Abstract
Multiplexed PCR, the amplification of multiple targets in a single reaction, presents a new set of challenges that further complicate more traditional PCR set-ups. These complications include a greater probability for non-specific amplicon formation and for imbalanced amplification of different targets, each of which can compromise quantification and detection of multiple targets. Despite these difficulties, multiplex PCR is frequently used in such applications as pathogen detection, RNA quantification, mutation analysis and now, next generation DNA sequencing. Herein, we investigate the utility of primers with one or two thermolabile 4-oxo-1-pentyl phosphotriester modifications in improving multiplex PCR performance. Initial endpoint and real-time analyses reveal a decrease in off-target amplification and subsequent increase in amplicon yield. Furthermore, the use of modified primers in multiplex set-ups revealed a greater limit of detection and more uniform amplification of each target as compared to unmodified primers. Overall, the thermolabile modified primers present a novel and exciting avenue in improving multiplex PCR performance.
Keywords: Multiplex PCR, PCR, Real-Time PCR, off-target amplification, DNA polymerase, primer dimer, Hot Start PCR
Introduction
The Polymerase Chain Reaction (PCR) is a commonly used molecular biology technique for the amplification of DNA. One variation of PCR is the amplification of multiple loci using two or more primer pairs known as multiplex PCR. Multiplex PCR, first used to amplify multiple loci in the human dystrophin gene [1], is now commonly used in mutation and polymorphism analysis [2, 3], real time PCR quantitative calculations [4], RNA detection [5], and recently DNA sequencing [6]. In such fields as molecular diagnostics, multiplex PCR is a valuable tool as it increases PCR capacity by increasing the number of targets detected in a single reaction, effectively lowering reagent costs [3].
Despite widespread use, inherent problems still plague multiplex PCR, such as preferential target amplification and non-specific amplifications, including primer dimer and mis-priming artifacts[6–10]. The extension of mis-priming and primer dimer complexes can subsequently consume essential PCR substrates such as the dNTPs and DNA polymerase from the desired amplification. This depletion of substrates not only lowers the specificity of the reaction but also can lower amplicon yield. In multiplex PCR, the problem of off-target amplification is compounded as the number of targets and as a result, the number of primer pairs is increased. Another common problem, preferential amplification of one target sequence over other target sequences, has been attributed to both PCR drift and selection [11]. PCR drift, which results from the stochastic interaction of PCR reagents especially in the early cycles of PCR, is difficult to adjust or correct due to the randomness of the variation and the irreproducibility of each fluctuation [12, 13]. On the other hand, PCR selection is a mechanism that inherently favors the amplification of certain targets because of specific properties of the target, the flanking sequences, or the whole genome. For example, GC content can often lower amplicon efficiencies due to the formation of secondary structures by the template [15, 16]. Additional sources for PCR bias have been linked to heteroduplex formation [14] and to preferential DNA rehybridization of more abundant amplicons in the later stage of PCR [17, 18].
As a result of the above-mentioned problems, optimization of each PCR component has become essential in attaining a robust multiplex PCR assay [19, 20]. Increased concentration of DNA polymerase and magnesium chloride as well as the use of other additives have resulted in improved amplification [2, 19, 21, 22]. In addition to basic optimizations and design, different methodologies have been developed, such as multi-step manipulations of the template DNA before PCR amplification which greatly increase the number of targets able to be amplified [23, 24]. Finally, “Hot Start” technologies centered around modifying the DNA polymerase to be inactive at the lower temperature set-up conditions have been demonstrated to improve multiplex PCR [25–27].
Much time and effort in multiplex PCR optimization is also placed into optimizing primer design and concentration [28–30]. Despite the important role that primers play in the success of the PCR reactions, primer modifications that prevent off-target amplification have not been investigated extensively. Recently, primers containing 4-oxo-1-pentyl (to be abbreviated OXP hereafter) phosphotriester modifications have demonstrated promise as a primer-based “Hot Start” solution in PCR [31]. The OXP modifications are introduced at the 3′ terminal phosphodiester linkage (termed single modified OXP) or at both the 3′ terminal and the penultimate phosphodiester linkages (termed double modified OXP) of the primers. The two types of primers differ in conversion rates to unmodified, viable primers as the time for half the double modified OXP primers to convert to unmodified primers is approximately twice as long as the single modified OXP primer [31]. The presence of thermolabile primer modifications has been shown to block DNA polymerase primer extension at low temperatures but release at a known rate at higher thermal cycling temperatures. Previous work has shown that in experiments where a single target has been amplified, the OXP modified primers reduced or prevented off-target amplification while enhancing amplicon yield [31]. Herein, the utility of OXP primers will be tested where more than one target is being detected in a multiplex PCR format.
Materials and Methods
Materials
Primers which contained either one or two 4-oxo-1-pentyl phosphotriester modifications to the 3′-terminal and penultimate internucleotide linkages (CleanAmp™ Turbo and CleanAmp™ Precision Primers, respectively) were prepared using the protocol described previously in Lebedev et. al [31]. Unmodified PCR primers and TaqMan® probes were prepared using standard methods. Bacteriophage Lambda genomic DNA used as the template was purchased from Roche Applied Science. Taq DNA polymerase was bought from Invitrogen and the dNTPs from New England Biolabs (NEB). “Hot Start” polymerases used were Platinum® Taq DNA Polymerase (Invitrogen), AmpliTaq® Gold DNA Polymerase (Applied BioSystems), DyNAzyme™ II Hot Start DNA Polymerase (New England Biolabs), HotStart-IT® DNA Polymerase (USB), and Multiplex PCR Kit (Qiagen). All polymerases were used with the manufacturers’ recommended 1X PCR buffer solution. The passive reference dye ROX used in real-time PCR experiments was purchased from Stratagene. Endpoint experiments were either conducted on a Perkin Elmer GeneAmp 9600® or a Perkin Elmer GeneAmp 2400® Thermal cycler. Real-time experiments were performed on a Stratagene MX3005P® QPCR System Instrument.
