Abstract
AIM: To establish a cell model harboring replicative clinical hepatitis B virus (HBV) isolates and evaluate its application in individualized selection of anti-HBV agents for chronic hepatitis B (CHB) patients.
METHODS: The full-length HBV genomic DNA from 8 CHB patients was amplified by polymerase chain reaction (PCR). All the patients were treated with lamivudine for at least seven months and finally became resistant to lamivudine. The amplified HBV DNA fragments were inserted into pHY106 vectors by Sap I digestion. The recombinant plasmids containing 1.1 copies of HBV genome were transiently transfected into Huh7 cell line, and the levels of HBsAg, HBeAg and intercellular HBV replicative intermediates were determined by ELISA and Southern blot analysis, respectively, with or without lamivudine and adefovir treatment. The antiviral treatment with adefovir was administered to the patients and analyzed in parallel.
RESULTS: A total of 25 independent HBV isolates were obtained from the sera of 8 patients, each patient had at least two isolates. One isolate from each individual was selected and subcloned into pHY106 vector, including 5 isolates with YVDD mutation and 3 isolates with YIDD mutation. All recombinant plasmids harboring HBV isolates were transfected into Huh7 cells. The results indicated that HBV genome carried in HBV replicons of clinical HBV isolates could effectively replicate and express in Huh7 cells. Adefovir, but not lamivudine, inhibited HBV replication both in vitro and in vivo, and in vitro inhibition was dose-dependent.
CONCLUSION: The novel method described herein enables individualized selection of anti-HBV agents in clinic and is useful in future studies of antiviral therapy for CHB.
Keywords: Hepatitis B virus, Chronic hepatitis B, Hepatitis B virus isolate, Antiviral agents
INTRODUCTION
Hepatitis B virus (HBV) infection may lead to acute liver disease, chronic active hepatitis, liver cirrhosis, and hepatocellular carcinoma (HCC). Over 350 million people worldwide are estimated to be infected chronically by HBV and are, therefore, at risk of liver failure, cirrhosis, or HCC[1–4]. The principal treatment for chronic hepatitis B (CHB) involves the use of interferon alpha (IFN-α) or nucleoside analogs.
Although the use of IFN-α and nucleoside analogs (such as lamivudine and adefovir) has improved the treatment of chronically infected HBV patients, an effective reduction in virus load is only observed in less than 40% of treated patients[5–10]. The molecular basis for resistance to antiviral therapy is not clearly defined. However, studies have suggested that HBV genome mutations may play a direct role in the development of resistance to antiviral agents[11–15].
In vitro analysis of clinical HBV isolates is difficult due to the lack of HBV cell culture model[16–19]. Fortunately, the lack of in vitro infectivity can be conquered by transfecting recombinant plasmids encoding over-length HBV genome into human liver cell lines. Following transfection, HBV pre-genomic and messenger RNA are transcribed from the plasmids and progeny virus is replicated and released from cells. The present study is to develop a more efficient method for selecting anti-HBV agents using a highly efficient HBV expression vector. It is supposed to be useful for future studies on resistance surveillance and novel drug discovery.
MATERIALS AND METHODS
Patients
Eight patients with CHB were selected according to the diagnostic criteria of CHB[20], and the infection of hepatitis A, C, E and G viruses, human immunodeficiency virus, cytomegalovirus and the use of alcohol were excluded. All patients were treated with lamivudine for more than seven months, and were found to be resistant to lamivudine treatment in the Department of Infectious Diseases of Union Hospital of Tongji Medical College, Huazhong University of Science and Technology, from January 2005 to August 2005. All the patients were HBsAg and HBeAg positive, with HBV DNA levels above 106 copies/mL and elevated serum ALT (≥ 1.5 times that of the upper normal limit) at the time of sample collection. Access to the materials complied with the Helsinki Declaration and approved by the local ethics committee, and the informed consents were obtained from all the patients.
Plasmids and bacteria
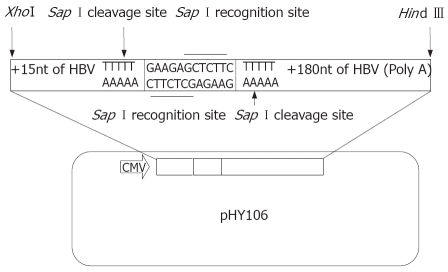
HBV cloning and expression vector pHY106 was kindly provided by Dr. Lu MJ (Institute of Virology, Duisburg-Essen University, Germany), which contains a cytomegalovirus (CMV) promoter upstream of a short, recombinant HBV sequence that allows the in-frame insertion of a full-length HBV genome by Sap I digestion (Figure 1). The HBV replicon, pHBV1.3, containing 1.3-fold full length genome of HBV (ayw subtype), was conducted in our laboratory as described previously[21]. Plasmid pUC19 and E.coli JM109 strain were maintained in our laboratory and stored at -80°C.
Figure 1.
The schematic map of HBV expression vector of pHY106.
Cell culture
Human hepatoblastoma cell line, Huh7, was maintained in MEM medium supplemented with 10% fetal bovine serum (FBS) at 37°C in a moist atmosphere containing 5% CO2.
