Abstract
ATP-powered AAA+ proteases degrade specific proteins in intracellular environments occupied by thousands of different proteins. These proteases operate as powerful molecular machines that unfold stable native proteins before degradation. Understanding how these enzymes choose the ‘right’ protein substrates at the ‘right’ time is key to understanding their biological function. Recently, proteomic approaches have identified numerous substrates for some bacterial enzymes and the sequence motifs responsible for recognition. Advances have also been made in elucidating the mechanism and impact of adaptor proteins in regulating substrate choice. Finally, recent biochemical dissection of the ATPase cycle and its coupling to protein unfolding has revealed fundamental operating principles of this important, ubiquitous family of molecular machines.
The AAA+ proteases
AAA+ proteases (the term AAA comes from ‘ATPase associated with cellular activities’) are multimeric machines that function with exquisite specificity in all domains of life to recognize, to unfold and to degrade proteins. Energy-dependent proteases differ widely in their complexity. For example, the 26S proteasome is built from >30 different types of protein, whereas a single type of subunit assembles to form the hexameric FtsH and Lon proteases. Nevertheless, ATP-dependent proteases share common architectural features [1-3].
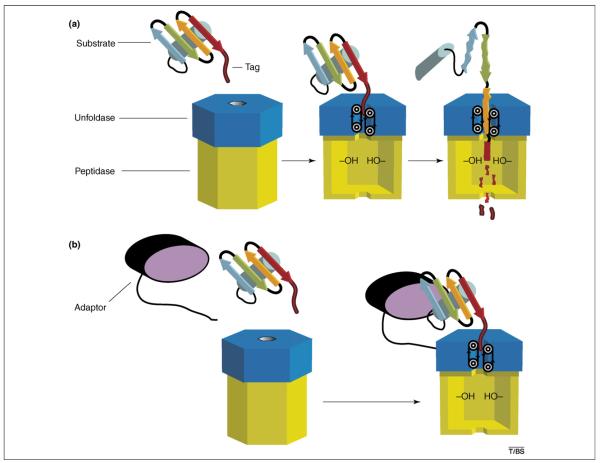
For example, the active sites that catalyze peptide bond cleavage are sequestered in a hollow interior chamber, typically constructed from rings of six or seven subunits or domains (Figure 1a). Substrates enter these degradation compartments through axial channels or portals that are too narrow to admit folded native proteins. This restriction prevents the undesirable cleavage of most cellular proteins but requires the coordinated enzymatic recognition, unfolding and translocation of correct substrates before degradation. Hexameric rings of subunits or domains, which belong to the AAA+ ATPase family, carry out these mechanical denaturation and translocation steps. Moreover, these proteins and related AAA+ enzymes often function to disassemble macromolecular complexes and to resolubilize protein aggregates [4,5]. Substrate recognition can be mediated directly by the AAA+ domains of ATP-dependent proteases or indirectly by additional domains or adaptor proteins (Figure 1b).
Figure 1.
Substrate recognition, unfolding and degradation by an AAA+ protease. (a) The protease consists of a hexameric AAA+ unfoldase (blue) and a compartmental peptidase (yellow). The initial recognition step shown here is mediated by residues in the central pore of the AAA+ ring, which bind to a peptide tag sequence attached to an otherwise native protein substrate. Repetitive conformational changes in the AAA+ ring, driven by ATP hydrolysis, translocate the tag and attached protein through the pore. This vectorial movement unfolds the protein and transports the denatured polypeptide into the degradation chamber of the peptidase. Active sites in the peptidase chamber cleave the unfolded substrate into short peptides, which are subsequently released. (b) Binding of protein substrates to AAA+ proteases can be assisted by adaptor proteins. In this example, the ternary recognition complex is stabilized by interactions between the adaptor (black and purple) and the substrate, between a flexible tail of the adaptor and the AAA+ ring of the protease, and between the substrate tag and the pore of the AAA+ ring. The ternary complex is more stable than the binary complex shown in (a), enabling more efficient degradation at low substrate concentrations. Some adaptor proteins are also degraded, whereas others are resistant to proteolysis and can therefore participate in many rounds of substrate delivery.
