Abstract
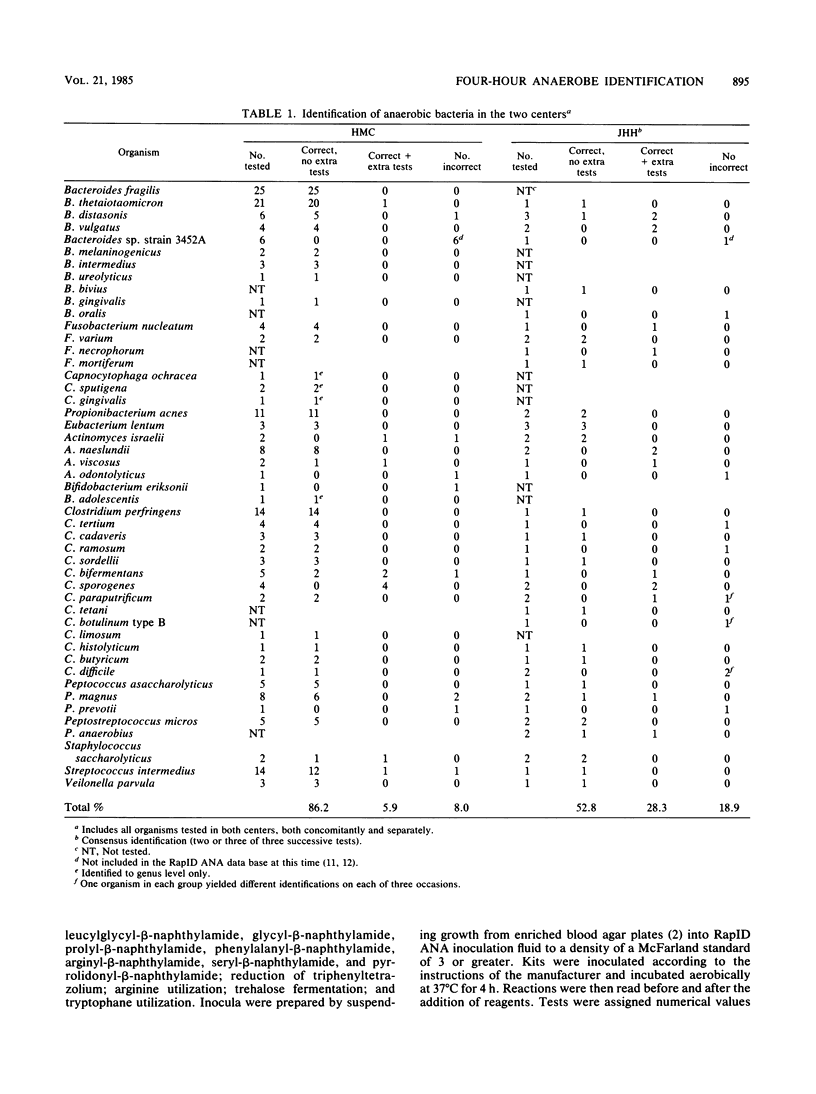
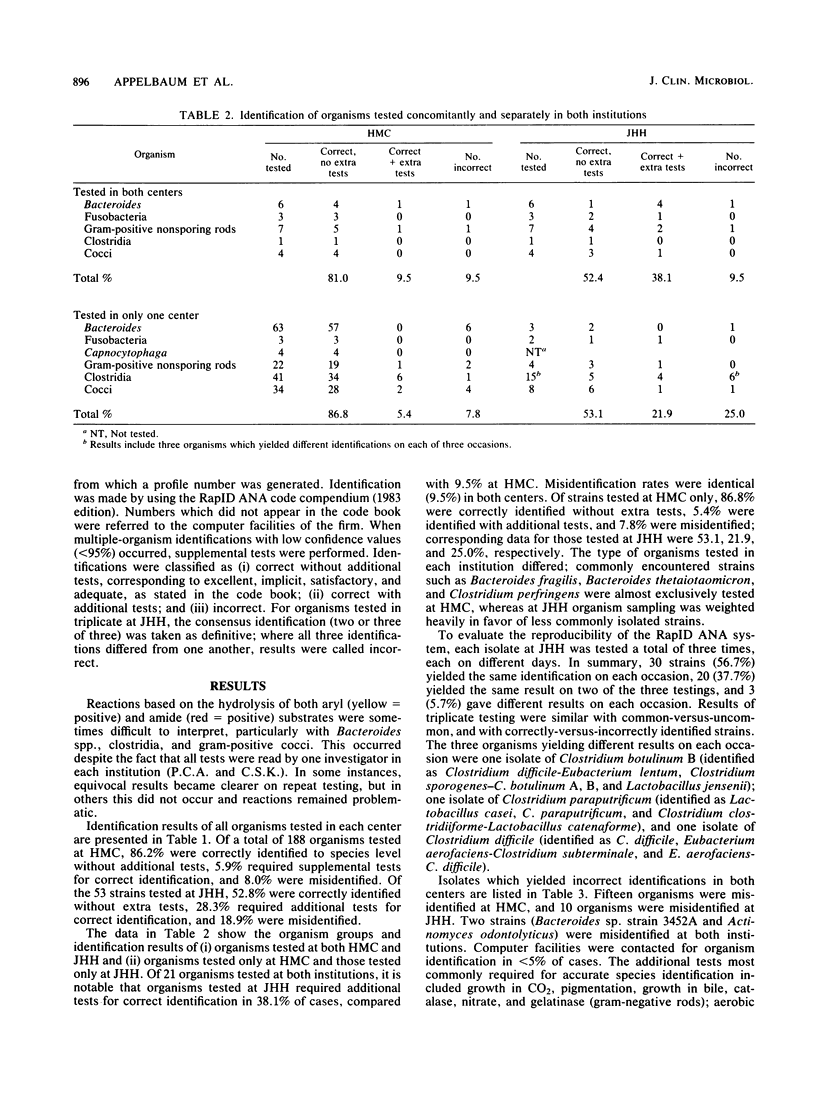
In this study, we evaluated the ability of a 4-h enzyme assay kit system, the RapID ANA method (Innovative Diagnostic Systems, Inc., Atlanta, Ga.) to accurately and reproducibly identify a spectrum of clinically significant anaerobic bacteria in two separate institutions. Additional tests were performed as required. Of a total of 188 organisms tested at Hershey Medical Center (HMC), 86.2% were correctly identified to species level without additional tests, 5.9% required extra tests for correct identification, and 8.0% were misidentified. Of 53 strains tested at Johns Hopkins Hospital (JHH), 52.8% were correctly identified without extra tests, 28.3% required extra tests for correct identification, and 18.9% were misidentified. Of 21 organisms tested at both institutions, those tested at JHH required additional tests for correct identification in 38.1% of cases, compared with 9.5% at HMC. Misidentification rates were identical (9.5%) in both centers. Of strains tested at HMC only, 86.8% were correctly identified without extra tests, 5.4% were identified with additional tests, and 7.8% were misidentified: corresponding data for JHH were 53.1, 21.9, and 25.0%, respectively. Of 53 strains tested in triplicate at JHH, 56.7% yielded the same result on each occasion, 37.7% were identical in two of three tests, and 5.7% gave different results on each of three occasions. Discrepancies between identification rates at HMC and JHH may be explained by differences in species tested (more commonly encountered species were tested at HMC) and interpretation of reactions by the two different readers. The RapID ANA method has the potential for rapid identification of clinically isolated anaerobes; however, accuracy and reproducibility may vary as a function of the specific laboratory setting.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Appelbaum P. C., Kaufmann C. S., Keifer J. C., Venbrux H. J. Comparison of three methods for anaerobe identification. J Clin Microbiol. 1983 Sep;18(3):614–621. doi: 10.1128/jcm.18.3.614-621.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edson R. S., Rosenblatt J. E., Lee D. T., McVey E. A., 3rd Recent experience with antimicrobial susceptibility of anaerobic bacteria: increasing resistance to penicillin. Mayo Clin Proc. 1982 Dec;57(12):737–741. [PubMed] [Google Scholar]
