Abstract
We have integrated single molecule fluorescence microscopy imaging into an optical tweezers set-up and studied the force extension behavior of individual DNA molecules in the presence of various YOYO-1 and YO-PRO-1 concentrations. The fluorescence modality was used to record fluorescent images during the stretching and relaxation cycle. Force extension curves recorded in the presence of either dye did not show the overstretching transition that is characteristic for bare DNA. Using the modified wormlike chain model to curve-fit the force extension data revealed a contour length increase of 6% and 30%, respectively, in the presence of YO-PRO-1 and YOYO-1 at 100 nM. The fluorescence images recorded simultaneously showed that the number of bound dye molecules increased as the DNA molecule was stretched and decreased again as the force on the complex was lowered. The binding constants and binding site sizes for YO-PRO-1 and YOYO-1 were determined as a function of the force. The rate of YO-PRO-1 binding and unbinding was found to be 2 orders of magnitude larger than that for YOYO-1. A kinetic model is proposed to explain this observation.
Introduction
Intercalation is a binding mode in which small molecules insert their planar aromatic moiety between two adjacent basepairs (bp) of double-stranded DNA (1). As a result of this binding mode the length of the DNA molecule increases, and may disturb processes such as transcription, replication, and DNA repair, which involve specific interactions between proteins and DNA (2). Some fluorescent intercalators are used in single DNA molecule fluorescence studies. Other intercalators, such as cisplatin, because of their specific property of interfering with DNA-protein interactions, are used in anticancer drug formulations (3). Understanding the interaction kinetics of these intercalating agents to double-stranded DNA is of utmost importance in the development of more specific and efficient drugs (4).
The dimer YOYO-1 (YOYO) and monomer YO-PRO-1 (YO) are oxazole yellow dyes (5–7), that bind to DNA through intercalation. They are used widely in single molecule fluorescence imaging of DNA (8–10). The dyes are virtually nonfluorescent in solution, but undergo a strong increase in fluorescence quantum yield of a few orders of magnitude when bound to DNA (5,6). YO is a mono-intercalator, which consists of only a single aromatic moiety. In the case of YOYO, which consists of two aromatic moieties connected with a linker, both moieties intercalate between bp leaving one position in between unoccupied (bis-intercalation) (10). The estimated binding constant for YOYO to DNA is 1010–1012 M−1. YO has a lower affinity (11).
Absorption spectroscopy, linear dichroism, and 1H NMR spectroscopy have been used to characterize the structure, binding mode, optical, and photophysical properties of the dye molecules, both free and bound to DNA (7,10,11). Single molecule techniques such as optical tweezers and atomic force microscopy have probed the mechanical properties of individual DNA molecules in interaction with YO and YOYO and other intercalating agents (12–15). These studies have shown that depending on the intercalator concentration the overstretching plateau, typically observed in stretching bare DNA, shortens, tilts, and disappears eventually. In a recent study by Vladescu et al. (16,17) the mechanical properties of DNA interacting with ethidium were studied. It was shown that the binding constant of the intercalating molecules increases from 4.6 × 105 M−1 at zero force to 5.5 × 107 M−1 at a force of 80 pN. Furthermore it was found that the size of the binding site of ethidium reduced from 2.4 bp to 1.7 bp.
In this study, we present a single molecule study using optical tweezers combined with fluorescence microscopy imaging in which we study the interaction of individual DNA molecules with the oxazole yellow dyes YO and YOYO. The mechanical properties of individual DNA molecules at various dye concentrations have been measured. From these force extension data we calculated fractional elongation and fitted this with a McGhee-von Hippel binding isotherm (18). This fit resulted in the binding constants and the binding site sizes as a function of force for both dye molecules.
For a 100 nM concentration of YO and YOYO we have simultaneously measured the force and the total fluorescence intensity and directly related the number of fluorescent molecules bound to the DNA to the force applied. These data showed clearly that the number of fluorophores increases as the force increases. They also showed that the kinetics of YOYO binding was relatively slow. Force relaxation experiments were carried out to get more detailed insight into these kinetics of dye molecules binding to DNA. The binding and unbinding rate of YOYO was two orders of magnitude lower than for YO. We propose a model that describes and explains the interaction kinetics measured for the intercalating oxazole yellow molecules.
