Abstract
The development of electrospray ionization mass spectrometry has been critical for the analyses of lipidomes from subcellular organelles. The myocardial nuclear lipidome likely has a key role in the molecular regulation of gene expression. In fact, recent studies have suggested that specific phospholipid classes bind and regulate specific transcription factors. The dynamic regulation of the myocardial nuclear lipidome may be critical in mediating long-term pathological responses to stresses such as ischemia, tachycardia and hypertension. In this brief review, the preparation of myocardial nuclei is discussed, and the resulting nuclear lipidome from rat and rabbit are shown as examples. The rabbit myocardial nuclear lipidome contains relatively more plasmenylcholine/phosphatidylcholine molecular species in comparison to that ratio observed in the rat myocardial nuclear lipidome. The composition of the rat myocardial nuclear choline glycerophospholipid pool was relatively enriched with molecular species containing arachidonic acid and docosahexaenoic acid in comparison to that in the rabbit myocardial nuclear choline glycerophospholipid pool. While the ethanolamine glycerophospholipids of the rabbit myocardial nuclei are enriched with arachidonic acid and plasmalogens, the ethanolamine glycerophospholipid profile from rat myocardial nuclei show less plasmalogen and more species containing docosahexaenoic acid. Last, significant differences in the ethanolamine glycerophospholipid molecular species were observed in the rabbit heart lipidomes from the nucleus and the mitochondria. Quantitation of these lipid species in hearts subjected to pathophysiological stresses may provide important information on the role of the myocardial nuclear lipidome on long-term cardiac cell function.
Keywords: electrospray ionization, myocardium, mass spectrometry, lipidome, lipidomics, mass spectrometry, mitochondria, nuclei, phospholipids, plasmalogen, sphingomyelin, subcellular organelles
2. Introduction
Over the past two decades, it has become ever-increasingly appreciated that individual subcellular lipidomes are regulated by specific lipases and contain unique constituents that are essential for their physiological and biochemical properties. Perhaps one of the most intensely studied tissues for lipid alterations has been the heart. The cardiac cell is exquisitely sensitive to changes in lipid constituents in specific subcellular membrane domains. For example, changes in the cardiac sarcolemmal phospholipid constituents, including alterations in plasmalogen and anionic phospholipid content, can have profound effects on sodium-calcium exchange protein enzymic activity (1,2). Further, intercalation of amphipathic metabolites of phospholipolysis and fatty acid catabolism (e.g., lysophosphatidylcholine and acylcarnitine) into cardiac sarcolemma have profound effects on the electrophysiological properties of heart cells (3,4). Oxidation of cardiolipin species in cardiac mitochondrial membranes is associated with release of cytochrome C and subsequent triggering of apoptosis (5,6). Nuclear and mitochondrial membranes have also been shown to be the targets of phospholipases activated during myocardial ischemia and reperfusion (7,8).
From a historical perspective, one of the first subcellular membrane lipidomic studies was performed by Gross in the mid-1980s with the characterization of the phospholipid constituents of canine sarcoplasmic reticulum, sarcolemma and mitochondrial membranes (9,10). These studies revealed the predominance of both choline and ethanolamine plasmalogen molecular species in the subcellular membrane pools of the canine heart. These early studies employed both GC analyses of aliphatic groups of HPLC-purified phospholipid classes, reversed phase HPLC analyses of individual molecular species as well as FAB-MS analyses of the HPLC-purified phospholipid species. The predominance of plasmalogens in the canine heart is similar to that in the human heart. Post and coworkers further elucidated the sidedness of the cardiac sarcolemma, which revealed the presence of plasmenylethanolamine species on the internal face of the cardiac sarcolemma (11).
The development of electrospray ionization mass spectrometry (ESI-MS) over the past 10–15 years has provided a powerful tool for the analyses of subcellular membrane lipidomes. One of the first studies employing ESI-MS was performed by Han and coworkers on erythrocyte plasma membranes (12). These studies demonstrated that the sensitivity of ESI-MS can be exploited to determine the molecular species of lipid components of subcellular membranes from minute quantities (e.g. erythrocyte plasma membranes from 25 nl of blood contain sufficient phospholipid for the delineation of their lipidome using ESI-MS). This study revealed the potential of this extraordinarily sensitive technique for determining lipidomes in membrane fractions that were previously difficult to assess due to limited amounts of isolated membranes.
In conclusion, advances in mass spectrometry now make it feasible to determine the lipidomes of individual subcellular membrane pools of cells and tissues. While it is difficult to assess changes in whole cell lipid composition during tissue stimulation or stress, it is quite possible that specific phospholipases are targeted to specific organelles, resulting in the loss of parent phospholipids and the generation of bioactive lipid products such as lysophospholipids, phosphatidic acid, diglycerides and arachidonic acid.
