zfh-1 expression. This would explain how Notch can be sufficient to induce zfh-1 expression (Fig. 4C), but work independently of Delta, Numb and Su(H) (Figs. 4E–H). Furthermore, since canonical Notch signaling is repressed in the EW neurons by Numb, a non-canonical pathway would explain how zfh-1 expression could be induced in the EW neurons, which we have shown becomes detectable in the eg289 mutant (Fig. 3B) and is necessary for EW maturation (Fig. 2). An alternative explanation of our results would be that all three mutations we have used maintain some residual level of canonical Notch signaling which is sufficient to induce zfh-1, but insufficient to induce apoptosis. Although a formal possibility we think this unlikely to be the case, since it’s doubtful that all three of our independent analyses would have suffered from leaky Notch signaling. In addition, if the distinguish between apoptosis and zfh-1 expression were solely dependent on levels of canonical Notch signaling one would expect variability in these mutants where some of the cells would have low enough Notch levels to both rescue cells from apoptosis and inhibit zfh-1 expression, but all rescued cells are consistently Zfh-1 positive. Therefore our results favor an interpretation where alternative mechanisms of Notch signaling induce zfh-1 expression and apoptosis.
We have shown that Eg efficiently suppress zfh-1 expression in NB 7-3 lineage (Figs. 3B,C) and other Zfh-1 expressing neurons when ectopically expressed (Fig. 3E). In a wild-type fly, the preferential loss of Eg in the GW by stage 16 (Fig. 1D) insures that detectable Zfh-1 is limited just to the GW motor neuron. Although the regulation of eg expression is unknown, we have previously shown that Eg can negatively regulate its own expression in the GW neuron (Lundell and Hirsh, 1998). It is also possible that eg expression might be reciprocally suppressed by Zfh-1. Since detectable Eg expression is limited to only four neuroblast lineages in the wild-type CNS, it may be necessary to globally restricted Eg expression throughout the CNS to allow for zfh-1 expression that is found in many lineages. In summary, the levels of zfh-1 expression in the NB 7-3 lineage are carefully regulated by both Notch and Eg.
The role of Notch in the NB 7-3 lineage is similar to asymmetric divisions of mesodermal progenitor cells in the heart where the cells that have active Notch signaling, also have Zfh-1 expression, and the Zfh-1 expression appears to be independent of the Numb/Notch pathway (Su et al., 1999). On the other hand, these results are fundamentally different from the aCC/pCC and RP2/RP2sib asymmetric neuronal lineages where Zfh-1 is expressed in the cell where Notch signaling is inactivated by Numb. In these lineages mutations in the canonical Notch signaling pathway lead to a de-repression and expansion of Zfh-1 expression (Skeath and Doe, 1998). Our results present a possible explanation for these lineage differences in that the variation could be due to multiple Notch mechanisms that induce zfh-1 expression and/or various cell specific regulators of zfh-1expression, similar to the function of Eg presented in this manuscript.
The role of Zfh-1 in motor neuron development
Zfh-1 has previously been shown to be expressed in most if not all motor neurons and has been implicated in the differentiation of motor neurons (Garces and Thor, 2006; Lai et al., 1991; Layden et al., 2006). Here we show that the GW motor neuron of the NB 7-3 lineage converts to an EW1 interneuron fate in the absence of Zfh-1 (Fig. 2D). Although a zfh-1 mutation has been shown to disrupt cell fates in mesodermal tissues (Broihier et al., 1998; Lai et al., 1993; Su et al., 1999), this is the first report of a neuronal conversion in cell fate.
When we examined the motor system in the zfh-12 null mutant more globally, we found that the motor neuron marker OK6 is completely disrupted and that both longitudinal and lateral FasII axon bundles show disorganization and defasciculation (Fig. 6). Layden et al. recently reported that a loss of function in zfh-1 prevents some motor axons from exiting the Drosophila (Layden et al., 2006). A mutation in zag-1, the C. elegans zfh-1 homolog, also shows errors in axon guidance, fasciculation and branching of both motor neurons and interneurons (Clark and Chiu, 2003; Wacker et al., 2003). Finally we have shown that ectopic Eg can suppress both zfh-1 and OK-6 expression throughout the CNS (Figs. 3E,7B,7D) and produce larvae with obvious locomotor defects (Fig. 7E). All of these results support the hypothesis that Zfh-1 has a critical role in motor neuron differentiation.