Primers design
Primers were designed according to Gunther’s method[22], P1 (1803-1821): 5’-CCGGAAAGCTTGAGCTCTTC-TTTTTCACCTCTGCCTAATCA-3’ and P2 (1841-1822): 5’-CCGGAAAGCTTGAGCTCTTCAAAAAGTTGCATGGTGCTGG-3’ for amplifying HBV full-length genome. The italics denoted Hind III, Sac I and Sap I sites for inserting HBV genome into pUC19 and PHY106.
Amplification of full-length HBV genomic DNA
HBV DNA was isolated from 200 μL sera using QIAamp Blood Kit (Qiagen Co., Germany) according to manufacturer’s instructions. Polymerase chain reaction (PCR) amplification of full-length HBV genomes was performed using primers P1 and P2 according to the methods of Gunther et al[22]. If the initial PCR reactions were negative, a second round of PCR was performed under identical conditions using 5 μL of 1:10 dilution of the first round reaction products as template.
Construction of recombinant plasmids harboring full-length HBV genome
Following PCR amplification, PCR product was purified using a QIAquick Gel Kit (Qiagen Co., Germany) according to manufacturer’s instructions. Then PCR products and plasmid pUC19 were digested with the restriction enzyme Sac I, and the 3.2 kb PCR products were inserted into the linear pUC19 vector (Figure 2). Recombinant plasmids were identified by restriction enzyme cleavage and termed as pUC19-HBV1.0.
Figure 2.
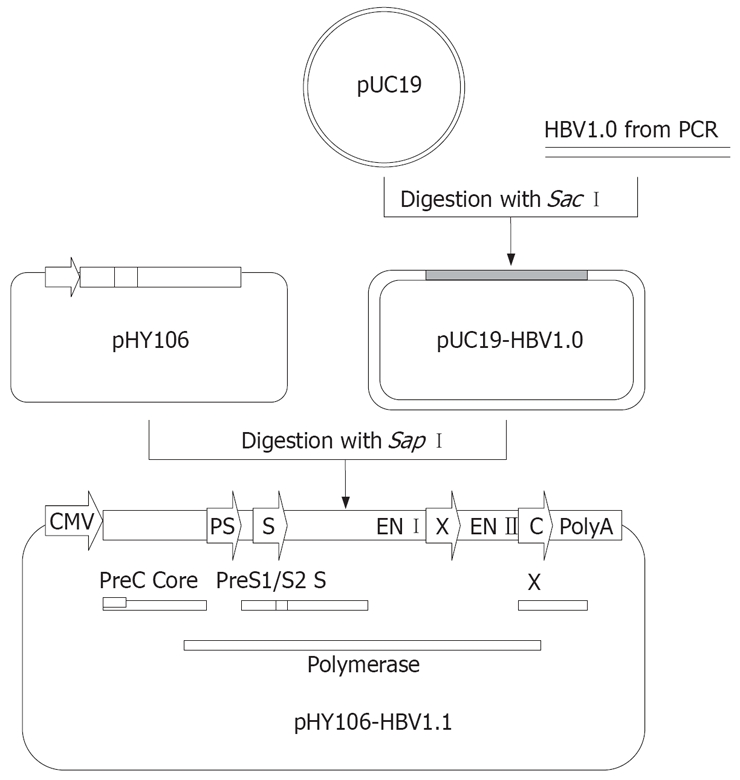
Construction of replicons of clinical HBV isolates using pHY106. Full-length HBV genomes from PCR amplication can be digested with Sap I and inserted into Sap I-digested pHY106 to produce HBV replicon. HBV open reading frames (ORFs) for the pre-core (PC), core, preS1 (PS1), preS2 (2) surface (S), polymerase, and X genes are indicated by square frame. Promoter sequences for the preS (PS), surface (S) and core (C) genes are indicated by arrows.
Construction of HBV replicons harboring 1.1-fold length HBV genome
pUC19-HBV1.0 was cleaved with the restriction enzyme Sap I, and a 3.2 kb fragment was recovered and subcloned into pHY106 vector (Figure 2). Recombinant plasmids were identified by Hind III and Nsi Idigestion and termed as pHY106-HBV1.1. HBV polymerase gene of pHY106-HBV1.1 was analyzed by sequencing.
Replication of clinical HBV replicon in Huh7 cell line
The plasmid pHY106-HBV1.1 was transiently transfected into Huh7 cells, and the replication of clinical HBV replicons was measured. Huh7 cells were seeded with 5 × 105/well and cultured in six-well plates. Sixteen hours later, the cells were transfected with 5 μg of plasmid pHY106-HBV1.1 using 5 μL lipofectamine reagent (Invitrogen Co., USA) according to the manufacturer’s instructions. Following transfection, cultures were fed with fresh media and incubated for 96 h. The levels of HBsAg, HBeAg and HBV DNA in supernatant were determined by ELISA and quantitative PCR everyday. Meanwhile, the total cellular DNA was isolated and HBV replicative intermediates were detected by Southern blot hybridization.