In the heart, we have previously shown that rat myocardial nuclear phospholipids are lost during ischemia/reperfusion and that this loss of lipids is partially blocked by an inhibitor of calcium-independent phospholipase A2 (8). Others have also shown that rat myocardial mitochondrial cardiolipin is oxidized during myocardial ischemia/reperfusion (6). It is well known that cardiac tissue is enriched in plasmalogens and the role of this phospholipid subclass in the heart is not fully understood. Interestingly, the enrichment of plasmalogens in human heart is considerably greater than that of the rat heart (11,13). In this review, we will briefly outline the issues in characterizing myocardial nuclear lipidomics and then compare the plasmalogen content in the rat and rabbit heart nuclei.
3. Complexity of myocardial subcellular lipidomics
The common animal models that are used for myocardial experimentation are mouse, rat and rabbit. These species contain four-chambered hearts with distinguishable right and left atria as well as right and left ventricles. To date, no lipidomic studies have thoroughly evaluated the lipidomics of these individual chambers. It should be appreciated that each of these chambers has tremendously different roles in cardiac physiology. The chamber with the largest mass is the left ventricle, which is responsible for pumping oxygenated blood through the arterial circulatory system to the body. In general, myocardial lipidomics is performed using tissue from either both ventricles, the left ventricle, or affected ventricular zones, such as those that are rendered ischemic.
It is also problematic to consider the disparate lipid composition of the heart. The heart is largely comprised of cardiac muscle cells, endothelial cells and smooth muscle cells. The majority of the mass of the heart is from the cardiac cells, but approximately 27% of the total nuclei in the heart are derived from cells other than the cardiac cell (14). Specific techniques have been developed that minimize the contribution of endothelial cells to the preparation of subcellular membrane fractions from heart cells (15).
Figure 1 is an electron micrograph of a small section of a cardiac cell that contains the cell’s nucleus. This micrograph demonstrates the abundance of myofibrils (MF) and mitochondria (M) in the heart. In comparison, each myocardial cell contains only one nucleus (N). Based on inspection of the ultrastructure of the heart, it is easy to follow that mitochondrial phospholipids are a predominant pool of phospholipids in the heart while the nuclear phospholipid pool is much smaller. Thus, the yield of cardiac mitochondria is much greater than that of cardiac nuclei. Prior to the availability of ESI-MS for lipidomic studies, it would have been virtually impossible to evaluate the nuclear lipidome by techniques such as FAB-MS.
Figure 1.

Electron micrograph of rat heart papillary muscle sample obtained from a isolated perfused rat heart. Original magnification is 9,900X. M, N and MF indicate mitochondria, nucleus and myofibrils, respectively.
Although specific techniques have been developed to specifically isolate subcellular organelles/membranes of the heart, it should be remembered that these organelles are derived from a heterogeneous organ containing functional compartments and multiple cell types. It will be of particular interest in the future to delineate the subcellular lipidomes of the different chambers in the heart. In particular, the atrial and ventricular chambers may have profound differences due to their significant differences in contractile properties.
4. Methodological considerations for the isolation of nuclei
Specific methods have been developed for the isolation of cardiac cell nuclei from whole hearts (14,15). In general, these techniques begin by perfusing the hearts with a crystalline buffer to remove blood-borne cells followed by homogenization and a series of centrifugations, as well as filtering through nylon sieve mesh (100 µm). Since phospholipases have been shown to translocate to myocardial nuclei, it is important to add phospholipase inhibitors such as quinacrine to buffers throughout preparation of nuclei. The isolated nuclei from these preparations should be assessed for purity by marker enzyme/protein analyses and stored in liquid nitrogen until further analyses.
4.1 Purity of subcellular organelle
The purity of the nuclear preparations is verified by immunoblotting with anti-lamin B1 in addition to comparisons with other subcellular markers (16). Figure 2 shows an immunoblot of subcellular markers. Lamin B1 is a marker of the nuclear envelope while SERCA2 ATPase is a marker of the sarcoplasmic reticulum. The absence of SERCA2 ATPase is of particular importance to document the purity of the preparations since the nuclear envelope is continuous with the sarcoplasmic reticulum.
Figure 2.

Immunoblot analyses of marker enzymes. Cytosol, particulate and nuclei were prepared from isolated perfused rat hearts. Cytosolic, particulate, and nuclear proteins were subjected to SDS-PAGE and immunoblot analyses using either anti-lamin B1, anti-SERCA2 ATPase or anti-GATA4 as the primary antibodies.