Endpoint PCR Experiments
All the triplex experiments contained 1X PCR buffer (20mM Tris (pH 8.4), 50mM KCl, 2.5mM MgCl2), 0.2mM dNTPs, and 1.25 U Taq DNA polymerase. Taq DNA polymerase and Platinum® Taq DNA Polymerase reactions with more than three targets contained 1X PCR buffer (20mM Tris (pH 8.4), 50mM KCl, 2.5mM MgCl2) with an additional 1.5mM MgCl2 (4mM total), 0.2mM dNTPs (0.4mM total), and 1.25 units of DNA polymerase (2.5 U total). An additional 40mM KCl (90mM final concentration) purchased from Fluka was added to only the single modified OXP primers and Taq DNA polymerase reaction. The corresponding addition of KCl to the Platinum® Taq reactions inhibited amplification and was therefore not included. Reactions were conducted in a single, thin walled 200μL tube with a total volume of 50μL. The primer concentration for all the reactions was 200nM. Primer sequences can be found in Table 1. Thermal cycling conditions included a 10 minute initial denaturation step at 95°C followed by 35 PCR cycles at 95°C for 40 seconds, 56°C for 30 seconds, 72°C for 2 minutes, and a final extension step at 72°C for 7 minutes. Lambda genomic DNA was used as the template in concentrations ranging from 50 copies to 500,000 copies. After PCR, 20μL of each sample was loaded in a 2% agarose E-gel cartridge (Invitrogen) and ran for 30 minutes. An image of the gel was then taken using an Alpha Innotech Corporation Multi Image Light Cabinet with CCD Camera and quantified using AlphaEaseFC™ software.
Table 1.
Primers and TaqMan® Probes used.
| Target Length (bp) |
Forward Primer (5′-3′) | Reverse Primer (5′-3′) | TaqMan® Probe (5′-3′) |
|---|---|---|---|
| 139 | ACGATCTGGTACGCAATAACG | AAGGCACGGGCTTCTTCC | - |
| 249 | GAACTGCGACTGGATAGG | TCCTCCTCTGTTGATTGTC | - |
| 330 | AAACAGGGTGTGGAAATC | AGTCTCTTTGGTCATTCG | - |
| 431 | CGTCGTGGCTCTAATTCC | GGCAATCAGTTCATCTTTCG | - |
| 533 | CAGGAGCTGGACTTTACTGATGC | CGGGATATCGACATTTCTGCACC | 6FAM-ACCGTGTGGCTTACCTGACCGCCG-BHQ1 |
| 600 | TCCGGATGCGGAGTCTTATCCGTG | GGCCTGTTATTAGCTCAGTAATGT | HEX-CGTTGCCGCGCTAACAGGCGC-BHQ1 |
| 715 | GTCACCGCCAGTTAATCC | ATCAGCACATCATCTTCAGG | - |
| 860 | CGATAGATAGCCACGGACTTC | GCTCTCGGAATATCAATGAAGG | - |
| 962 | ATCAGAAACGAACGCATCATCAAGT | GCCTCGCATATCAGGAAGCAC | Alexa Fluor 647-CTTCCTTCGGCAACGGCGGCAGAAA-BHQ2 |
Real-time PCR Experiments
All conditions are similar as an endpoint triplex reaction except the addition of ROX passive reference dye (30nM) and TaqMan® probe (100nM). Reactions were monitored using one of the four channels: FAM, HEX, Cy-5, and ROX. Thermal cycling conditions included a 10 minute initial denaturation step at 95°C followed by 35 cycles at 95°C for 40 seconds, 56°C for 30 seconds, 72°C for 2 minutes. Lambda genomic DNA was used as the template in concentrations ranging from 50 copies to 500,000 copies.
Results
In these studies, PCR primers containing one or two thermolabile OXP modifications were assessed for their potential benefit in improving multiplex PCR performance. Experiments included the PCR evaluation of modified primers in triplex experiments, with endpoint and real-time detection. Further studies compared this approach to other Hot Start DNA polymerases and investigated expansion of the triplex studies into experiments with more targets.
Evaluation of single-plex and triplex PCR experiments
Three separate targets, 533bp, 600bp, 962bp in length (L533, L600, and L962, respectively), found in Lambda genomic DNA were amplified either in three separate reactions or collectively in a single tube multiplex PCR. In these conditions, the relative performance of three types of primers was tested: primers with no modifications, primers with a single OXP modification, and primers with a double OXP modification with analysis by agarose gel electrophoresis.