Here, we focus on prokaryotic examples of the widespread family of AAA+ proteases (especially ClpXP and ClpAP) and their well-characterized adaptor proteins (SspB and RssB, and ClpS). In particular, we highlight recent developments that have advanced our understanding of the mechanisms by which the AAA+ proteases and their adaptors function.
Recognition through peptide ‘tags’
One mechanism that enables ATP-dependent proteases to recognize specific substrates involves binding to unstructured peptide sequences, which are typically located at the N-terminal or C-terminal end of a target protein. Recognition of ‘degradation tags’ can be the sole determinant of targeted proteolysis. For example, one quality control system in Escherichia coli adds the ‘ssrA tag’ sequence (AANDENYALAA) to the C terminus of proteins for which biosynthesis cannot be completed normally (e.g. because the mRNA is broken and therefore lacks a stop codon), thereby targeting the tagged, incomplete protein for degradation by proteases such as ClpXP and ClpAP [1,6]. These enzymes consist of hexameric ring ATPases (ClpX or ClpA) and a double-ring peptidase (ClpP). A pore through the center of the ClpX or ClpA ring binds to the ssrA tag, enabling ATP-dependent translocation of the tag sequence to drive unfolding of the attached protein and transport of the denatured polypeptide into ClpP for degradation [1,7,8] (Figure 1a).
Degradation of ssrA-tagged proteins by ClpXP and ClpAP illustrates a simple strategy that can be used when rapid destruction is the desired biological goal. For example, the synthesis of some proteins is highly induced during DNA damage but these proteins are deleterious once the damage has been repaired [9,10]. Thus, it is beneficial to remove them from the cell as soon as possible. The damage-response protein RecN is synthesized with a C-terminal sequence similar to that of the ssrA tag. As a consequence, ClpXP continually degrades RecN. When the rate of RecN synthesis is sufficiently high, a moderate steady-state level of RecN is maintained. As soon as damage-induced expression ceases, however, degradation by ClpXP causes a rapid decline in the intracellular amount of RecN [11]. This type of ‘hard-coded’ instability of stress-response proteins is likely to be common, helping to ensure that the proteome can rapidly return to the pre-stressed state.
Regulated exposure of degradation tags
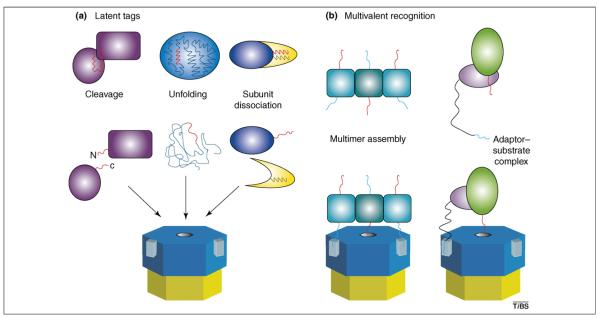
Degradation tags for ATP-dependent proteases can be created or exposed by an endoproteolytic cleavage event (Figure 2a). For example, proteolytic clipping of the LexA repressor or the transmembrane regulator RseA at a single site produces N-terminal fragments that are subsequently degraded by ClpXP [12,13]. In these proteins, a combination of the new α-carboxyl group and the increased accessibility of the proximal ssrA-like sequence creates a degradation tag that is recognized by ClpXP. Temporal control of the initial cleavage event enables these regulatory proteins to exist stably in the cell under normal conditions, but to be eradicated as soon as the appropriate signal for endoproteolysis is received. For example, LexA autoproteolysis is stimulated by RecA bound to single-stranded DNA that accumulates when replication cannot proceed past a site of damage [14]. Interestingly, in numerous situations, protein degradation is required both to initiate a specific cellular stress response and to rebalance the proteome after stress during the return to normal physiology.
Figure 2.
Mechanisms of regulated substrate recognition. (a). When degradation tags (red) are part of the folded structure of a protein or complex, they can be inaccessible to AAA+ proteases. Such ‘hidden’ tags can be revealed by internal cleavage (left), unfolding (center), or complex dissociation (right). In these cases, protein turnover can be regulated by the reactions that lead to exposure of the degradation tag. (b) Macromolecular assembly can be required for efficient substrate recognition and degradation, when monomeric recognition signals are weak. Left, trimerization of a protein places a degradation tag (red) and two tethering peptides (cyan) in the proper geometry to interact simultaneously with the pore and tethering sites (light blue rectangles) of an AAA+ protease. Multivalent binding of this type can be orders of magnitude stronger than any of the individual binding reactions. Right, heteromeric assembly of an adaptor containing a tethering signal and a substrate containing a degradation tag is required for efficient substrate recognition.
