Summary of recent advances
A major breakthrough in understanding the bacterial cell cycle is the discovery that bacteria exhibit a high degree of intracellular organization. Chromosomal loci and many protein complexes are positioned at particular subcellular sites. In this review, we examine recently discovered control mechanisms that make use of dynamically localized protein complexes to orchestrate the Caulobacter crescentus cell cycle. Protein localization, notably of signal transduction proteins, chromosome partition proteins, and proteases, serves to coordinate cell division with chromosome replication and cell differentiation. The developmental fate of daughter cells is decided before completion of cytokinesis, via the early establishment of cell polarity by the distribution of activated signaling proteins, bacterial cytoskeleton and landmark proteins.
Introduction
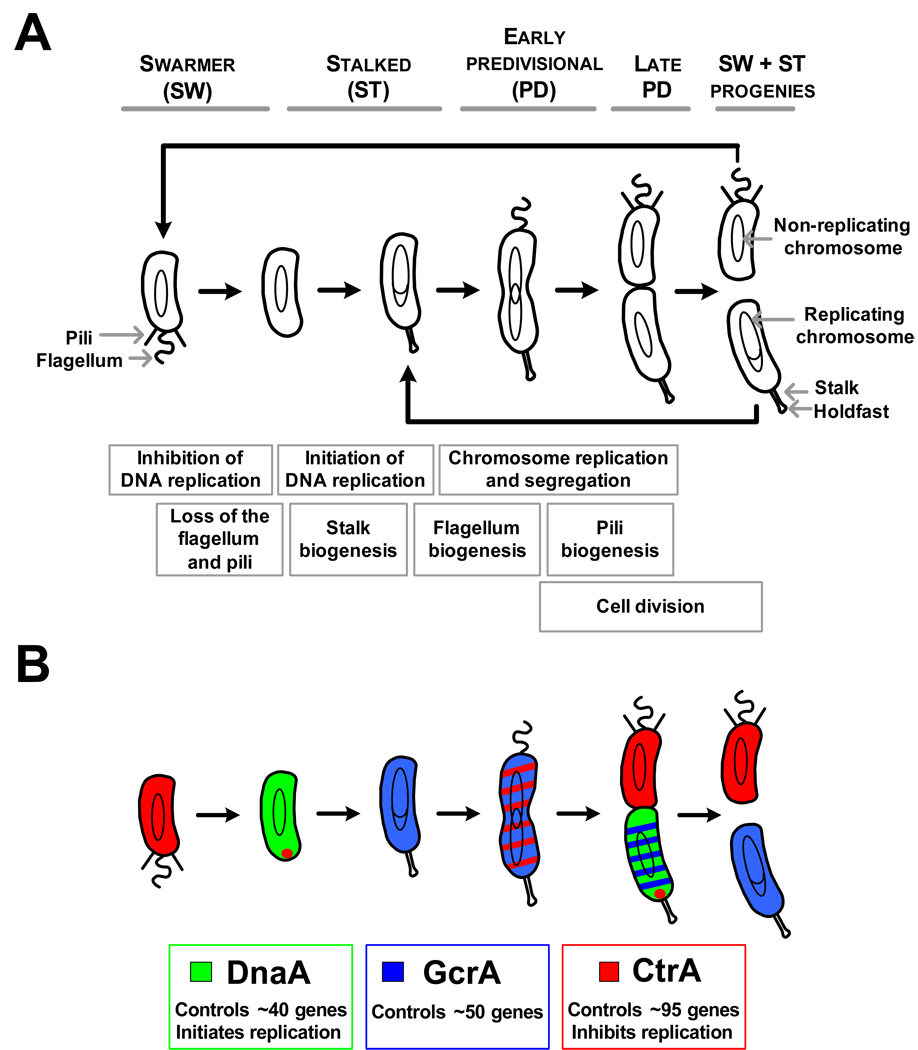
The bacterial cell is now known to be highly organized and to use cytoskeletal proteins to facilitate dynamic cellular processes and cell shape. Examples of both protein complexes and chromosomal loci have been shown to have specific cellular addresses. To address the question of how dynamic subcellular positioning contributes to the regulatory mechanisms that control the cell cycle, the α-proteobacterium Caulobacter crescentus has been the focus of extensive investigation. Caulobacter divides asymmetrically in each cell cycle, yielding a flagellated swarmer cell and a sessile stalked cell, each exhibiting different morphological features and cell fates (Fig.1A). The motile swarmer cells are incapable of initiating chromosome replication and cell division until they differentiate into stalked cells. During the swarmer-to-stalked cell differentiation, which is the bacterial equivalent of the eukaryotic G1-to-S transition, Caulobacter loses its polar flagellum, pili and chemoreceptors, and builds a stalk, capped with a holdfast, at the cell pole previously occupied by the flagellum. The stalked cell, generated as a result of cell division or by differentiation of a swarmer cell, immediately initiates chromosome replication. Pre-divisional cells construct a new flagellum and pili at the cell pole opposite the stalk. The cytoplasm of the pre-divisional cell partitions significantly before cell separation [1,2]. The two compartments formed in the late pre-divisional cell express different genetic programs, leading to the distinct cell fates of the two progeny cells.