The role of Zfh-1 in interneuron development
We were surprised to find that the zfh-12 mutant allele also affects maturation of the EW interneurons. The Zfh-1 antibody does not detect protein in the EW interneurons (Fig 1. C), except when Eg suppression is removed (Fig. 3B). These results suggest that, in a wild-type fly, Zfh-1 is expressed in the EW neurons at low undetectable levels.
The zfh-12 allele disrupts expression of the terminal products, serotonin and corazonin and also causes defasciculation and ectopic branching of the EW axons (Fig. 2). In C. elegans, a mutation in zag-1 disrupts the development of the HSN serotonergic neurons (Clark and Chiu, 2003). The HSN neurons show aberrant axon projections, branch extensively and downregulate tryptophan hydroxylase the rate limiting enzyme in serotonin biosynthesis. Ectopic expression of zfh-1 leads to aberrant projections of the EW axons (Fig. 5B and Layden et al., 2006). We have previously reported a similar mutant phenotype for eg289 (Lundell and Hirsh, 1998), which we have shown leads to ectopic zfh-1 expression (Fig. 3B). Therefore although Zfh-1 is required for fasciculation and maturation of the EW interneurons, the level of Zfh-1 in these cells must be carefully maintained by Eg suppression to prevent detrimental effects that can be induced by excessive Zfh-1.
The role of Zfh-1 and Notch in axonal guidance of NB 7-3 progeny
Because the GW motor neuron and the EW interneurons have very different axon projections we asked whether Zfh-1 expression or Notch signaling might be responsible for this variation. We found dramatic ipsilateral rerouting of EW axons when Notch is overexpressed (Fig. 5C). Notch appears to have a fundamental role in differentiating between ipsilateral and contralateral projections. Presumably the unrestricted canonical Notch signaling in the GW neuron would direct an ipsilateral projection, whereas Numb blocks this function in the EW neurons allowing contralateral projections. Notch signaling has been reported to direct axonal guidance in other lineages of Drosophila (Crowner et al., 2003; Giniger, 1998; Giniger et al., 1993; Skeath and Doe, 1998)
We observe that ectopic Zfh-1 can misdirect EW axons away from the midline (Fig. 5B), as previously reported (Layden et al., 2006). However, we also found that axons that project toward the midline can be abruptly repelled when expressing ectopic Zfh-1 (Fig. 5B). This suggests that ectopic Zfh-1 might make the cells more sensitive to a midline repulsive cue, such as Slit or Netrin. This could occur by either inducing the expression of a member of the Roundabout (Robo) family, the Slit receptor (reviewed in Couch and Condron, 2002; Dickson, 2002) or UNC5 a repulsive Netrin receptor (Keleman and Dickson, 2001). Alternatively Zfh-1 might downregulate Commissureless (Comm), an intracellular protein that induces Robo degradation. Robo, Comm and Slit have been shown to be involved in the midline crossing of the EW neurons (Couch et al., 2004). We have previously observed that eg289 can also prevent midline crossing of the EW interneurons (Lundell and Hirsh, 1998).
Therefore, ectopic expression of both Notch and Zfh-1 can prevent midline crossing in the NB 7-3 lineage. This may not be surprising since we have shown that Notch can induce zfh-1 expression. The observation that ectopic Notch signaling produces a more severe phenotype than ectopic Zfh-1 suggests that the role of Notch in guidance of NB 7-3 axons is probably more complex than simply inducing zfh-1 expression. The specific epistatic relationship of Notch and Zfh-1 with respect to the growth of the GW axon remains to be determined.