Cryptic degradation signals can also become accessible to an AAA+ protease as a consequence of protein unfolding (Figure 2a). For example, native green fluorescent protein is not degraded by ClpAP unless it bears an appended degradation tag, but unfolded green fluorescent protein is rapidly proteolyzed without the need for this additional tag [15,16]. In the cell, protein unfolding and subsequent degradation could occur as a consequence of heat shock, changes in pH or redox environment, the depletion of a folding partner or chaperone, or the incorporation of amino acid analogs. Dissociation of protein—protein, protein—DNA or protein—RNA complexes could also expose degradation signals that would otherwise be inaccessible (see, for example, Ref. [17])(Figure 2a).
Distinct types of peptide degradation tags can target intracellular substrates for degradation by ClpXP [18]. In each class, the degradation signals are short and only a few key residues seem to mediate recognition by ClpX. Having various degradation signals might facilitate independent regulation of the proteolysis of different types of substrate (see later). Degradation tags for AAA+ proteases other than ClpXP have not been characterized extensively, but the examples that are known suggest a common use of exposed peptides in which a few crucial residues mediate specific recognition [19-22].
Adaptor-mediated recognition of substrates
Adaptor proteins often regulate proteolysis, either by facilitating or preventing the degradation of specific substrates [1,23]. Some adaptors enhance degradation by tethering substrates to an AAA+ protease, thereby increasing the ‘effective’ local concentrations of these molecules (Figure 2b). For example, SspB uses flexible tails to bind ClpX and a peptide-binding groove to bind some ClpXP substrates including ssrA-tagged proteins and RseA [24-27]. This adaptor-mediated tethering facilitates efficient degradation at low concentrations in which the substrate alone would not bind strongly to ClpXP.
SspB is not required for ClpXP-mediated proteolysis of RseA or ssrA-tagged substrates, but it increases the degradation of low concentrations of these substrates approximately tenfold [13,24]. Interestingly, SspB enhances ClpXP-mediated degradation of substrates with ‘weak’ degradation tags to an even greater extent. For example, when the C terminus of the ssrA tag is changed from Leu-Ala-Ala (LAA) to Arg-Ala-Ser (DAS), ClpX binding is weakened considerably. At low substrate concentrations, ClpXP degrades DAS-tagged proteins >100-fold more slowly than when SspB is present [28]. Indeed, DAS-tagged substrates can be stably expressed in E. coli when SspB concentrations are low, but induction of SspB results in rapid degradation by ClpXP and clearance of the DAS-tagged proteins from the cell. This synthetic system provides a simple illustration of how adaptor-mediated tethering of a substrate to an AAA+ protease can be required to achieve biologically meaningful rates of degradation.
Additional examples in which tethering seems to be required for efficient degradation by ClpXP or related bacterial enzymes include the adaptor—substrate pairs MecA—ComK, RssB—σS and UmuD—UmuD’ [1,23]. Similarly, in eukaryotes polyubiquitin chains tether covalently attached substrates to the 26S proteasome [2]. In this system, the enzymes that add polyubiquitin to specific proteins carry out the initial reactions required for targeted degradation [2]. As in bacteria, however, degradation requires both proteasomal recognition of additional peptide signals in the substrate and ubiquitin-mediated tethering [29].
ClpS is an adaptor protein for the ClpAP protease that alters substrate preferences positively and negatively [30,31]. Binding of ClpS to ClpA inhibits the binding and/or degradation of ssrA-tagged substrates but activates the binding and/or degradation of ‘N-end rule substrates’ — a class of unstable proteins that have a specific amino acid at their N termini (F, W, L, R or K) responsible for their rapid turnover. How ClpS inhibits the recognition of some substrates, while acting as a molecular matchmaker for new substrates is not understood; however, ClpS clearly provides an important example of how a single adaptor molecule can reprogram the recognition properties of an ATP-dependent protease, inhibiting ‘default’ recognition mechanisms and establishing new substrate-binding specificities.