Southern blot analysis of HBV replicative intermediates
Five μg of total DNA of transfected cells was separated on 1.5% agarose gels, and blot onto Hybond-N nylon membranes (Amersham Bioscience Inc, UK) using standard Southern blot procedures. Membranes were hybridized with a 32P-labeled full-length HBV DNA fragment and viral DNA was analyzed using a storage phosphor system (Cyclone, Packard Bioscience Co.).
Antiviral treatment in Huh7 cells tranfected with clinical HBV replicons
Plasmid pHY106-HBV1.1 was transiently transfected into Huh7 cells as described above. The next day, cells were fed with fresh medium containing 0.01, 0.1, 1 and 10 μmol/L of adefovir (Gilead Sciences, USA) and 0.01, 0.1, 1 and 10 μmol/L of lamivudine, respectively, for 72 h, after which intracellular HBV replicative intermediates were isolated and quantified as described above.
Statistical analysis
Data were presented as mean ± SD error of the mean (SEM). Statistical significance of differences between the measured parameters was determined by Mann-Whitney’s test. Statistical analyses were performed using the SPSS13.0 software. The values were statistically significant at P < 0.05.
RESULTS
Amplification and cloning of full-length clinical HBV isolates
Using the method of Gunther et al, full-length HBV DNA was amplified from the sera of 6 patients following a single round of PCR. For the other two cases, a second round of PCR was performed to amplify HBV full-length genome successfully. In general, it was more difficult to amplify full-length HBV genome from patients with low levels of viremia. The PCR products were cloned into pUC19 vector, and a total of 25 independent HBV clones were obtained from the sera of the 8 patients and each patient had at least two clones.
Construction of HBV replicons harboring 1.1-fold length HBV genome
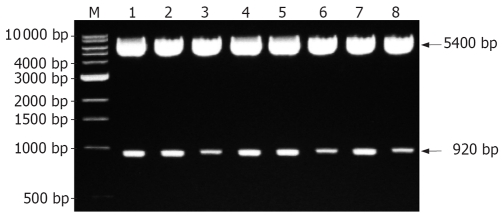
One clone was selected from each patient, and was subcloned into pHY106 vector. The directions of inserted clones were identified by digestion with restriction enzyme Hind III and Nsi I (Nsi I is a single restriction site at nt1064 of HBV genome). If HBV clones were forwardly inserted, the recombinant plasmid should be cleaved into two fragments with a molecular weight of 5400 bp and 920 bp (Figure 3). Polymerase gene of 5 HBV isolates showed YVDD mutation and the other 3 isolates showed YIDD mutation.
Figure 3.
Identification of recombinant expression plasmids by restriction endonuclease digestion analysis. M: 1 kb DNA ladder marker; Lane 1-8: Recombinant plasmids digested with Hind III and Nsi I.
Replication of clinical HBV replicons in Huh7 cell line
Eight of the HBV isolates cloned into the PHY106 vector were analyzed for their ability to replicate in vitro. Recombinant plasmids were transfected into Huh7 cells and allowed to replicate for 96 h, after which HBsAg, HBeAg and HBV DNA were detected, and intracellular replicative intermediates were extracted and analyzed by Southern blot hybridization. A marked variation was observed in the levels of HBsAg, HBeAg, HBV DNA and intracellular replicative intermediates among individual clones (Tables 1 and 2, Figure 4). The HBsAg and HBeAg levels rapidly reached to a plateau within 72 h. Further incubation did not increase the concentration of HBsAg and HBeAg in the culture supernatants. There was also a consistency of the HBsAg, HBeAg and HBV DNA levels in supernatants (Tables 1 and 2).
Table 1.
