Abstract
There is increasing evidence that UVA radiation, which makes up ∼95% of the solar UV light reaching the Earth's surface and is also commonly used for cosmetic purposes, is genotoxic. However, in contrast to UVC and UVB, the mechanisms by which UVA produces various DNA lesions are still unclear. In addition, the relative amounts of various types of UVA lesions and their mutagenic significance are also a subject of debate. Here, we exploit atomic force microscopy (AFM) imaging of individual DNA molecules, alone and in complexes with a suite of DNA repair enzymes and antibodies, to directly quantify UVA damage and reexamine its basic mechanisms at a single-molecule level. By combining the activity of endonuclease IV and T4 endonuclease V on highly purified and UVA-irradiated pUC18 plasmids, we show by direct AFM imaging that UVA produces a significant amount of abasic sites and cyclobutane pyrimidine dimers (CPDs). However, we find that only ∼60% of the T4 endonuclease V-sensitive sites, which are commonly counted as CPDs, are true CPDs; the other 40% are abasic sites. Most importantly, our results obtained by AFM imaging of highly purified native and synthetic DNA using T4 endonuclease V, photolyase, and anti-CPD antibodies strongly suggest that CPDs are produced by UVA directly. Thus, our observations contradict the predominant view that as-yet-unidentified photosensitizers are required to transfer the energy of UVA to DNA to produce CPDs. Our results may help to resolve the long-standing controversy about the origin of UVA-produced CPDs in DNA.
Introduction
Ultraviolet (UV) radiation spans the range of wavelengths between 200 and 400 nm and is divided into three groups: UVC (200–290 nm), UVB (290–320 nm), and UVA (320–400 nm). The biological effects of UVC and UVB have been studied extensively, and it has been generally concluded that both types of UV light may directly and indirectly damage DNA, contributing to various types of skin cancer (1–8). The main DNA lesions generated by UVC and UVB include direct products of photochemical reactions within DNA, such as cyclobutane pyrimidine dimers (CPDs) and 6-4 lesions (5,8–11). Other types of damage include single- and double-strand breaks (SSBs and DSBs, respectively), and numerous modified bases, such as 8-oxo-guanine, thymine glycol, 5,6-dihydrothymine, and cytosine photohydrate (5,12–15). All of these DNA alterations are well characterized chemically and have been precisely quantified for various absorbed doses of UV (2,3,16).
On the other hand, the biological effects of UVA have been studied only fairly recently (6–8,10,13,16–20), even though it is the predominant UV radiation to which humans are exposed. The initial results suggest strong mutagenic properties of this ever-present radiation (5–8,10,13,16,21–24). However, the distribution and accurate fractions of various DNA lesions attributed to UVA radiation are still unknown, and the various results obtained in different studies are a subject of a debate (6–8,20,24,25).
Recently, Mouret et al. (7) and Douki et al. (10) postulated that UVA-induced CPDs are the main promutagenic DNA lesions. However, the mechanism by which they are generated remains unclear (7,24,26). In earlier works by Kielbassa et al. (13), Kuluncsics et al. (20), and Perdiz et al. (25), UVA was proposed to generate CPDs directly. However, based on the most recent analysis of the types of CPDs induced by UVA, and the apparent lack of 6-4 photoproducts among UVA-produced lesions, other investigators (5,7,10,16) support the notion that these CPDs are produced indirectly. The putative mechanism would involve a triplet energy transfer of UVA that must be absorbed by an as-yet-unidentified photosensitizer (7,16,27). However, this view seems to downplay the important fact that the absorbance of UVA light by DNA, although believed to be small, is not insignificant (26), and experiments with UVA light typically involve very large radiation doses. In our opinion, the lack of an understanding of the fundamental mechanism by which UVA generates CPDs warrants new studies in this area.
Because of the extraordinary complexity of the intracellular environment, it is difficult to examine the mechanism of UVA damage to DNA in vivo. Therefore, to clarify the origin of UVA-induced CPDs, we simplified the system to be studied and examined isolated and dialyzed DNA that was purified of any possible photosensitizers. There are many methods that can be used to detect and quantify DNA damage (2,7–11,28–32). These methods typically involve fairly harsh DNA degradation, processing, staining, and labeling procedures, which by themselves may obscure or even alter the DNA damage. It should be also noted that in some of the earlier works on UVA damage to DNA, T4 endonuclease V was used as a gold standard for detection and/or calibration of these lesions (10,13,20,25,32). However, this enzyme detects not only CPDs but also abasic sites. Thus, it is possible that the number of CPDs reported in studies that exploited T4 endonuclease V may have been significantly overestimated. In the work presented here, we addressed this issue by directly determining the fraction of UVA-induced T4 endonuclease V-sensitive sites that correspond to abasic sites. To identify and accurately quantify UVA damage in this work, we used a novel direct single-molecule approach (33,34). This method involves atomic force microscopy (AFM) imaging of individual DNA molecules (35–41) and DNA complexes with photodamage-specific enzymes and antibodies (see Materials and Methods). AFM detection of DNA damage has several advantages over other methods in that harsh DNA treatment and degradation are avoided, extremely small amounts of DNA and protein material are needed for observation, and DNA and proteins do not need to be labeled or stained. Of importance, AFM imaging of photolyase and antibodies allows for direct detection of the location of each lesion. To the best of our knowledge, this work shows the first direct images of individual anti-CPD antibodies on DNA captured by AFM.
Materials and Methods
Materials
The enzymes and antibody used for UVA damage detection are summarized in Table 1 (42). pUC18 (2686 basepairs) was isolated from Escherichia coli and purified using the Qiafilter plasmid maxi kit (Qiagen, Valencia, CA). Poly(dA)-poly(dT) was purchased from Sigma-Aldrich (St. Louis, MO). DNA was dialyzed in ultrapure Millipore water or buffer using a Slide-A-Lyzer mini dialysis unit from Pierce Biotechnology (Rockford, IL) following its normal dialysis protocol. The molecular weight cutoff of the membrane of the chosen dialysis tube was 10K to remove all possible small-molecule chemicals, such as salts and possible chromophores. Tris-EDTA buffer 100× concentrate, sodium chloride solution (∼5 M), and magnesium chloride (1 M) were purchased from Sigma-Aldrich.
Table 1.