Figure 1. The Caulobacter cell cycle and its regulation by master regulators.
(A) Schematic of the Caulobacter cell cycle. Cell cycle events are shown in boxes that are aligned with the cell cycle model to show when these events take place during the cell cycle.
(B) Schematic of the differential accumulation of the three master regulators during the Caulobacter cell cycle. Red indicates CtrA, green indicates DnaA, and blue indicates GcrA. The approximate number of genes included in the regulon of each master regulator is also indicated.
In this review, we focus on the transcriptional, posttranscriptional, and signal transduction mechanisms controlling Caulobacter cell cycle progression that integrate the dynamic localization of protein complexes and chromosomal loci involved in these processes.
Spatially confined master regulators control cell cycle progression
Progression of the Caulobacter cell cycle requires a coordinated succession of genetic events that control chromosome replication, polar differentiation, and cell division. The time of duration of each of these events is shown in Fig.1A. Global analysis of mRNA levels during the Caulobacter cell cycle using whole genome microarrays, showed that the expression of about 15% of the genes is cell cycle-regulated [3]. Three essential master transcriptional regulators have been identified, which control the temporal and spatial expression of approximately 200 cell cycle-regulated genes in Caulobacter (Fig.1B). The CtrA response regulator directly controls the transcription of 95 genes, and in swarmer cells binds to and silences the chromosomal origin of replication [4–7]. The DnaA protein, well known to function as an essential replication initiator in nearly all bacteria studied, was recently shown to regulate the transcription of a minimum of 40 genes during the Caulobacter cell cycle [8,9]. Finally, GcrA regulates the transcription of about 50 genes [10]. The three master regulators oscillate out-of-phase temporally and spatially, so that each cell cycle-regulated gene is activated at its time of function (Fig.1B) [10,11]. Caulobacter has developed robust mechanisms to strictly control the accumulation, and sometimes the activity, of each of these master regulators (Fig.2A) [10–13]. The best understood of these regulatory pathways is the temporally and spatially-regulated activation and degradation of the CtrA protein.
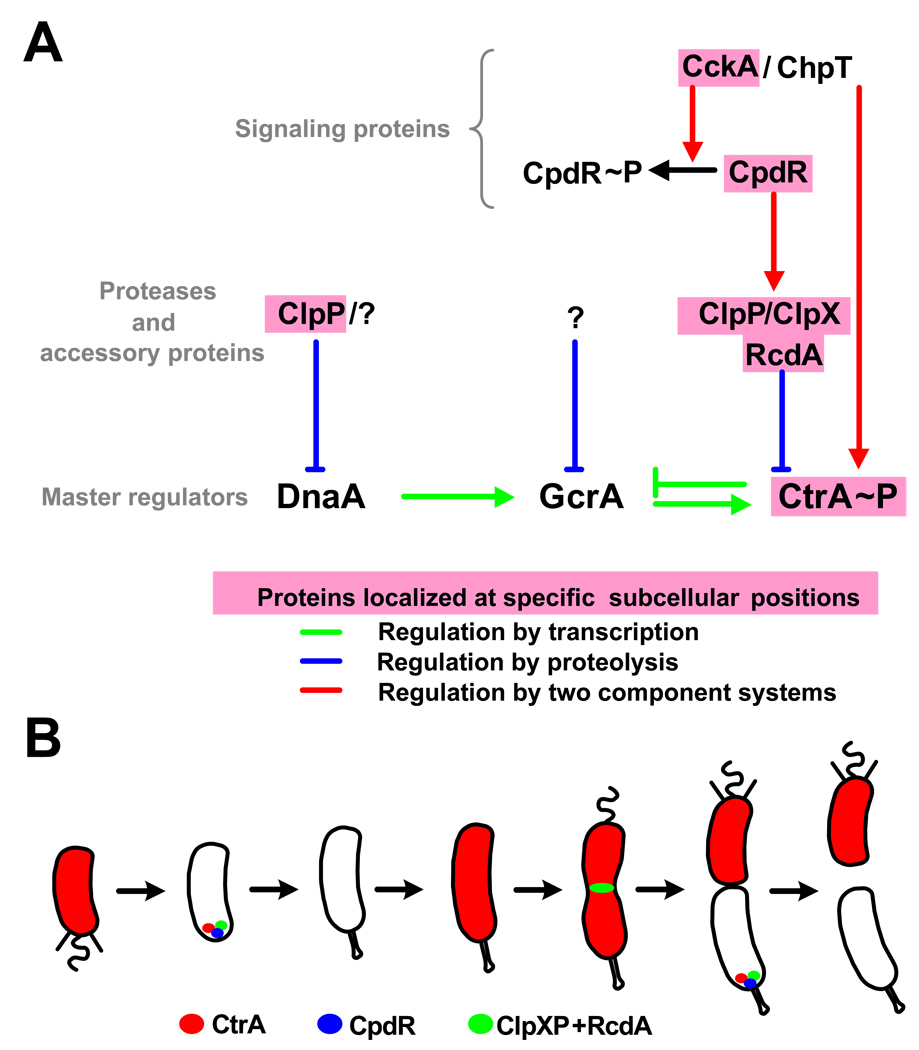
Figure 2. Spatial regulation of CtrA proteolysis.