The role of canonical and non-canonical Notch signaling in NB 7-3 lineage
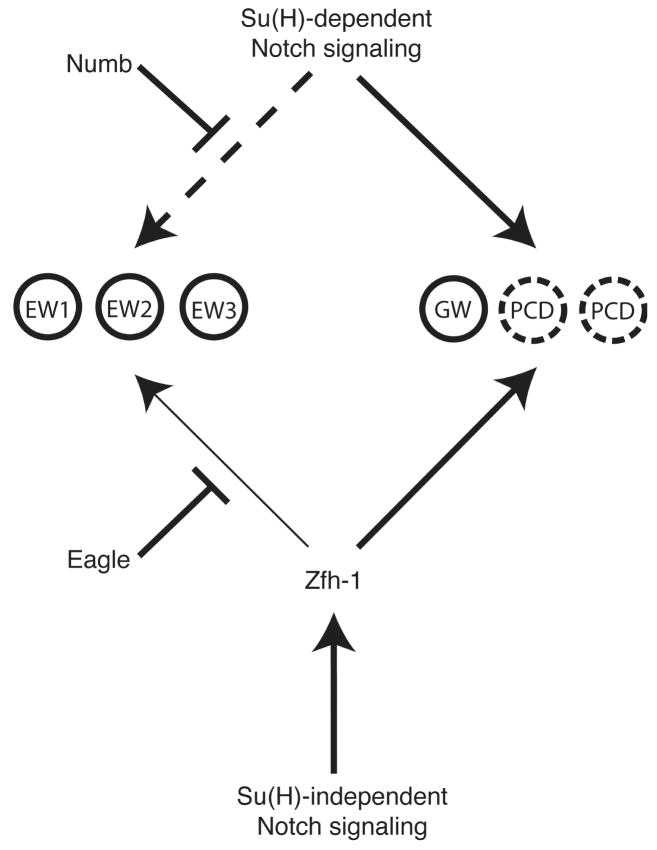
Our results suggest a model where both canonical and non-canonical Notch signaling have a role in development of the NB 7-3 lineage (Fig. 8). We propose that canonical/Su(H)-dependent Notch signaling is required for inducing apoptosis and ipsilateral axonal guidance, whereas non-canonical/Su(H)-independent Notch signaling is responsible for inducing zfh-1 expression.
Fig. 8. A model for the regulation of NB 7-3 development by Notch, Eagle and Zfh-1.
Su(H)-dependent Notch signaling is active in the GW motor neuron and apoptotic sister cells (dashed circles) but is inhibited by Numb in the EW interneurons (dashed arrow). Zfh-1 is expressed at high levels in the GW motor neuron and apoptotic cells but is limited in expression in the EW interneurons by Eg (thin arrow). The results presented in this manuscript suggest that Zfh-1 may be induced by Su(H)-independent Notch signaling. Thus both Su(H)-dependent and Su(H)-independent Notch signaling have specific functions within the lineage.
In EW interneurons, Su(H)-dependent Notch signaling is blocked by the asymmetric distribution of Numb; this prevents both apoptosis (Karcavich and Doe, 2005; Lundell et al., 2003) and ipsilateral axonal projection (Fig. 5E). Zfh-1 is induced in the EW interneurons by Su(H)-independent Notch signaling but Eg counteracts this pathway keeping Zfh-1 at very low (undetectable) levels (Fig. 3B). The low level of Zfh-1 maintained in the EW interneurons is critical for axon fasciculation and terminal differentiation (Figs. 2F,H).
The GW motor neuron not only has Su(H)-dependent Notch signaling, but the preferential loss of Eg expression in GW (Fig. 1D) might also activate the Su(H)-independent Notch signaling pathway. Both of these pathways could contribute to the high levels of Zfh-1 unique to the GW neuron. The high concentration of Zfh-1 specifies the GW motor neuron cell fate (Fig. 2C) and contributes to determining the ipsilateral projection of its axon (Fig. 5B and Layden et al., 2006).
Apoptosis within the NB 7-3 lineage is clearly driven by canonical Notch signaling since a numb mutant induces cell death (Karcavich and Doe, 2005; Lundell et al., 2003) and Dl−, Su(H)− or UAS-numb alleles are all capable of rescuing the cells from cell death (Fig. 4E–H). Despite the fact that the apoptotic cells express Zfh-1 (Fig. 4A), apoptosis is not dependent on Zfh-1 since these cells still die in zfh-12 mutant flies (Fig. 2C).
