Abstract
Innate immunity is part of the antiviral response. Interferon (IFN)-beta plays a leading role in this system. To investigate the influence of hepatitis C virus (HCV) on innate immunity, we examined the effect of viral proteins on IFN-beta induction. HepG2 cells were co-transfected with plasmids for seven HCV proteins (core protein, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) and the IFN-beta promoter luciferase. Toll-like receptor (TLR) 3 and Toll/IL-1 receptor domain-containing adapter inducing IFN-beta (TRIF) play key roles in dsRNA-mediated activation of interferon regulatory factor (IRF)-3 and IFN-beta; therefore, the participation of TLR3/TRIF in NS5B-mediated IFN induction was examined. Among seven HCV proteins, only NS5B, a viral RNA-dependent RNA polymerase (RdRp), activated the IFN-beta promoter. However, mutant NS5B without RdRp activity or template/primer association did not activate the IFN-beta promoter. Activation of the IFN-beta promoter by NS5B required the positive regulatory domain III, a binding sequence for IRF-3. Moreover, IRF-3 was phosphorylated by NS5B. Both inhibition of TLR3 expression by small interfering RNA and expression of the dominant negative form of TRIF significantly reduced NS5B-induced activation of IFN-beta. Of the six other HCV proteins, NS4A, NS4B, and NS5A efficiently inhibited this activation. HCV NS5B is a potent activator of the host innate immune system, possibly through TLR3/TRIF and synthesis of dsRNA. Meanwhile, NS4A, NS4B, and NS5A block IFN-beta induction by NS5B, which may contribute toward the persistence of this virus.
Keywords: HCV, Persistent infection, Innate immunity, Toll-like receptor, Toll/IL-1 receptor domain-containing adapter inducing IFN-beta
Introduction
Hepatitis C virus (HCV) is a major causative agent of chronic hepatitis, cirrhosis, and hepatocellular carcinoma (HCC) throughout the world [1–3]. HCV is a positive-sense, ssRNA virus, consisting of an approximately 10-kb genome containing a large open reading frame, encoding a polyprotein precursor of 3010–3033 amino acids and an untranslated region at the 5′ and 3′ ends. The putative organization of the HCV genome includes the 5′-untranslated region, 3–4 structural proteins (core, E1, and E2/p7), six non-structural (NS) proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B), and the 3′-untranslated region [4, 5]. NS5B is a viral RNA-dependent RNA polymerase (RdRp) with the GDD motif, which is highly conserved among RdRp [6, 7].
Viral infection induces an innate immune response, including the production of interferon (IFN) and other cytokines [8]. Initially, Toll-like receptor (TLR) 3 recognizes dsRNA, a replicative intermediate generated in the course of viral RNA replication, and triggers activation of the innate immune system [9]. TLR3 then activates IFN regulatory factor (IRF)-3 through the adapter protein Toll/Interleukin-1 receptor domain containing adapter inducing IFN-beta (TRIF) [10–12]. Phosphorylated and dimerized IRF-3 upregulates the IFN-beta promoter [13, 14]. Finally, IFN-beta binds to its cell surface type 1 receptor, activates the Janus kinase and signal transducer of transcription (JAK-STAT) pathway, and induces the IFN regulated genes responsible for the antiviral response [15].
Persistent HCV infection probably results from disruption of host antiviral immunity [16–18]. Recently, it was reported that HCV NS3/4A protein blocks the phosphorylation and effector actions of IRF-3 [19, 20]. However, the influence of HCV on innate immunity has not been elucidated. Therefore, in this study, we analyzed the effect of HCV proteins on the innate immune system, especially induction of IFN-beta.
Materials and methods
Cell culture
HepG2 (human hepatoblastoma) cells were obtained from RIKEN cell bank (Tsukuba, Japan). Cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% fetal bovine serum. Cells were cultured in 100-mm tissue culture dishes (IWAKI Glass, Chiba, Japan) at 37°C in humidified air containing 5% CO2, and passaged at intervals of 3 or 4 days.
Construction of plasmids expressing HCV proteins
Plasmids expressing HCV proteins were constructed as described previously [21]. Briefly, HCV core, NS2, NS3, NS4A, NS4B, NS5A, and NS5B derived from genotype 1b were cloned into the Xho1 site of pCXN2 (kindly provided by J. Miyazaki, Osaka University, Japan), a mammalian expression vector with β-actin-based CAG promoter and SV40 origin. Expression of these HCV proteins was confirmed by Western blotting [21].
N-terminal hemagglutinin (HA)-tagged NS5B protein expression plasmid (pCXN2-HA-NS5B) was constructed using the Gateway cloning system (Invitrogen, Carlsbad, CA, USA). First, an HA (YPYDVPDYA) tag was attached to the N-terminus of the NS5B gene by PCR. Then, additional sequences required for the recombination reaction, attB1 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCT-3′) and attB2 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGT-3′), were attached by PCR to the HA-NS5B fragment. The resultant DNA fragment, attB1-HA-NS5B-attB2, was inserted into a donor vector, pDONR201 (Invitrogen), by recombination, creating the entry clone pDONR-HANS5B. Next, the HA-NS5B fragment was transferred into the pCXN2 vector by recombination, creating pCXN2-HA-NS5B. Expression of the HA-tagged NS5B protein was confirmed by Western blotting using both anti-NS5B mouse monoclonal antibodies (kindly provided by I. Fuke, Osaka University, Japan) and anti-HA (Y-11) rabbit polyclonal antibodies (Santa Cruz, CA, USA).