Materials and Methods
The optical tweezers instrument has been described previously (15). For the fluorescence microscope, a 10 mW laser with a wavelength of 488 nm (model 161C-01; Spectra Physics, Mountain View, CA) was used. The laser beam was expanded in one direction using a cylindrical lens (f = 40 mm), such that the line-shaped illumination pattern had dimensions of 0.3 × 3.5 μm2 in the focal plane of the objective. A scanning mirror (AE1000; General Scanning, Watertown, MA) was added to scan the line-shaped beam at a frequency of 12.5 Hz. Maximum amplitude of the movement of the beam in the focal plane was 40 μm, which was sufficient to illuminate the full length of extended λ-DNA. This was realized by imaging the center of the scanning mirror onto the back aperture of the objective using two lenses in a 4f lens configuration (19). A 45° dichroic mirror, which reflects from 400–700 nm and transmits from 800–1100 nm, combines the laser beam for optical trapping and the excitation laser beam for fluorescence. The fluorescence emission is collected by the objective and separated from the excitation light using a second dichroic mirror, which reflects below 495 nm and transmits above 505 nm. Fluorescence images were acquired with an EMCCD camera (iXon DV887-BV; Andor, Belfast, Northern Ireland).
Bacteriophage λ-DNA with a contour length of 16.4 μm was obtained from New England BioLabs (Ipswich, MA). The 12 bp single-stranded DNA overhangs were biotinylated with the use of Klenow DNA polymerase and biotinylated nucleotides (Sigma, St. Louis, MO) (15). Single DNA molecules were attached between two streptavidin-coated 2.6-μm beads (Bangs Laboratories, Fishers, IN) in a flow cell, as described elsewhere (15). One bead was trapped in the optical trap and a second bead was attached to a glass micropipette. The flow cell was mounted on a piezo-stage (P-561.3CD; Physik Instruments, Karlsruhe, Germany). The stage moves the micropipette bead with respect to the bead in the trap, which results in an extension or relaxation of the DNA molecule. Experiments were carried out in buffer solution that contains 10 mM Tris-HCl, pH 7.5, 1 mM ethylene-diaminetetraacetic acid supplemented with 150 mM NaCl, and 0.05% NaN3. After successful attachment of a single DNA molecule between two beads, the buffer was replaced by an identical buffer supplemented with various concentrations of YO or YOYO (Invitrogen-Molecular Probes, Carlsbad, CA). The dye-buffer was bubbled with nitrogen gas for 1 h to remove dissolved oxygen to reduce the occurrence of DNA strand breaks during the stretching of the molecule (20,21).
Force spectroscopy experiments were carried out at a rate of 3 μm/s if no fluorescent images were recorded. In the experiments in which the fluorescence was recorded simultaneously, a rate of 1 μm/s was used; fluorescence images were acquired at 10 Hz, in the presence of 100 nM YOYO and YO. The fluorescence intensities were obtained from the images by integrating for every frame the light intensity in a fixed-width rectangle with a length equal to the DNA length. This rectangle enclosed the entire DNA molecule at all extensions. An identical rectangle was selected in a dark region, in which the total intensity was used for background correction. Fluorescence kymographs were constructed from the sequence of images, as described in Waterman-Storer et al. (22).
Results and Discussion
Interaction of YOYO with DNA
Force spectroscopy was carried out on individual DNA molecules in the absence and presence of various concentrations of YOYO (100, 200, 500, and 1000 nM). Fig. 1 A shows the force extension/relaxation curves. In the absence of dye molecules, the overstretching transition was observed at ∼65 pN and hysteresis was observed between 17 and 24 μm during relaxation. The contour length (16.4 ± 0.2 μm), persistence length (52 ± 4 nm), and stretch modulus (1100 ± 100 pN) were determined from the force extension data with the help of the modified worm-like chain model (MWLC) (23). These results are in agreement with published results (24,25). Fig. 1 A shows the force extension curves of DNA in the presence of various concentrations of YOYO. These force extension curves were remarkably different from the force extension curve of a bare dsDNA molecule. The overstretching transition was not observed with YOYO at these concentrations up to 110 pN, and the contour length increased as function of the dye concentration in the surrounding buffer (Table 1 and Fig. S1 A in the Supporting Material). Fig. 1 B presents force extension curves of DNA-YOYO complex at 100 nM YOYO pulled at different speeds. These curves have also been curve-fitted with the MWLC and the results are presented in Table 1. These changes were qualitatively similar to those observed by Sischka et al. (14), who carried out force spectroscopy of DNA molecules at a YOYO concentration of 1 μM. Different amounts of hysteresis were observed in the force extension curve when recorded in the presence of YOYO, but these were of an entirely different nature as observed in the absence of the dye, in agreement with earlier reports (14,15). The dependence of the force extension curve on the pulling rate suggests that DNA in the presence of YOYO is not in equilibrium during stretching and relaxation and that the binding kinetics of YOYO to DNA is slow with respect to the time of the experiment.