Other adaptors also downregulate degradation. For example, the PinA protein of bacteriophage T4 binds to the Lon protease and prevents the degradation of some substrates [32]. ClpAP-mediated degradation of ssrA-tagged substrates can be inhibited by SspB binding to the ssrA tag, which masks sequences required for ClpA recognition [33]. In principle, any binding event that masks a degradation tag or a tethering signal protein could prevent or diminish degradation. Similarly, binding of other molecules to the protease sites that recognize these tethering sequences or degradation tags will also inhibit degradation. For example, the peptide sequences that facilitate SspB- and UmuD-mediated tethering to ClpX compete with each other for ClpX binding [34]. Thus, expression of one adaptor protein can enhance the degradation of cognate substrates and, at the same time, slow the degradation of substrates that use other adaptors.
Multiple weak signals: combinatorial control and evolution
As we have seen, AAA+ proteases frequently recognize multiple signals on a substrate and/or an associated adaptor. By themselves, these signals might be too weak for efficient degradation. Coupling weak recognition signals creates stronger binding but, more importantly, facilitates combinatorial regulation.
For example, two requisite signals might be recognized only after assembly of a protein multimer or aggregate (Figure 2b). Alternatively, a protein modification such as phosphorylation could strengthen or weaken a particular signal or unmask an otherwise cryptic signal. Evolutionary creation of signals is also facilitated if only a few unstructured residues need to be specified. In addition, if many different sequences can mediate the weak binding affinities needed, then it might be easier to craft signals that can be masked or unmasked by protein folding or the association—dissociation of molecular partners.
AAA+ proteases are powerful protein unfolding machines
After substrate recognition by AAA+ proteases, native substrates must still be unfolded, translocated into the degradation chamber and ultimately proteolyzed. Enzymes such as ClpAP, ClpXP and the 26S proteasome can denature protein substrates that have exceptionally high thermodynamic, kinetic and mechanical stability [15,16,35-38]. How do the AAA+ portions of these enzymes carry out ATP-fueled unfolding?
Enzymatic trapping of globally denatured substrates has been ruled out as a general mechanism, because AAA+ enzymes unfold and degrade some substrates more than a million times faster than their rates of spontaneous solution unfolding [15,16,35]. Moreover, binding of some substrates to AAA+ proteases is not, by itself, destabilizing [16,37].
Do the AAA+ rings or associated domains trap locally unfolded regions of substrates? This model (akin to a ‘brownian ratchet’ whereby movement is generated by thermal fluctuations trapped by a vectoral process) is more difficult to disprove rigorously but seems highly unlikely. All proteins undergo local unfolding reactions, but these partially denatured structures are extremely short-lived and could occur anywhere in a substrate. Thus, it is improbable that a transiently unfolded segment of a substrate would encounter and be trapped by a specific site in an AAA+ protease before refolding. This mechanism seems even more unlikely when one considers that some AAA+ proteases degrade hundreds of different natural substrates in addition to artificial substrates constructed with proteins that could not have coevolved with these proteases [15,16,18,35-38].
Enzyme-induced protein denaturation almost certainly involves the application of a mechanical force. For example, translocation of a degradation tag through the center of an AAA+ hexamer could generate force by trying to pull an attached native protein through this small opening (Figure 3). This ‘pulling’ model explains how a single degradation signal, such as the ssrA tag, could mediate enzymatic denaturation of various structurally unrelated proteins, and is also appealing because the same cycle of conformational changes in the enzyme would be sufficient to drive translocation and denaturation [1]. In addition, this type of model is supported both by experiments that show that substrates are unfolded more readily when their degradation tags are attached to relatively ‘weak’ regions of local structure and vice versa and by studies demonstrating strong correlations between translocation and denaturation activities [36-39].
Figure 3.