NS5B acts as an RdRp and has the GDD motif (aa 317–319), which is highly conserved among RdRps in RNA viruses. Mutation of G (aa 317) to V in the GDD motif is reported to abolish RdRP activity [22]. HCV NS5B also acts as a cooperative RNA-binding protein that is required for viral RNA synthesis. The mutation of Y (aa 276) to A in the template/primer association site abolishes RNA-binding activity and RdRP activity [22]. Mutagenesis in the substitutions pCXN2-HA-NS5Bm1 (G317V) and pCXN2-HA-NS5Bm2 (Y276A) was generated by the Quikchange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). The primers used for G317V were 5′-GCTCGTGAACGTAGACGACCTTGTC-3′ and 5′-GACAAGGTCGTCTACGTTCACGAGC-3′. The primers used for Y276A were 5′-GAACTGCGGTGCTCGCCGGTGCCGC-3′ and 5′-GCGGCACCGGCGGAGCACCGCAGTTC-3′. Mutagenesis PCR was performed using pDONR-HA-NS5B as a template, according to the manufacturer’s instructions. Each mutated entry clone was transferred into pCXN2 vector by recombination. Expression of these mutated NS5B proteins was confirmed by Western blotting using anti-HA antibodies.
Reporter plasmids for luciferase assay
The promoter region of the human IFN-beta gene (−125 to +19), derived from p125-luc (kindly provided by T. Taniguchi, University of Tokyo, Japan), was transferred to a position upstream of the luciferase gene in the pGL3-basic vector using the Gateway cloning system. The constructed plasmid, designated pGL3-p125 luc, containing the Photinus pyralis (firefly) luciferase reporter gene driven by a basic promoter element (TATA box) plus 5′-flanking region of the IFN-beta promoter gene, was used as reporter plasmid. pRL-TK, expressing Renilla reniformis (seapansy) luciferase driven by the herpes simplex virus thymidine kinase promoter, was used to monitor and standardize the efficacy of transfection (Promega, Madison, WI, USA). The IFN-beta promoter contains four positive regulatory domains (PRDs). Among these, PRD I and III are known to be binding sequences for IRF-3 [8]. To identify the site responsible for NS5B-induced activation of IFN-beta promoter, we constructed three reporter plasmids containing deletion mutants of the IFN-beta promoter: pGL3-p125 PRD IV luc (bp −97 to +19), pGL3-p125 PRD III luc (bp −84 to +19), and pGL3-p125 PRD I luc (bp −71 to +19). All cloned plasmids were purified using an Endofree plasmid kit (Qiagen, Heiden, Germany).
Inhibition of TLR3 expression by RNA interference
We used small interfering RNA (siRNA) to inhibit the expression of TLR3. The sequence of TLR3 siRNA used in this study was obtained from a previous study [12]. We cloned this sequence into pSilencer 2.1-U6 hygro siRNA expression vector (Ambion, Austin, TX, USA).
Expression plasmids of TRIF and its dominant negative form
A recent study showed that TRIF activates the IFN-beta promoter, and a dominant negative form of TRIF blocks the response of TLR3 to dsRNA [11]. We used plasmids expressing human full length TRIF (pEFBOS-myc-TRIF, 712 amino acids) and TRIF dominant negative form (pCMV-myc-TRIF DN) that harbors only the TIR domain (amino acids 380–541; both kindly provided by S. Akira, Osaka University, Japan).
Transfection, luciferase assays, and reagents
Approximately 4 × 105 HepG2 cells were placed in a six-well tissue culture plate (IWAKI Glass) 24 h before transfection. Using the Effectene transfection reagent (Qiagen), 0.2 μg reporter plasmid (0.18 μg pGL3-p125 luc and 0.02 μg pRL-TK) were transiently co-transfected into cells with a total of 0.4 μg plasmid expressing HCV proteins. Thirty-six hours after transfection, whole cell lysates were examined for luciferase activity (PicaGene Dual Seapansy system; Toyo Ink, Japan) with a luminometer (Lumat LB9507; EG&G Berthold). Synthetic dsRNA [poly(I:C)] was used for the positive control of IFN-beta promoter activation at 100 μg/ml in culture medium. Absolute firefly luciferase activity was normalized for transfection efficiency on the basis of seapansy–luciferase activity. The luciferase activity of the cells transfected with the reporter plasmid plus pCXN2 was set as 1.0, and the experimental luciferase activity was compared to this standard value. All experiments were performed at least in duplicate.
Western blotting
The antibodies used were anti-IRF3 rabbit polyclonal antibody (Santa Cruz), anti-HA (Y-11) rabbit polyclonal antibody, anti-NS5B mouse monoclonal antibody, and anti-rabbit and anti-mouse immunoglobulin horseradish peroxidase-conjugated secondary antibodies (Amersham Pharmacia Biotech, Piscataway, NJ, USA). Cell lysates, prepared using SDS lysis buffer (62.5 mM Tris pH 6.8, 2% SDS, 10% glycerol, 50 mM DTT, and 0.1% BPB) or NP-40 lysis buffer [15], were separated by SDS-PAGE and transferred by electrophoresis to polyvinylidene difluoride membrane that was incubated with each primary antibody. Immunoreactive bands were visualized by the use of appropriate secondary antibodies and a Western blotting detection system (ECL Advance; Amersham Pharmacia Biotech).