Figure 1.
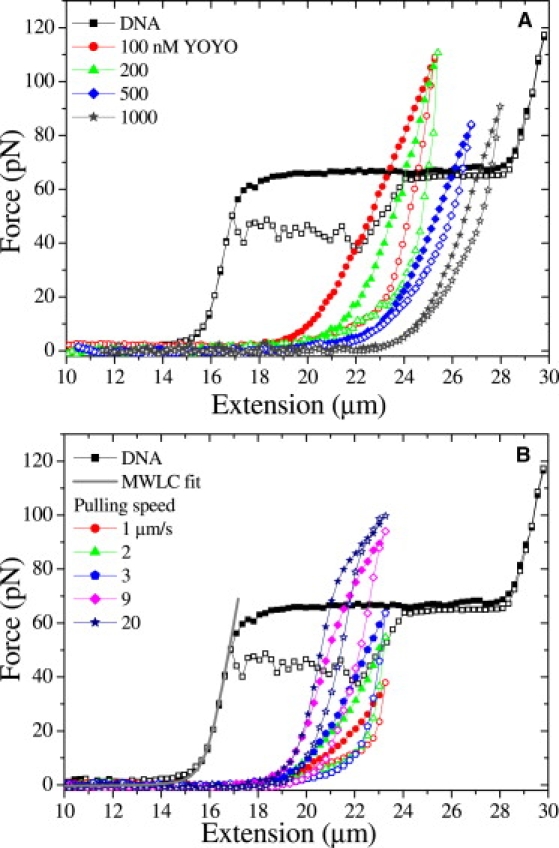
(A) Force extension curves of a single λ-DNA in the presence of various concentrations of YOYO molecules in the surrounding buffer pulled at 3 μm/s. (B) Force extension curves of DNA-YOYO complex in the presence of 100 nM YOYO pulled at different pulling speeds. Solid and open symbols present extension and relaxation respectively. The curve fitting MWLC model to the extension part of the λ-DNA gray line.
Table 1.
Parameters obtained by curve fitting the extension part by the MWLC model to the force extension of the DNA-YOYO complex at various concentrations of YOYO and at 100 nM YOYO pulled at different pulling speeds
| Concentration YOYO (nM) | Persistence length (nm) | Contour length (μm) | Stretch modulus (pN) |
|---|---|---|---|
| 0 | 52.00 | 16.40 | 1100 |
| 100 | 16.70 | 21.32 | 577 |
| 200 | 12.90 | 22.97 | 783 |
| 500 | 14.90 | 24.67 | 786 |
| 1000 | 30.10 | 25.85 | 986 |
| Pulling speed (μm/s) | |||
| 1 | 8.80 | 22.38 | 415 |
| 2 | 14.40 | 21.37 | 443 |
| 3 | 16.70 | 21.32 | 577 |
| 9 | 25.60 | 20.36 | 943 |
| 20 | 32.10 | 20.29 | 1120 |
The actual fits are presented in Fig. S1.
Fig. 2 A shows a number of fluorescence images taken out of a video sequence recorded during the stretching and relaxation of DNA in the presence of 100 nM YOYO. The total fluorescence intensity of the DNA-dye complex is determined from these images (see Materials and Methods) and plotted as a function of force (Fig. 2 B). It is evident from these data that the fluorescence intensity of the complex increases with force. The intensity is ∼3 times higher at a force of 55 pN than at a force of 5 pN. The fluorescence intensity returns to its original value after a full cycle, indicating full reversibility. The hysteresis observed in this plot again indicates that the DNA-YOYO complex is not in equilibrium with the free YOYO in the surrounding buffer during the stretching and relaxation cycle. Fig. 2 C shows the results in a kymograph.
Figure 2.