Thus, our model suggests that canonical and non-canonical Notch signaling have specific functions within the NB 7-3 lineage. It remains to be determined how Su(H)-independent Notch signaling activates Zfh-1 expression, how Eg can block this process and how the intracellular components of two different Notch signaling mechanisms are linked or resolved in the GW motor neuron. Furthermore, Wingless (wg) has been proposed as a ligand for Notch that can antagonize Su(H)-independent Notch signaling (reviewed in Martinez Arias et al., 2002). A wg mutation result in a loss of detectable NB 7-3 progeny (Deshpande et al., 2001). Examination of the relationship of Notch and Wnt signaling in the NB 7-3 lineage should provide further insight into the crosstalk between these pathways.
EXPERIMENTAL METHODS
Drosophila Stocks
The following fly lines were used in this work: eg-lacZ (eg289), eg-Gal4 (egmz360), UAS-eg (from G. Technau); UAS-Pon-GFP, numb1 (from C. Doe); UAS-NACT (Notch with a deleted extracellular domain but intact transmembrane and intracellular domains), UAS-numb (from Y. N. Jan); OK6-Gal4 (from B. Zhang); zfh-12 (a protein null allele from Z.C. Lai); DlRevF10SerRX82 (a lethal null allele, where both mutations are deletions), Su(H)1(a lethal null allele, where the molecular lesion is unknown), Elav-Gal4, UAS-GFP, UAS-zfh-1, UAS-GAP-GFP, UAS-p35, UAS-mCD8-GFP and Canton S (from the Bloomington stock center).
Immunohistochemistry
Embryonic and larval CNS were dissected, fixed in 4% paraformaldehyde and subsequently incubated with primary and secondary antisera as previously described (Lundell and Hirsh, 1994). Primary antibodies were: rabbit anti-Eg (1:2000, G. Technau), guinea pig anti-Hb (1:600, East Asian Distribution Center for Segmentation Antibodies), rabbit anti-Ddc (1:50, M. Lundell), rat anti-Ddc (1:200, M. Lundell), rabbit anti-Crz (1:200, C. Doe), rabbit anti-Zfh-1 (1:400, R. Lehman), guinea pig anti-Zfh-1 (1:450, Z. C. Lai), rat anti-5-HT (1:100, Accurate Chemical), mouse anti-FasII monoclonal1D4 (1:5, Developmental Studies Hybridoma Bank) and rat anti-lacZ (1:5000, C. Doe). All Texas Red, FITC and Cy5 secondary antibodies were from Jackson Laboratories and used at 1/200. Images were obtained using a BioRad 1024 laser-scanning microscope and edited with Adobe Photoshop 7.0.
Larval Locomotor Assay
Larvae were subjected to a 12-hour light/dark cycle. The assay was done two hours before the light-to-dark transition time. Third-instar larvae were randomly sampled and placed on a Petri dish (8.5 × 1.4 cm) that was coated with 10 ml of nonnutritive 0.8% agar. After two minutes to allow for acclimatization, larvae locomotion was tested for three minutes. To measure the trail distance, the Petri dish was placed on top of a light box that had a marked grid (5×5mm). The number of grid squares the larva crossed in three minutes was counted. At least 20 larvae of each genotype were tested.
Statistical Analysis
Statistical analysis was performed using SAS software (SAS Institute, Inc.). The larval locomotor assay was analyzed using analysis of variance (ANOVA) followed by Tukey’s Studentized Range Test to compare larval activity means in four genotypes: Canton S, OK6-Gal4, UAS-eg and OK6-Gal4/UAS-eg.