(A) A diagram of the regulatory pathways controlling the synthesis, the stability, and the activation of master transcriptional regulators. Protein names highlighted in pink correspond to proteins that localize at specific sub-cellular positions. (B) Model of the cell cycle-dependent localization of proteins involved in the degradation of the CtrA global regulator [14,15,16].
Localized signaling protein and protease complexes control the time and place of CtrA activation and proteolysis
The removal of active CtrA (CtrA-P) during the swarmer-to-stalked cell transition and in the stalked compartment of the late pre-divisional cell is essential for the initiation of DNA replication [12]. Once replication has initiated, CtrA again accumulates to allow its functions as a transcriptional regulator. Thus, the timing of CtrA proteolysis must be tightly controlled. CtrA is degraded by the essential ClpXP ATP-dependent protease (Fig.2A&B) [14,15]. Because ClpXP is present throughout the cell cycle [15], but CtrA is only degraded during a narrow time frame in stalked cells, a mechanism must be operative to control the timing of CtrA proteolysis by ClpXP. Surprisingly, the proteolysis of CtrA requires the transient localization of the ClpXP protease to the stalked pole of the cell. A localization factor, RcdA, in complex with CtrA, interacts with ClpXP at the stalked cell pole, providing a cell localization mechanism to control the degradation of CtrA at the correct time in the cell cycle (Fig.2B) [14]. It was further shown that a single domain response regulator, CpdR, is required to localize the ClpXP protease to the stalked pole [16]. During the swarmer-to-stalked cell transition and in the stalked compartment of the late pre-divisional cell, CpdR is not in the phosphorylated state. Only unphosphorylated CpdR localizes to the stalked pole and promotes the same polar localization of the ClpXP/RcdA complex to eliminate CtrA. Once phosphorylated, CpdR~P accumulates in the flagellated compartment of the predivisional cell, it loses its polar localization and consequently its ability to promote CtrA degradation. CpdR is phosphorylated by the cell cycle-regulated CckA histidine kinase, via the ChpT phosphotransferase [17]. Because the CckA/ChpT pathway also phosphorylates CtrA [17,18], it has the dual function of directly activating CtrA while preventing its degradation [16].
Like CtrA, the McpA chemoreceptor (that is positioned at the cell pole) is also degraded by the polar ClpXP complex during the G1-to-S transition, in a CpdR-dependent manner [16]. Yet, the proteolysis of McpA does not require RcdA [14]. It is also interesting to note the dynamic nature of ClpXP/RcdA localization: the ClpXP/RcdA complex leaves the cell pole following the initiation of DNA replication and transiently localizes to the division plane in pre-divisional cells (Fig.2B) [14], but its localization at that site does not require CpdR [16]. The ClpXP/RcdA complex at the division plane may function in the disassembly of the FtsZ division machinery upon completion of cytokinesis. These observations suggest a novel aspect of the role of the three dimensional deployment of complex regulatory networks to control protein degradation during the Caulobacter cell cycle.