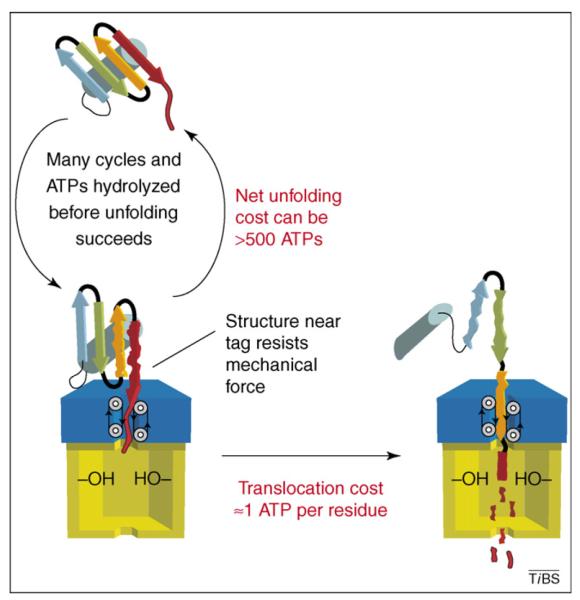
The cost of degradation. For denatured substrates and native substrates that are easily unfolded, the cost of degradation is determined mainly by the cost of translocation. For some substrates of ClpXP, this cost is ∼1 ATP molecule for each residue degraded [37]. Thus, ∼100 ATP molecules would be hydrolyzed during degradation of a 100-residue protein. For native substrates that are difficult to denature, the cost of degradation can be much higher. When the native structure of the substrate is sufficiently stable to resist mechanical unfolding by the protease, the substrate is usually released [40]. In these instances, many cycles of binding, attempted unfolding and release can occur before enzymatic denaturation is successful, and the energetic cost of unfolding alone can exceed the hydrolysis of 500 molecules of ATP [37].
Degradation can be expensive
The most detailed studies of the energetic price of degradation have been carried out on ClpXP [37]. Depending on the substrate, the cost of enzymatic denaturation of a single protein can range from the hydrolysis of fewer than 20 ATP molecules to that of >500 ATP molecules (Figure 3). Translocation costs an additional 0.3–1 ATP molecules per amino acid. Thus, the net cost of degradation is extraordinarily high for some substrates and substantial for all substrates. What accounts for the enormous variability in the cost of denaturation of different protein substrates?
ATP hydrolysis must power cyclical conformational changes in the hexameric AAA+ ring, which in turn drive unfolding and translocation. Moreover, the all-or-none nature of domain unfolding means that proteins cannot be unraveled in a piece-by-piece manner. Thus, each ATP cycle must represent a new unfolding attempt. But if numerous ATP molecules are hydrolyzed before denaturation occurs, then most unfolding attempts must be unsuccessful. Indeed, hard-to-denature substrates are clearly bound and released many times by ClpXP before they are successfully unfolded [40]. Why, then, would the nth denaturation attempt be successful when all previous efforts failed? Because neither the enzyme nor the substrate has any memory of previous encounters, random factors must alter the probability of denaturation. In a population of otherwise identical molecules, stochastic thermal fluctuations continually cause transient structural distortions. If these deformations weaken structure near the degradation tag at the same time that an AAA+ enzyme tugs on this tag, then the probability of protein unfolding will increase.
At first glance, this relentless ‘try and try again’ mechanism seems inelegant and energetically wasteful, sometimes using more energy to degrade a protein than was originally used for its biosynthesis. Remember, however, that AAA+ proteases must denature an eclectic assortment of proteins with markedly different structures and stabilities, and thus evolution cannot optimize activity for any single substrate. Pulling on a peptide tag represents a simple and universal mechanism that might represent the best compromise. Furthermore, because substrates are released when unfolding is unsuccessful [40], an AAA+ enzyme will not become jammed when a hyperstable substrate resists denaturation. Lastly, this release mechanism ensures that when an AAA+ protease encounters a mixture of substrates, only some of which are denatured easily, it will preferentially degrade the ‘easy’ substrates, thereby maximizing energetic efficiency.
Operational dissection of the AAA+ machine is underway
The hexameric ATPases are the engines of AAA+ proteases. As yet, we do not understand how these molecular machines engage the degradation tags of substrates during denaturation or interact with translocating polypeptides. Moreover, the nucleotide-dependent conformational changes that drive substrate translocation and unfolding have not been elucidated. Nevertheless, we are beginning to understand some aspects of these reactions. For example, loops that line the central pore of the AAA+ hexamer are clearly involved in translocation and in the recognition of some substrates [7,8,41,42]. Different structures of HslUV show nucleotide-dependent changes in loop conformations [43-45]. Although the functional relevance of some of these structures is unclear, related loop movements could drive polypeptide translocation through the pore.