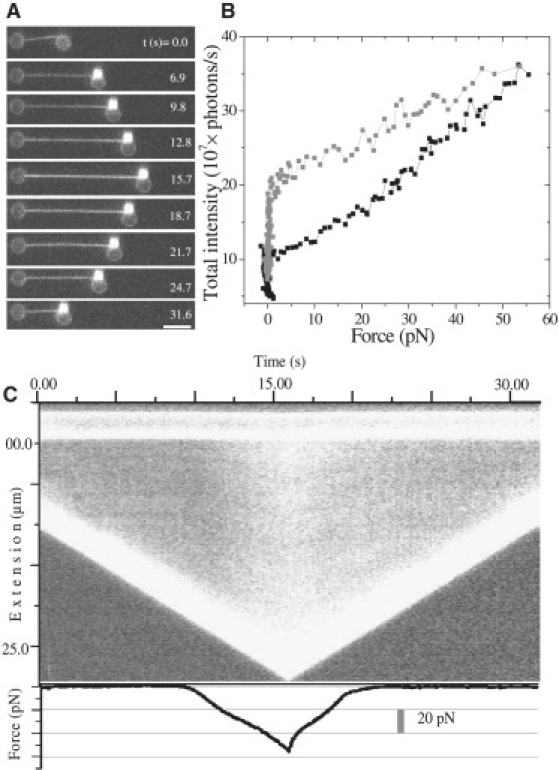
(A) Fluorescence images taken from a sequence recorded during the extension and relaxation of the DNA-YOYO complex in the presence of 100 nM YOYO (pulling speed = 1 μm/s, scale bar = 5 μm). (B) Total fluorescence intensity plotted as a function of force (black squares, extension; gray squares, relaxation). (C) Kymograph created from the sequence of images recorded simultaneously while carrying out force spectroscopy, where the horizontal axis represents time, and the vertical axis is extension. The two bright lines in the kymograph represent the streptavidin coated polystyrene beads at either end of the DNA molecules. The force on the complex is plotted as a function of time in the plot below the kymograph.
The fluorescence intensity is linearly proportional to the number of YOYO molecules when one assumes a single, intercalative binding mode of YOYO with DNA that contributes significantly to the fluorescence (26). The results observed here can be explained by assuming a binding constant that is force dependent. The hysteresis observed in the force extension curves measured at different pulling rates is the result of the relatively low binding and unbinding rates. The rates are too low to achieve equilibrium during the stretching and relaxation cycle, even at a pulling rate as low as 1 μm/s.
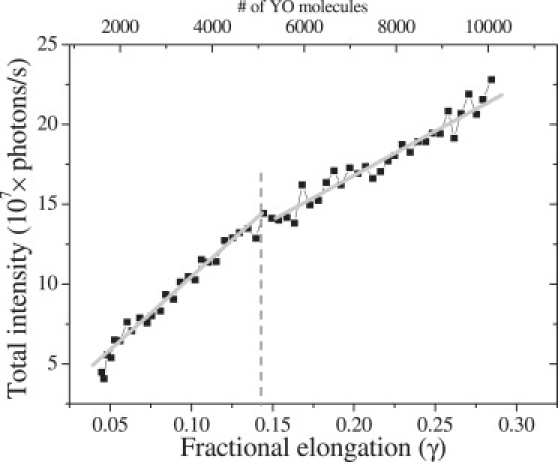
Apart from the fluorescence intensity, the number of intercalated dye molecules can also be extracted from the molecule's elongation. Fractional elongation γ of a DNA molecule due to the intercalation of dye molecules is defined in Eq. 1 (17):
| (1) |
Here xcomplex(F) and xDNA(F) represents the length of DNA at force F, respectively in the presence and absence of YOYO/YO. In Fig. 3 the fractional elongation is plotted versus the fluorescence intensity, and a linear relationship was indeed established. Because each intercalating YOYO molecule increases the contour length of DNA by 0.884 nm (10), one can calculate the number of molecules bound to DNA using Eq. 2:
| (2) |
Figure 3.
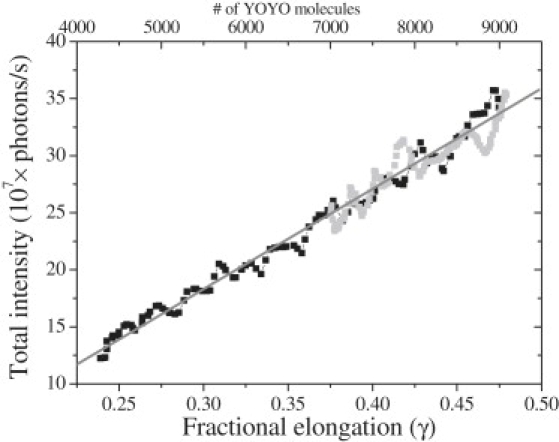
Total fluorescence intensity versus the fractional elongation obtained during the stretching (black squares) and relaxation (gray squares) of a DNA molecule at 1 μm/s in 100 nM YOYO. The number of YOYO molecules (top x axis) calculated using Eq. 2. Solid gray line presents the linear fit to the data.