Statistical analysis
Data are expressed as the mean ± SD. Statistical analysis was done using Student’s t-test. P < 0.05 was considered to be statistically significant.
Results
HCV NS5B protein activates IFN-beta promoter
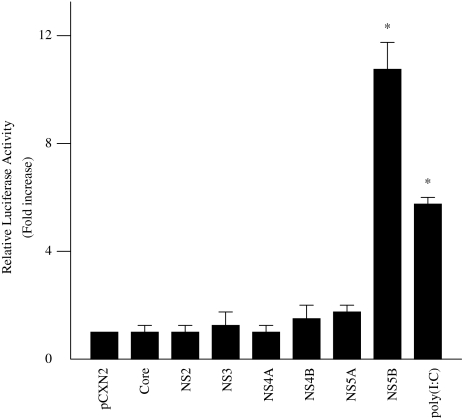
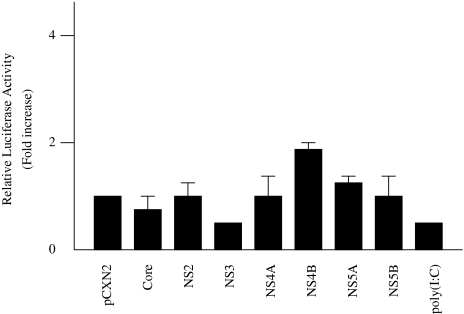
To determine the effect of HCV proteins on the induction of IFN-beta, HepG2 cells were co-transfected with pGL3-p125 luc, containing the luciferase reporter gene driven by the IFN-beta promoter, and each of seven HCV protein expression plasmids. Thirty-six hours later, the cells were assayed for luciferase activity. Relative firefly luciferase activity of PGL3-p125 luc/pCXN2-NS5B-transfected cell lysates was 10.8 times higher than that of PGL3-p125 luc/pCXN2-transfected cell lysates. The other HCV proteins, however, did not influence IFN-beta promoter activity (Fig. 1).
Fig. 1.
Effect of HCV proteins on IFN-beta promoter activity. HepG2 cells were co-transfected with 0.18 μg pGL3-p125luc, 0.02 μg PRL-TK, and 0.4 μg expression plasmids for each of seven HCV proteins. As a positive control, 100 μg/ml poly(I:C) were added to the culture medium. Thirty-six hours after transfection, cells were harvested and luciferase activity was measured. The normalized luciferase activities were calculated by assigning a value of 1 to the levels of activity of cells that were transfected with the empty vector. *P < 0.05 compared with the activity of the cells with the empty vector
RdRp activity of NS5B is required for the activation of IFN-beta
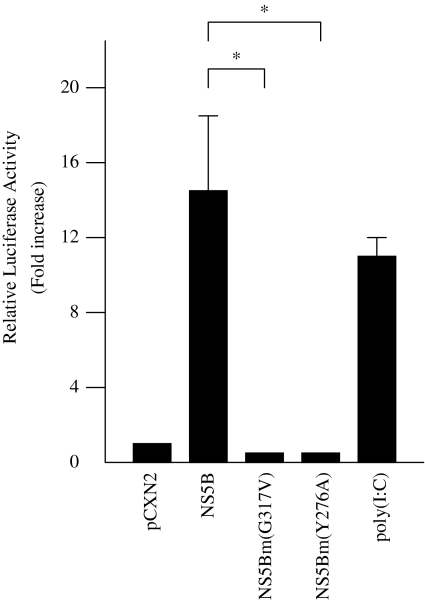
HCV NS5B is an RdRp and a cooperative RNA-binding protein that is required for viral replication and RNA synthesis. To examine whether activation of IFN-beta by NS5B protein was mediated via RdRp activity, substitution mutations were generated in the GDD motif (G317V) and template/primer association site (Y276A) of NS5B. Expression of NS5B G317V mutant protein did not activate the IFN-beta promoter, suggesting that NS5B protein induces IFN-beta via its RdRp activity (Fig. 2). Expression of NS5B Y276A also did not activate the IFN-beta promoter, suggesting that a putative template association with NS5B is necessary to activate the promoter (Fig. 2).
Fig. 2.