It is also becoming clear that AAA+ hexamers function in an asymmetric manner. For example, although the ClpX hexamer contains six subunits of identical sequence, some of these subunits do not bind ATP, some bind ATP tightly and some bind ATP weakly [46]. Some structures of the HslU homohexamer also show three classes of subunit [43].
The six subunits of ClpX can be covalently linked without disrupting function, enabling enzyme variants with different numbers of active or inactive subunits to be constructed and studied [39]. Linked enzymes with only a single subunit capable of hydrolyzing ATP have weak activity, suggesting that nucleotide hydrolysis in one subunit is sufficient to power the conformational changes required for degradation. Enzymes with two wild-type subunits on opposing sides of the hexamer are substantially more active, degrading substrates at a third of the rate and with the same thermodynamic efficiency as the fully wild-type hexamer. The locations of the two active subunits in the hexameric ring is an important determinant of activity, as are the abilities of neighboring subunits to mimic ‘empty’ or ‘ATP-bound’ conformations. These results show that subunit—subunit communication and cooperation have important roles in determining translocation and denaturation activity. Given that some AAA+ machines, including the proteasome, function with non-identical subunits, it makes sense that different subunits in homomeric AAA+ rings also have distinct functional roles [4,46].
The properties of linked ClpX variants suggest that ATP hydrolysis occurs in a sequential but not strictly ordered manner [39]. In other words, hydrolysis of ATP in one subunit of the hexameric ring is not always followed by hydrolysis in an adjacent subunit, in the next nearest neighbor, or in the opposite subunit. ClpX hydrolyzes ATP faster when it is translocating an unfolded substrate than when it is attempting to denature a native substrate; thus, it is possible that the hydrolysis activity of any ATP-bound subunit in contact with a translocating substrate is stimulated. Because unfolded polypeptides have highly irregular properties and shapes, this mechanism could lead to hydrolysis that occurs without any regular sequence or pattern but instead drives conformational changes in subunits best positioned to carry out the next step of translocation.
Future perspectives
Spectacular advances in our understanding of ATP-dependent proteases have been made in the past decade. Questions about biological regulation and enzymatic mechanism can now be posed and tackled in increasing molecular detail. Much remains, however, to be learned.
For example, in most cases we do not know the rules of engagement well enough to predict from sequence or structure whether a specific protein will be a substrate for a given ATP-dependent protease. As a consequence, only a small fraction of the complete substrate repertoire for any ATP-dependent protease is known. In addition, our current level of structural information about AAA+ proteases needs to be expanded to include structures of enzymes that have engaged or are translocating protein substrates to increase our understanding of the mechanisms by which they function. How ATP-fueled proteases function in concert with other molecular chaperones in the cell to ensure protein quality control is also poorly understood and many adaptor molecules remain to be identified. Clearly, important and exciting challenges lie ahead for the coming decade.
Acknowledgements
We thank present and past members of our laboratories for discussions. Work in the authors’ laboratories was supported by grants AI-15706, AI-16892 and GM-049224, and by the Howard Hughes Medical Institute (HHMI). T.A.B. is an employee of the HHMI.