Enzymes and antibody used for UVA damage detection
| Enzyme or antibody for damage detection | Supplier | Specific lesions these enzymes can detect | Enzyme's activity (42) |
|---|---|---|---|
| E. coli endonuclease IV | New England Biolabs | AP site base paired with adenine | 100% |
| 5,6-dihydrothymine | <10% | ||
| T4 endonuclease V | New England Biolabs and Epicentre | CPDs | 100% |
| AP site base paired with adenine | 100% | ||
| E. coli endonuclease III | Trevigen | thymine glycol | 100% |
| AP site base paired with adenine | 100% | ||
| 5,6-dihydrothymine | <10% | ||
| E. coli Fpg formamidopyrimidine DNA glycosylase | Trevigen | 8 oxoguanine base paired with a cytosine or guanine | 100% |
| AP site base paired with adenine | <10% | ||
| 5,6-dihydrothymine | <10% | ||
| 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine | |||
| E. coli photolyase | Trevigen | cis-syn CPDs | |
| Antithymine-dimer antibody, clone KTM53 | Kamiya Biomedical | Thymine dimers | |
UVA irradiation
For broadband UVA, irradiations were performed at the peak wavelength of 365 nm using a high-intensity UVA lamp (model B-100A) from UVP (Upland, CA). The intensity of UV light was measured by a UVX radiometer from UVP and read a value of ∼450 W/m2 at the distance of 10 cm, where the DNA was irradiated. For narrowband UVA, a bandpass filter was inserted between the UVA source and sample to cut off both sides of the UVA spectrum (43). The high-transmission UV bandpass filter (model XHQA365; Asahi Spectra, Torrance, CA) had a central wavelength of 365 nm, a full width at half-maximum of 10 nm, and a minimum transmission of 70%. Then 50 μL of a 40 μg/mL supercoiled pUC18 DNA solution in different buffers were put into an open NMR tube exposed to UVA light at room temperature for different durations. NMR tubes were cut to 1 cm length and cleaned by 12 M hot HCl for 30 min before use. NMR tubes were used instead of plastic tubes to reduce a possible reaction of the tube material to the radiation, causing the release of possible DNA damaging products (20).
Endonuclease enzyme treatment of DNA
Irradiated or control supercoiled pUC18 plasmids (5.64 nM or 10 μg/mL, respectively) were incubated with different kinds of enzymes to a total volume of 40 μL at 37°C for 30–60 min in their preferred reaction buffer, as suggested by the enzyme provider. Control experiments showed that when the concentration of the enzymes was twice that suggested by the manufacturer, the number of lesions detected was only slightly higher as compared to the number of lesions detected with the manufacturer-suggested amount of enzyme. Thus, to make sure that all of the lesions generated were detected, we used concentrations of the enzymes that were five times higher than suggested by the manufacturers. After enzyme incubation, all of the solutions were diluted by DNA buffer (10 mM Tris HCl, 1 mM EDTA, and 100 mM NaCl) to the final DNA concentration of 0.5–1 μg/mL and then deposited on 1-(3-aminopropyl)silatrane-functionalized mica (APS-mica) (44) for imaging.
Photolyase treatment of DNA
DNA (10 μg/mL) was incubated with photolyase (2 μg/mL) in 1× REC buffer 14 (20 mM Tris-HCl, pH = 7.8; 1 mM EDTA; 1 mM DTT; and 50 mM NaCl) at a total volume of 20 μL. The incubation was performed for 30 min at room temperature in darkness to prevent the photolyase from repairing damage and disassociating from the damage sites. The solution was then diluted with the addition of 10 mM Tris HCl, 1 mM EDTA, and 5–10 mM MgCl2 buffer to a final DNA concentration of 0.5–1 μg/mL. The sample was then deposited onto a freshly cleaved mica surface immediately for AFM imaging.
Antithymine-dimer antibody treatment of DNA
DNA (10 μg/mL) was incubated with an antithymine dimer (0.5–1.0 μg/mL) in PBS buffer at a total volume of 20 μL. The incubation was performed for 30 min at 37°C. Then the solution was diluted by PBS buffer again to the final DNA concentration of 0.5–1 μg/mL. The sample was then deposited onto an APS-mica surface immediately for AFM imaging.
Immobilization of DNA molecules for AFM imaging
For experiments in which the imaging buffer did not have MgCl2, APS-mica was used for the binding of DNA molecules. APS-mica was prepared as described by Shlyakhtenko et al. (44). A drop of 10–50 μL of DNA solution (DNA concentration of 0.5–1 μg/mL) was deposited on the APS-mica surface at room temperature for 3 min. The sample was rinsed and air-dried before imaging.
AFM imaging
Images were taken using a Nanoscope IIIa MultiMode scanning probe microscope (Veeco Instruments, Santa Barbara, CA) using Tapping mode with an E scanner. RTESP probes (Veeco) were used for imaging in air. The spring constant of the AFM cantilevers was 20–80 N/m and their resonance frequency was 275–316 kHz. All images were collected at a scan rate of 2.0–3.0 Hz, a scan resolution of 512 × 512 pixels, and scan sizes of 1000–5000 nm. In each experiment, 24–36 AFM images were captured and more than 500 DNA molecules were analyzed to determine the fractions of supercoiled, circular, and linear molecules. The results are expressed as the mean ± SD for each fraction.
Quantification and verification of DNA damage from AFM images
Supercoiled DNA is a good model for studying DNA damage because its configuration is highly sensitive to environmental conditions and structural modifications, such as SSB and DSB. It responds to different buffer or salt conditions by undergoing large topological changes (33,45,46), such as switching freely between supercoiled and circular forms. One SSB will relax its superhelical structure to a circular form permanently. Similarly, one DSB will open the circular plasmid to a linear form. Plasmid relaxation will occur spontaneously if SSBs are generated by the UV light. In addition, typical UV-produced damage sites will be converted to SSBs by means of damage-specific endonucleases. All of these topological or length variations of DNA are easy to resolve using AFM imaging (44,47–53). Our criteria for differentiating between intact and damaged DNA are described in detail elsewhere (33). Briefly, pUC18 molecules with more than five supercoiled nodes were considered intact, whereas molecules with five or fewer nodes indicated relaxed DNA. The number of various lesions was quantified by a Poisson distribution (54,55) based on percentages of supercoiled and relaxed DNA. The average number of lesions per molecule, P, can be obtained from P = −ln[f(P,0)], where f(P,0) is the fraction of supercoiled (intact) plasmids (34). Using this method, we can directly detect and quantify UVA damage to supercoiled DNA by visualizing and counting individual intact and damaged plasmids captured by AFM imaging. To remove bias and verify the accuracy of damage quantification, we performed numerous blind tests in which the person who analyzed the AFM images did not know which sample they originated from. We also verified our AFM assay by performing gel electrophoresis (33,34) (see Fig. S3 and Table S1 in the Supporting Material).