4.2 Selective isolation of cardiac cell nuclei from whole heart and other considerations
To specifically determine that nuclei are derived from cardiac cells when isolated from whole hearts, it is necessary to assess specific markers of cardiac cell nuclei. As shown in Figure 2, the nuclear preparation is highly enriched with nuclei as shown by the enrichment of lamin B1, and it is specifically enriched with cardiac cell nuclei since it is enriched with GATA4. GATA4 is a transcription factor that is found specifically in cardiac cell nuclei. Comparisons of the amounts of GATA4 (cardiac cell marker) to GATA2 (non-cardiac cell marker) can be used to show the relative enrichment of cardiac and non-cardiac cell nuclei (8,17,18).
The use of marker enzyme analyses for the purity of the nuclear preparations is particularly important in comparisons of nuclear phospholipids in hearts subjected to intense stress such as ischemia, ischemia/reperfusion and infarction. Under these conditions, the subcellular integrity could vary considerably, resulting in disparate isolation and purity of subcellular fractions. It is particularly important to determine the purity of the nuclear preparations by enzyme marker analyses under these conditions prior to assessing alterations in lipid content by mass spectrometry.
5. Recent developments in nuclear membrane lipidomics
Recently, the precise definition of nuclear lipid has been reviewed (19). The nuclear matrix is surrounded by a nuclear envelope that is contiguous with the endoplasmic reticulum of cells. In our studies, we have isolated intact nuclei containing both nuclear membrane and nuclear matrix. Contamination of the endoplasmic reticulum is minimal as shown by the absence of the endoplasmic reticulum marker, SERCA2 ATPase. Several studies have suggested that up to 6% of the cellular phospholipid content is present in the nuclear matrix (20,21). This percentage is likely much lower in the cardiac cell due to the abundance of mitochondria in these cells. It is important however to consider, in future experiments, the dissociation of the lipid content of the nuclear envelope and that of the matrix in cardiac cell nuclei. The possible dynamic changes of nuclear matrix lipid in the cardiac cell and the impact of these lipids on transcription will be important to determine. In fact, many acidic phospholipids have been implicated as important regulators of nuclear signaling. Coupled with this putative role of acidic phospholipids as regulators of transcription are the findings of signal transduction pathways leading to the translocation of phospholipases to the nucleus (22–25).
6. Examples of myocardial nuclear lipidomics
As previously stated, the subcellular lipidomes can vary considerably within a cell and can vary between animal species. In these examples of myocardial lipidomes, we will compare and contrast the choline and ethanolamine glycerophospholipids of the myocardial nuclei from the rat and rabbit. Also within one species, the rabbit, we will compare the ethanolamine glycerophospholipid molecular species present in the nuclear lipidome to that of the mitochondrial lipidome.
6.1 Comparison of rat and rabbit cardiac cell nuclear lipidomes
Figure 3 and Figure 4 show ESI-MS spectra in the positive ion mode from rabbit and rat myocardial nuclei, respectively. The ions that are labeled are the sodium adducts of the choline glycerophospholipid molecular species, as well as those of sphingomyelin. In Figure 3, molecular ions at m/z 764.7, 766.7, 788.7, 790.7 and 816.7 are plasmenylcholine molecular species. These plasmalogens are not present if the sample subjected to ESI-MS is first subjected to a 45 sec treatment with HCl vapors. It is also important to note that plasmenylcholine molecular species are identified more readily by neutral loss scanning of 59.1 compared to neutral loss scanning of 183.1 (26–28). Thus, triple quadrupole analyses and differential appearance of plasmalogens under these two neutral loss scanning modes can be used to identify plasmalogen molecular species. In contrast to rabbit myocardial nuclei, the rat myocardial nuclei are relatively void of plasmalogen molecular species (Figure 4). Rat myocardial nuclear lipid extracts were also subjected to HCl vapor treatment, but no changes in the spectra were observed (data not shown). Additionally, there is little plasmalogen detected in the neutral loss scanning of 59.1 mode. Two other striking differences are also observed in these positive ion scans of rat myocardial nuclei in comparison to rabbit heart nuclei. The choline glycerophospholipid profile from rat heart nuclei also showed that this pool contains more arachidonic acid (20:4) compared to that from the rabbit heart nuclei analyses. For example, molecular ions at m/z 832.7 and 804.6 are the sodiated adducts of 18:0–20:4 and 16:0–20:4 phosphatidylcholine molecular species, respectively. Furthermore 18:0–22:6 choline glycerophospholipid (m/z 856.7) is observed in the profile from rat nuclei (this species was not observed in profiles from rabbit nuclei, data not shown). Also, from these profiles it appears that rat myocardial nuclei contain relatively more sphingomyelin (molecular ions at m/z 725.6 and 753.6) compared to that of the rabbit. These are the sodiated adducts of 16:0 and 18:0 sphingomyelin, respectively.