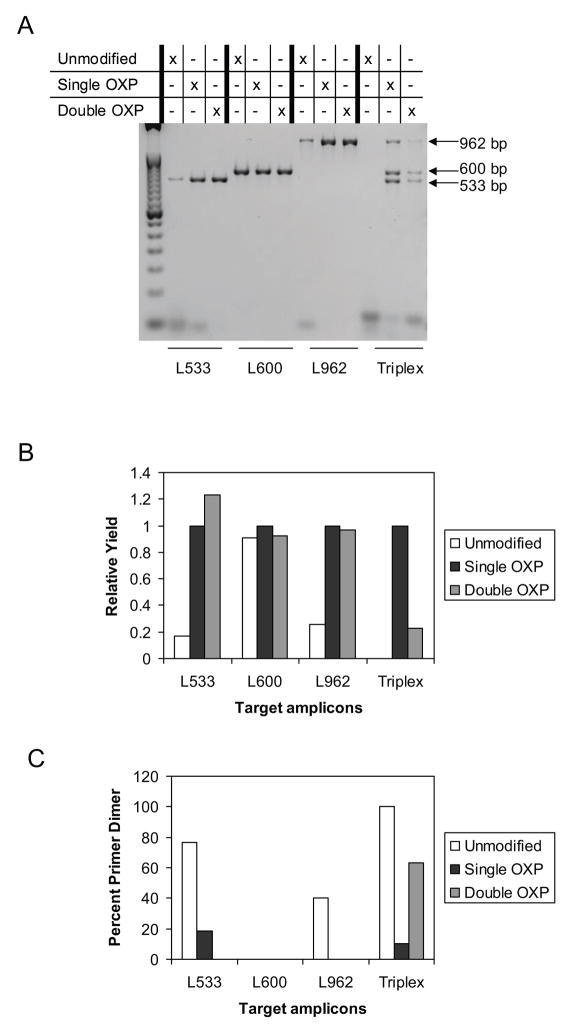
All three targets were found to amplify individually with varying yields in set-ups that contained both the unmodified and modified primers (Fig. 1). For reactions containing the unmodified primers, primer dimer formation complicated the amplification of L533 and L962 while reactions which employed the single and double OXP modified primers provided similar amplicon yields with no detectable primer dimer formation. However, when all three targets were amplified together in multiplex PCR (Fig. 1), the set-up with unmodified primers did not amplify any of the three targets and showed an increase in primer dimer formation. In contrast, tri-plex reactions containing modified primers amplified all three targets, with the double modified OXP primers exhibiting lower amplicon yield than the single modified OXP primers. The single modified OXP primers provided the best solution for this multiplex PCR reaction as they afforded the greatest amplicon yield, with reduced off-target amplicon formation.
Figure 1.
Comparison of the PCR performance of unmodified, single modified and double modified OXP primers in the amplification of three individual targets separately and collectively from 500 copies of Lambda genomic DNA. A. Agarose gel image of individual amplifications of three separate targets and a multiplex amplification of the three targets in a single reaction. B. Relative amplicon yield by densitometry integration of agarose gel. For each of the target amplicons, the yield of single modified OXP primers was normalized to 1.0. C. Percent primer dimer found by densitometry integration.
Comparison of unmodified, single modified OXP, and double modified OXP primers at different template concentrations
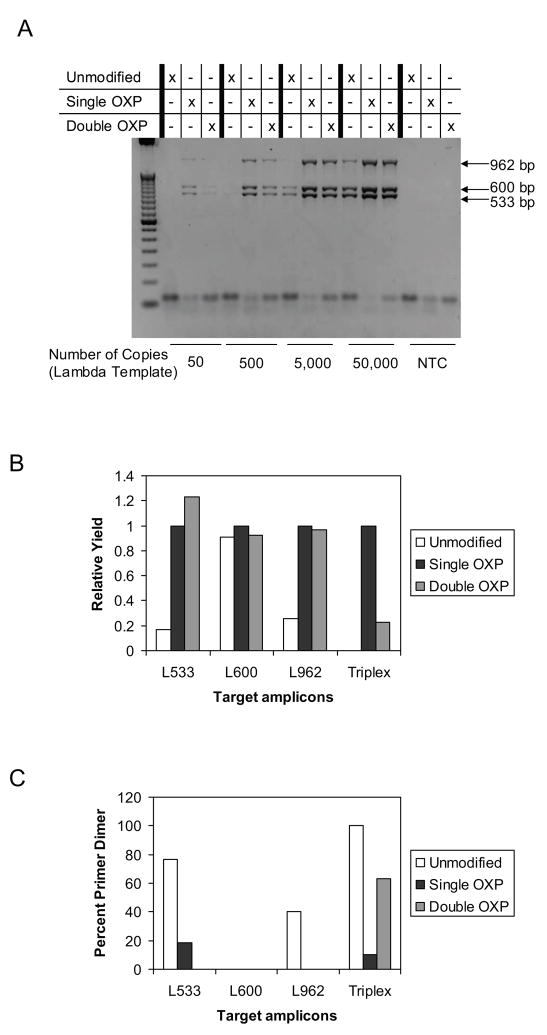
To test the level of sensitivity of the primers, the same triplex reaction set-up from the previous experiment with unmodified, single and double modified OXP primers was employed, using input amounts of Lambda genomic DNA template ranging from 50 to 50,000 copies (Fig. 2). Agarose gel analysis of reaction products reveals that the unmodified primers were unable to amplify all three targets at lower template concentrations. At 5,000 copies, reactions with unmodified primers successfully detected all 3 targets, however, the overall yield was still much lower than the corresponding reactions with single or double modified OXP primers. Although the relative amplicon yield for reactions with unmodified primers was found to increase as template concentration increased, the overall yield remained below the analogous reactions with single and double modified OXP primers. The use of double modified OXP primers in a triplex reaction was found to have slightly lower amplicon yield compared to the single OXP primers across all template concentrations, albeit sensitivity was not compromised to as great of an extent as for unmodified primers. The single modified OXP primers amplified most efficiently at all template concentrations and provided the greatest limit of detection at template concentrations as low as 50 copies.