Using this value, the fluorescence emission per YOYO molecule is calculated to be (41.7 ± 0.8) × 103 photons/s at 0.4 kW/cm2 power in the focal plane.
To acquire quantitative information on the kinetics of the YOYO-DNA interaction, DNA was stretched to different preset extensions with a pulling rate of 3 μm/s in the presence of 100 nM YOYO. Subsequently the force was recorded whereas the extension remained constant. Fig. 4 A shows force relaxation results for different constant extensions (21.5, 22.5, 23.5, 24.5, and 25.5 μm). The force drops exponentially to a lower value (fitted curves are presented in Fig. S2). In all cases the force dropped to 58.8 ± 5.0% of its initial value independent of the initial force. Changes in the pulling rate, however, did have an effect on this relative drop in force. When pulling rates were varied from 1 to 30 μm/s, the drop in force was observed to increase linearly for pulling rates up to 5 μm/s. At higher pulling rates this relative drop in force remained constant at 67.8 ± 0.5% (Fig. S3).
Figure 4.
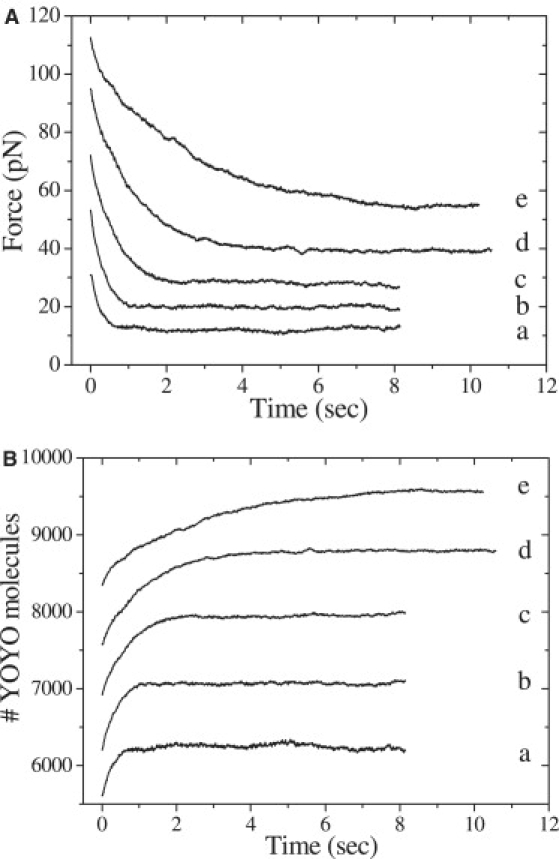
(A) Force as a function of time recorded as the DNA molecule is left to equilibrate after being pulled to different extensions at 3 μm/s pulling speed in the presence of 100 nM YOYO. Curves a–e represent constant extension of 21.5, 22.5, 23.5, 24.5, and 25.5 μm, respectively. (B) The number of YOYO molecules bound to DNA as function of time during force relaxation calculated using Eq. 2.
The force as a function of time (Fig. 4 A) was fitted with a single exponential function (Fig. S2) to acquire a characteristic time constant τFR, which is presented in Table 2. The exponential decrease in force reflects the binding of YOYO molecules to restore equilibrium. The larger extension of the complex leads to a larger initial force and a larger τFR. The number of bound YOYO molecules during the force relaxation was calculated using Eq. 2 and plotted as a function of time (Fig. 4 B). These curves were fitted using a single exponential function, which resulted in time constants that were in agreement with the τFR.
Table 2.
Characteristic time constant as function of the extension, obtained by fitting the force relaxation curves in Fig. 4A with a single-exponential function
| Extension (μm) | 21.5 | 22.5 | 23.5 | 24.5 | 25.5 |
| Time constant τFR (s) | 0.23 | 0.33 | 0.64 | 1.13 | 2.54 |
The DNA-YOYO complex was pulled to the present extension at 3 μm/s pulling speed. Fitted curves are presented in Fig. S2.