Figure 3.

Electrospray ionization mass spectrometric analyses of rabbit myocardial nuclear choline glycerophospholipids. Myocardial nuclei were isolated from rabbit ventricles. Nuclei were extracted by Bligh-Dyer extraction and the organic extract was subsequently subjected to ESI-MS. For all spectra, samples were directly infused into the electrospray ionization source at a flow rate of 3 microliters per minute. The top spectrum was acquired in the positive-ion mode directly from a lipid extract that was diluted to less than 20 pmol of total lipids per microliter (4/1, methanol/chloroform) with the addition of approximately 10 pmol NaOH per microliter to the lipid solution. The same samples were also subjected to tandem mass spectrometry with neutral loss (NL) scanning of 59.1 or 183.1 as indicated to specifically identify phospholipids containing the choline polar head group. Spectra were also acquired from lipid extract that was first dried and treated for 45 sec with HCl vapors prior to preparation for mass spectrometry (HCl treated). All mass spectral traces are displayed after normalization to the base peak in each individual spectrum.
Figure 4.

Electrospray ionization mass spectrometric analyses of rat myocardial nuclear choline glycerophospholipids. Myocardial nuclei were isolated from rat ventricles. Nuclei were extracted by Bligh-Dyer extraction and the organic extract was subsequently subjected to ESI-MS. For all spectra, samples were directly infused into the electrospray ionization source at a flow rate of 3 microliters per minute. The top spectrum was acquired in the positive-ion mode directly from a lipid extract that was diluted to less than 20 pmol of total lipids per microliter (4/1, methanol/chloroform) with the addition of approximately 10 pmol NaOH per microliter to the lipid solution. The same samples were also subjected to tandem mass spectrometry with neutral loss (NL) scanning of 59.1 or 183.1 as indicated to specifically identify phospholipids containing the choline polar head group. All mass spectral traces are displayed after normalization to the base peak in each individual spectrum.
Figure 5 and Figure 6 show ESI-MS spectra in the negative ion mode from rabbit and rat myocardial nuclei, respectively. Rabbit myocardial nuclei are enriched with plasmenylethanolamine molecular species in comparison to phosphatidylethanolamine molecular species. Ethanolamine glycerophospholipids were confirmed by precursor ion scanning of 196 in the negative ion mode (27). Pretreatment of lipid extracts with HCl vapors removed plasmalogen molecular species from the spectra. Additionally, rabbit myocardial nuclear ethanolamine glycerophospholipids were highly enriched with arachidonic acid which can be readily identified by precursor ion scanning of 303.3. In comparison to the rabbit myocardial nuclei, less plasmenylethanolamine was present in the rat myocardial nuclei (Fig. 6). Additionally, in comparison to rabbit myocardial nuclei, the ethanolamine glycerophospholipids of rat myocardial nuclei contained molecular species containing substantial amounts of docosahexaenoic acid (identified by precursor ion scanning of 327.4). Taken together, analyses of the phospholipid profiles from rat myocardial nuclear choline and ethanolamine glycerophospholipid pools reveal fewer plasmalogen molecular species, but more molecular species containing arachidonic acid and docosahexaenoic acid in comparison to rabbit.
Figure 5.