Effect of HCV NS5B mutated proteins on IFN-beta promoter activity. HepG2 cells were co-transfected with 0.18 μg pGL3-p125luc, 0.02 μg PRL-TK, and 0.4 μg pCXN2-HA-NS5B, pCXN2-HA-NS5Bm1 (G317V), or pCXN2-HA-NS5Bm2 (Y276A). As a positive control, 100 μg/ml poly(I:C) were added to the culture medium. Thirty-six hours after transfection, cells were harvested and luciferase activity was measured. The normalized luciferase activities were calculated by assigning a value of 1 to the levels of activity of cells that were transfected with the empty vector. *P < 0.05
Mapping the region of the IFN-beta promoter responsible for activation by NS5B protein
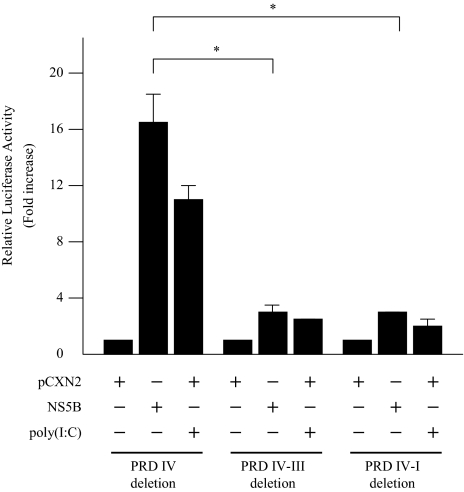
The IFN-beta promoter contains four positive regulatory domains, and PRD III is known as a binding sequence for IRF-3. To identify the site responsible for NS5B-induced activation of the IFN-beta promoter, three luciferase reporter plasmids with different lengths of IFN-beta promoter region were co-transfected with pCXN2-HA-NS5B. For comparison, poly(I:C) was added to the incubation medium at a concentration of 100 μg/ml. The NS5B response/luciferase activity ratio was reduced markedly when the PRD III site was deleted (Fig. 3). The poly(I:C) response/luciferase activity ratio was also reduced when the PRD III site was deleted. These results suggest that the PRD III site (bp −97 to −84) of the IFN-beta promoter was the major element responsible for NS5B-induced upregulation of IFN-beta.
Fig. 3.
Identification of the IFN-beta promoter region responsible for activation by HCV NS5B protein. Three luciferase reporter plasmids with different lengths of IFN-beta promoter region, deleting PRD IV (bp −97 to +19), PRD III (bp −84 to +19), or PRD I (bp −71 to +19), were co-transfected with pCXN2-HA-NS5B. The normalized luciferase activities were calculated by assigning a value of 1 to the levels of activity of cells that were transfected with the empty vector. *P < 0.05
IRF-3 was phoshorylated by NS5B expression
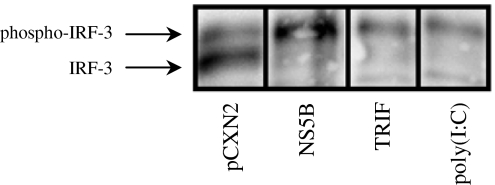
Viral infection induces phosphorylation of IRF-3 serine residues, which then induces a comformational change, leading to IRF-3 dimerization and subsequent nuclear translocation [13]. Phosphorylated IRF-3 is known to migrate slower in SDS-PAGE than the non-phosphorylated form [14]. Therefore, we examined the effect of NS5B on IRF-3 phosphorylation. The intensity of slow-migrating IRF-3 was increased by NS5B expression at 36 h after transfection (Fig. 4). The slow-migrating form was also increased by poly(I:C) treatment or TRIF expression, both of which are known to activate IRF-3 by phosphorylation (Fig. 4). These results show that NS5B can phosphorylate IRF-3, leading to induction of IFN-beta.
Fig. 4.
Activation of IRF-3 by HCV NS5B protein. HepG2 cells were transfected with 0.4 μg pCXN2-HA-NS5B or 0.4 μg pEF-BOS myc TRIF. As a positive control, 100 μg/ml poly(I:C) were added to the culture medium. Thirty-six hours after transfection, whole cell lysates were prepared using NP-40 lysis buffer and analyzed by Western blotting using anti-IRF-3 rabbit polyclonal antibody. Slow-migrating bands resent phospho-IRF-3
HCV NS5B activation of IFN-beta through TLR3 and TRIF
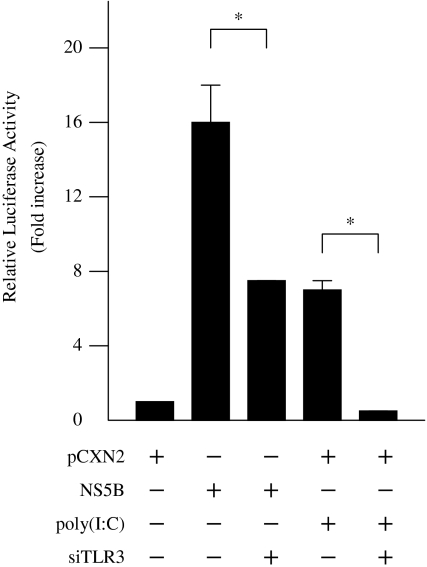
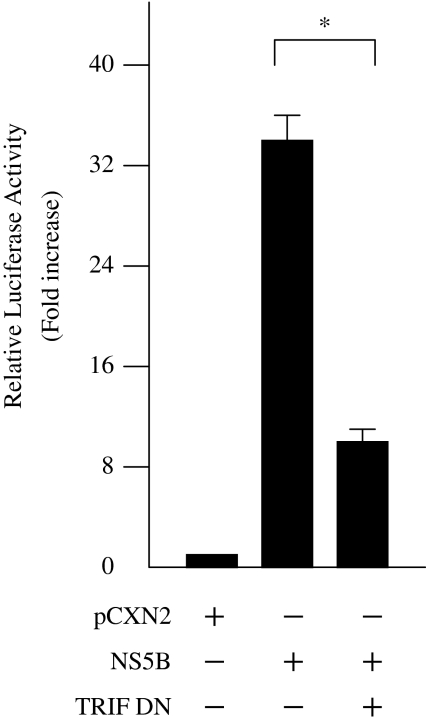
To examine whether NS5B-induced activation of IFN-beta is mediated by TLR3, HepG2 cells were co-transfected with pCXN2/pCXN2-HA-NS5B, pGL3-p125 luc, and siRNA-TLR3 expression vectors. Inhibition of TLR3 expression by siRNA significantly decreased NS5B-induced IFN-beta activation to 45.6% (Fig. 5). To confirm the effect of siRNA on TLR3 expression, we added poly(I:C), a specific TLR3 ligand, to HepG2 cells transfected with pCXN2 and pGL3-p125 luc. Activation of the IFN-beta promoter by poly(I:C) was also significantly inhibited by induction of TLR3 siRNA (Fig. 5). TRIF was recently shown to be a TLR3 adapter protein and a potent inducer of IRF3, leading to activation of the IFN-beta promoter. To establish whether activation of the IFN-beta pathway by NS5B protein was mediated through TRIF, HepG2 cells were co-transfected with pCXN2/pCXN2-HA-NS5B, pGL3-p125 luc, and pCMV-myc TRIF DN, which expresses the dominant negative form of TRIF. Expression of the dominant negative TRIF significantly reduced NS5B-induced IFN-beta activation to 30.4% in HepG2 cells (Fig. 6). These results suggest that NS5B protein activates the IFN-beta pathway through TLR3 and TRIF.