Regulated intramembrane proteolysis (Rip) controls the functions of a cell polarity determinant
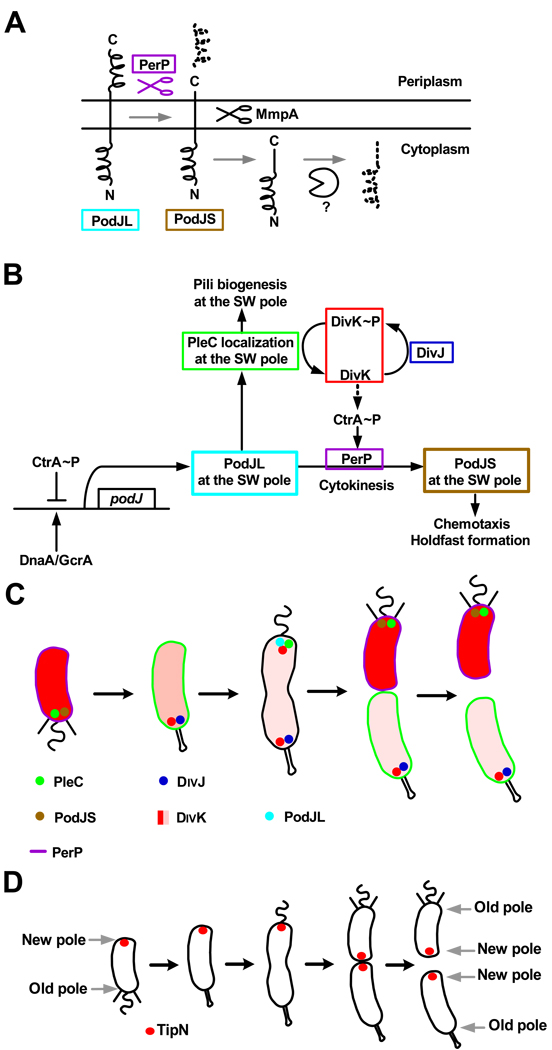
Another well understood, but different, example of a spatially-regulated proteolytic event during the Caulobacter cell cycle occurs when the PodJ polarity determinant is cleaved to a shorter form, thus modulating its functions (Fig.3A&B) [19–21]. The trans-membrane protein PodJL is synthesized in early pre-divisional cells, where it localizes at the incipient swarmer pole to recruit factors required for pili biogenesis, including the PleC histidine kinase (Fig.3B&C) [22–24]. Single molecule visualization of PleC-GFP in living cells indicated that PleC is positioned at the cell pole by a diffusion/capture mechanism [25], suggesting that PleC is captured by a complex positioned at that pole at a specific time in the cell cycle. As the cell divides, the periplasmic domain of PodJL is cleaved by the periplasmic protease PerP [21] . This proteolytic event gives rise to a truncated form (PodJS) that stays localized at the flagellated pole of the future swarmer progeny cell. PodJS is required for chemotaxis and holdfast formation in swarmer cells, presumably by recruiting polar factors required for those processes [19,23]. The expression of the perP gene is activated by a signal transduction pathway in response to the completion of cytokinesis [21]. An important protein in this pathway is the DivK single domain response regulator. DivK is phosphorylated by the DivJ kinase at the stalked pole and dephosphorylated by PleC functioning as a phosphatase at the flagellated pole [26]. Before compartmentalization of the cytoplasm, the DivK protein shuttles rapidly between the two poles of the pre-divisional cell [27]. Upon completion of cytokinesis, the activities of PleC and DivJ are physically separated, resulting in differential levels of DivK and DivK~P in the two compartments of the late pre-divisional cell: DivK~P in the stalked compartment and DivK in the swarmer compartment. It has been proposed that DivK~P indirectly promotes the degradation of CtrA in the stalked cell [28]. Because perP transcription is activated by CtrA [21], perP expression is confined to the swarmer cell, resulting in the coupling of the completion of cytokinesis to perP expression and therefore PodJL cleavage into PodJS. In turn, PodJS is cleaved within the cytoplasmic membrane by the trans-membrane metalloprotease MmpA, belonging to the Rip family of proteases [20]. Following this second cleavage event, PodJ gets rapidly degraded during the swarmer-to-stalked cell transition by an unknown protease, thereby maintaining asymmetry in the predivisional cell.
Figure 3. Regulation and localization of proteins involved in the polarization of Caulobacter cells.
(A) A schematic diagram of the sequential proteolytic processing of PodJ, first by the periplasmic protease PerP, then by the Rip metalloprotease MmpA, and finally by an unknown cytoplasmic protease [21]. (B) A schematic of the regulatory pathway controlling the polarity determinant PodJ. SW pole indicates the flagellated pole of the pre-divisional cell. PodJL is synthesized in early pre-divisional cells, where it localizes at the SW pole and recruits PleC. Compartmentalization, monitored in part by PleC, activates the expression of the PerP protease. PerP cleaves PodJL into PodJS. The polar functions controlled by PodJL and PodJS are indicated [20–24]. (C) Model of the cell cycle-dependent localization of proteins involved in polar in PodJ processing and polar organelles development. (D) Model of the cell cycle-dependent localization of the polarity determinant TipN. The new pole of the cell is marked by TipN throughout the cell cycle [29,30].