Results and Discussion
UVA produces a variety of lesions in highly purified DNA
As our model system for investigating UVA damage, we chose supercoiled DNA because its configuration is highly sensitive to structural modifications such as SSB and DSB, to which it responds with large topological changes that are easily identified with AFM imaging (33,45,46,50) (see Materials and Methods). Since UVA is expected to produce SSB and DSB, these lesions can be detected directly. In addition, CPDs (7,10,17,23,24) and other oxidative lesions can be easily converted into SSBs with the use of appropriate endonucleases (e.g., T4 endonuclease V for CPDs; see Table 1 for the enzymes used for damage detection). The supercoiled DNA plasmid pUC18, which we used in this study, has been extracted from E. coli cells. Thus, it might have certain impurities that could potentially mediate energy transfer to DNA. The purpose of dialysis is thus to remove these impurities before the DNA is treated with UVA (see Materials and Methods).
The AFM image in Fig. 1 A shows the UVA damage detected directly without using any enzyme, and Fig. 1 C shows the distribution of the different plasmid configurations. After ∼1.3 MJ/m2 of UVA irradiation at 365 ± 5 nm (see Materials and Methods), 88.2 ± 1.9% of DNA remained supercoiled, and 11.1 ± 1.0% of plasmid relaxed to a circular form. According to a Poisson distribution (see Materials and Methods), the number of SSBs developed is 0.125 SSB/pUC18, i.e., 46.7 ± 1.5 lesions/Mbp. By subtracting the number of background damage of 14.7 ± 0.4 lesions/Mbp (see Fig. S1), we calculated the number of new SSB lesions to be 32.0 ± 1.9 SSB/Mbp per 1.3 MJ/m2 of UVA radiation, i.e., 21.2 ± 1.5 SSB/Mbp/MJ/m2. We similarly quantified other lesions by converting them into SSBs using specific enzymes (Table 1). Typically, these DNA enzymes target multiple lesions. Here, we list only their primary targets that we aimed to detect. Thus, we used E. coli endonuclease IV to detect apurinic/apyrimidinic (AP) sites, T4 endonuclease V for CPDs and AP sites, E. coli endonuclease III for oxidized pyrimidines and AP sites, and E. coli Fpg for oxidized purines. The results obtained by incubating UVA-irradiated pUC18 with these enzymes are summarized in Fig. 1 E (see also Fig. S2).
Figure 1.
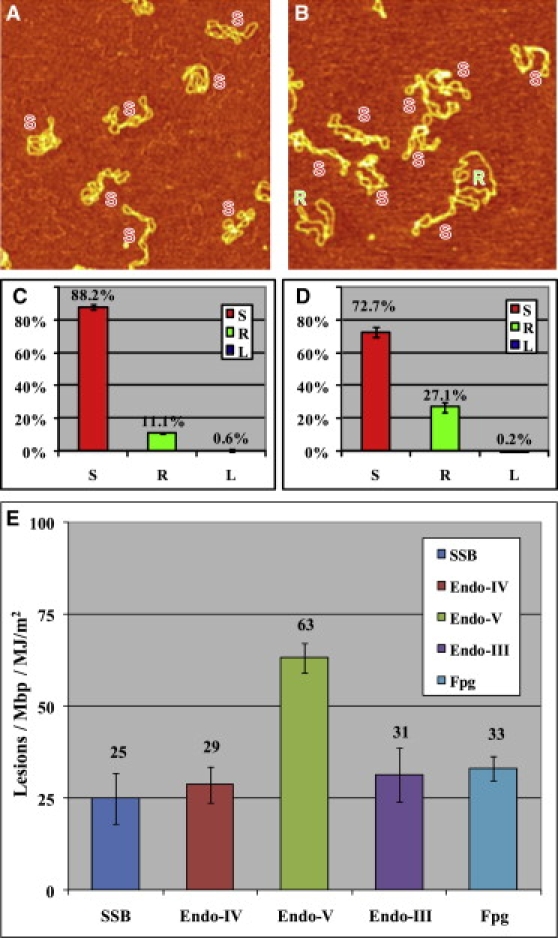
AFM images on APS-mica (48) of different pUC18 DNAs that were subjected to 1.3 MJ/m2 UVA radiation and different enzyme treatments before imaging. DNA was dialyzed in 10 mM Tris-HCl, 1 mM EDTA, and 100 mM NaCl buffer, and irradiated in the same solution by UVA. After that, the sample was diluted back to a suitable buffer for different enzyme incubations: (A) no enzyme treatment as control, (B) T4 endonuclease V. The scan size in all the images is 1 × 1 μm2. (C and D) Histograms of the occurrence of various configurations of pUC18 plasmids determined from the AFM images, such as these shown in A and B. Color code: red, supercoiled DNA (S); green, relaxed circular plasmids (R); blue, linear DNA (L). The error bars in the figures represent the SD. Each histogram is based on 600–1000 DNA molecules from 30–36 AFM images. (E) Histogram summarizes the number of different lesions/Mbp/MJ/m2 after UVA irradiation and specific enzyme treatments. The values shown in the histogram represent averages from two to five separate experiments.
T4 endonuclease V detects CPDs produced directly by UVA
T4 endonuclease V is commonly used to detect CPDs in UV-irradiated DNA (13,20,25,32). However, we note that T4 endonuclease V has both DNA glycosylase and AP-lyase activity, which means that it has the same endonucleolytic activity as E. coli endonuclease IV to cleave the phosphodiester bond at AP sites (56). Thus, it is possible that some or even all of the T4 endonuclease V-sensitive sites may actually be AP sites, not CPDs. Therefore, to determine the actual number of CPDs generated by UVA, in a separate experiment we first treated the dialyzed and irradiated DNA by E. coli endonuclease IV to relax all the AP sites (28.7 ± 4.8 lesions/Mbp/MJ/m2), and then incubated the same sample with T4 endonuclease V, which resulted in the total number of lesions being 70.9 ± 7.3 lesions/Mbp/MJ/m2. The results are shown in Fig. 2. After subtracting all the lesions detected by E. coli endonuclease IV treatment, we determined 70.9–28.7 = 42.2 lesions/Mbp/MJ/m2. Thus, the real number of CPD lesions generated by UVA is ∼67% of the sites detected by T4 endonuclease V alone. Of importance, in previous studies of UVA damage, the AP-lyase activity of T4 endonuclease V was ignored, which could have resulted in a significant overestimation of the number of CPDs generated by UVA adding more confusion to the interpretation of UVA damage. Yet, from our results it is clear that in the absence of photosensitizers, UVA directly generated a significant number of CPDs as detected by a combined action of E. coli endonuclease IV and T4 endonuclease V. The detailed mechanism underlying the formation of these CPDs by UVA may be quite complex and involve some intra-DNA energy transfer, and warrants further studies (2,3). The above results were independently reproduced by gel electrophoresis measurements as shown in Fig. S3 and Table S1.