Fig. 5.
HCV NS5B activation of IFN-beta promoter through TLR3. HepG2 cells were co-transfected with pCXN2-HA-NS5B, pGL3-p125luc, and siRNA TLR3 expression vectors. The normalized luciferase activities were calculated by assigning a value of 1 to the levels of activity of cells that were transfected with the empty vector. *P < 0.05
Fig. 6.
HCV NS5B activation of IFN-beta promoter through TRIF. HepG2 cells were co-transfected with pCXN2-HA-NS5B, pGL3-p125 luc, and PCMV-myc TRIF DN. The normalized luciferase activities were calculated by assigning a value of 1 to the levels of activity of cells that were transfected with the empty vector. *P < 0.05
HCV NS4A, NS4B, and NS5A inhibit NS5B-induced activation of IFN-beta
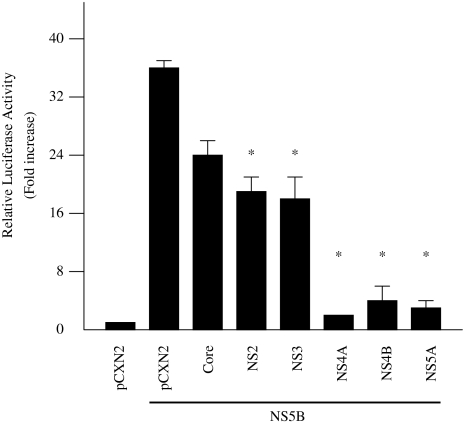
To examine the effect of other HCV proteins on NS5B-induced activation of IFN-beta, HepG2 cells were co-transfected with pCXN2/pCXN2-HA-NS5B, pGL3-p125 luc, and expression plasmids for each of the six HCV proteins, core protein, NS2, NS3, NS4A, NS4B, and NS5A. Of these six HCV proteins, NS5A efficiently inhibited NS5B-induced IFN-beta activation to 5.3% (Fig. 7). Meanwhile, NS3 inhibited this activation to 50.5%. NS4A and NS4B also inhibited IFN-beta activation (Fig. 7). A deletion mutant form of NS5A that lacks NS5B binding sites did not efficiently inhibit NS5B-induced IFN-beta activation (data not shown).
Fig. 7.
Inhibition of HCV NS5B-induced activation of IFN-beta by HCV proteins. HepG2 cells were co-transfected with pCXN2-HA-NS5B, pGL3-p125 luc, and expression plasmids for the six HCV proteins, core protein, NS2, NS3, NS4A, NS4B, and NS5A. The normalized luciferase activities were calculated by assigning a value of 1 to the levels of activity of cells that were transfected with the empty vector. *P < 0.05 compared with the activity of the cells with the vector plus NS5B
HCV NS4A, NS4B, and NS5A do not inhibit TRIF-induced activation of IFN-beta
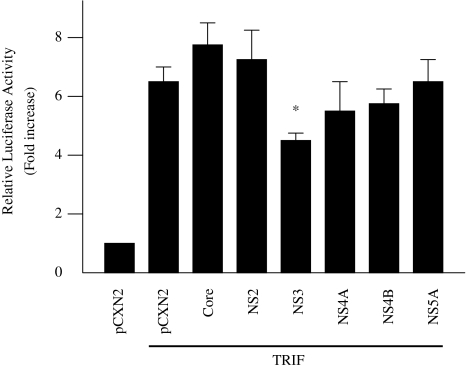
To examine the effect of HCV proteins on TRIF-induced activation of IFN-beta, similar experiments were conducted using pEFBOS-myc-TRIF and expression plasmids for the six HCV proteins, core protein, NS2, NS3, NS4A, NS4B, and NS5A. Of these six proteins, NS3 inhibited the TRIF-induced IFN-beta activation to 68.1%, but NS4A, NS4B, and NS5A had almost no effect (Fig. 8). These results suggest that NS4A, NS4B, and NS5A can inhibit IFN-beta activation upstream of TRIF.