HBsAg and HBeAg levels of clinical HBV isolates in culture supernatants (mean ± SD)
|
24 h |
48 h |
72 h |
96 h |
|||||
| Isolates | HBsAg | HBeAg | HBsAg | HBeAg | HBsAg | HBeAg | HBsAg | HBeAg |
| 1 | 0.236 ± 0.028 | 0.158 ± 0.018 | 0.792 ± 0.055 | 0.435 ± 0.039 | 1.892 ± 0.168 | 1.276 ± 0.138 | 1.725 ± 0.258 | 1.212 ± 0.237 |
| 2 | 0.225 ±0.019 | 0.149 ± 0.014 | 0.646 ± 0.043 | 0.453 ± 0.044 | 1.737 ± 0.132 | 1.321 ± 0.215 | 1.722 ± 0.633 | 1.121 ± 0.153 |
| 3 | 0.214 ± 0.038 | 0.152 ± 0.015 | 0.756 ± 0.075 | 0.421 ± 0.054 | 1.654 ± 0.156 | 1.232 ± 0.342 | 1.563 ± 0.742 | 1.133 ± 0.217 |
| 4 | 0.195 ± 0.022 | 0.122 ± 0.012 | 0.533 ± 0.057 | 0.349 ± 0.033 | 1.278 ± 0.132 | 1.133 ± 0.142 | 1.236 ± 0.571 | 1.123 ± 0.342 |
| 5 | 0.189 ± 0.028 | 0.112 ± 0.013 | 0.636 ± 0.062 | 0.352 ± 0.024 | 0.996 ± 0.114 | 0.982 ± 0.211 | 1.006 ± 0.263 | 1.012 ± 0.411 |
| 6 | 0.188 ± 0.033 | 0.132 ± 0.013 | 0.537 ± 0.075 | 0.384 ± 0.042 | 0.987 ± 0.132 | 0.952 ± 0.156 | 0.945 ± 0.154 | 0.943 ± 0.125 |
| 7 | 0.179 ± 0.018 | 0.122 ± 0.012 | 0.639 ± 0.031 | 0.298 ± 0.027 | 1.139 ± 0.163 | 0.892 ± 0.142 | 0.997 ± 0.218 | 0.885 ± 0.218 |
| 8 | 0.236 ± 0.043 | 0.155 ± 0.015 | 0.836 ± 0.061 | 0.475 ± 0.039 | 1.754 ± 0.174 | 1.312 ± 0.111 | 1.825 ± 0.318 | 1.212 ± 0.323 |
| Positive | 0.336 ± 0.048 | 0.217 ± 0.028 | 0.956 ± 0.078 | 0.552 ± 0.066 | 1.868 ± 0.218 | 1.202 ± 0.324 | 1.922 ± 0.525 | 1.189 ± 0.305 |
| Negative | 0.046 ± 0.012 | 0.032 ± 0.011 | 0.044 ± 0.018 | 0.040 ± 0.015 | 0.047 ± 0.020 | 0.038 ± 0.012 | 0.042 ± 0.013 | 0.039 ± 0.018 |
Plasmid pHBV 1.3 is positive control; Plasmid pHY106 is negative control.
Table 2.
Viral loads in supernatants of Huh7 cells transfected with HBV replicon (copies/mL)
| Replicons |
Transfected time (h) |
|||
| 24 | 48 | 72 | 96 | |
| 1 | 5.6 × 102 | 2.5 × 103 | 3.6 × 105 | 3.9 × 105 |
| 2 | 5.2 × 102 | 2.0 × 103 | 2.8 × 105 | 2.7 × 105 |
| 3 | 4.5 × 102 | 1.8 × 103 | 2.4 × 105 | 2.6 × 105 |
| 4 | < 102 | 1.1 × 103 | 7.5 × 104 | 9.7 × 104 |
| 5 | < 102 | 1.2 × 103 | 8.2 × 104 | 9.1 × 104 |
| 6 | < 102 | 1.5 × 103 | 7.8 × 104 | 8.9 × 104 |
| 7 | < 102 | 1.3 × 103 | 9.5 × 104 | 1.2 × 105 |
| 8 | 5.5 × 102 | 2.3 × 103 | 2.8 × 105 | 2.9 × 105 |
| pHBV1.3 | 6.5 × 102 | 2.8 × 103 | 2.5 × 105 | 2.7 × 105 |
| pHY106 | < 102 | < 102 | < 102 | < 102 |
pHBV1.3 is positive control; pHY106 is negative control.
Figure 4.
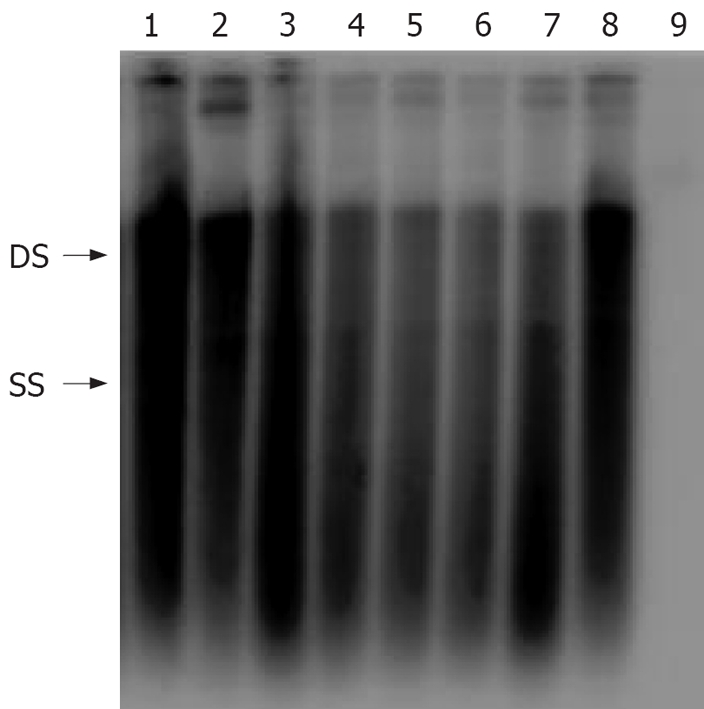
Analysis of intracellular replication of clinical HBV isolates. HBV isolates were cloned into the vector pHY106 and transfected into Huh2 cells. Seventy-two hours after transfection, intracellular replicative intermediates were isolated and analyzed by Southern blot. Double-stranded (DS) and single-stranded (SS) intermediates are indicated. Lane 1-8: eight replicons of clinical HBV isolates; Lane 9: pHY106 negative control.