Acknowledgments
This work was supported by a NIH/MBRS/SCORE grant GM 08194 to M.J.L. We thank C. Doe, Y. N. Jan, Z.C. Lai, G. Technau, B. Zhang and the Bloomington stock center for provision of fly stocks. We thank C. Doe, R. Lehman, Z.C. Lai, G. Technau, the Iowa Hybridoma Bank and East Asian Distribution Center for provision of antibodies. We thank K.W. Lee for assistance with the behavioral assay. We thank E. Wheeler and P. Mueller for their comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aberle H, Haghighi AP, Fetter RD, McCabe BD, Magalhaes TR, Goodman CS. wishful thinking encodes a BMP type II receptor that regulates synaptic growth in Drosophila. Neuron. 2002;33:545–558. doi: 10.1016/s0896-6273(02)00589-5. [DOI] [PubMed] [Google Scholar]
- Bassez G, Camand OJ, Cacheux V, Kobetz A, Dastot-Le Moal F, Marchant D, Catala M, Abitbol M, Goossens M. Pleiotropic and diverse expression of ZFHX1B gene transcripts during mouse and human development supports the various clinical manifestations of the “Mowat-Wilson” syndrome. Neurobiol Dis. 2004;15:240–250. doi: 10.1016/j.nbd.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Bossing T, Udolph G, Doe CQ, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev Biol. 1996;179:41–64. doi: 10.1006/dbio.1996.0240. [DOI] [PubMed] [Google Scholar]
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Broihier HT, Moore LA, Van Doren M, Newman S, Lehmann R. zfh-1 is required for germ cell migration and gonadal mesoderm development in Drosophila. Development. 1998;125:655–666. doi: 10.1242/dev.125.4.655. [DOI] [PubMed] [Google Scholar]
- Budnik V, Wu CF, White K. Altered branching of serotonin-containing neurons in Drosophila mutants unable to synthesize serotonin and dopamine. J Neurosci. 1989;9:2866–2877. doi: 10.1523/JNEUROSCI.09-08-02866.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SG, Chiu C. C. elegans ZAG-1, a Zn-finger-homeodomain protein, regulates axonal development and neuronal differentiation. Development. 2003;130:3781–3794. doi: 10.1242/dev.00571. [DOI] [PubMed] [Google Scholar]
- Couch J, Condron B. Axon guidance: Comm hither, Robo. Curr Biol. 2002;12:R741–742. doi: 10.1016/s0960-9822(02)01253-8. [DOI] [PubMed] [Google Scholar]
- Couch JA, Chen J, Rieff HI, Uri EM, Condron BG. robo2 and robo3 interact with eagle to regulate serotonergic neuron differentiation. Development. 2004;131:997–1006. doi: 10.1242/dev.00962. [DOI] [PubMed] [Google Scholar]
- Crowner D, Le Gall M, Gates MA, Giniger E. Notch steers Drosophila ISNb motor axons by regulating the Abl signaling pathway. Curr Biol. 2003;13:967–972. doi: 10.1016/s0960-9822(03)00325-7. [DOI] [PubMed] [Google Scholar]
- Deshpande N, Dittrich R, Technau GM, Urban J. Successive specification of Drosophila neuroblasts NB 6-4 and NB 7-3 depends on interaction of the segment polarity genes wingless, gooseberry and naked cuticle. Development. 2001;128:3253–3261. doi: 10.1242/dev.128.17.3253. [DOI] [PubMed] [Google Scholar]
- Dickson BJ. Molecular mechanisms of axon guidance. Science. 2002;298:1959–1964. doi: 10.1126/science.1072165. [DOI] [PubMed] [Google Scholar]
- Dittrich R, Bossing T, Gould AP, Technau GM, Urban J. The differentiation of the serotonergic neurons in the Drosophila ventral nerve cord depends on the combination function of the zinc finger proteins Eagle and Huckebein. Development. 1997;124:2515–2525. doi: 10.1242/dev.124.13.2515. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development. 1992;116:855–863. doi: 10.1242/dev.116.4.855. [DOI] [PubMed] [Google Scholar]