Fig. 8.
Effect of HCV proteins on TRIF-induced activation of IFN-beta. HepG2 cells were co-transfected with pEF-BOS-myc-TRIF, pGL3-p125 luc, and expression plasmids for the six HCV proteins, core protein, NS2, NS3, NS4A, NS4B, and NS5A. The normalized luciferase activities were calculated by assigning a value of 1 to the levels of activity of cells that were transfected with the empty vector. *P < 0.05 compared with the activity of the cells with the vector plus TRIF
HCV NS4A, NS4B, and NS5A partially inhibit dsRNA-induced activation of IFN-beta
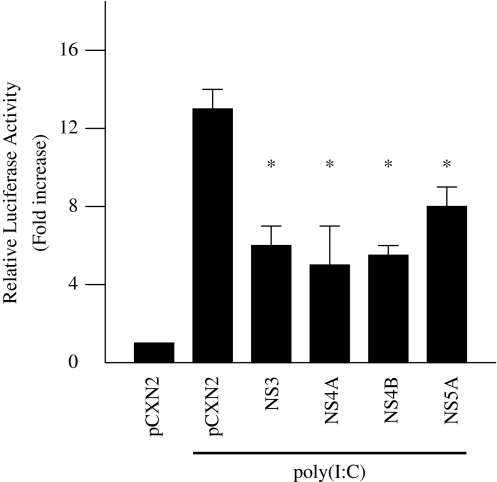
TLR3 recognizes dsRNA, a replicative intermediate generated in the course of RNA virus replication, and triggers activation of host innate immunity. To examine the effect of other HCV proteins on dsRNA-induced activation of IFN-beta, HepG2 cells were co-transfected with pGL3-p125 luc and expression plasmids for four HCV proteins, NS3, NS4A, NS4B, and NS5A, and treated with poly(I:C) at 100 μg/ml. NS4A, NS4B, and NS5A inhibited dsRNA-induced activation of IFN-beta by 50% (Fig. 9). This result suggests that NS4A, NS4B, and NS5A can partially inhibit dsRNA-induced activation of IFN-beta upstream of TLR3.
Fig. 9.
Inhibition of dsRNA-induced activation of IFN-beta by HCV proteins. HepG2 cells were co-transfected with pGL3-p125 luc with the expression plasmids for the four HCV proteins, NS3, NS4A, NS4B, and NS5A, and treated with 100 μg/ml poly(I:C). The normalized luciferase activities were calculated by assigning a value of 1 to the levels of activity of cells that were transfected with the empty vector. *P < 0.05 compared with the activity of the cells with the vector plus poly(I:C) treatment
Discussion
Viral infection induces host innate immunity, which can immediately eliminate most viruses. In contrast, HCV infection often leads to long-term persistence, and consequently causes chronic hepatitis, cirrhosis, and HCC [16, 17]. However, the influence of HCV infection on host innate immunity has not been elucidated. In this study, we analyzed the effect of HCV proteins on innate immunity. We demonstrated that HCV NS5B induces intrinsic IFN-beta, through TLR3 and TRIF, by its RdRp activity, and HCV NS4A, NS4B, and NS5A specifically inhibit this NS5B-induced activation of IFN-beta.
Among seven HCV proteins tested, we found that NS5B could activate IFN-beta in HepG2 cells. NS5B acts as an RdRp in HCV replication, and mutant NS5B, lacking RdRp activity, had no effect on IFN-beta promoter activation. Therefore, NS5B activates IFN-beta via its RdRp activity. The IFN-beta promoter contains four positive regulatory domains. We constructed three luciferase reporter plasmids with different lengths of the IFN-beta promoter region. We found that NS5B-induced activation of IFN-beta was markedly reduced when the PRD III site was deleted. This suggests that PRD III, which is known to be an IRF-3 binding sequence, was the major element responsible for NS5B-induced activation of IFN-beta.
TLR3 recognizes dsRNA and activates IFN-beta through TRIF and IRF-3. In our study, inhibition of TLR3 expression by siRNA significantly reduced NS5B-induced activation of IFN-beta. Expression of the dominant negative form of TRIF also significantly reduced NS5B-induced activation of IFN-beta. HCV is an ssRNA virus that generates a dsRNA intermediate during replication. The fact that NS5B protein activated IFN-beta by its RdRp activity supports the hypothesis that NS5B synthesizes non-specific dsRNA, without an HCV specific template, which could be recognized by TLR3 and lead to IFN-beta activation. NS5B is reported to exhibit little binding specificity to various RNA templates and carries out RNA synthesis using several RNAs from both HCV and non-HCV sources [23–26].
We found that NS4A, NS4B, and NS5A are able to inhibit NS5B-induced activation of IFN-beta. These three HCV proteins partially inhibited dsRNA-induced activation of IFN-beta. In contrast, they showed no effect on TRIF-induced activation of IFN-beta. These results indicate that NS4A, NS4B, and NS5A may block synthesis of non-specific dsRNA by NS5B and recognition of dsRNA by TLR3. Our findings suggest that NS4A, NS4B, and NS5A block synthesis of non-specific dsRNA generated by NS5B, through formation of a replication complex including NS5B. HCV NS proteins form a complex on cytoplasmic membranes that catalyzes replication of the viral genome [27]. The formation of multi-protein complexes requires specific interactions between NS proteins [28]. NS5A is reported to interact with NS5B, modulating its RdRp activity [29], and is critical for HCV RNA replication [30]. There may be another mechanism where NS4A, NS4B, and NS5A block TLR3 recognition of dsRNA. It is possible that NS4A, NS4B, and NS5A bind TLR3 and inhibit the interaction between dsRNA and TLR3, resulting in the inhibition of IFN-beta activation. HCV NS4A, NS4B, and NS5A may modulate the function of NS5B or block TLR3 recognition of dsRNA, leading to the inhibition of IFN-beta activation.