References
- 1.Sauer RT, et al. Sculpting the proteome with AAA+ proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmidt M, et al. Proteasome-associated proteins: regulation of a proteolytic machine. Biol. Chem. 2005;386:725–737. doi: 10.1515/BC.2005.085. [DOI] [PubMed] [Google Scholar]
- 3.Ito K, Akiyama Y. Cellular functions, mechanism of action, and regulation of FtsH protease. Annu. Rev. Microbiol. 2005;59:211–231. doi: 10.1146/annurev.micro.59.030804.121316. [DOI] [PubMed] [Google Scholar]
- 4.Bukau B, et al. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 5.Burton BM, Baker TA. Remodeling protein complexes: insights from the AAA+ unfoldase ClpX and Mu transposase. Protein Sci. 2005;14:1945–1954. doi: 10.1110/ps.051417505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Withey JH, Friedman DI. A salvage pathway for protein structures: tmRNA and trans-translation. Annu. Rev. Microbiol. 2003;57:101–123. doi: 10.1146/annurev.micro.57.030502.090945. [DOI] [PubMed] [Google Scholar]
- 7.Siddiqui SM, et al. Role of the processing pore of the ClpX AAA+ ATPase in the recognition and engagement of specific protein substrates. Genes Dev. 2004;18:369–374. doi: 10.1101/gad.1170304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinnerwisch J, et al. Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell. 2005;121:1029–1041. doi: 10.1016/j.cell.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 9.Mizusawa S, Gottesman S. Protein degradation in Escherichia coli: the lon gene controls the stability of SulA protein. Proc. Natl. Acad. Sci. U. S. A. 1983;80:358–362. doi: 10.1073/pnas.80.2.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez M, et al. Subunit-specific degradation of the UmuD/D’ heterodimer by the ClpXP protease: the role of trans recognition in UmuD’ stability. EMBO J. 2000;19:5251–5258. doi: 10.1093/emboj/19.19.5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neher SB, et al. Proteomic profiling of ClpXP substrates after DNA damage reveals extensive instability within SOS regulon. Mol. Cell. 2006;22:193–204. doi: 10.1016/j.molcel.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Neher SB, et al. Latent ClpX-recognition signals ensure LexA destruction after DNA damage. Genes Dev. 2003;17:1084–1089. doi: 10.1101/gad.1078003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flynn JM, et al. Modulating substrate choice: the SspB adaptor delivers a regulator of the extracytoplasmic-stress response to the AAA+ protease ClpXP for degradation. Genes Dev. 2004;18:2292–2301. doi: 10.1101/gad.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bell CE. Structure and mechanism of Escherichia coli RecA ATPase. Mol. Microbiol. 2005;58:358–366. doi: 10.1111/j.1365-2958.2005.04876.x. [DOI] [PubMed] [Google Scholar]
- 15.Weber-Ban EU, et al. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature. 1999;401:90–93. doi: 10.1038/43481. [DOI] [PubMed] [Google Scholar]
- 16.Hoskins JR, et al. Protein binding and unfolding by the chaperone ClpA and degradation by the protease ClpAP. Proc. Natl. Acad. Sci. U. S. A. 2000;97:8892–8897. doi: 10.1073/pnas.97.16.8892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levchenko I, et al. ClpX and MuB interact with overlapping regions of Mu transposase: implications for control of the transposition pathway. Genes Dev. 1997;11:1561–1572. doi: 10.1101/gad.11.12.1561. [DOI] [PubMed] [Google Scholar]
- 18.Flynn JM, et al. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell. 2003;11:671–683. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- 19.Ishii Y, et al. Regulatory role of C-terminal residues of SulA in its degradation by Lon protease in Escherichia coli. J. Biochem. (Tokyo) 2000;127:837–844. doi: 10.1093/oxfordjournals.jbchem.a022677. [DOI] [PubMed] [Google Scholar]
- 20.Burton RE, et al. Nucleotide-dependent substrate recognition by the AAA+ HslUV protease. Nat. Struct. Mol. Biol. 2005;12:245–251. doi: 10.1038/nsmb898. [DOI] [PubMed] [Google Scholar]
- 21.Shah IM, Wolf RE., Jr Sequence requirements for Lon-dependent degradation of the Escherichia coli transcription activator SoxS: identification of the SoxS residues critical to proteolysis and specific inhibition of in vitro degradation by a peptide comprised of the N-terminal 21 amino acid residues. J. Mol. Biol. 2006;357:718–731. doi: 10.1016/j.jmb.2005.12.088. [DOI] [PubMed] [Google Scholar]
- 22.Hoskins JR, et al. Substrate recognition by the ClpA chaperone component of ClpAP protease. J. Biol. Chem. 2000;275:35361–35367. doi: 10.1074/jbc.M006288200. [DOI] [PubMed] [Google Scholar]
- 23.Dougan DA, et al. AAA+ proteins and substrate recognition, it all depends on their partner in crime. FEBS Lett. 2002;529:6–10. doi: 10.1016/s0014-5793(02)03179-4. [DOI] [PubMed] [Google Scholar]