Antiviral treatment in Huh7 cells tranfected with clinical HBV replicons
A cell-based antiviral assay was used to test the lamivudine and adefovir susceptibility of the 8 clinical isolates that replicated efficiently in cell culture. The results indicated that adefovir but not lamivudine, efficiently inhibited the replication of clinical HBV isolates in vitro, furthermore, the inhibition effect depended upon the concentration of adefovir. The viral replication was completely inhibited in the transfected cells treated with 10 μmol/L adefovir (Figure 5).
Figure 5.
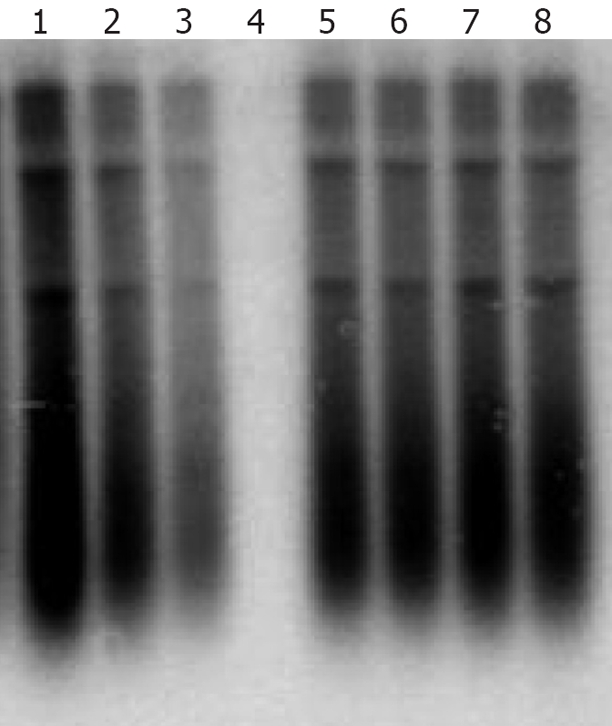
Antiviral susceptibility of an HBV isolate cloned from a patient with lamivudine resistance. An HBV isolate amplified from the sera of patients failing in lamivudine therapy was cloned into the pHY106 vector and analyzed in vitro. Lane 1-4: transfected cells treated with 0.01, 0.1, 1 and 10 μmol/L of adefovir; Lane 5-8: transfected cells treated with 0.01, 0.1, 1 and 10 μmol/L of lamivudine.
Antiviral treatment in vivo
All chronic HBV infected patients who had resistance to lamivudine were treated with adefovir for 6 mo. These patients were sensitive to adefovir therapy, and their HBV viral load in sera dropped at least 103-105 copies/mL. Together with the results of antiviral test of adefovir in vitro as described above, it indicated that antiviral susceptibility of clinical HBV isolates was well coincident between in vivo and in vitro.
DISCUSSION
HBV contains a 3.2-kb, circular, double-stranded DNA genome and causes acute and chronic hepatitis B, cirrhosis, and eventually HCC[23]. Investigation of the expression and replication of the HBV genome as well as the full viral life cycle is hampered by the lack of an in vitro tissue culture system in which HBV is propagated[16–19]. The HBV cell model system has been the subject of many studies in the last four decades. Unfortunately, no study has reported the identification of an appropriate tissue culture system to propagate HBV. This has greatly hindered the progress of HBV studies in many fields, such as virology, molecular biology, immunology and antiviral therapy. In an attempt to overcome this obstacle, several researchers transducted HBV genome into human hepatocyte via plasmids, in which HBV gene could replicate, express, and even assemble infectious virions[21,24–26]. Because the HBV genome is very dense and genes overlap with each other, HBV genome was inserted into plasmids in a head-to-tail linkage manner.
Recently, many groups have transfected over-length HBV genome into hepatocytes for viral replication and anti-HBV studies[21,24–26]. It is well known that the longest HBV transcript is a 3.5 kb mRNA; therefore, several plasmids carrying HBV multiple genomes were constructed, such as 2.0 copies and 1.3 copies of HBV genome, these over-length HBV genome contains complete HBV replication units, such as Enh I, Enh II, DR1, DR2, the transcription origin site of viral pre-genome, promoters and ORFs. Although these artificially constructed HBV replicons played an important role in anti-HBV studies, they were not suitable for the individualized selection of antiviral agents. Recently many scientists working in HBV fields considered that HBV replicated as quasi-species in vivo and individual species are likely to vary[27,28]. HBV genome mutation is mostly caused by immune and drug pressures, for example, CHB patients treated with lamivudine, when mutation appeared in polymerase gene of HBV, will be resistant to lamivudine therapy. As a result, HBV replicon can be used for the individualized selection of antiviral agents in vitro based on clinical HBV isolates.