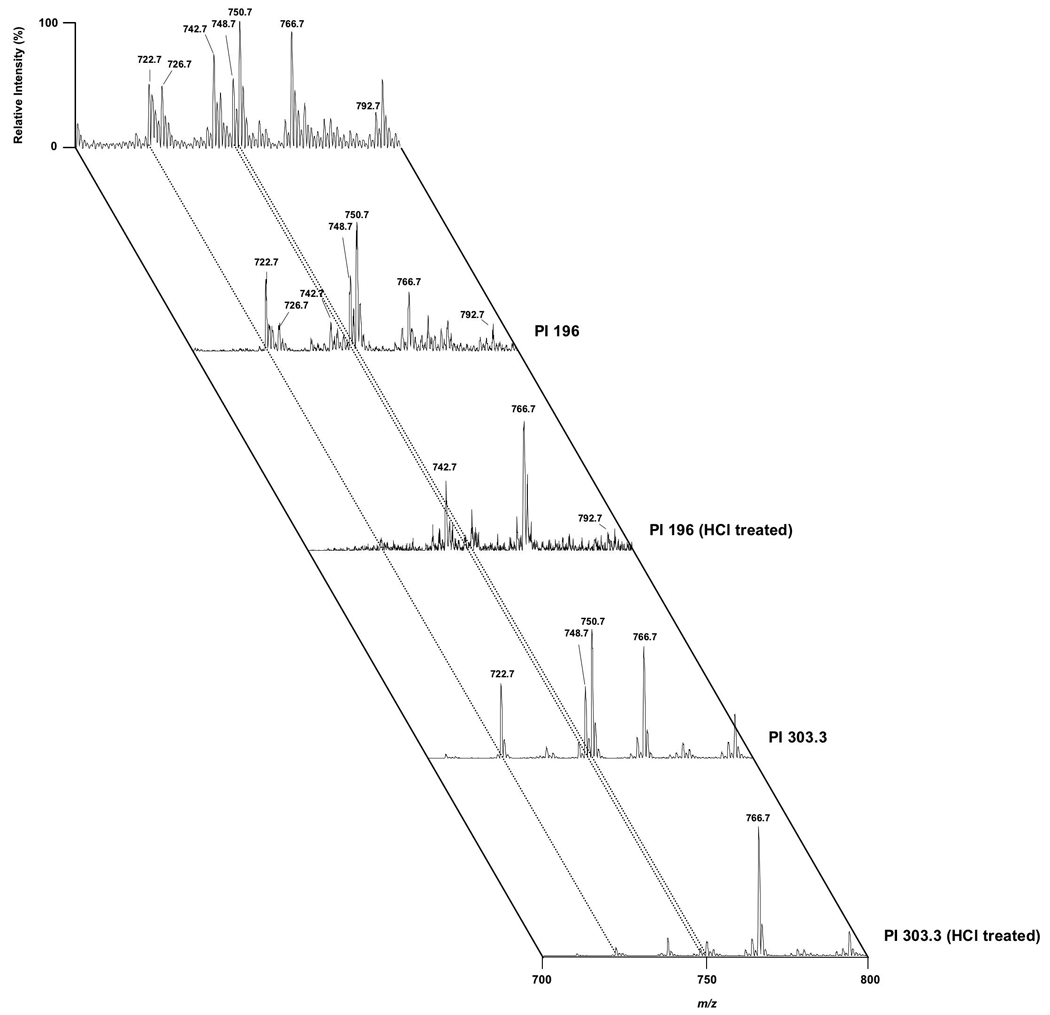
Electrospray ionization mass spectrometric analyses of rabbit myocardial nuclear ethanolamine glycerophospholipids. Myocardial nuclei were isolated from rabbit ventricles. Nuclei were extracted by Bligh-Dyer extraction and the organic extract was subsequently subjected to ESI-MS. For all spectra, samples were directly infused into the electrospray ionization source at a flow rate of 3 microliters per minute. The top spectrum was acquired in the negative-ion mode directly from a lipid extract that was diluted to less than 20 pmol of total lipids per microliter (4/1, methanol/chloroform) with the addition of approximately 10 pmol NaOH per microliter to the lipid solution. The same samples were also subjected to tandem mass spectrometry with precursor ion scanning of either 196, 279.2, 283.2, 303.3 or 327.4 as indicated to specifically identify phospholipids containing the phosphoethanolamine polar head group, esterified linoleic acid, esterified stearic acid, esterified arachidonic acid or esterified docosahexaenoic acid, respectively. Spectra were also acquired from lipid extract that was first dried and treated for 45 sec with HCl vapors prior to preparation for mass spectrometry (HCl treated). All mass spectral traces are displayed after normalization to the base peak in each individual spectrum.
Figure 6.

Electrospray ionization mass spectrometric analyses of rat myocardial nuclear ethanolamine glycerophospholipids. Myocardial nuclei were isolated from rat ventricles. Nuclei were extracted by Bligh-Dyer extraction, and the organic extract was subsequently subjected to ESI-MS. For all spectra, samples were directly infused into the electrospray ionization source at a flow rate of 3 microliters per minute. The top spectrum was acquired in the negative-ion mode directly from a lipid extract that was diluted to less than 20 pmol of total lipids per microliter (4/1, methanol/chloroform) with the addition of approximately 10 pmol NaOH per microliter to the lipid solution. The same samples were also subjected to tandem mass spectrometry with precursor ion scanning of either 196, 279.2, 283.2, 303.3 or 327.4 as indicated to specifically identify phospholipids containing the phosphoethanolamine polar head group, esterified linoleic acid, esterified stearic acid, esterified arachidonic acid or esterified docosahexaenoic acid, respectively. Spectra were also acquired from lipid extract that was first dried and treated for 45 sec with HCl vapors prior to preparation for mass spectrometry (HCl treated). All mass spectral traces are displayed after normalization to the base peak in each individual spectrum.