Figure 2.
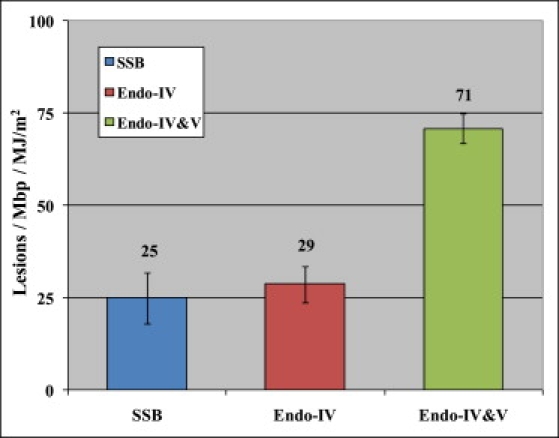
Histogram summarizes the actual CPD number/Mbp/MJ/m2 of UVA-irradiated DNA by combining E. coli endonuclease IV and T4 endonuclease V enzyme treatments. The values shown in the histogram represent averages from two to five separate experiments.
CPDs are not generated by the UVB tail of the UV lamp spectrum
To verify that CPDs were indeed generated by light at UVA wavelengths and not the UVB tail of the UV lamp spectrum, we repeated the same experiments without the narrowband UVA filter, which attenuates the intensity of transmitted UVB by a factor of 105 (see above). The number of T4 endonuclease V-sensitive sites increased after the filter was removed, as shown in Fig. 3. At 365 ± 5 nm, filter transmission was ∼70%. We calculated the slope of both lines and found that generation rate of T4 endonuclease-sensitive sites without and with the filter were 90 ± 3 sites/Mbp and 61 ± 2 sites/Mbp, respectively, i.e., a 68% difference, consistent with filter transmission. We determined that in the absence of the filter, the number of T4 endonuclease-sensitive sites generated within the same exposure time increased only by 47% and not by a factor of 105. Along the same line, in another experiment (data not shown) without the filter, we combined E. coli endonuclease IV with T4 endonuclease V treatment and determined that ∼60% of the T4 endonuclease V-sensitive sites were true CPDs, confirming the results from our experiment with the filter and ruling out the possibility that those CPDs were generated by UVB light.
Figure 3.
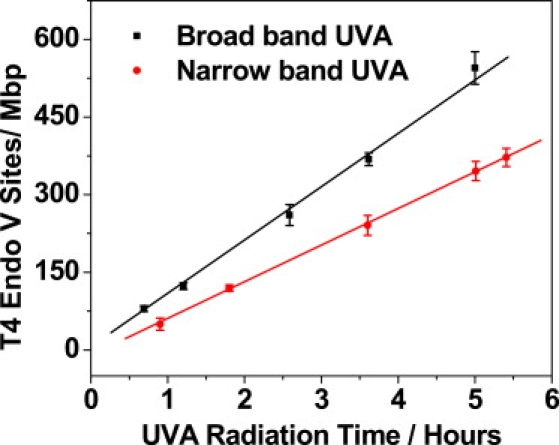
T4 endonuclease V-sensitive sites generated by broadband (without filter) and narrowband (with filter) UVA irradiation as a function of radiation time.
More CPDs are produced by UVA irradiation of DNA in pure water
To further test whether other chemicals present in the DNA solution (such as the constituents of the buffer) may somehow participate in CPD generation by UVA, we extensively dialyzed pUC18 supercoiled plasmid against ultrapure Millipore water (see Materials and Methods and Fig. S4) before subjecting the DNA to UVA radiation. The use of a covalently closed supercoiled plasmid for this study has a clear advantage over linear DNA because pUC18 strands cannot fully separate during the dialyses. Although pUC18 temporarily assumes a circular form in pure water, the plasmid remains intact and recoils to a supercoiled structure when transferred to the original buffer. Thus, this treatment does not introduce any unexpected damage (Fig. S4). According to the Poisson distribution, the background damage for this intact DNA when suspended in pure water was 18.9 ± 0.4 lesions/Mbp. Fig. 4 summarizes the results obtained by irradiating pUC18 in pure water and compares the different lesions with those obtained when irradiation was done in the buffer (see also Fig. S5).
Figure 4.
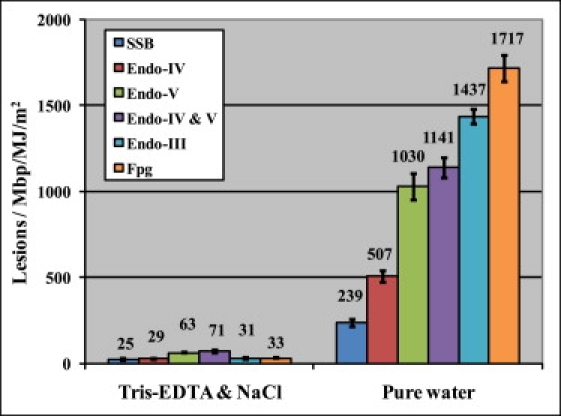
Histograms show the different lesions after UVA irradiation in 10 mM Tris-HCl, 1 mM EDTA, and 100 mM NaCl, and ultrapure Millipore water. DNA was treated by different enzymes at their preferred buffer condition and then diluted by regular imaging buffer (10 mM Tris-HCl, 1 mM EDTA, and 100 mM NaCl) before deposition onto APS-mica.
We note that the number of all oxidative lesions generated in pure water (the sum is 3154.4 ± 123.3 lesions/Mbp/MJ/m2) increased almost 50 times as compared to the total oxidative damage generated when DNA was irradiated in a regular buffer (10 mM Tris-HCl, 1 mM EDTA, and 100 mM NaCl). At the same time, the number of CPDs as measured by the combined action of endonuclease IV and T4 endonuclease V increased 15 times. The significant increase in the oxidative damage likely reflects the partial denaturation of DNA in pure water, The higher number of CPDs generated in pure water is consistent with earlier studies of UVB damage, which suggested that the increased mobility of nucleobases in the partially denatured state increases the efficiency of CPD formation (20), and with the results of a recent study of thymine dimerization in single-stranded DNA (57). Thus, the results of DNA irradiation in pure water reinforce the conjecture that CPDs are generated as a result of a direct absorption of UVA by DNA.