A birth scar protein marks the new cell pole
The biogenesis of organelles such as flagella and pili at the cell pole at specific times in the cell cycle is a hallmark of the Caulobacter cell cycle. What are the mechanisms that direct the synthesis of new structures at a specific site on the cell? An indication of how this might occur has recently been provided by the discovery of a new protein, TipN, that serves as a landmark for the biogenesis of these polar structures [29,30]. TipN is located at the division plane in late pre-divisional cells and is retained at the new pole of each of the two progeny cells after the completion of division (Fig.3D). As the progeny cells proceed through the cell cycle, TipN relocates to the new division site, so that TipN will again mark the new pole at the next division. TipN is necessary for the assembly of the flagellum at the correct pole of the cell in early pre-divisional cells, via the localization of the TipF protein at the new pole [29]. Flagella are assembled at ectopic locations when TipN is absent, and new flagella assemble along the lateral side of the cell when TipN is overproduced, as if new cell poles were created [29,30]. Other phenotypes associated with a tipN deletion include mislocalization of the division plane, and mislocalization of PleC and the pilus secretion protein CpaE [30]. Thus, TipN provides a positional cue to orient the polarity axis of the cell.
A cytoskeletal protein polarizes the cell
The actin homolog MreB contributes to Caulobacter cell polarity. MreB forms a dynamically contracting and expanding spiral-like structures that lines the inner surface of the cytoplasmic membrane [31,32]. MreB, which is essential for viability, affects cell shape as well as the dynamic positioning of developmental regulators, such as PleC and DivJ, to the correct cell pole: PleC to the swarmer pole and DivJ to the stalked pole [31]. When MreB is depleted, prior to a change is cell shape, the polar localization of PleC and DivJ are lost. This effect is reversible in that upon re-synthesis of MreB both PleC and DivJ are localized to cell poles. However, these polar proteins become randomly distributed to either cell pole. Their polar specificity is lost [31]. Furthermore, cells recovering from MreB depletion exhibit branching morphology and possess ectopic poles [33]. Thus, the spatial organization of MreB actin contributes to the establishment of cell polarity and protein localization in Caulobacter. Since the dynamic reorganization of MreB in pre-divisional cells is affected in a tipN deletion mutant, it is likely that TipN is upstream of MreB in regulating cell polarity [30].
Spatial control of the division site placement
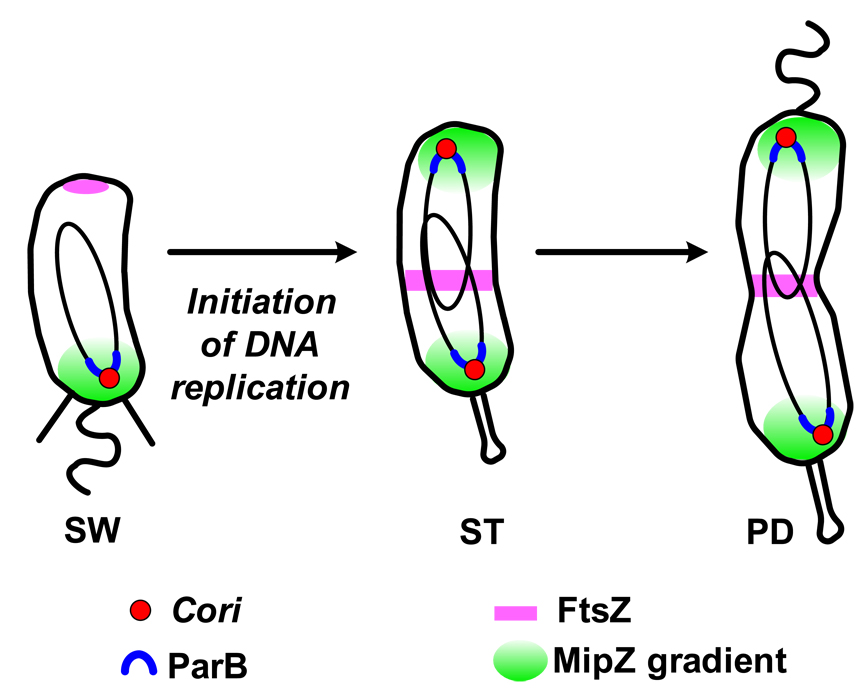
In Caulobacter, cytokinesis initiates shortly after entry into S phase. It starts with the repositioning of tubulin-like FtsZ protein, left at the new cell pole following the previous cell division, to a position close to mid-cell (Fig.4) [34]. How is FtsZ directed to a site at mid-cell? This problem in spatial dynamics makes use of a newly discovered protein, MipZ, that coordinates chromosome dynamics with cytokinesis [34]. In swarmer cells, the single circular chromosome is oriented such that the origin sequence resides at the pole bearing the flagellum and the terminus resides at the opposite pole [35]. Upon the initiation of replication in the new stalked cell, a copy of the origin sequence moves rapidly to the opposite cell pole [36]. The mechanism that directs FtsZ monomers to mid-cell depends on the timing and movement of the replicated origin to the cell pole. MipZ is an ATPase that forms a complex with the partitioning protein ParB at the origin region of the chromosome (Fig.4). Upon initiation of chromosome replication and segregation, duplicated origins maintain the ParB and MipZ complex. MipZ forms a gradient with its highest concentration peaking at the polar position of the origins and decreasing toward mid-cell. Because MipZ is an inhibitor of FtsZ polymerization [34], FtsZ ring formation occurs at the cell position with the lowest concentration of MipZ at midcell. This mechanism also provides a checkpoint to prevent FtsZ ring assembly before the onset of chromosomal replication. Upon completion of cytokinesis but prior to cell separation, the stalked compartment, but not the swarmer compartment, continues to grow yielding progeny of different sizes.