6.2 Comparison of rabbit cardiac cell nuclear and mitochondrial ethanolamine glycerophospholipid molecular species
Figure 7 shows ESI-MS spectra in the negative ion mode from rabbit myocardial mitochondria. In comparison to the ethanolamine glycerophospholipids of the rabbit myocardial nuclei, the mitochondrial pool contains less plasmalogen. In fact, the ion intensity of 18:0–18:2 phosphatidylethanolamine (m/z 742.6) in the mitochondrial pool is of similar magnitude to that of the plasmenylethanolamine molecular species containing arachidonic acid (m/z 722.5, 748.6 and 750.6). The predominant ethanolamine glycerophospholipid molecular species present in rabbit myocardial mitochondria is18:0–0:4 phosphatidylethanolamine (Fig. 7). In comparison, the rabbit myocardial nuclei (Fig. 5) contains more 18:0–20:4 plasmenylethanolamine (m/z 750.7) compared to 18:0–20:4 phosphatidylethanolamine (m/z 766.7). An abundant cardiolipin ion at m/z 723.6 is observed in the spectra from the myocardial mitochondrial lipids (Fig. 7). This is the double-charged ion of tetra-18:2 cardiolipin.
Figure 7.

Electrospray ionization mass spectrometric analyses of rabbit myocardial mitochondrial ethanolamine glycerophospholipids. Myocardial mitochondria were isolated from rabbit ventricles. Mitochondria were extracted by Bligh-Dyer extraction and the organic extract was subsequently subjected to ESI-MS. For all spectra, samples were directly infused into the electrospray ionization source at a flow rate of 3 microliters per minute. The top spectrum was acquired in the negative-ion mode directly from a lipid extract that was diluted to less than 20 pmol of total lipids per microliter (4/1, methanol/chloroform) with the addition of approximately 10 pmol NaOH per microliter to the lipid solution. The same samples were also subjected to tandem mass spectrometry with precursor ion scanning of either 196, 279.2, 283.2, 303.3 or 327.4 as indicated to specifically identify phospholipids containing the phosphoethanolamine polar head group, esterified linoleic acid, esterified stearic acid, esterified arachidonic acid or esterified docosahexaenoic acid, respectively. Spectra were also acquired from lipid extract that was first dried and treated for 45 sec with HCl vapors prior to preparation for mass spectrometry (HCl treated). All mass spectral traces are displayed after normalization to the base peak in each individual spectrum.
6.3 Discussion of myocardial nuclear lipidomics
The results reported herein are consistent with the phospholipid profiles that have previously been described by our laboratory for rat myocardial nuclear lipids (8). This previous study is, to date, the only extensive analyses of myocardial nuclear phospholipid profiles. The present data expand these findings by comparing the rat myocardial nuclear phospholipid profiles to that of rabbit myocardial nuclei.
The rat and rabbit nuclear phospholipids contain molecular species that contain both arachidonic acid and docosahexaenoic acid. This is consistent with one other study that demonstrated a selective incorporation of radiolabeled arachidonic acid into the nuclear phospholipid pool of metabolically-poisoned cardiac muscle cells (29). In these studies electron microscopic autoradiography was performed. However the specific phospholipid pools that incorporated arachidonic acid under metabolically-poisoned conditions were not determined. With the sensitivity of ESI-MS techniques, it would be of great interest to determine this pool using stable isotope labeled fatty acids. In contrast, others have shown in cell culture that polyunsaturated fatty acids are predominantly esterified to inositol glycerophospholipids in the nucleus while choline glycerophospholipids are comprised predominantly of molecular species containing saturated fatty acids (19,20). It is possible that the disparate phospholipid profiles of rat and rabbit nuclei compared to nuclei from neuroblastoma cells is due to differentiation of cell culture lines compared to primary tissue, differences in neural and myocardial tissue and differences in animal species.
7. Summary and perspective
Subcellular lipidomes represent an exciting area of investigation. The specific lipids in subcellular organelles in many instances have critical physiological roles that ultimately impact the function of the entire cell, tissue and organism. Within the nuclear lipidome, the lipidomes of the nuclear envelope and the nuclear matrix remain to be elucidated. The nuclear envelope lipids may represent an important pool of precursor lipids that are transported to the nuclear matrix, as well as targets for extranuclear phospholipases. Our past and current studies have elucidated the myocardial nuclear lipidome. Nuclei possess many choline and ethanolamine glycerophospholipid molecular species, and rabbit myocardial nuclei contain abundant plasmalogen species in both of these pools. Future studies will be directed toward delineating the phospholipid classes and molecular species in the nuclear matrix from cardiac cells and determining how they are dynamically altered under the severe stresses that can be placed on the heart such as ischemia/reperfusion, tachycardia and hypertrophy.