Photolyase detects CPDs directly produced by UVA in purified and dialyzed pUC18
So far, we have used the combined action of endonuclease IV and T4 endonuclease V to detect UVA-generated CPDs. To use an independent method for CPD detection, we irradiated pUC18 plasmids in pure water and incubated them with E. coli photolyase, an enzyme that specializes in direct photoreactivation of CPDs and (6-4) lesions (58–60). In this reaction, photolyase uses the energy of visible light to break the bonds between dimers and restore DNA integrity. Here, we incubated the UVA-irradiated DNA with photolyase in the dark to promote its binding to DNA and prevent photoreactivation. Photolyase (MW = 54 KDa) is large enough to be directly visualized by AFM imaging (61). Before AFM imaging, the DNA was suspended in a buffer that contained 5 mM Mg and no Na (33). Under these conditions, supercoiled plasmids are forced to assume a circular form with no supercoiled strand crossings, although they remain covalently closed. This DNA structure allowed us to image and count photolyase particles on DNA more accurately. The AFM image in Fig. 5 A shows photolyase bound onto 1 MJ/m2 UVA-irradiated pUC18 plasmids, and Fig. 5 B shows the image of untreated DNA after incubation with the enzyme. We counted the number of photolyase particles on pUC18 molecules directly from AFM images and plotted their frequency histograms together with the Poisson distribution fits (green curves in Fig. 5, C and D). After subtracting the number of enzyme molecules bound nonspecifically to the DNA, we estimate 0.98 photolyase-sensitive sites per plasmid at 1 MJ/m2, i.e., 365 CPD/Mbp/MJ/m2.
Figure 5.
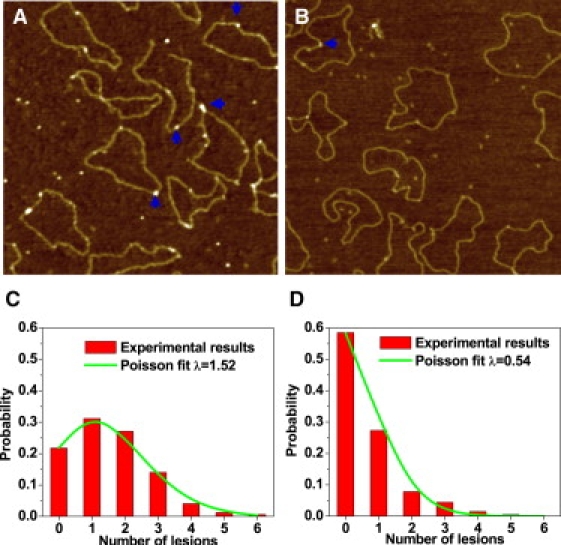
AFM images show photolyases binding to the CPD sites of pUC18 (some of them marked by blue arrows) with (A) 1 MJ/m2 UVA radiation and (B) no UVA radiation. Irradiation was performed on dialyzed plasmids suspended in pure water. The scan size in all the images is 1 × 1 μm2. (C and D) Histograms show the distribution of photolyase on pUC18 molecules as shown in A and B. The curves show the Poisson distribution fits, which give the average damage λ = 1.52/plasmid for the UVA-radiated DNA and 0.54 for control DNA, respectively.
Thus, the number of photolyase-sensitive sites is close to but somewhat lower than the number of CPDs determined from the combined results of T4 endonuclease V and E. coli endonuclease IV treatments (634 lesions/Mbp/MJ/m2). We conclude that, consistent with the results obtained by means of specific endonucleases, the photolyase assay detects a significant amount of CPDs produced by UVA in the absence of any photosensitizers. This result additionally supports the hypothesis that UVA can produce CPDs by direct action on DNA.
Photolyase and antibodies detect CPDs produced directly by UVA in synthetic homopolynucleotide
To further test whether UVA directly generates CPDs in DNA without possible exposure to cellular photosensitizers, we irradiated, in a Tris-EDTA buffer, a synthetic double-stranded homopolynucleotide, poly(dA)-poly(dT), with 6 MJ/m2 UVA at 365 ± 5 nm, and incubated it with photolyase and with anti-CPD antibodies (see Materials and Methods). Fig. 6 shows the AFM images, which clearly reveal numerous protein particles on the irradiated DNA proving that CPDs have indeed been formed. To the best of our knowledge, Fig. 6 C shows for the first time direct images of individual anti-CPD antibodies on DNA captured by AFM. The numbers of CPD sites determined from these and similar images were (after subtracting nonspecifically bound proteins): for poly(dA)-poly(dT) 78 ± 12 CPD/Mbp/MJ/m2 as detected by photolyase, and 68 ± 5 CPD/Mbp/MJ/m2 as detected by antithymine-dimer antibodies. These numbers of CPDs are higher than the number of CPDs detected by the combined action of T4 endonuclease V and endonuclease IV on pUC18 molecules in the same buffer (42 CPD/Mbp/MJ/m2). This result is understandable because poly(dA)-poly(dT) has more TT neighbors per unit length that can potentially form TT dimers than does pUC18.
Figure 6.
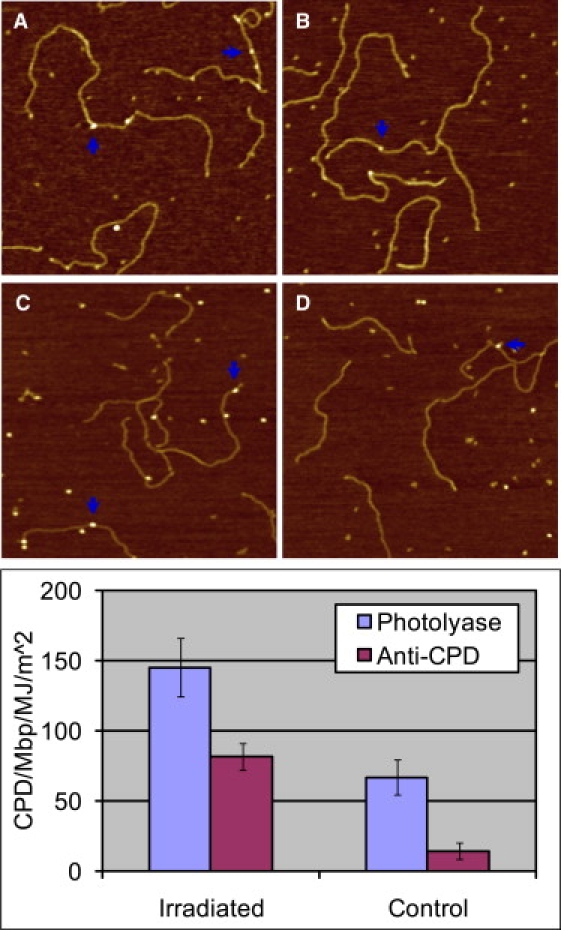
(A and B) AFM images show photolyases binding to the CPD sites of poly(dA)-poly(dT) with (A) 6 MJ/m2 UVA radiation, and (B) no UVA radiation. (C and D) AFM images show antithymine-dimer antibodies binding to the CPD sites of poly(dA)-poly(dT) with (C) 6 MJ/m2 UVA radiation, and (D) no UVA radiation as control. The scan size in all the images is 1 × 1 μm2. The histograms compare the lesions detected by photolyase and antithymine-dimer antibody on UVA-irradiated poly(dA)-poly(dT) and intact poly(dA)-poly(dT).