Figure 2.
Comparison of unmodified, single modified, and double modified OXP primers in the amplification of a range of Lambda genomic template DNA ranging from 50 to 50,000 copies. A. Agarose gel for amplification products using unmodified, single modified, and double modified OXP primers. B. Relative yield of all three targets using densitometry measurements where the error bars represent the standard deviation of a single experiment done in triplicate. Yield values for all three targets were combined. For each of the target amplicons, the yield of single modified OXP primers was normalized to 1.0. C. Relative primer dimer percentages by densitometry measurements.
Real-time PCR analysis of unmodified, single modified OXP, and double modified OXP primers over a wide range of template concentrations
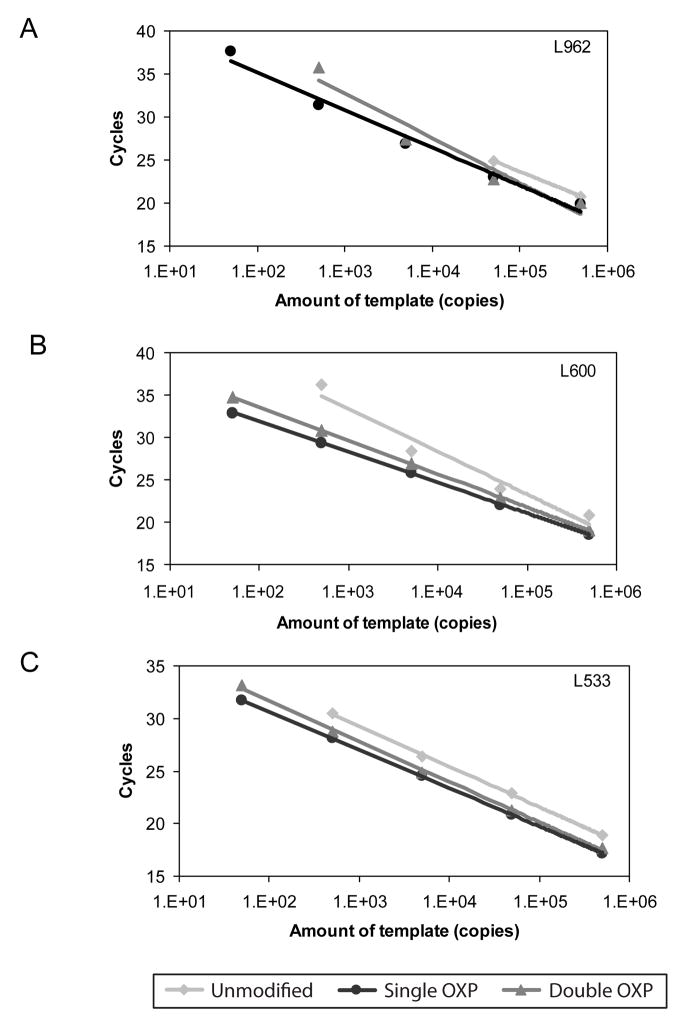
A similar triplex experiment was performed in real-time PCR, a much more sensitive and thorough method to analyze PCR efficiency, using TaqMan probe detection (Fig. 3). The resultant standard curves from the amplification of 50 to 50,000 copies of Lambda genomic DNA revealed that for all three targets, the set-up with single modified OXP primers had the lowest Ct (cycle threshold). At lower template concentrations, reactions containing the unmodified primers did not amplify the amplicons in 40 cycles, findings which correlate well with the previous endpoint findings. For reactions with the double modified OXP primers, Ct values at high template concentrations were similar to reactions with the single modified OXP primers. However, as template concentration was decreased, the Ct values for reactions with double modified OXP primers began to deviate from linearity or were absent, suggesting a decrease in PCR amplification efficiency relative to single modified OXP primers. Overall, these results correlate with previous results in endpoint analysis and demonstrate that single modified OXP primers exhibit better PCR efficiency and specificity even at low template concentrations.
Figure 3.
Triplex Real-time PCR experiments using TaqMan® probe detection of 50 to 50,000 copies of Lambda genomic template DNA. Standard curve for the A. L533, B. L600, and C. L962 targets from a triplex reaction comparing the performance of unmodified, single modified OXP and double modified OXP primers. The resultant data for each standard curve follows.