In this study, we established a novel method to assess in vitro replication of clinical HBV isolate and investigated its application for selecting antiviral agents for CHB patients. We employed pHY106 vector to construct HBV replicons of clinical isolates. The pHY106 contains a CMV promoter, followed by a 15 nucleotide HBV sequence encoding the pre-core initiation site (plus the next two amino acids of the pre-core protein), a short heterologous linker sequence that contains two Sap I sites, and finally a 180 nucleotide region that encodes the carboxy terminus of the HBV X gene (five amino acids) and the polyadenylation signal for HBV mRNA. Following digestion of pHY106 and full-length HBV genomes with Sap I (Figure 2), HBV genomes can be inserted in-frame, resulting in the generation of replication-competent HBV clones, 95% of which are derived from patient virus (Figure 2). The CMV promoter upstream of the pre-core initiation site allows efficient transcription of the 3.5 kb pre-genomic RNA after transfection of liver cell lines. The minimal 5’ and 3’ sequence derived from the wild type HBV strain in pHY106 is highly conserved among published HBV isolates. Thus, HBV isolates cloned into the pHY106 vector should be representative of patient virus with little or no genotypic changes.
We obtained 8 clinical HBV replicons from CHB patients with lamivudine resistance, and found that these replicons were not susceptive to lamivudine treatment in vitro because of YMDD mutation of polymerase gene. Sequence analysis of clinical HBV isolates with established drug resistant mutations will provide useful information for selecting the existing or novel antiviral agents or the combination of antiviral agents for further treatment. At least four major mutational patterns are associated with lamivudine resistance (rtM204I, rtL180M + rtM204I, rtL180M + M204V, and rtV173L + rtL180M + M204V)[29]. Many mutations enhance viral replication, e.g., the pre-core mutation G1896A has been shown to greatly enhance the replication capacity of HBV encoding the rtM204I (but not rtM204V) lamivudine resistant mutation[30]. Adefovir demonstrated an excellent therapeutic effect for CHB patients who failed in lamivudine therapy[31–35]. Together with the results of antiviral treatment of adefovir in vitro and in vivo, our results confirmed that adefovir inhibited the replication of HBV with lamivudine resisitance in vitro and in vivo, and indicated that antiviral susceptibility of clinical HBV isolates was well coincident between in vivo and in vitro. Therefore, the antiviral treatment in vitro will provide useful information for individualized selection of antiviral agents in vivo.
Several antiviral agents including lamivudine, adefovir and entacavir are available for CHB treatment, and several novel antiviral agents for CHB treatment are being developed. In vitro analysis of drug susceptibility based on clinical HBV isolates is useful for personalized selection of the existing or novel antiviral agents or combinations of antiviral agents for treatment. However, there are still some problems to be resolved. Firstly, HBV replicates as quasi-species in vivo, and the replication capacity of individual species are likely to vary; therefore the predominant HBV isolate should be selected in this study. Secondly, the replication of HBV in vitro is significantly different from that of in vivo, and the influence of the difference between in vitro and in vivo on antiviral agent selection should be considered. Finally, despite the use of a high fidelity DNA polymerase for HBV genome amplification, artificial mutation may occur in HBV genome, and the replication of HBV isolates may be affected.
Our study described a novel strategy and method for developing an in vitro system for selection of antiviral agents based on clinical HBV isolates, and evaluated the application in individualized selection of antiviral agents for chronic hepatitis B patients. The results indicated that in vitro analysis of drug susceptibility based on clinical HBV isolates is useful for individualized selection of antiviral agents for patients with CHB.
COMMENTS
Background
Hepatitis B virus (HBV) is the leading cause of chronic hepatitis. Antiviral treatment of chronic hepatitis B (CHB) relies on interferon alpha and nucleoside analogs that inhibit activity of HBV polymerase. However, resistance of HBV to antiviral nucleoside analogs, caused by mutation of HBV polymerase gene, has become a major clinical problem. Moreover, the lack of reliable in vitro infection system and convenient models has hindered the new treatment options. Therefore, establishment of new strategies for studying in vitro the drug susceptibility of clinical isolates of HBV is extremely important for antiviral therapy for CHB patients.
Research frontiers
The phenotypic assays have first been developed for monitoring drug susceptibility of human immunodeficiency virus viral populations. Some of them are used currently in clinical practice to monitor drug resistance. As compared with the human immunodeficiency virus phenotypic assay, the development and clinical use of the HBV phenotypic assay are still at an early stage. This study demonstrated the relevance of this type of assay for longitudinal monitoring of the drug susceptibility status of the viral population isolated from clinical samples.
Innovations and breakthroughs
This study describes a novel strategy and method for the cloning of HBV genomes isolated from CHB patients into plasmidic vectors and the basis of another phenotypic assay capable of assessing HBV drug susceptibility in vitro and evaluates the effects of antiviral agents in circulating viral isolates from chronic HBV.
Applications
This work may help determine the drug susceptibility of HBV quasi-species to antivirals and select antiviral agents for CHB patients.
Peer review
In this study the authors established a novel method to assess in vitro replication of clinical HBV isolate and investigated its application for selecting antiviral agents for CHB patients. It will be very useful in future studies of antiviral therapy for CHB.