Acknowledgements
This research was supported by NIH grants HL 74214 (DAF), RR19232 (DAF) and Grant-in-Aid 0650044Z (DAF) from the American Heart Association.
Abbreviations
- ESI
electrospray ionization
- FAB
fast atom bombardment
- GC
gas chromatography
- HPLC
high performance liquid chromatography
- MS
mass spectrometry
- NL
neutral loss
- PI
precursor ion
References
- 1.Hale CC, Ebeling EG, Hsu FF, Ford DA. The selective activation of the cardiac sarcolemmal sodium-calcium exchanger by plasmalogenic phosphatidic acid produced by phospholipase D. FEBS Letters. 1998;422:247–251. doi: 10.1016/s0014-5793(98)00017-9. [DOI] [PubMed] [Google Scholar]
- 2.Ford DA, Hale CC. Plasmalogen and anionic phospholipid dependence of the cardiac sarcolemmal sodium-calcium exchanger. FEBS Letters. 1996;394:99–102. doi: 10.1016/0014-5793(96)00930-1. [DOI] [PubMed] [Google Scholar]
- 3.Corr PB, Snyder DW, Cain ME, Crafford WA, Jr, Gross RW, Sobel BE. Electrophysiological effects of amphiphiles on canine purkinje fibers. Implications for dysrhythmia secondary to ischemia. Circ Res. 1981;49:354–363. doi: 10.1161/01.res.49.2.354. [DOI] [PubMed] [Google Scholar]
- 4.Katz AM, Messineo FC. Lipid-membrane interactions and the pathogenesis of ischemic damage in the myocardium. Circ Res. 1981;48:1–16. doi: 10.1161/01.res.48.1.1. [DOI] [PubMed] [Google Scholar]
- 5.Vlasova II, Tyurin VA, Kapralov AA, Kurnikov IV, Osipov AN, Potapovich MV, Stoyanovsky DA, Kagan VE. Nitric oxide inhibits peroxidase activity of cytochrome c.cardiolipin complex and blocks cardiolipin oxidation. J Biol Chem. 2006;281:14554–14562. doi: 10.1074/jbc.M509507200. [DOI] [PubMed] [Google Scholar]
- 6.Petrosillo G, Di Venosa N, Ruggiero FM, Pistolese M, D'Agostino D, Tiravanti E, Fiore T, Paradies G. Mitochondrial dysfunction associated with cardiac ischemia/reperfusion can be attenuated by oxygen tension control. Role of oxygen-free radicals and cardiolipin. Biochim Biophys Acta. 2005;1710:78–86. doi: 10.1016/j.bbabio.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Williams SD, Gottlieb RA. Inhibition of mitochondrial calcium-independent phospholipase A2 (iPLA2) attenuates mitochondrial phospholipid loss and is cardioprotective. Biochem J. 2002;362:23–32. doi: 10.1042/0264-6021:3620023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams SD, Hsu FF, Ford DA. Electrospray ionization mass spectrometry analyses of nuclear membrane phospholipid loss after reperfusion of ischemic myocardium. J Lipid Res. 2000;41:1585–1595. [PubMed] [Google Scholar]
- 9.Gross RW. Identification of plasmalogen as the major phospholipid constituent of cardiac sarcoplasmic reticulum. Biochemistry. 1985;24:1662–1668. doi: 10.1021/bi00328a014. [DOI] [PubMed] [Google Scholar]
- 10.Gross RW. High plasmalogen and arachidonic acid content of canine myocardial sarcolemma: a fast atom bombardment mass spectroscopic and gas chromatography-mass spectroscopic characterization. Biochemistry. 1984;23:158–165. doi: 10.1021/bi00296a026. [DOI] [PubMed] [Google Scholar]
- 11.Post JA, Verkleij AJ, Roelofsen B, Op de Kamp JA. Plasmalogen content and distribution in the sarcolemma of cultured neonatal rat myocytes. FEBS Letters. 1988;240:78–82. doi: 10.1016/0014-5793(88)80343-0. [DOI] [PubMed] [Google Scholar]
- 12.Han X, Gross RW. Electrospray ionization mass spectroscopic analysis of human erythrocyte plasma membrane phospholipids. Proc Nat Acad Sci USA. 1994;91:10635–10639. doi: 10.1073/pnas.91.22.10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hazen SL, Hall CR, Ford DA, Gross RW. Isolation of a human myocardial cytosolic phospholipase A2 isoform. Fast atom bombardment mass spectroscopic and reverse-phase high pressure liquid chromatography identification of choline and ethanolamine glycerophospholipid substrates. J Clin Invest. 1993;91:2513–2522. doi: 10.1172/JCI116487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackowski G, Liew CC. Fractionation of rat ventricular nuclei. Biochem J. 1980;188:363–373. doi: 10.1042/bj1880363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liew CC, Jackowski G, Ma T, Sole MJ. Nonenzymatic separation of myocardial cell nuclei from whole heart tissue. Am J Physiol. 1983;244:C3–C10. doi: 10.1152/ajpcell.1983.244.1.C3. [DOI] [PubMed] [Google Scholar]