- Doherty D, Feger G, Younger-Shepherd S, Jan LY, Jan YN. Delta is a ventral to dorsal signal complementary to Serrate, another Notch ligand, in Drosophila wing formation. Genes Dev. 1996;10:421–434. doi: 10.1101/gad.10.4.421. [DOI] [PubMed] [Google Scholar]
- Fortini ME, Lai Z, Rubin GM. The Drosophila zfh-1 and zfh-2 genes encode novel proteins containing both zinc-finger and homeodomain motifs. Mechs of Dev. 1991;34:113–122. doi: 10.1016/0925-4773(91)90048-b. [DOI] [PubMed] [Google Scholar]
- Fuwa TJ, Hori K, Sasamura T, Higgs J, Baron M, Matsuno K. The first deltex null mutant indicates tissue-specific deltex-dependent Notch signaling in Drosophila. Mol Genet Genomics. 2006;275:251–263. doi: 10.1007/s00438-005-0087-3. [DOI] [PubMed] [Google Scholar]
- Garces A, Thor S. Specification of Drosophila aCC motoneuron identity by a genetic cascade involving even-skipped, grain and zfh1. Development. 2006;133:1445–1455. doi: 10.1242/dev.02321. [DOI] [PubMed] [Google Scholar]
- Giniger E. A role for Abl in Notch signaling. Neuron. 1998;20:667–681. doi: 10.1016/s0896-6273(00)81007-7. [DOI] [PubMed] [Google Scholar]
- Giniger E, Jan LY, Jan YN. Specifying the path of the intersegmental nerve of the Drosophila embryo: a role for Delta and Notch. Development. 1993;117:431–440. doi: 10.1242/dev.117.2.431. [DOI] [PubMed] [Google Scholar]
- Guo M, Jan LY, Jan YN. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron. 1996;17:27–41. doi: 10.1016/s0896-6273(00)80278-0. [DOI] [PubMed] [Google Scholar]
- Harper JA, Yuan JS, Tan JB, Visan I, Guidos CJ. Notch signaling in development and disease. Clin Genet. 2003;64:461–472. doi: 10.1046/j.1399-0004.2003.00194.x. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Rudloff E, Campos-Ortega JA. The pattern of proliferation of the neuroblasts in the wild-type embryo of Drosophila melanogaster. Roux’s Arch Dev Biol. 1987;196:473–485. doi: 10.1007/BF00399871. [DOI] [PubMed] [Google Scholar]
- Hayward P, Brennan K, Sanders P, Balayo T, DasGupta R, Perrimon N, Martinez Arias A. Notch modulates Wnt signalling by associating with Armadillo/beta-catenin and regulating its transcriptional activity. Development. 2005;132:1819–1830. doi: 10.1242/dev.01724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi Y, Moribe H, Takagi T, Sekido R, Kawakami K, Kikutani H, Kondoh H. Impairment of T cell development in deltaEF1 mutant mice. J Exp Med. 1997;185:1467–1479. doi: 10.1084/jem.185.8.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashijima S, Shishido E, Matsuzaki M, Saigo K. Eagle, a member of the steroid-receptor gene superfamily, is expressed in a subset of neuroblasts and regulates the fate of their putative progeny in the Drosophila CNS. Development. 1996;122:527–536. doi: 10.1242/dev.122.2.527. [DOI] [PubMed] [Google Scholar]
- Hori K, Fostier M, Ito M, Fuwa TJ, Go MJ, Okano H, Baron M, Matsuno K. Drosophila deltex mediates suppressor of Hairless-independent and late-endosomal activation of Notch signaling. Development. 2004;131:5527–5537. doi: 10.1242/dev.01448. [DOI] [PubMed] [Google Scholar]
- Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- Karcavich R, Doe CQ. Drosophila neuroblast 7-3 cell lineage: a model system for studying programmed cell death, Notch/Numb signaling, and sequential specification of ganglion mother cell identity. J Comp Neurol. 2005;481:240–251. doi: 10.1002/cne.20371. [DOI] [PubMed] [Google Scholar]
- Keleman K, Dickson BJ. Short- and long-range repulsion by the Drosophila Unc5 netrin receptor. Neuron. 2001;32:605–617. doi: 10.1016/s0896-6273(01)00505-0. [DOI] [PubMed] [Google Scholar]