Supported by The National Natural Science Foundation of China, No. 30271170, and the Ph.D. Program Fund of Chinese Ministry of Education, No. 20070487152
Peer reviewer: Vasiliy I Reshetnyak, MD, PhD, Professor, Scientific Research Institute of General Reanimatology, 25-2, Petrovka Str., 107031, Moscow, Russia
S- Editor Li DL L- Editor Ma JY E- Editor Yin DH
References
- 1.Maddrey WC. Hepatitis B: an important public health issue. J Med Virol. 2000;61:362–366. doi: 10.1002/1096-9071(200007)61:3<362::aid-jmv14>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 2.Lai CL, Ratziu V, Yuen MF, Poynard T. Viral hepatitis B. Lancet. 2003;362:2089–2094. doi: 10.1016/S0140-6736(03)15108-2. [DOI] [PubMed] [Google Scholar]
- 3.Villeneuve JP. The natural history of chronic hepatitis B virus infection. J Clin Virol. 2005;34 Suppl 1:S139–S142. doi: 10.1016/s1386-6532(05)80024-1. [DOI] [PubMed] [Google Scholar]
- 4.Cougot D, Neuveut C, Buendia MA. HBV induced carcinogenesis. J Clin Virol. 2005;34 Suppl 1:S75–S78. doi: 10.1016/s1386-6532(05)80014-9. [DOI] [PubMed] [Google Scholar]
- 5.Farrell GC, Teoh NC. Management of chronic hepatitis B virus infection: a new era of disease control. Intern Med J. 2006;36:100–113. doi: 10.1111/j.1445-5994.2006.01027.x. [DOI] [PubMed] [Google Scholar]
- 6.Loomba R, Liang TJ. Novel approaches to new therapies for hepatitis B virus infection. Antivir Ther. 2006;11:1–15. [PubMed] [Google Scholar]
- 7.Craxi A, Antonucci G, Camma C. Treatment options in HBV. J Hepatol. 2006;44:S77–S83. doi: 10.1016/j.jhep.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 8.Zoulim F. Entecavir: a new treatment option for chronic hepatitis B. J Clin Virol. 2006;36:8–12. doi: 10.1016/j.jcv.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 9.Liu CJ, Lai MY, Chao YC, Liao LY, Yang SS, Hsiao TJ, Hsieh TY, Lin CL, Hu JT, Chen CL, et al. Interferon alpha-2b with and without ribavirin in the treatment of hepatitis B e antigen-positive chronic hepatitis B: a randomized study. Hepatology. 2006;43:742–749. doi: 10.1002/hep.21100. [DOI] [PubMed] [Google Scholar]
- 10.Leemans WF, Flink HJ, Janssen HL, Niesters HG, Schalm SW, de Man RA. The effect of pegylated interferon-alpha on the treatment of lamivudine resistant chronic HBeAg positive hepatitis B virus infection. J Hepatol. 2006;44:507–511. doi: 10.1016/j.jhep.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 11.Das K, Xiong X, Yang H, Westland CE, Gibbs CS, Sarafianos SG, Arnold E. Molecular modeling and biochemical characterization reveal the mechanism of hepatitis B virus polymerase resistance to lamivudine (3TC) and emtricitabine (FTC) J Virol. 2001;75:4771–4779. doi: 10.1128/JVI.75.10.4771-4779.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buti M, Rodriguez-Frias F, Jardi R, Esteban R. Hepatitis B virus genome variability and disease progression: the impact of pre-core mutants and HBV genotypes. J Clin Virol. 2005;34 Suppl 1:S79–S82. doi: 10.1016/s1386-6532(05)80015-0. [DOI] [PubMed] [Google Scholar]
- 13.Liaw YF. The current management of HBV drug resistance. J Clin Virol. 2005;34 Suppl 1:S143–S146. doi: 10.1016/s1386-6532(05)80025-3. [DOI] [PubMed] [Google Scholar]
- 14.Tong S. Mechanism of HBV genome variability and replication of HBV mutants. J Clin Virol. 2005;34 Suppl 1:S134–S138. doi: 10.1016/s1386-6532(05)80023-x. [DOI] [PubMed] [Google Scholar]
- 15.Pawlotsky JM. The concept of hepatitis B virus mutant escape. J Clin Virol. 2005;34 Suppl 1:S125–S129. doi: 10.1016/s1386-6532(05)80021-6. [DOI] [PubMed] [Google Scholar]
- 16.Paran N, Geiger B, Shaul Y. HBV infection of cell culture: evidence for multivalent and cooperative attachment. EMBO J. 2001;20:4443–4453. doi: 10.1093/emboj/20.16.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, Ding X, Zhang Y, Bo X, Zhang M, Wang S. Fibronectin is essential for hepatitis B virus propagation in vitro: may be a potential cellular target? Biochem Biophys Res Commun. 2006;344:757–764. doi: 10.1016/j.bbrc.2006.03.204. [DOI] [PubMed] [Google Scholar]
- 18.Guha C, Mohan S, Roy-Chowdhury N, Roy-Chowdhury J. Cell culture and animal models of viral hepatitis. Part I: hepatitis B. Lab Anim (NY) 2004;33:37–46. doi: 10.1038/laban0704-37. [DOI] [PubMed] [Google Scholar]