- 16.Albert CJ, Ford DA. Identification of specific nuclear protein kinase C isozymes and accelerated protein kinase C-dependent nuclear protein phosphorylation during myocardial ischemia. FEBS Letters. 1998;438:32–36. doi: 10.1016/s0014-5793(98)01264-2. [DOI] [PubMed] [Google Scholar]
- 17.Dorfman DM, Wilson DB, Bruns GA, Orkin SH. Human transcription factor GATA-2. Evidence for regulation of preproendothelin-1 gene expression in endothelial cells. J Biol Chem. 1992;267:1279–1285. [PubMed] [Google Scholar]
- 18.Molkentin JD, Kalvakolanu DV, Markham BE. Transcription factor GATA-4 regulates cardiac muscle-specific expression of the alpha-myosin heavy-chain gene. Molec Cell Biol. 1994;14:4947–4957. doi: 10.1128/mcb.14.7.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunt AN. Dynamic lipidomics of the nucleus. J Cell Biochem. 2006;97:244–251. doi: 10.1002/jcb.20691. [DOI] [PubMed] [Google Scholar]
- 20.Hunt AN, Clark GT, Attard GS, Postle AD. Highly saturated endonuclear phosphatidylcholine is synthesized in situ and colocated with CDP-choline pathway enzymes. J Biol Chem. 2001;276:8492–8499. doi: 10.1074/jbc.M009878200. [DOI] [PubMed] [Google Scholar]
- 21.Vann LR, Wooding FB, Irvine RF, Divecha N. Metabolism and possible compartmentalization of inositol lipids in isolated rat-liver nuclei. Biochem J. 1997;327:569–576. doi: 10.1042/bj3270569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raben DM, Baldassare JJ. Phospholipid metabolism and nuclear envelope signaling. Adv Enz Regul. 2000;40:97–123. doi: 10.1016/s0065-2571(99)00023-0. [DOI] [PubMed] [Google Scholar]
- 23.Baldassare JJ, Jarpe MB, Alferes L, Raben DM. Nuclear translocation of RhoA mediates the mitogen-induced activation of phospholipase D involved in nuclear envelope signal transduction. J Biol Chem. 1997;272:4911–4914. doi: 10.1074/jbc.272.8.4911. [DOI] [PubMed] [Google Scholar]
- 24.Su X, Dowhan W. Translational regulation of nuclear gene COX4 expression by mitochondrial content of phosphatidylglycerol and cardiolipin in Saccharomyces cerevisiae. Molec Cell Biol. 2006;26:743–753. doi: 10.1128/MCB.26.3.743-753.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stallings JD, Tall EG, Pentyala S, Rebecchi MJ. Nuclear translocation of phospholipase C-delta1 is linked to the cell cycle and nuclear phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 2005;280:22060–22069. doi: 10.1074/jbc.M413813200. [DOI] [PubMed] [Google Scholar]
- 26.Han X, Gross RW. Structural determination of picomole amounts of phospholipids via electrospray ionization tandem mass spectrometry. J Am Soc Mass Spectrom. 1995;6:1202–1210. doi: 10.1016/1044-0305(95)00568-4. [DOI] [PubMed] [Google Scholar]
- 27.Brugger B, Erben G, Sandhoff R, Wieland FT, Lehmann WD. Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc Nat Acad Sci USA. 1997;94:2339–2344. doi: 10.1073/pnas.94.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu FF, Bohrer A, Turk J. Formation of lithiated adducts of glycerophosphocholine lipids facilitates their identification by electrospray ionization tandem mass spectrometry. J Am Soc Mass Spectrom. 1998;9:516–526. doi: 10.1016/S1044-0305(98)00012-9. [DOI] [PubMed] [Google Scholar]
- 29.Miyazaki Y, Gross RW, Sobel BE, Saffitz JE. Selective turnover of sarcolemmal phospholipids with lethal cardiac myocyte injury. Am J Physiol. 1990;259:C325–C331. doi: 10.1152/ajpcell.1990.259.2.C325. [DOI] [PubMed] [Google Scholar]