Our results indicate that HCV NS5B protein activates host innate immunity and induces intrinsic IFN-beta. However, HCV cannot be eliminated by the host immune system and often leads to long-term persistence. Persistent infection probably results from disruption of the host antiviral response or HCV protein-induced alteration of signaling pathways in the host immune response. A recent study showed that HCV NS3/4A protein blocks activation of IRF-3 induced by TRIF expression or Sendai virus infection [19]. The blocking of IRF-3 activation by HCV NS3/4A protein is considered one of the causes of persistent infection. In our study, NS3 partially inhibited both NS5B- and TRIF-induced activation of IFN-beta. In contrast, NS4A, NS4B, and NS5A efficiently inhibited NS5B-induced, but not TRIF-induced, activation of IFN-beta. These results suggest that NS3 blocks the IFN pathway downstream of TRIF, and NS4A, NS4B, and NS5A may inhibit NS5B function or recognition of dsRNA by TLR3. In other words, HCV has a dual strategy against host innate immunity, which may contribute to the higher rate of persistent infection compared to other viruses.
Recently, significant advances have been made in developing cell culture systems for HCV [31, 32]. The HCV subgenomic replicon system is based on the self-replication of HCV RNA in a transfected human hepatoma cell line. The HCV replicon is known to be propagated only in Huh7 cells. In our study, NS5B and other HCV proteins did not influence IFN-beta promoter activity in Huh7 cells. Poly(I:C) treatment also did not activate IFN-beta in Huh7 cells (Fig. 10). These results indicate that innate immunity may be disrupted in Huh7 cells. This could be one of the mechanisms allowing replication of HCV replicon system in Huh7 cells.
Fig. 10.
Effect of HCV proteins on IFN-beta promoter activity in Huh7 cells. Huh7 cells were co-transfected with 0.18 μg pGL3-p125luc, 0.02 μg PRL-TK, and 0.4 μg expression plasmids for each of seven HCV proteins. As a positive control, 100 μg/ml poly(I:C) were added to the culture medium. The normalized luciferase activities were calculated by assigning a value of 1 to the levels of activity of cells that were transfected with the empty vector
In conclusion, HCV NS5B potentially activates host innate immunity through TLR3, TRIF, and IRF-3 by its RdRp activity. Meanwhile, HCV NS4A, NS4B, and NS5A specifically inhibit NS5B-induced activation of IFN-beta. NS4A, NS4B, and NS5A combine with NS3 to act against the host antiviral response, which may contribute to the higher rate of persistent infection seen with HCV.
Acknowledgement
This work was supported in part by Health Science Research Grants for Medical Frontier Strategy Research from the Ministry of Health, Labor, and Welfare of Japan, and by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
References
- 1.Kuo G, Choo QL, Alter HJ, Gintick GL, Redeker AG, Purcell RH, et al. An assay for circulating antibodies to a major etiologic virus of human non-A non-B hepatitis. Science 1989;244:362–4. [DOI] [PubMed]
- 2.Saito M, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kichi S, et al. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA 1990;87:6547–9. [DOI] [PMC free article] [PubMed]
- 3.Shiratori Y, Shiina S, Imamura M, Kato N, Kanai F, Okudaira T, et al. Characteristic difference of hepatocellular carcinoma between hepatitis B- and C- viral infection in Japan. Hepatology 1995;22:1027–33. [DOI] [PubMed]
- 4.Choo QL, Richman KH, Han JH, Berger K, Lee C, Dong C, et al. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA 1991;88:2451–5. [DOI] [PMC free article] [PubMed]
- 5.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Shimotohno K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci USA 1991;88:5547–51. [DOI] [PMC free article] [PubMed]
- 6.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J 1989;8:3867–74. [DOI] [PMC free article] [PubMed]
- 7.Behrens SE, Tomei L, De Francesco R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J 1996;15:12–22. [PMC free article] [PubMed]
- 8.Du W, Maniatis T. An ATF/CREB binding site protein is required for virus induction of the human interferon-beta gene. Proc Natl Acad Sci USA 1992;89:2150–4. [DOI] [PMC free article] [PubMed]
- 9.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol 2001;2:675. [DOI] [PubMed]
- 10.Fitzgerald KA, Palsson-McDermott EM, Bowie AG, Jefferies C, Mansell AS, Brady G et al. Mal (MyD88-adaptor like) is required for Toll-like receptor-4 signal transduction. Nature 2001;413:78. [DOI] [PubMed]