- Lai ZC, Fortini ME, Rubin GM. The embryonic expression patterns of zfh-1 and zfh-2, two Drosophila genes encoding novel zinc-finger homeodomain proteins. Mech Dev. 1991;34:123–134. doi: 10.1016/0925-4773(91)90049-c. [DOI] [PubMed] [Google Scholar]
- Lai ZC, Rushton E, Bate M, Rubin GM. Loss of function of the Drosophila zfh-1 gene results in abnormal development of mesodermally derived tissues. Proc Natl Acad Sci U S A. 1993;90:4122–4126. doi: 10.1073/pnas.90.9.4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon T, Hayward P, Brennan K, Wirtz-Peitz F, Sanders P, Zecchini V, Friday A, Balayo T, Martinez Arias A. Notch receptor encodes two structurally separable functions in Drosophila: a genetic analysis. Dev Dyn. 2006;235:998–1013. doi: 10.1002/dvdy.20735. [DOI] [PubMed] [Google Scholar]
- Layden MJ, Odden JP, Schmid A, Garces A, Thor S, Doe CQ. Zfh1, a somatic motor neuron transcription factor, regulates axon exit from the CNS. Dev Biol. 2006;291:253–263. doi: 10.1016/j.ydbio.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Le Borgne R, Bardin A, Schweisguth F. The roles of receptor and ligand endocytosis in regulating Notch signaling. Development. 2005;132:1751–1762. doi: 10.1242/dev.01789. [DOI] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Lundell MJ, Chu-LaGraff Q, Doe CQ, Hirsh J. The engrailed and huckebein genes are essential for development of serotonin neurons in the Drosophila CNS. Mol Cell Neurosci. 1996;7:46–81. doi: 10.1006/mcne.1996.0004. [DOI] [PubMed] [Google Scholar]
- Lundell MJ, Hirsh J. Temporal and spatial development of serotonin and dopamine neurons in the Drosophila CNS. Dev Biol. 1994;165:385–396. doi: 10.1006/dbio.1994.1261. [DOI] [PubMed] [Google Scholar]
- Lundell MJ, Hirsh J. eagle is required for the specification of serotonin neurons and other neuroblast 7-3 progeny in the Drosophila CNS. Development. 1998;125:463–472. doi: 10.1242/dev.125.3.463. [DOI] [PubMed] [Google Scholar]
- Lundell MJ, Lee HK, Perez E, Chadwell L. The regulation of apoptosis by Numb/Notch signaling in the serotonin lineage of Drosophila. Development. 2003;130:4109–4121. doi: 10.1242/dev.00593. [DOI] [PubMed] [Google Scholar]
- Martinez Arias A, Zecchini V, Brennan K. CSL-independent Notch signalling: a checkpoint in cell fate decisions during development? Curr Opin Genet Dev. 2002;12:524–533. doi: 10.1016/s0959-437x(02)00336-2. [DOI] [PubMed] [Google Scholar]
- Maruhashi M, Van De Putte T, Huylebroeck D, Kondoh H, Higashi Y. Involvement of SIP1 in positioning of somite boundaries in the mouse embryo. Dev Dyn. 2005;234:332–338. doi: 10.1002/dvdy.20546. [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Maruhashi M, Van De Putte T, Kondoh H, Huylebroeck D, Higashi Y. Complementary expression pattern of Zfhx1 genes Sip1 and deltaEF1 in the mouse embryo and their genetic interaction revealed by compound mutants. Dev Dyn. 2006;235:1941–1952. doi: 10.1002/dvdy.20799. [DOI] [PubMed] [Google Scholar]
- Moore LA, Broihier HT, Van Doren M, Lehmann R. Gonadal mesoderm and fat body initially follow a common developmental path in Drosophila. Development. 1998;125:837–844. doi: 10.1242/dev.125.5.837. [DOI] [PubMed] [Google Scholar]
- Muraoka O, Ichikawa H, Shi H, Okumura S, Taira E, Higuchi H, Hirano T, Hibi M, Miki N. Kheper, a novel ZFH/deltaEF1 family member, regulates the development of the neuroectoderm of zebrafish (Danio rerio) Dev Biol. 2000;228:29–40. doi: 10.1006/dbio.2000.9909. [DOI] [PubMed] [Google Scholar]
- Novotny T, Eiselt R, Urban J. Hunchback is required for the specification of the early sublineage of neuroblast 7-3 in the Drosophila central nervous system. Development. 2002;129:1027–1036. doi: 10.1242/dev.129.4.1027. [DOI] [PubMed] [Google Scholar]