The number of CPDs produced by UVA in vitro exceeds the number of CPDs produced in vivo
Finally, we compare the number of CPDs generated by UVA in isolated DNA that we determined using our AFM methodology with the number of CPDs determined in previous studies, in which cells and skin tissues were subjected to prolonged UVA radiation (6–8). The latter number varies among different systems and studies, and ranges from 1.5 to 18 CPD/Mbp/MJ/m2 (5–8,10,22,23). Thus, the number of CPDs generated in cellular DNA by UVA is lower than the number of CPDs generated in isolated DNA (42.2 CPDMbp/MJ/m2). Since our results strongly suggest that UVA generates CPDs in isolated DNA directly, it is possible that some, if not all, cellular CPDs are also generated by UVA directly. However, it is also possible that the protection of cellular DNA against direct UVA damage is so high that some photosensitizers are still required to account for the numbers of CPDs generated in vivo. Further studies to clarify these difficult issues are warranted.
Conclusions
By executing endonuclease-driven supercoiled plasmid relaxation assays on the AFM imaging platform, and AFM visualization of photolyase and anti-CPD antibodies on UVA-irradiated native and synthetic DNA, we were able to directly detect and quantify a variety of DNA lesions. We found that only ∼60% of T4 endonuclease V-sensitive sites are true CPDs; the other 40% are abasic sites. Our results show that, similarly to UVC and UVB, UVA can directly produce a significant amount of CPDs when dialyzed DNA is irradiated in a buffer, and even more CPDs when DNA is irradiated in pure water. The amount of CPDs produced in vitro was actually greater than the amount of CPDs generated by the same UVA dose in vivo, suggesting that some, if not all, cellular CPDs are also produced by UVA directly.
Acknowledgments
We are grateful to Dr. Aziz Sancar and Dr. Paul Modrich for their comments on the manuscript. This work was funded by grants from the National Science Foundation and National Institutes of Health to P.E.M. and R.L.C.
Supporting Material
References
- 1.Setlow R.B. Wavelengths in sunlight effective in producing skin cancer—theoretical analysis. Proc. Natl. Acad. Sci. USA. 1974;71:3363–3366. doi: 10.1073/pnas.71.9.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morrison H. John Wiley & Sons; New York: 1990. Bioorganic Photochemistry, Vol. 1, Photochemistry and the Nucleic Acids. [Google Scholar]
- 3.Friedberg E.C., Walker G.C., Siede W. ASM Press; Washington, D.C.: 1995. DNA Repair and Mutagenesis. [Google Scholar]
- 4.Sinha R.P., Hader D.P. UV-induced DNA damage and repair: a review. Photochem. Photobiol. Sci. 2002;1:225–236. doi: 10.1039/b201230h. [DOI] [PubMed] [Google Scholar]
- 5.Cadet J., Sage E., Douki T. Ultraviolet radiation-mediated damage to cellular DNA. Mutat. Res. 2005;571:3–17. doi: 10.1016/j.mrfmmm.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 6.Kozmin S., Slezak G., Reynaud-Angelin A., Elie C., de Rycke Y. UVA radiation is highly mutagenic in cells that are unable to repair 7,8-dihydro-8-oxoguanine in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2005;102:13538–13543. doi: 10.1073/pnas.0504497102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mouret S., Baudouin C., Charveron M., Favier A., Cadet J. Cyclobutane pyrimidine dimers are predominant DNA lesions in whole human skin exposed to UVA radiation. Proc. Natl. Acad. Sci. USA. 2006;103:13765–13770. doi: 10.1073/pnas.0604213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Besaratinia A., Synold T.W., Chen H.H., Chang C., Xi B.X. DNA lesions induced by UV A1 and B radiation in human cells: comparative analyses in the overall genome and in the p53 tumor suppressor gene. Proc. Natl. Acad. Sci. USA. 2005;102:10058–10063. doi: 10.1073/pnas.0502311102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Setlow R.B., Carrier W.L. Pyrimidine dimers in ultraviolet-irradiated DNA's. J. Mol. Biol. 1966;17:237–254. doi: 10.1016/s0022-2836(66)80105-5. [DOI] [PubMed] [Google Scholar]
- 10.Douki T., Reynaud-Angelin A., Cadet J., Sage E. Bipyrimidine photoproducts rather than oxidative lesions are the main type of DNA damage involved in the genotoxic effect of solar UVA radiation. Biochemistry. 2003;42:9221–9226. doi: 10.1021/bi034593c. [DOI] [PubMed] [Google Scholar]
- 11.Setlow R.B. Cyclobutane-type pyrimidine dimers in polynucleotides. Science. 1966;153:379–386. doi: 10.1126/science.153.3734.379. [DOI] [PubMed] [Google Scholar]
- 12.Wehner J., Horneck G. Effects of vacuum UV and UVC radiation on dry Escherichia-coli plasmid Puc19. 1. Inactivation, lacz(−) mutation-induction and strand breaks. J. Photochem. Photobiol. B. 1995;28:77–85. doi: 10.1016/1011-1344(94)07089-7. [DOI] [PubMed] [Google Scholar]
- 13.Kielbassa C., Roza L., Epe B. Wavelength dependence of oxidative DNA damage induced by UV and visible light. Carcinogenesis. 1997;18:811–816. doi: 10.1093/carcin/18.4.811. [DOI] [PubMed] [Google Scholar]
- 14.Folkard M., Prise K.M., Vojnovic B., Brocklehurst B., Michael B.D. Critical energies for ssb and dsb induction in plasmid DNA by vacuum-UV photons: an arrangement for irradiating dry or hydrated DNA with monochromatic photons. Int. J. Radiat. Biol. 2000;76:763–771. doi: 10.1080/09553000050028913. [DOI] [PubMed] [Google Scholar]
- 15.Folkard M., Prise K.M., Turner C.J., Michael B.D. The production of single strand and double strand breaks in DNA in aqueous solution by vacuum UV photons below 10 eV. Radiat. Prot. Dosimetry. 2002;99:147–149. doi: 10.1093/oxfordjournals.rpd.a006746. [DOI] [PubMed] [Google Scholar]
- 16.Cadet J., Courdavault S., Ravanat J.L., Douki T. UVB and UVA radiation-mediated damage to isolated and cellular DNA. Pure Appl. Chem. 2005;77:947–961. [Google Scholar]
- 17.Tyrrell R.M. Induction of pyrimidine dimers in bacterial DNA by 365 nm radiation. Photochem. Photobiol. 1973;17:69–73. doi: 10.1111/j.1751-1097.1973.tb06334.x. [DOI] [PubMed] [Google Scholar]