- 24.Levchenko I, et al. A specificity-enhancing factor for the ClpXP degradation machine. Science. 2000;289:2354–2356. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- 25.Wah DA, et al. Flexible linkers leash the substrate-binding domain of SspB to a peptide module that stabilizes delivery complexes with the AAA+ ClpXP protease. Mol. Cell. 2003;12:355–363. doi: 10.1016/s1097-2765(03)00272-7. [DOI] [PubMed] [Google Scholar]
- 26.Levchenko I, et al. Structure of a delivery protein for a AAA+ protease in complex with a peptide degradation tag. Mol. Cell. 2003;12:365–372. doi: 10.1016/j.molcel.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 27.Levchenko I, et al. Versatile modes of peptide recognition by the AAA+ adaptor protein SspB. Nat. Struct. Mol. Biol. 2005;12:520–525. doi: 10.1038/nsmb934. [DOI] [PubMed] [Google Scholar]
- 28.McGinness KE, et al. Engineering controllable protein degradation. Mol. Cell. 2006;22:701–707. doi: 10.1016/j.molcel.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 29.Prakash S, et al. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat. Struct. Mol. Biol. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- 30.Dougan DA, et al. ClpS, a substrate modulator of the ClpAP machine. Mol. Cell. 2002;9:673–683. doi: 10.1016/s1097-2765(02)00485-9. [DOI] [PubMed] [Google Scholar]
- 31.Erbse A, et al. ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature. 2006;439:753–756. doi: 10.1038/nature04412. [DOI] [PubMed] [Google Scholar]
- 32.Hilliard JJ, et al. PinA inhibits ATP hydrolysis and energy-dependent protein degradation by Lon protease. J. Biol. Chem. 1998;273:524–527. doi: 10.1074/jbc.273.1.524. [DOI] [PubMed] [Google Scholar]
- 33.Flynn JM, et al. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10584–10589. doi: 10.1073/pnas.191375298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neher SB, et al. Distinct peptide signals in the UmuD and UmuD’ subunits of UmuD/D’ mediate tethering and substrate processing by the ClpXP protease. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13219–13224. doi: 10.1073/pnas.2235804100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim YI, et al. Dynamics of substrate denaturation and translocation by the ClpXP degradation machine. Mol. Cell. 2000;5:639–648. doi: 10.1016/s1097-2765(00)80243-9. [DOI] [PubMed] [Google Scholar]
- 36.Lee C, et al. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol. Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- 37.Kenniston JA, et al. Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of an AAA+ degradation machine. Cell. 2003;114:511–520. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
- 38.Kenniston JA, et al. Effects of local protein stability and the geometric position of the substrate degradation tag on the efficiency of ClpXP denaturation and degradation. J. Struct. Biol. 2004;146:130–140. doi: 10.1016/j.jsb.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 39.Martin A, et al. Rebuilt AAA+ motors reveal operating principles for ATP-fueled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- 40.Kenniston JA, et al. Partitioning between unfolding and release of native domains during ClpXP degradation determines substrate selectivity and partial processing. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1390–1395. doi: 10.1073/pnas.0409634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamada-Inagawa T, et al. Conserved pore residues in the AAA protease FtsH are important for proteolysis and its coupling to ATP hydrolysis. J. Biol. Chem. 2003;278:50182–50187. doi: 10.1074/jbc.M308327200. [DOI] [PubMed] [Google Scholar]
- 42.Park E, et al. Role of the GYVG pore motif of HslU ATPase in protein unfolding and translocation for degradation by HslV peptidase. J. Biol. Chem. 2005;280:22892–22898. doi: 10.1074/jbc.M500035200. [DOI] [PubMed] [Google Scholar]
- 43.Bochtler M, et al. The structures of HsIU and the ATP-dependent protease HsIU–HsIV. Nature. 2000;403:800–805. doi: 10.1038/35001629. [DOI] [PubMed] [Google Scholar]
- 44.Sousa MC, et al. Crystal and solution structures of an HslUV protease-chaperone complex. Cell. 2000;103:633–643. doi: 10.1016/s0092-8674(00)00166-5. [DOI] [PubMed] [Google Scholar]
- 45.Wang J, et al. Nucleotide-dependent conformational changes in a protease-associated ATPase HslU. Structure. 2001;9:1107–1116. doi: 10.1016/s0969-2126(01)00670-0. [DOI] [PubMed] [Google Scholar]
- 46.Hersch GL, et al. Asymmetric interactions of ATP with the AAA+ ClpX6 unfoldase: allosteric control of a protein machine. Cell. 2005;121:1017–1027. doi: 10.1016/j.cell.2005.05.024. [DOI] [PubMed] [Google Scholar]