- Postigo AA. Opposing functions of ZEB proteins in the regulation of the TGFbeta/BMP signaling pathway. Embo J. 2003;22:2443–2452. doi: 10.1093/emboj/cdg225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postigo AA, Ward E, Skeath JB, Dean DC. zfh-1, the Drosophila homologue of ZEB, is a transcriptional repressor that regulates somatic myogenesis. Mol Cell Biol. 1999;19:7255–7263. doi: 10.1128/mcb.19.10.7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roegiers F, Jan YN. Asymmetric cell division. Curr Opin Cell Biol. 2004;16:195–205. doi: 10.1016/j.ceb.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Rothe M, Nauber U, Jäckle H. Three hormone receptor-like Drosophila genes encode an identical DNA- binding finger. Embo J. 1989;8:3087–3094. doi: 10.1002/j.1460-2075.1989.tb08460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal S, Narayanan R, Consoulas C, Ramaswami M. Evidence for cell autonomous AP1 function in regulation of Drosophila motor-neuron plasticity. BMC Neurosci. 2003;4:20. doi: 10.1186/1471-2202-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid A, Chiba A, Doe CQ. Clonal analysis of Drosophila embryonic neuroblasts: neural cell types, axon projections and muscle targets. Development. 1999;126:4653–4689. doi: 10.1242/dev.126.21.4653. [DOI] [PubMed] [Google Scholar]
- Schweisguth F. Notch signaling activity. Curr Biol. 2004;14:R129–138. [PubMed] [Google Scholar]
- Skeath JB, Doe CQ. Sanpodo and Notch act in opposition to Numb to distinguish sibling neuron fates in the Drosophila CNS. Development. 1998;125:1857–1865. doi: 10.1242/dev.125.10.1857. [DOI] [PubMed] [Google Scholar]
- Su MT, Fujioka M, Goto T, Bodmer R. The Drosophila homeobox genes zfh-1 and even-skipped are required for cardiac-specific differentiation of a numb-dependent lineage decision. Development. 1999;126:3241–3251. doi: 10.1242/dev.126.14.3241. [DOI] [PubMed] [Google Scholar]
- Sykes PA, Condron BG. Development and sensitivity to serotonin of Drosophila serotonergic varicosities in the central nervous system. Dev Biol. 2005;286:207–216. doi: 10.1016/j.ydbio.2005.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi T, Moribe H, Kondoh H, Higashi Y. DeltaEF1, a zinc finger and homeodomain transcription factor, is required for skeleton patterning in multiple lineages. Development. 1998;125:21–31. doi: 10.1242/dev.125.1.21. [DOI] [PubMed] [Google Scholar]
- Van de Putte T, Maruhashi M, Francis A, Nelles L, Kondoh H, Huylebroeck D, Higashi Y. Mice lacking ZFHX1B, the gene that codes for Smad-interacting protein-1, reveal a role for multiple neural crest cell defects in the etiology of Hirschsprung disease-mental retardation syndrome. Am J Hum Genet. 2003;72:465–470. doi: 10.1086/346092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker I, Schwarz V, Hedgecock EM, Hutter H. zag-1, a Zn-finger homeodomain transcription factor controlling neuronal differentiation and axon outgrowth in C. elegans. Development. 2003;130:3795–3805. doi: 10.1242/dev.00570. [DOI] [PubMed] [Google Scholar]
- Weinmaster G, Kopan R. A garden of Notch-ly delights. Development. 2006;133:3277–3282. doi: 10.1242/dev.02515. [DOI] [PubMed] [Google Scholar]
- Wilkin MB, Baron M. Endocytic regulation of Notch activation and down-regulation (review) Mol Membr Biol. 2005;22:279–289. doi: 10.1080/09687860500129778. [DOI] [PubMed] [Google Scholar]
- Yoshimoto A, Saigou Y, Higashi Y, Kondoh H. Regulation of ocular lens development by Smad-interacting protein 1 involving Foxe3 activation. Development. 2005;132:4437–4448. doi: 10.1242/dev.02022. [DOI] [PubMed] [Google Scholar]