Figure 4. Integration of the spatial deployment of the replication origin and the mid-cell placement of FtsZ cytokinetic ring in Caulobacter.
In swarmer cells (SW), the single origin of replication (Cori) is located at the flagellated cell pole and the FtsZ tubulin is at the opposite pole. After the initiation of replication, one copy of the Cori moves to the opposite end of the cell. The ParB partition protein in complex with the MipZ ATPase binds to the origin regions that are then localized at the two cell poles in the stalked cells. MipZ, an inhibitor of FtsZ polymerization, is distributed in a bipolar gradient with the highest concentrations at the poles and lowest at mid-cell. As soon as MipZ reaches the cell pole, FtsZ is displaced from that pole and moves to mid-cell, thereby marking the site of the cell division plane [34]. SW, swarmer cell; ST, stalked cell; PD, pre-divisional cell.
Conclusions
The development of powerful imaging technologies has opened new doors to the investigation of the spatial localization of protein complexes and chromosomal loci in living bacterial cells. Cellular events can now be followed in time and space, leading to an understanding that the dynamic three dimensional organization of the bacterial cell plays a fundamental role in the mechanisms that control the cell cycle. It is now clear that many regulatory events take place at specific subcellular locations during the bacterial cell cycle. In some cases, signaling and regulatory proteins are not localized at a specific subcellular location, but instead in a specific cell compartment when the cell is about to divide. Temporal and spatial control is used to coordinate chromosome replication and segregation with cell division and polar organelle development.
Although it has been established that the bacterial cell is highly organized and that this organization contributes to the regulation of cell cycle events, many questions remain unanswered. An outstanding issue is an understanding of the mechanisms used to localize proteins to specific sites in the bacterial cells. There are examples of localization mechanisms involving diffusion/capture [25,37] and targeted membrane insertion [38,39] in bacterial cells. The challenge now is to understand how the signaling, landmark, and cytoskeletal proteins described here are involved in protein or DNA localization at specific subcellular positions within the cell. New cytoskeletal structures have recently been visualized in Caulobacter cells [40] and their role in dynamic cellular processes is being explored. Also under investigation are the possibilities that localized proteins recognize specific membrane lipid domains [41], and that mRNA can be localized in bacterial cells to direct the synthesis of proteins at specific cellular sites, as seen in eukaryotic cells [42].
Acknowledgements
We are very grateful to all of the members of the Shapiro and the McAdams labs, and particularly to Antonio Iniesta, for helpful discussions when writing this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and annotations
• of special interest
•• of outstanding interest
- 1.Judd EM, Ryan KR, Moerner WE, Shapiro L, McAdams HH. Fluorescence bleaching reveals asymmetric compartment formation prior to cell division in Caulobacter. Proc Natl Acad Sci U S A. 2003;100:8235–8240. doi: 10.1073/pnas.1433105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Judd EM, Comolli LR, Chen JC, Downing KH, Moerner WE, McAdams HH. Distinct constrictive processes, separated in time and space, divide Caulobacter inner and outer membranes. J Bacteriol. 2005;187:6874–6882. doi: 10.1128/JB.187.20.6874-6882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laub MT, McAdams HH, Feldblyum T, Fraser CM, Shapiro L. Global analysis of the genetic network controlling a bacterial cell cycle. Science. 2000;290:2144–2148. doi: 10.1126/science.290.5499.2144. [DOI] [PubMed] [Google Scholar]