- 19.Tang H, Raney AK, McLachlan A. Replication of the wild type and a natural hepatitis B virus nucleocapsid promoter variant is differentially regulated by nuclear hormone receptors in cell culture. J Virol. 2001;75:8937–8948. doi: 10.1128/JVI.75.19.8937-8948.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chinese Society of Hepatology and Chinese Society of infectious Diseases, Chinese Medical Association. The guideline of prevention and treatment for chronic hepatitis B. Zhonghua Ganzangbing Zazhi. 2005;13:881–891. [PubMed] [Google Scholar]
- 21.Lu YP, Wang BJ, Dong JH, Liu Z, Guan SH, Lu MJ, Yang DL. Construction and Characterization of a Hepatitis B Virus Replicon. Virologica Sinica. 2007;22:8–13. [Google Scholar]
- 22.Gunther S, Li BC, Miska S, Kruger DH, Meisel H, Will H. A novel method for efficient amplification of whole hepatitis B virus genomes permits rapid functional analysis and reveals deletion mutants in immunosuppressed patients. J Virol. 1995;69:5437–5444. doi: 10.1128/jvi.69.9.5437-5444.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sells MA, Chen ML, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yaginuma K, Shirakata Y, Kobayashi M, Koike K. Hepatitis B virus (HBV) particles are produced in a cell culture system by transient expression of transfected HBV DNA. Proc Natl Acad Sci USA. 1987;84:2678–2682. doi: 10.1073/pnas.84.9.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang PL, Althage A, Chung J, Chisari FV. Hydrodynamic injection of viral DNA: a mouse model of acute hepatitis B virus infection. Proc Natl Acad Sci USA. 2002;99:13825–13830. doi: 10.1073/pnas.202398599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu H, Peng M, Qing Y, Ling N, Lan Y, Liang Z, Cai D, Li Y, Ren H. A Quasi species of the pre-S/S gene and mutations of enhancer II/core promoter/pre-C in mothers and their children infected with hepatitis B virus via mother-to-infant transmission. J Infect Dis. 2006;193:88–97. doi: 10.1086/498573. [DOI] [PubMed] [Google Scholar]
- 28.Burda MR, Gunther S, Dandri M, Will H, Petersen J. Structural and functional heterogeneity of naturally occurring hepatitis B virus variants. Antiviral Res. 2001;52:125–138. doi: 10.1016/s0166-3542(01)00177-2. [DOI] [PubMed] [Google Scholar]
- 29.Westland CE, Yang H, Delaney WE 4th, Wulfsohn M, Lama N, Gibbs CS, Miller MD, Fry J, Brosgart CL, Schiff ER, et al. Activity of adefovir dipivoxil against all patterns of lamivudine-resistant hepatitis B viruses in patients. J Viral Hepat. 2005;12:67–73. doi: 10.1111/j.1365-2893.2005.00578.x. [DOI] [PubMed] [Google Scholar]
- 30.Chen RY, Edwards R, Shaw T, Colledge D, Delaney WE 4th, Isom H, Bowden S, Desmond P, Locarnini SA. Effect of the G1896A precore mutation on drug sensitivity and replication yield of lamivudine-resistant HBV in vitro. Hepatology. 2003;37:27–35. doi: 10.1053/jhep.2003.50012. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki F, Kumada H, Nakamura H. Changes in viral loads of lamivudine-resistant mutants and evolution of HBV sequences during adefovir dipivoxil therapy. J Med Virol. 2006;78:1025–1034. doi: 10.1002/jmv.20658. [DOI] [PubMed] [Google Scholar]
- 32.Zeng M, Mao Y, Yao G, Wang H, Hou J, Wang Y, Ji BN, Chang CN, Barker KF. A double-blind randomized trial of adefovir dipivoxil in Chinese subjects with HBeAg-positive chronic hepatitis B. Hepatology. 2006;44:108–116. doi: 10.1002/hep.21225. [DOI] [PubMed] [Google Scholar]
- 33.Schildgen O, Sirma H, Funk A, Olotu C, Wend UC, Hartmann H, Helm M, Rockstroh JK, Willems WR, Will H, et al. Variant of hepatitis B virus with primary resistance to adefovir. N Engl J Med. 2006;354:1807–1812. doi: 10.1056/NEJMoa051214. [DOI] [PubMed] [Google Scholar]
- 34.Marcellin P, Chang TT, Lim SG, Tong MJ, Sievert W, Shiffman ML, Jeffers L, Goodman Z, Wulfsohn MS, Xiong S, et al. Adefovir dipivoxil for the treatment of hepatitis B e antigen-positive chronic hepatitis B. N Engl J Med. 2003;348:808–816. doi: 10.1056/NEJMoa020681. [DOI] [PubMed] [Google Scholar]
- 35.Hadziyannis SJ, Tassopoulos NC, Heathcote EJ, Chang TT, Kitis G, Rizzetto M, Marcellin P, Lim SG, Goodman Z, Wulfsohn MS, et al. Adefovir dipivoxil for the treatment of hepatitis B e antigen-negative chronic hepatitis B. N Engl J Med. 2003;348:800–807. doi: 10.1056/NEJMoa021812. [DOI] [PubMed] [Google Scholar]