- George W. L., Kirby B. D., Sutter V. L., Citron D. M., Finegold S. M. Gram-negative anaerobic bacilli: Their role in infection and patterns of susceptibility to antimicrobial agents. II. Little-known Fusobacterium species and miscellaneous genera. Rev Infect Dis. 1981 May-Jun;3(3):599–626. doi: 10.1093/clinids/3.3.599. [DOI] [PubMed] [Google Scholar]
- Hansen S. L., Stewart B. J. Comparison of API and Minitek to Center for Disease Control methods for the biochemical characterization of anaerobes. J Clin Microbiol. 1976 Sep;4(3):227–231. doi: 10.1128/jcm.4.3.227-231.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson C. W., Cassorla R., Martin W. J. API and Minitek systems in identification of clinical isolates of anaerobic gram-negative bacilli and Clostridium species. J Clin Microbiol. 1979 Jul;10(1):14–18. doi: 10.1128/jcm.10.1.14-18.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway Y., Dankert J. Identification of anaerobes on the Minitek System, compared to a conventional system. Zentralbl Bakteriol Orig A. 1979;245(3):324–331. [PubMed] [Google Scholar]
- Karachewski N. O., Busch E. L., Wells C. L. Comparison of PRAS II, RapID ANA, and API 20A systems for identification of anaerobic bacteria. J Clin Microbiol. 1985 Jan;21(1):122–126. doi: 10.1128/jcm.21.1.122-126.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard G. L., Whaley D. N., Dowell V. R., Jr Comparison of media in the Anaerobe-Tek and Presumpto plate systems and evaluation of the Anaerobe-Tek system for identification of commonly encountered anaerobes. J Clin Microbiol. 1982 Dec;16(6):1066–1072. doi: 10.1128/jcm.16.6.1066-1072.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore H. B., Sutter V. L., Finegold S. M. Comparison of three procedures for biochemical testing of anaerobic bacteria. J Clin Microbiol. 1975 Jan;1(1):15–24. doi: 10.1128/jcm.1.1.15-24.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stargel D., Thompson F. S., Phillips S. E., Lombard G. L., Dowell V. R., Jr Modification of the Minitek Miniaturized Differentiation System for characterization of anaerobic bacteria. J Clin Microbiol. 1976 Mar;3(3):291–301. doi: 10.1128/jcm.3.3.291-301.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr S. E., Thompson F. S., Dowell V. R., Jr, Balows A. Micromethod system for identification of anaerobic bacteria. Appl Microbiol. 1973 May;25(5):713–717. doi: 10.1128/am.25.5.713-717.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]