L533: Unmodified primers: Y = −3.842*LOG(X) + 38.79, Eff. = 82.1%, RSq:0.999, Single OXP primers: Y = −3.638*LOG(X) + 37.93, Eff. = 88.3%, RSq:1.000, Double OXP primers: Y = −3.839*LOG(X) + 39.34, Eff. = 82.2%, RSq:0.998
L600: Unmodified primers: Y = −5.664*LOG(X) + 49.60, Eff. = 50.2%, RSq:0.929, Single OXP primers: Y = −3.603*LOG(X) + 39.06, Eff. = 89.5%, RSq:1.000, Double OXP primers: Y = −3.742*LOG(X) + 39.91, Eff. = 85.0%, RSq:0.996
L962: Unmodified primers: Y = −4.049*LOG(X) + 41.88, Eff. = 76.6%, RSq:1.000, Single OXP primers: Y = −4.924*LOG(X) + 46.12, Eff. = 59.6%, RSq:0.963, Double OXP primers: Y = −5.538*LOG(X) + 49.59, Eff. = 51.6%, RSq:0.959
Evaluation of single modified OXP primers in comparison to and in combination with other “Hot Start” Technologies
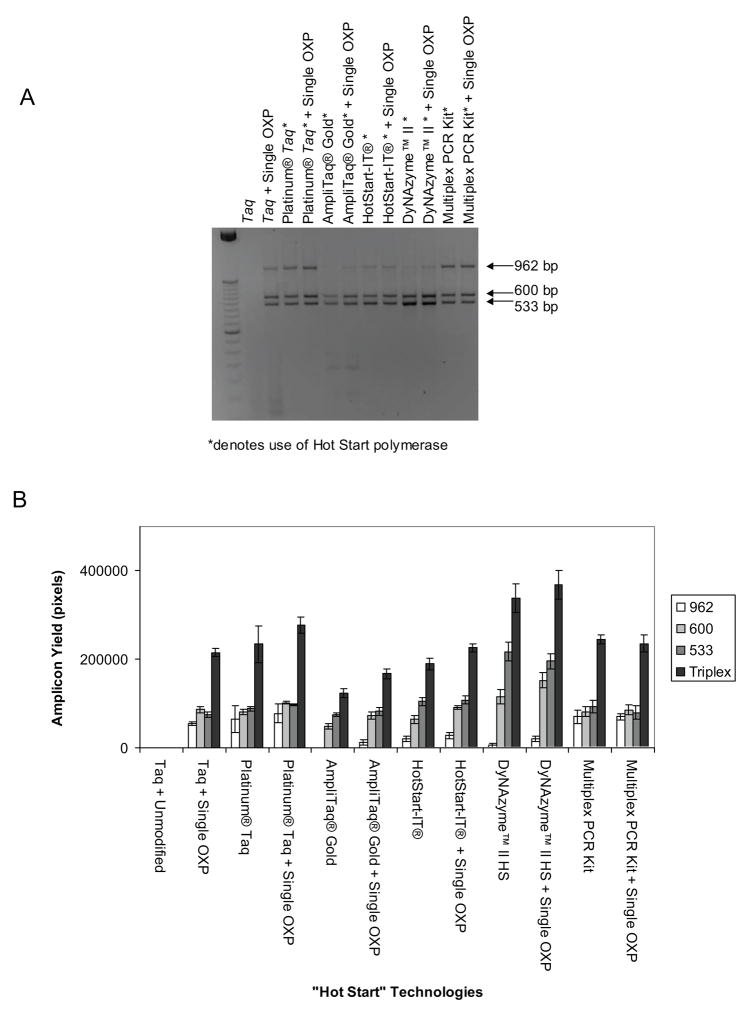
Single modified OXP primers were compared to other “Hot Start” technologies for amplicon yield. The single modified OXP primers were also combined with the same “Hot Start” technologies to see if there was further improvement (Fig. 4). Most of these “Hot Start” approaches include modifications to the DNA polymerase which prevent extension at the low set-up temperatures and resume normal DNA extension at higher stringency, elevated temperatures. The single modified OXP primers, Platinum® Taq, and Qiagen’s Multiplex PCR kit all displayed similar efficiency in amplifying each individual target and similar overall yields. However, while the smaller L533 and L600 products were amplified well, the longer L962 proved difficult to amplify for some of the modified DNA polymerases such as AmpliTaq® Gold, HotStart-IT®, and DyNAzyme™ II Hot Start Polymerase. The combination of single modified OXP primers with the existing “Hot Start” approaches did not dramatically improve reaction performance relative to reactions with unmodified primers. However, there were indications of a slight increase in amplicon yield for all three targets for all OXP-modified primer combinations with the exception of the Qiagen Multiplex PCR kit. Overall, OXP-modified primers provided comparable and in some cases improved performance to other Hot Start technologies, with the promise for improvements when used in combination with these technologies.
Figure 4.
Single modified OXP primers in comparison and combination with other “Hot Start” technologies. A. Triplex PCR reaction at 500 copies template DNA with single modified OXP primers and other modified DNA polymerases. The modified DNA polymerases were assayed with either unmodified or single modified OXP primers. B. Densitometry integration of each individual target and the total yield (sum of all three targets) of each triplex reaction, where the error bars represent the standard deviation of two independent experiments performed in duplicate.
