Abstract
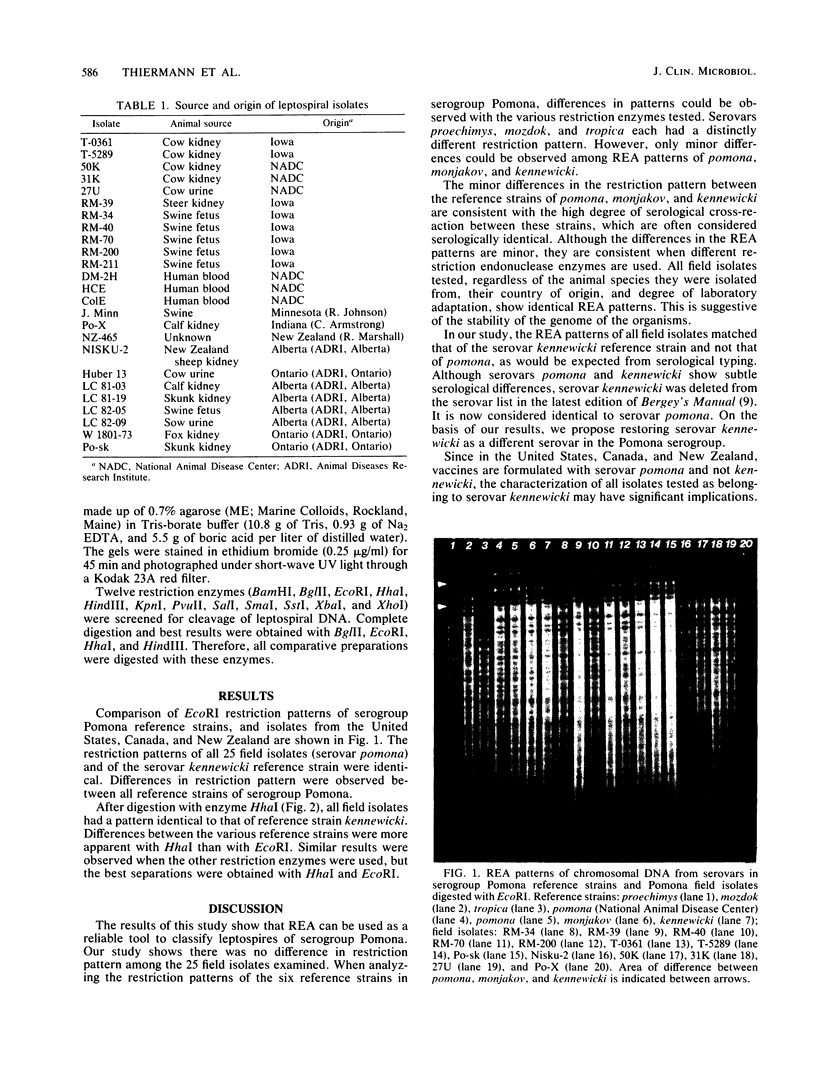
The genomes of leptospiral field isolates belonging to serogroup Pomona were analyzed and compared with those of type strains by cleavage with restriction endonucleases. This new classification method shows differences among these organisms not indicated by the conventional serological typing method. No differences were observed among isolates from the United States, Canada, and New Zealand. Although all isolates selected for this study had been serologically typed as belonging to serovar pomona, the restriction endonuclease analysis indicates that they belong to serovar kennewicki. kennewicki, a serovar of North American origin, has recently been eliminated from the official serovar list because it was found to be indistinguishable from serovar pomona by the serological method.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chang A., Faine S., Williams W. T. Cross-reactivity of the axial filament antigen as a criterion for classification of Leptospira. Aust J Exp Biol Med Sci. 1974 Jun;52(Pt 3):549–568. doi: 10.1038/icb.1974.53. [DOI] [PubMed] [Google Scholar]
- Green S. S., Goldberg H. S., Blenden D. C. Enzyme patterns in the study of leptospira. Appl Microbiol. 1967 Sep;15(5):1104–1113. doi: 10.1128/am.15.5.1104-1113.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haapala D. K., Rogul M., Evans L. B., Alexander A. D. Deoxyribonucleic acid base composition and homology studies of Leptospira. J Bacteriol. 1969 May;98(2):421–428. doi: 10.1128/jb.98.2.421-428.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward G. S., Frenkel N., Roizman B. Anatomy of herpes simplex virus DNA: strain differences and heterogeneity in the locations of restriction endonuclease cleavage sites. Proc Natl Acad Sci U S A. 1975 May;72(5):1768–1772. doi: 10.1073/pnas.72.5.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull R. A., Gill R. E., Hsu P., Minshew B. H., Falkow S. Construction and expression of recombinant plasmids encoding type 1 or D-mannose-resistant pili from a urinary tract infection Escherichia coli isolate. Infect Immun. 1981 Sep;33(3):933–938. doi: 10.1128/iai.33.3.933-938.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall R. B., Wilton B. E., Robinson A. J. Identification of Leptospira serovars by restriction-endonuclease analysis. J Med Microbiol. 1981 Feb;14(1):163–166. doi: 10.1099/00222615-14-1-163. [DOI] [PubMed] [Google Scholar]
- Paul P. S., Mengeling W. L., Pirtle E. C. Differentiation of pseudorabies (Aujeszky's disease) virus strains by restriction endonuclease analysis. Arch Virol. 1982;73(2):193–198. doi: 10.1007/BF01314727. [DOI] [PubMed] [Google Scholar]
- Robinson A. J., Ramadass P., Lee A., Marshall R. B. Differentiation of subtypes within Leptospira interrogans serovars Hardjo, Balcanica and Tarassovi, by bacterial restriction-endonuclease DNA analysis (BRENDA). J Med Microbiol. 1982 Aug;15(3):331–338. doi: 10.1099/00222615-15-3-331. [DOI] [PubMed] [Google Scholar]
- Studdert M. J., Simpson T., Roizman B. Differentiation of respiratory and abortigenic isolates of equine herpesvirus 1 by restriction endonucleases. Science. 1981 Oct 30;214(4520):562–564. doi: 10.1126/science.6270790. [DOI] [PubMed] [Google Scholar]