- 4.Quon KC, Marczynski GT, Shapiro L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 5.Quon KC, Yang B, Domian IJ, Shapiro L, Marczynski GT. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc Natl Acad Sci U S A. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laub MT, Chen SL, Shapiro L, McAdams HH. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc Natl Acad Sci U S A. 2002;99:4632–4637. doi: 10.1073/pnas.062065699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reisenauer A, Quon K, Shapiro L. The CtrA response regulator mediates temporal control of gene expression during the Caulobacter cell cycle. J Bacteriol. 1999;181:2430–2439. doi: 10.1128/jb.181.8.2430-2439.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hottes AK, Shapiro L, McAdams HH. DnaA coordinates replication initiation and cell cycle transcription in Caulobacter crescentus. Mol Microbiol. 2005;58:1340–1353. doi: 10.1111/j.1365-2958.2005.04912.x. [DOI] [PubMed] [Google Scholar]
- 9.Gorbatyuk B, Marczynski GT. Physiological consequences of blocked Caulobacter crescentus dnaA expression, an essential DNA replication gene. Mol Microbiol. 2001;40:485–497. doi: 10.1046/j.1365-2958.2001.02404.x. [DOI] [PubMed] [Google Scholar]
- 10.Holtzendorff J, Hung D, Brende P ReisenauerA, Viollier PH, McAdams HH, Shapiro L. Oscillating global regulators control the genetic circuit driving a bacterial cell cycle. Science. 2004;304:983–987. doi: 10.1126/science.1095191. [DOI] [PubMed] [Google Scholar]
- 11. Collier J, Murray SR, Shapiro L. DnaA couples DNA replication and the expression of two cell cycle master regulators. Embo J. 2006;25:346–356. doi: 10.1038/sj.emboj.7600927. This article reports that GcrA transcription is directly activated by DnaA and that the DnaA/GcrA/CtrA regulatory cascade drives the forward progression of the Caulobacter cell cycle
- 12.Domian IJ, Quon KC, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 13.Gorbatyuk B, Marczynski GT. Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus. Mol Microbiol. 2005;55:1233–1245. doi: 10.1111/j.1365-2958.2004.04459.x. [DOI] [PubMed] [Google Scholar]
- 14. McGrath PT, Iniesta AA, Ryan KR, Shapiro L, McAdams HH. A dynamically localized protease complex and a polar specificity factor control a cell cycle master regulator. Cell. 2006;124:535–547. doi: 10.1016/j.cell.2005.12.033. This study shows that the ClpXP protease is dynamically localized in Caulobacter cells and that the RcdA protein is needed for the localization and degradation of CtrA by ClpXP at the cell pole.
- 15.Jenal U, Fuchs T. An essential protease involved in bacterial cell-cycle control. Embo J. 1998;17:5658–5669. doi: 10.1093/emboj/17.19.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Iniesta AA, McGrath PT, Reisenauer A, McAdams HH, Shapiro L. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc Natl Acad Sci U S A. 2006;103:10935–10940. doi: 10.1073/pnas.0604554103. The identification of the new single domain response regulator CpdR is reported in this paper. CpdR is phosphorylated by the CckA kinase. The unphosphorylated form of CpdR directs the polar localization of the ClpXP/RcdA/CtrA complex, facilitating CtrA degradation.
- 17. Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, Ryan KR, Laub MT. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature. 2006;444:899–904. doi: 10.1038/nature05321. This paper identifies a new phosphotransferase that transfers phosphate from the CckA histidine kinase to both CtrA and to CpdR, thus activating CtrA while preventing its degradation.
- 18.Jacobs C, Ausmees N, Cordwell SJ, Shapiro L, Laub MT. Functions of the CckA histidine kinase in Caulobacter cell cycle control. Mol Microbiol. 2003;47:1279–1290. doi: 10.1046/j.1365-2958.2003.03379.x. [DOI] [PubMed] [Google Scholar]
- 19.Lawler ML, Larson DE, Hinz AJ, Klein D, Brun YV. Dissection of functional domains of the polar localization factor PodJ in Caulobacter crescentus. Mol Microbiol. 2006;59:301–316. doi: 10.1111/j.1365-2958.2005.04935.x. [DOI] [PubMed] [Google Scholar]
- 20.Chen JC, Viollier PH, Shapiro L. A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol Microbiol. 2005;55:1085–1103. doi: 10.1111/j.1365-2958.2004.04443.x. [DOI] [PubMed] [Google Scholar]
- 21. Chen JC, Hottes AK, McAdams HH, McGrath PT, Viollier PH, Shapiro L. Cytokinesis signals truncation of the PodJ polarity factor by a cell cycle-regulated protease. Embo J. 2006;25:377–386. doi: 10.1038/sj.emboj.7600935. These authors demonstrate that the PodJ polarity determinant is cleaved by the periplasmic PerP protease, whose expression is activated by CtrA in swarmer cells after cytokinesis, thereby coupling organelle biogenesis with cytokinesis.