Increasing multiplex PCR reaction from three to nine targets
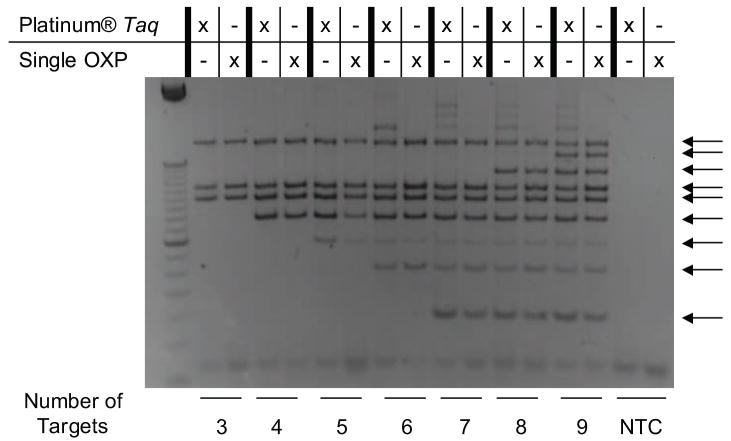
In order to further test the limits of multiplex PCR with single OXP modified primers, the number of targets from Lambda genomic DNA was increased from three to nine by sequentially adding additional primer pairs (Fig. 5). In the same experiment, the single OXP modified primers were compared to Platinum® Taq, a “Hot Start” approach which performed comparably well amplifying the triplex targets (Fig. 4). Although increasing the number of primer pairs could cause greater potential for primer self-hybridization and concomitant primer dimer formation, the single OXP modified primers were still able to efficiently amplify each target with similar efficiency as Platinum® Taq. However, reactions that employed Platinum® Taq were prone to increased off-target amplification of longer non-specific targets as the number of targets increased. Though the maximum number of targets able to be amplified using the OXP modified primers has still yet to be determined, nine targets can be robustly amplified when single OXP modified primers are employed.
Figure 5.
Determining upper limit of targets for single modified OXP primers in multiplex PCR. The number of targets amplified was increased from three to nine targets sequentially with Platinum® Taq and modified OXP primers with 500 copies of Lambda genomic template DNA.
Discussion
Multiplex PCR is a powerful tool which allows for the detection of multiple targets in a single reaction. Not only does it provide an economical alternative for higher-priced reagents, it has also opened research possibilities in such areas as mutation and polymorphism analysis [3], bacterial species identification [32], and relative quantification of RNA [4]. Multiplex PCR faces two key problems: non-specific product formation and preferential amplification. The prevention of off-target amplification is essential for robust product formations in multiplex PCR [2]. As demonstrated earlier (Fig. 1), an increase in the number of primer pairs in a reaction further increases spurious product formation when unmodified primers are employed, which sequesters other essential PCR components and limits the overall efficiency of amplicon formation. Herein, we have demonstrated that the introduction of OXP modifications into PCR primers can effectively improve the performance of multiplex set-ups, with single modified OXP primers providing a greater level of detection (Figs. 2, 3) and improved real time performance (Fig. 3) in relation to reactions containing unmodified primers. When compared to “Hot Start” DNA polymerases, single modified OXP primers provide both robust and uniform amplicon yields similar to the top performing “Hot Start” technologies; Platinum® Taq and Qiagen’s Multiplex PCR kit. Furthermore, the combination of the single modified OXP primers with other “Hot Start” approaches demonstrated some promise for slight improvements in amplicon yield (Fig. 4).
The ability to detect at least nine individual targets cleanly from genomic DNA is promising as Platinum® Taq, with increased primers pairs, appeared to exhibit greater off target amplification. From recent literature, multiplex PCR has been found to be limited to 10–20 targets when amplifying from genomic DNA [33, 34]. To circumvent this limitation and to accommodate higher multiplexing, many new methodologies have been developed, which include ligating adaptor sequences to endonuclease treated genomic DNA, followed by PCR amplification with a single primer pair [23, 24]. Though these methods have been successful, they require much manipulation before the PCR amplification. Further studies into determining the upper limit of detectable targets using single modified OXP primers could prove to be valuable, as it may allow for higher multiplexing directly from genomic DNA, especially in combination with the above-mentioned methodologies.
One unexpected advantage of the use of OXP modified primers in multiplexed reactions is the unbiased amplification of all targets. Though PCR drift is unavoidable in any multiplex reaction in principle, PCR selection can be modulated in order to encourage equal amplification of targets. From the comparison of single modified OXP primers to several of the “Hot Start” DNA polymerases, the longer 962 base pair target was amplified with lower efficiency for the modified DNA polymerases. The reduced efficiency for longer target formation indicates a potential length dependent effect when using those polymerases. The reactions with single modified OXP primers did not experience such an effect, as all three targets were amplified equally. Perhaps the rate of conversion from single modified OXP primers to viable unmodified primers during thermal cycling conditions provides the correct balance between too abundant or too scarce unmodified primer concentrations for unbiased amplification. This observation would imply that a controlled release of viable primers into the reaction is extremely important in preventing preferential amplification in multiplex PCR.
The kinetics of the removal of the OXP group(s) must also be sufficient to satisfy the number of unmodified primers molecules needed to anneal to the target of interest. If the kinetics of release of unmodified primer is too fast, the resultant excess of unmodified primers will have a greater likelihood of annealing to each other or other non-specific sites along the template. On the other hand, much like the results seen from studies with the double modified OXP primers, if the kinetics are too slow, the PCR efficiency will be compromised by limiting primer molecules relative to target, leading to less product formed [31]. Based on our findings, there appears to a balance between excess unmodified primers, where off-target amplifications predominate the reaction and limiting modified primers where the concentration of available primer and template is below the Km of the enzyme [35]. The single OXP modified primer seemingly is able to remain between both extremes and therefore promote efficient amplification by release of the correct balance of unmodified primer into the reaction.