- 11.Yamamoto M, Sato S, Mori K, Hoshino K, Takeuchi O, Takeda K, et al. A novel toll/IL-1 receptor domain-containing adaptor that preferentially activates the IFN-beta promoter in the toll-like receptor signaling. J Immunol 2002;169:6669–72. [DOI] [PubMed]
- 12.Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nature Immunol 2003;4:161–7. [DOI] [PubMed]
- 13.Suhara W, Yoneyama M, Iwamura T, Yoshimura S, Tamura K, Namiki H, et al. Analysis of virus-induced homomeric and heteromeric protein associations between IRF-3 and coactivator CBP/p300. J Biochem 2000;128:301–7. [DOI] [PubMed]
- 14.Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. Direct triggering of the type I interferon system by virus infection: activation of transcription factor complex containing IRF-3 and CBP/p300. EMBO J 1998;17:1087–95. [DOI] [PMC free article] [PubMed]
- 15.Servant MJ, ten Oever B, LePage C, Conti L, Gessani S, Julkunen I, et al. Identification of distinct signaling pathways leading to the phosphorylation of interferon regulatory factor 3. J Biol Chem 2001;276:355–63. [DOI] [PubMed]
- 16.Cerny A, Chisari FV. Pathogenesis of hepatitis C: immunological features of hepatic injury and viral persistence. Hepatology 1999;30:595–601. [DOI] [PubMed]
- 17.Disson O, Haouji D, Desagher S, Loesch K, Hahne M, Kremer EJ, et al. Impaired clearance of virus-infected hepatocytes in transgenic mice expressing the hepatitis C virus polyprotein. Gastroenterology 2004;126:859–72. [DOI] [PubMed]
- 18.Keskinen P, Nyqvist M, Sareneva T, Pirhonen J, Melen K, Julkunen I. Impaired antiviral response in human hepatoma cells. Virology 1999;263:364–75. [DOI] [PubMed]
- 19.Foy E, Li K, Wang C, Sumpter R Jr, Ikeda M, Lemon SM, et al. Regulation of interferon regulatory factor-3 by hepatitis C virus serine protease. Science 2003;300:1145–8. [DOI] [PubMed]
- 20.Otsuka M, Kato N, Moriyama M, Taniguchi H, Wang Y, Dharel N, et al. Interaction between the HCV NS3 protein and the host TBK1 protein leads to inhibiton of cellular antiviral responses. Hepatology 2005;41:1004–12. [DOI] [PubMed]
- 21.Kato N, Yoshida H, Ono-Nita SK, Kato J, Goto T, Otsuka M, et al. Activation of intracellular signaling by hepatitis B and C viruses: C-viral core is the most potent signal inducer. Hepatology 2000;32:405–12. [DOI] [PubMed]
- 22.Qin W, Yamashita T, Shirota Y, Ling Y, Wei W, Murakami S. Mutational analysis of the structure and functions of hepatitis C virus RNA-dependent RNA polymerase. Hepatology 2001;33:728–37. [DOI] [PubMed]
- 23.Vo NM, Oh JW, Lai MMC. Identification of RNA ligands that bind hepatitis C virus polymerase selectively and inhibit its RNA synthesis from the natural viral RNA templates. Virology 2003;307:301–16. [DOI] [PubMed]
- 24.Lohmann V, Ross A, Körner F, Koch JO, Bartenshlager R. Biochemical and kinetic analysis of NS5B RNA-dependent RNA polymerase of the hepatitis C virus. Virology 1998;249:108–18. [DOI] [PubMed]
- 25.Oh JW, Ito T, Lai MM. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full length viral RNA. J Virol 1999;73:7694–702. [DOI] [PMC free article] [PubMed]
- 26.Zhong W, Ferrari E, Lesburg CA, Maag D, Ghosh SK, Cameron CE,et al. Template/primer requirements and single nucleotide incorporation by hepatitis C virus nonstructural protein 5B polymerase. J Virol 2000;74:9134–43. [DOI] [PMC free article] [PubMed]
- 27.Mottola G, Cardinali G, Ceccacci A, Trozzi C, Batholomew L, Torrisi MR, et al. Hepatitis C virus nonstructural proteins are localized in a modified endoplasmic reticulum of cells expressing viral subgenomic replicons. Virology 2002;293:31–43. [DOI] [PubMed]
- 28.Dimitrova M, Imbert I, Kieny MP, Schuster C. Protein-protein interactions between hepatitis C virus nonstructural proteins. J Virol 2003;77:5401–14. [DOI] [PMC free article] [PubMed]
- 29.Shirota Y, Luo H, Qin W, Kaneko S, Yamashita T, Kobayashi K, et al. Hepatitis C virus (HCV) NS5A binds RNA-dependent RNA polymerase (RdRp) NS5B and modulates RNA-dependent RNA polymerase activity. J Biol Chem 2002;277:11149–55. [DOI] [PubMed]
- 30.Shimakami T, Hijikata M, Luo H, Ma YY, Kaneko S, Shimotohno K, et al. Effect of interaction between hepatitis C virus NS5A and NS5B on hepatitis C virus RNA replication with the hepatitis C virus replicon. J Virol 2004;78:2738–48. [DOI] [PMC free article] [PubMed]
- 31.Lohmann V, Körner F, Koch JO, Herian U, Theilmann L, Bartenshlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 1999;285:110–3. [DOI] [PubMed]
- 32.Lohmann V, Körner F, Dobeierzewska A, Bartenshlager R. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J Virol 2001;75:1437–49. [DOI] [PMC free article] [PubMed]