- 18.Peak M.J., Peak J.G., Webb R.B. Inactivation of transforming DNA by ultraviolet-light. 3. Further observations on effects of 365 nm radiation. Mutat. Res. 1973;20:143–148. doi: 10.1016/0027-5107(73)90184-x. [DOI] [PubMed] [Google Scholar]
- 19.Webb R.B., Brown M.S. Action spectra for oxygen-dependent and independent inactivation of Escherichia-coli-WP2S from 254-nm to 460-nm. Photochem. Photobiol. 1979;29:407–409. doi: 10.1111/j.1751-1097.1979.tb07068.x. [DOI] [PubMed] [Google Scholar]
- 20.Kuluncsics Z., Perdiz D., Brulay E., Muel B., Sage E. Wavelength dependence of ultraviolet-induced DNA damage distribution: involvement of direct or indirect mechanisms and possible artefacts. J. Photochem. Photobiol. B. 1999;49:71–80. doi: 10.1016/S1011-1344(99)00034-2. [DOI] [PubMed] [Google Scholar]
- 21.Peak M.J., Peak J.G. Solar-ultraviolet-induced damage to DNA. Photodermatol. 1989;6:1–15. [PubMed] [Google Scholar]
- 22.Douki T., Court M., Sauvaigo S., Odin F., Cadet J. Formation of the main UV-induced thymine dimeric lesions within isolated and cellular DNA as measured by high performance liquid chromatography-tandem mass spectrometry. J. Biol. Chem. 2000;275:11678–11685. doi: 10.1074/jbc.275.16.11678. [DOI] [PubMed] [Google Scholar]
- 23.Courdavault S., Baudouin C., Charveron M., Favier A., Cadet J. Larger yield of cyclobutane dimers than 8-oxo-7,8-dihydroguanine in the DNA of UVA-irradiated human skin cells. Mutat. Res. 2004;556:135–142. doi: 10.1016/j.mrfmmm.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 24.Mitchell D. Revisiting the photochemistry of solar UVA in human skin. Proc. Natl. Acad. Sci. USA. 2006;103:13567–13568. doi: 10.1073/pnas.0605833103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perdiz D., Grof P., Mezzina M., Nikaido O., Moustacchi E. Distribution and repair of bipyrimidine photoproducts in solar UV-irradiated mammalian cells—possible role of Dewar photoproducts in solar mutagenesis. J. Biol. Chem. 2000;275:26732–26742. doi: 10.1074/jbc.M001450200. [DOI] [PubMed] [Google Scholar]
- 26.Sutherland J.C., Griffin K.P. Absorption-spectrum of DNA for wavelengths greater than 300-nm. Radiat. Res. 1981;86:399–410. [PubMed] [Google Scholar]
- 27.Hiraku Y., Ito K., Hirakawa K., Kawanishi S. Photosensitized DNA damage and its protection via a novel mechanism. Photochem. Photobiol. 2007;83:205–212. doi: 10.1562/2006-03-09-IR-840. [DOI] [PubMed] [Google Scholar]
- 28.Sutherland B.M., Bennett P.V., Sidorkina O., Laval J. Clustered DNA damages induced in isolated DNA and in human cells by low doses of ionizing radiation. Proc. Natl. Acad. Sci. USA. 2000;97:103–108. doi: 10.1073/pnas.97.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sutherland B.M., Georgakilas A.G., Bennett P.V., Laval J., Sutherland J.C. Quantifying clustered DNA damage induction and repair by gel electrophoresis, electronic imaging and number average length analysis. Mutat. Res. 2003;531:93–107. doi: 10.1016/j.mrfmmm.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 30.Collins A.R. The comet assay for DNA damage and repair—principles, applications, and limitations. Mol. Biotechnol. 2004;26:249–261. doi: 10.1385/MB:26:3:249. [DOI] [PubMed] [Google Scholar]
- 31.Brendler-Schwaab S., Hartmann A., Pfuhler S., Speit G. The in vivo comet assay: use and status in genotoxicity testing. Mutagenesis. 2005;20:245–254. doi: 10.1093/mutage/gei033. [DOI] [PubMed] [Google Scholar]
- 32.Sutherland B.M., Bennett P.V., Conlon K., Epling G.A., Sutherland J.C. Quantitation of Supercoiled DNA cleavage in nonradioactive DNA—application to ionizing-radiation and synthetic endonuclease cleavage. Anal. Biochem. 1992;201:80–86. doi: 10.1016/0003-2697(92)90176-8. [DOI] [PubMed] [Google Scholar]
- 33.Jiang Y., Ke C.H., Mieczkowski P.A., Marszalek P.E. Detecting ultraviolet damage in single DNA molecules by atomic force microscopy. Biophys. J. 2007;93:1758–1767. doi: 10.1529/biophysj.107.108209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ke C., Jiang Y., Mieczkowski P.A., Muramoto G.G., Chute J.P. Nanoscale detection of ionizing radiation damage to DNA by atomic force microscopy. Small. 2008;4:288–294. doi: 10.1002/smll.200700527. [DOI] [PubMed] [Google Scholar]
- 35.Hansma H.G., Laney D.E., Bezanilla M., Sinsheimer R.L., Hansma P.K. Applications for atomic-force microscopy of DNA. Biophys. J. 1995;68:1672–1677. doi: 10.1016/S0006-3495(95)80343-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lyubchenko Y.L., Shlyakhtenko L.S. Visualization of supercoiled DNA with atomic force microscopy in situ. Proc. Natl. Acad. Sci. USA. 1997;94:496–501. doi: 10.1073/pnas.94.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guthold M., Bezanilla M., Erie D.A., Jenkins B., Hansma H.G. Following the assembly of RNA-polymerase DNA complexes in aqueous-solutions with the scanning force microscope. Proc. Natl. Acad. Sci. USA. 1994;91:12927–12931. doi: 10.1073/pnas.91.26.12927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shao Z.F., Mou J., Czajkowsky D.M., Yang J., Yuan J.Y. Biological atomic force microscopy: what is achieved and what is needed. Adv. Phys. 1996;45:1–86. [Google Scholar]
- 39.Hansma H.G. Surface biology of DNA by atomic force microscopy. Annu. Rev. Phys. Chem. 2001;52:71–92. doi: 10.1146/annurev.physchem.52.1.71. [DOI] [PubMed] [Google Scholar]
- 40.Hansma H.G., Vesenka J., Siegerist C., Kelderman G., Morrett H. Reproducible imaging and dissection of plasmid DNA under liquid with the atomic force microscope. Science. 1992;256:1180–1184. doi: 10.1126/science.256.5060.1180. [DOI] [PubMed] [Google Scholar]
- 41.Lindsay S.M., Thundat T., Nagahara L., Knipping U., Rill R.L. Images of the DNA double helix in water. Science. 1989;244:1063–1064. doi: 10.1126/science.2727694. [DOI] [PubMed] [Google Scholar]
- 42.New England Biolab. DNA Repair Glycosylases on Various Damaged Bases. Available at: http://www.neb.com/nebecomm/tech_reference/modifying_enzymes/dna_repair_damaged_bases.asp.