- 22.Hinz AJ, Larson DE, Smith CS, Brun YV. The Caulobacter crescentus polar organelle development protein PodJ is differentially localized and is required for polar targeting of the PleC development regulator. Mol Microbiol. 2003;47:929–941. doi: 10.1046/j.1365-2958.2003.03349.x. [DOI] [PubMed] [Google Scholar]
- 23.Viollier PH, Sternheim N, Shapiro L. Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins. Proc Natl Acad Sci U S A. 2002;99:13831–13836. doi: 10.1073/pnas.182411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Viollier PH, Sternheim N, Shapiro L. A dynamically localized histidine kinase controls the asymmetric distribution of polar pili proteins. Embo J. 2002;21:4420–4428. doi: 10.1093/emboj/cdf454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deich J, Judd EM, McAdams HH, Moerner WE. Visualization of the movement of single histidine kinase molecules in live Caulobacter cells. Proc Natl Acad Sci U S A. 2004;101:15921–15926. doi: 10.1073/pnas.0404200101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wheeler RT, Shapiro L. Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol Cell. 1999;4:683–694. doi: 10.1016/s1097-2765(00)80379-2. [DOI] [PubMed] [Google Scholar]
- 27.Matroule JY, Lam H, Burnette DT, Jacobs-Wagner C. Cytokinesis monitoring during development; rapid pole-to-pole shuttling of a signaling protein by localized kinase and phosphatase in Caulobacter. Cell. 2004;118:579–590. doi: 10.1016/j.cell.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 28.Hung DY, Shapiro L. A signal transduction protein cues proteolytic events critical to Caulobacter cell cycle progression. Proc Natl Acad Sci U S A. 2002;99:13160–13165. doi: 10.1073/pnas.202495099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH. Bacterial birth scar proteins mark future flagellum assembly site. Cell. 2006;124:1025–1037. doi: 10.1016/j.cell.2006.01.019. The identification of the TipN and TipF is reported in this paper. TipN is a birth scar protein that always localizes at the new cell pole and is necessary for the correct localization of the flagellum assembly factor TipF.
- 30. Lam H, Schofield WB, Jacobs-Wagner C. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell. 2006;124:1011–1023. doi: 10.1016/j.cell.2005.12.040. This article, reports the simultaneous identification of TipN. It also shows that TipN is required for the correct localization of PleC, CpaE, and MreB.
- 31.Gitai Z, Dye N, Shapiro L. An actin-like gene can determine cell polarity in bacteria. Proc Natl Acad Sci U S A. 2004;101:8643–8648. doi: 10.1073/pnas.0402638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim SY, Gitai Z, Kinkhabwala A, Shapiro L, Moerner WE. Single molecules of the bacterial actin MreB undergo directed treadmilling motion in Caulobacter crescentus. Proc Natl Acad Sci U S A. 2006;103:10929–10934. doi: 10.1073/pnas.0604503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wagner JK, Galvani CD, Brun YV. Caulobacter crescentus Requires RodA and MreB for Stalk Synthesis and Prevention of Ectopic Pole Formation. J Bacteriol. 2005;187:544–553. doi: 10.1128/JB.187.2.544-553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–162. doi: 10.1016/j.cell.2006.05.038. This study uses genetic, cell biological and biochemical experiments to demonstrate the role of a new protein, MipZ, involved in the spatial control of the division site placement . MipZ coordinates chromosome dynamics with cytokinesis in Caulobacter.
- 35.Jensen RB, Shapiro L. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc Natl Acad Sci U S A. 1999;96:10661–10666. doi: 10.1073/pnas.96.19.10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, Shapiro L. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci U S A. 2004;101:9257–9262. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rudner DZ, Pan Q, Losick RM. Evidence that subcellular localization of a bacterial membrane protein is achieved by diffusion and capture. Proc Natl Acad Sci U S A. 2002;99:8701–8706. doi: 10.1073/pnas.132235899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brandon LD, Goehring N, Janakiraman A, Yan AW, Wu T, Beckwith J, Goldberg MB. IcsA, a polarly localized autotransporter with an atypical signal peptide, uses the Sec apparatus for secretion, although the Sec apparatus is circumferentially distributed. Mol Microbiol. 2003;50:45–60. doi: 10.1046/j.1365-2958.2003.03674.x. [DOI] [PubMed] [Google Scholar]
- 39.Rubio A, Jiang X, Pogliano K. Localization of translocation complex components in Bacillus subtilis: enrichment of the signal recognition particle receptor at early sporulation septa. J Bacteriol. 2005;187:5000–5002. doi: 10.1128/JB.187.14.5000-5002.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Briegel A, Dias DP, Li Z, Jensen RB, Frangakis AS, Jensen GJ. Multiple large filament bundles observed in Caulobacter crescentus by electron cryotomography. Mol Microbiol. 2006;62:5–14. doi: 10.1111/j.1365-2958.2006.05355.x. Electron cryotomography was used to visualize new cytoskeletal structures in Caulobacter cells.
- 41.Mileykovskaya E, Dowhan W. Role of membrane lipids in bacterial division-site selection. Curr Opin Microbiol. 2005;8:135–142. doi: 10.1016/j.mib.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 42.Czaplinski K, Singer RH. Pathways for mRNA localization in the cytoplasm. Trends Biochem Sci. 2006;31:687–693. doi: 10.1016/j.tibs.2006.10.007. [DOI] [PubMed] [Google Scholar]