The use of OXP modified primers in multiplex PCR shows great promise in a number of different applications. The enhancement in the lower limit of detection provided by single modified OXP primers in multiplex PCR could prove to be useful in situations where template is limited. For example, many multiplex PCR assays identifying bacterial and viral pathogens lack sensitivity mainly due to difficulty of isolating the template [2, 36]. Perhaps the most exciting result afforded by the use of OXP modified primers is the unbiased amplification of each target. This advantage is necessary when trying to quantify DNA or RNA levels, as the amplifications and product yield would be a more accurate representation of initial template sample and not an artifact of PCR drift or selection [12]. At the RNA level, quantification of expression levels by multiplex PCR with OXP modified primers could act as a corollary and perhaps even replace traditional protein quantification assays such as ELISA. On the DNA level, OXP modified primers could greatly assist researchers studying copy number variation by reducing artificial PCR influences in each assay [37].
Acknowledgments
We thank E. Hidalgo Ashrafi, R. Hogrefe, T. Le, A. Lebedev, J. Yee, G. Zon for helpful suggestions and for critical reading of the manuscript. Funding for this work was provided by National Institutes of Health (NIH awarding numbers 1R43GM072177-01A1, 2R44GM072177-02, and 5R44GM072177-03).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chamberlain JS, Gibbs RA, Ranier JE, Nguyen PN, Caskey CT. Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res. 1988;16:11141–56. doi: 10.1093/nar/16.23.11141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elnifro EM, Ashshi AM, Cooper RJ, Klapper PE. Multiplex PCR: optimization and application in diagnostic virology. Clin Microbiol Rev. 2000;13:559–70. doi: 10.1128/cmr.13.4.559-570.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Lellis L, Curia MC, Veschi S, Aceto GM, Morgano A, Cama A. Methods for routine diagnosis of genomic rearrangements: multiplex PCR-based methods and future perspectives. Expert Rev Mol Diagn. 2008;8:41–52. doi: 10.1586/14737159.8.1.41. [DOI] [PubMed] [Google Scholar]
- 4.Sherlock J, Cirigliano V, Petrou M, Tutschek B, Adinolfi M. Assessment of diagnostic quantitative fluorescent multiplex polymerase chain reaction assays performed on single cells. Ann Hum Genet. 1998;62:9–23. doi: 10.1046/j.1469-1809.1998.6210009.x. [DOI] [PubMed] [Google Scholar]
- 5.Zou S, Stansfield C, Bridge J. Identification of new influenza B virus variants by multiplex reverse transcription-PCR and the heteroduplex mobility assay. J Clin Microbiol. 1998;36:1544–8. doi: 10.1128/jcm.36.6.1544-1548.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Porreca GJ, Zhang K, Li JB, Xie B, Austin D, Vassallo SL, LeProust EM, Peck BJ, Emig CJ, Dahl F, Gao Y, Church GM, Shendure J. Multiplex amplification of large sets of human exons. Nat Methods. 2007;4:931–6. doi: 10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 7.Brownie J, Shawcross S, Theaker J, Whitcombe D, Ferrie R, Newton C, Little S. The elimination of primer-dimer accumulation in PCR. Nucleic Acids Res. 1997;25:3235–41. doi: 10.1093/nar/25.16.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puskas LG, Bottka S. Reduction of mispriming in amplification reactions with restricted PCR. Genome Res. 1995;5:309–11. doi: 10.1101/gr.5.3.309. [DOI] [PubMed] [Google Scholar]
- 9.Shigemori Y, Mikawa T, Shibata T, Oishi M. Multiplex PCR: use of heat-stable Thermus thermophilus RecA protein to minimize non-specific PCR products. Nucleic Acids Res. 2005;33:e126. doi: 10.1093/nar/gni111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou Q, Russell M, Birch DE, Raymond J, Bloch W. Prevention of pre-PCR mis-priming and primer dimerization improves low-copy-number amplifications. Nucleic Acids Res. 1992;20:1717–23. doi: 10.1093/nar/20.7.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagner A, Blackstone N, Cartwright P, Dick M, Misof B, Snow P, Wagner GP, Bartels J, Murtha M, Pendleton J. Surveys of gene families using polymerase reaction: PCR selection and PCR drift. Syst Biol. 1994;43:250–261. [Google Scholar]
- 12.Polz MF, Cavanaugh CM. Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol. 1998;64:3724–30. doi: 10.1128/aem.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mutter GL, Boynton KA. PCR bias in amplification of androgen receptor alleles, a trinucleotide repeat marker used in clonality studies. Nucleic Acids Res. 1995;23:1411–8. doi: 10.1093/nar/23.8.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanagawa T. Bias and artifacts in multitemplate polymerase chain reactions (PCR) J Biosci Bioeng. 2003;96:317–23. doi: 10.1016/S1389-1723(03)90130-7. [DOI] [PubMed] [Google Scholar]