- 43.Woollons A., Kipp C., Young A.R., Petit-Frere C., Arlett C.F. The 0.8% ultraviolet B content of an ultraviolet A sunlamp induces 75% of cyclobutane pyrimidine dimers in human keratinocytes in vitro. Br. J. Dermatol. 1999;140:1023–1030. doi: 10.1046/j.1365-2133.1999.02899.x. [DOI] [PubMed] [Google Scholar]
- 44.Shlyakhtenko L.S., Gall A.A., Filonov A., Cerovac Z., Lushnikov A. Silatrane-based surface chemistry for immobilization of DNA, protein-DNA complexes and other biological materials. Ultramicroscopy. 2003;97:279–287. doi: 10.1016/S0304-3991(03)00053-6. [DOI] [PubMed] [Google Scholar]
- 45.Cherny D.I., Jovin T.M. Electron and scanning force microscopy studies of alterations in supercoiled DNA tertiary structure. J. Mol. Biol. 2001;313:295–307. doi: 10.1006/jmbi.2001.5031. [DOI] [PubMed] [Google Scholar]
- 46.Vologodskii A.V., Cozzarelli N.R. Conformational and thermodynamic properties of supercoiled DNA. Annu. Rev. Biophys. Biomol. Struct. 1994;23:609–643. doi: 10.1146/annurev.bb.23.060194.003141. [DOI] [PubMed] [Google Scholar]
- 47.Bussiek M., Mucke N., Langowski J. Polylysine-coated mica can be used to observe systematic changes in the supercoiled DNA conformation by scanning force microscopy in solution. Nucleic Acids Res. 2003;31:e137. doi: 10.1093/nar/gng137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shlyakhtenko L.S., Miloseska L., Potaman V.N., Sinden R.R., Lyubchenko Y.L. Intersegmental interactions in supercoiled DNA: atomic force microscope study. Ultramicroscopy. 2003;97:263–270. doi: 10.1016/S0304-3991(03)00051-2. [DOI] [PubMed] [Google Scholar]
- 49.Murakami M., Hirokawa H., Hayata I. Analysis of radiation damage of DNA by atomic force microscopy in comparison with agarose gel electrophoresis studies. J. Biochem. Biophys. Methods. 2000;44:31–40. doi: 10.1016/s0165-022x(00)00049-x. [DOI] [PubMed] [Google Scholar]
- 50.Pang D., Berman B.L., Chasovskikh S., Rodgers J.E., Dritschilo A. Investigation of neutron-induced damage in DNA by atomic force microscopy: experimental evidence of clustered DNA lesions. Radiat. Res. 1998;150:612–618. [PubMed] [Google Scholar]
- 51.Pang D., Rodgers J.E., Berman B.L., Chasovskikh S., Dritschilo A. Spatial distribution of radiation-induced double-strand breaks in plasmid DNA as resolved by atomic force microscopy. Radiat. Res. 2005;164:755–765. doi: 10.1667/rr3425.1. [DOI] [PubMed] [Google Scholar]
- 52.Boichot S., Fromm M., Cunniffe S., O'Neill P., Labrune J.C. Investigation of radiation damage in DNA by using atomic force microscopy. Radiat. Prot. Dosimetry. 2002;99:143–145. doi: 10.1093/oxfordjournals.rpd.a006745. [DOI] [PubMed] [Google Scholar]
- 53.Psonka K., Brons S., Heiss M., Gudowska-Nowak E., Taucher-Scholz G. Induction of DNA damage by heavy ions measured by atomic force microscopy. J. Phys. Condens. Matter. 2005;17:S1443–S1446. [Google Scholar]
- 54.Lobachevsky P.N., Karagiannis T.C., Martin R.F. Plasmid DNA breakage by decay of DNA-associated auger electron emitters: approaches to analysis of experimental data. Radiat. Res. 2004;162:84–95. doi: 10.1667/rr3187. [DOI] [PubMed] [Google Scholar]
- 55.Sachs R.K., Ponomarev A.L., Hahnfeldt P., Hlatky L.R. Locations of radiation-produced DNA double strand breaks along chromosomes: a stochastic cluster process formalism. Math. Biosci. 1999;159:165–187. doi: 10.1016/s0025-5564(99)00019-x. [DOI] [PubMed] [Google Scholar]
- 56.Gruskin E.A., Lloyd R.S. The DNA scanning mechanism of T4 endonuclease-V—effect of NaCl concentration on processive nicking activity. J. Biol. Chem. 1986;261:9607–9613. [PubMed] [Google Scholar]
- 57.Schreier W.J., Schrader T.E., Koller F.O., Gilch P., Crespo-Hernandez C.E. Thymine dimerization in DNA is an ultrafast photoreaction. Science. 2007;315:625–629. doi: 10.1126/science.1135428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 2003;103:2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 59.Kao Y.-T., Saxena C., Wang L., Sancar A., Zhong D. Inaugural article: direct observation of thymine dimer repair in DNA by photolyase. Proc. Natl. Acad. Sci. USA. 2005;102:16128–16132. doi: 10.1073/pnas.0506586102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sancar G.B., Sancar A. Purification and characterization of DNA photolyases. Methods Enzymol. 2006;408:121–156. doi: 10.1016/S0076-6879(06)08009-8. [DOI] [PubMed] [Google Scholar]
- 61.van Noort J., Orsini F., Eker A., Wyman C., de Grooth B. DNA bending by photolyase in specific and non-specific complexes studied by atomic force microscopy. Nucleic Acids Res. 1999;27:3875–3880. doi: 10.1093/nar/27.19.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