- 15.Dutton CM, Paynton C, Sommer SS. General method for amplifying regions of very high G+C content. Nucleic Acids Res. 1993;21:2953–4. doi: 10.1093/nar/21.12.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki MT, Giovannoni SJ. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–30. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathieu-Daude F, Welsh J, Vogt T, McClelland M. DNA rehybridization during PCR: the ‘Cot effect’ and its consequences. Nucleic Acids Res. 1996;24:2080–6. doi: 10.1093/nar/24.11.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurata S, Kanagawa T, Magariyama Y, Takatsu K, Yamada K, Yokomaku T, Kamagata Y. Reevaluation and reduction of a PCR bias caused by reannealing of templates. Appl Environ Microbiol. 2004;70:7545–9. doi: 10.1128/AEM.70.12.7545-7549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henegariu O, Heerema NA, Dlouhy SR, Vance GH, Vogt PH. Multiplex PCR: critical parameters and step-by-step protocol. Biotechniques. 1997;23:504–11. doi: 10.2144/97233rr01. [DOI] [PubMed] [Google Scholar]
- 20.Edwards MC, Gibbs RA. Multiplex PCR: advantages, development, and applications. PCR Methods Appl. 1994;3:S65–75. doi: 10.1101/gr.3.4.s65. [DOI] [PubMed] [Google Scholar]
- 21.McElhinney LM, Cooper RJ, Morris DJ. Multiplex polymerase chain reaction for human herpesvirus-6, human cytomegalovirus, and human beta-globin DNA. J Virol Methods. 1995;53:223–33. doi: 10.1016/0166-0934(95)00019-q. [DOI] [PubMed] [Google Scholar]
- 22.Holger Engel. In: Office EP, editor. 2004. [Google Scholar]
- 23.Dahl F, Gullberg M, Stenberg J, Landegren U, Nilsson M. Multiplex amplification enabled by selective circularization of large sets of genomic DNA fragments. Nucleic Acids Res. 2005;33:e71. doi: 10.1093/nar/gni070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fredriksson S, Baner J, Dahl F, Chu A, Ji H, Welch K, Davis RW. Multiplex amplification of all coding sequences within 10 cancer genes by Gene-Collector. Nucleic Acids Res. 2007;35:e47. doi: 10.1093/nar/gkm078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kellogg DE, Rybalkin I, Chen S, Mukhamedova N, Vlasik T, Siebert PD, Chemchik A. TaqStart Antibody™: “Hot Start” PCR Facilitated by a Neutralizing Monoclonal Antibody Directed Against Taq DNA Polymerase. Biotechniques. 1994;16:1134–1137. [PubMed] [Google Scholar]
- 26.Kermekchiev MB, Tzekov A, Barnes WM. Cold-sensitive mutants of Taq DNA polymerase provide a hot start for PCR. Nucleic Acids Res. 2003;31:6139–47. doi: 10.1093/nar/gkg813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moretti T, Koons B, Budowle B. Enhancement of PCR amplification yield and specificity using AmpliTaq Gold DNA polymerase. Biotechniques. 1998;25:716–22. [PubMed] [Google Scholar]
- 28.Heredia A, Soriano V, Weiss SH, Bravo R, Vallejo A, Denny TN, Epstein JS, Hewlett IK. Development of a multiplex PCR assay for the simultaneous detection and discrimination of HIV-1, HIV-2, HTLV-I and HTLV-II. Clin Diagn Virol. 1996;7:85–92. doi: 10.1016/s0928-0197(96)00255-3. [DOI] [PubMed] [Google Scholar]
- 29.Dieffenbach CW, Lowe TM, Dveksler GS. General concepts for PCR primer design. PCR Methods Appl. 1993;3:S30–7. doi: 10.1101/gr.3.3.s30. [DOI] [PubMed] [Google Scholar]
- 30.Nicodeme P, Steyaert JM. Selecting optimal oligonucleotide primers for multiplex PCR. Proc Int Conf Intell Syst Mol Biol. 1997;5:210–3. [PubMed] [Google Scholar]
- 31.Lebedev AV, Paul N, Yee J, Timoshchuk VA, Shum J, Miyagi K, Kellum J, Hogrefe RI, Zon G. Hot start PCR with heat-activatable primers: a novel approach for improved PCR performance. Nucleic Acids Res. 2008;36:e131. doi: 10.1093/nar/gkn575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Regan E, McCabe E, Burgess C, McGuinness S, Barry T, Duffy G, Whyte P, Fanning S. Development of a real-time multiplex PCR assay for the detection of multiple Salmonella serotypes in chicken samples. BMC Microbiol. 2008;8:156. doi: 10.1186/1471-2180-8-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Broude NE, Zhang L, Woodward K, Englert D, Cantor CR. Multiplex allele-specific target amplification based on PCR suppression. Proc Natl Acad Sci U S A. 2001;98:206–11. doi: 10.1073/pnas.98.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Syvanen AC. Toward genome-wide SNP genotyping. Nat Genet. 2005;37(Suppl):S5–10. doi: 10.1038/ng1558. [DOI] [PubMed] [Google Scholar]
- 35.Johnson KA. Conformational coupling in DNA polymerase fidelity. Annu Rev Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 36.Mackay IM, Arden KE, Nitsche A. Real-time PCR in virology. Nucleic Acids Res. 2002;30:1292–305. doi: 10.1093/nar/30.6.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Komura D, Shen F, Ishikawa S, Fitch KR, Chen W, Zhang J, Liu G, Ihara S, Nakamura H, Hurles ME, Lee C, Scherer SW, Jones KW, Shapero MH, Huang J, Aburatani H. Genome-wide detection of human copy number variations using high-density DNA oligonucleotide arrays. Genome Res. 2006;16:1575–84. doi: 10.1101/gr.5629106. [DOI] [PMC free article] [PubMed] [Google Scholar]