Interaction of YO with DNA
Force spectroscopy was carried out on DNA in the presence of various concentrations of YO, and the force extension behavior is presented in Fig. 5 A. The overstretching transition in DNA was not observed in the presence of the mono-intercalator YO at these concentrations up to 100 pN. Force extension curves were curve fitted with the MWLC model (Fig. S4) and the parameters obtained are presented in Table 3. The contour length of the DNA increases as the YO concentration increases. Between the stretching and relaxation curve no hysteresis was observed. At 100 nM YO in the enthalpic stretching regime, the force extension curve does reveal a change in slope from 13.9 pN/μm to 9.4 pN/μm at extensions beyond 18 μm (Fig. 5 B). This change in slope is also observed for 200 nM, but disappears at higher YO concentrations (Fig. 5 A). A similar change of slope was also observed in Vladescu et al. (17). Varying the pulling rate from 1 to 20 μm/s did not lead to significant differences in force extension behavior (Fig. 5 B), indicating that the binding of YO to DNA is in equilibrium at all times during the experiment. From this we conclude that the binding and unbinding rates of YO with DNA are larger than those for YOYO interacting with DNA.
Figure 5.
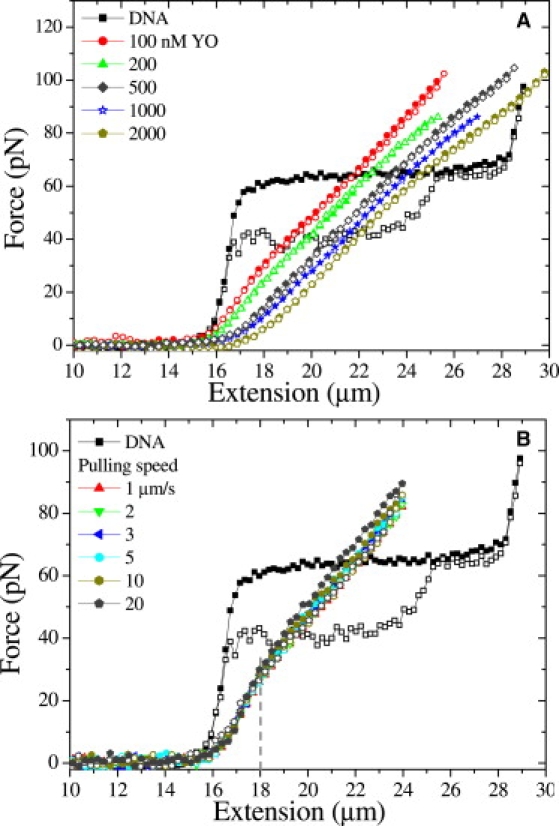
(A) Force extension curves of single λ-DNA in the absence and in the presence of various YO concentrations. (B) Force extension curves of DNA-YO complex at 100 nM YO pulled at different pulling speeds. Solid and open symbols present extension and relaxation respectively. The dotted line indicates the change in slope.
Table 3.
Parameters obtained by curve fitting the extension part by the MWLC model to the force extension of the DNA-YO complex at various concentrations of YO
| Concentration YO (nM) | Persistence length (nm) | Contour length (μm) | Stretch modulus (pN) |
|---|---|---|---|
| 0 | 52.00 | 16.4 | 1100 |
| 100 | 48.00 | 17.2 | 587 |
| 200 | 46.10 | 17.3 | 457 |
| 500 | 28.20 | 17.6 | 187 |
| 1000 | 23.30 | 18.2 | 196 |
| 2000 | 19.90 | 18.5 | 200 |
The actual fits are presented in Fig. S4.
Simultaneous force and intensity measurements have been carried out at a concentration of 100 nM YO. Fig. 6 A presents nine fluorescent microscopy images taken from the sequence at different points during the experiment. In Fig. 6 B the fluorescence intensity as a function of force is plotted. Furthermore, all fluorescence data are compiled into a kymograph, as presented in Fig. 6 C. As has been observed for YOYO (Fig. 2), the intensity of the DNA-YO complex also increases as the force is increased. The increase however for YO is larger than YOYO as the fluorescence intensity was five times higher at 55 pN compared to the intensity at 5 pN. The fluorescence intensity as a function of the extension reveals also a slight change in slope around an extension of 18 μm (Fig. S5), which coincides with the change in slope observed in the force extension curve (Fig. 5 B).
Figure 6.
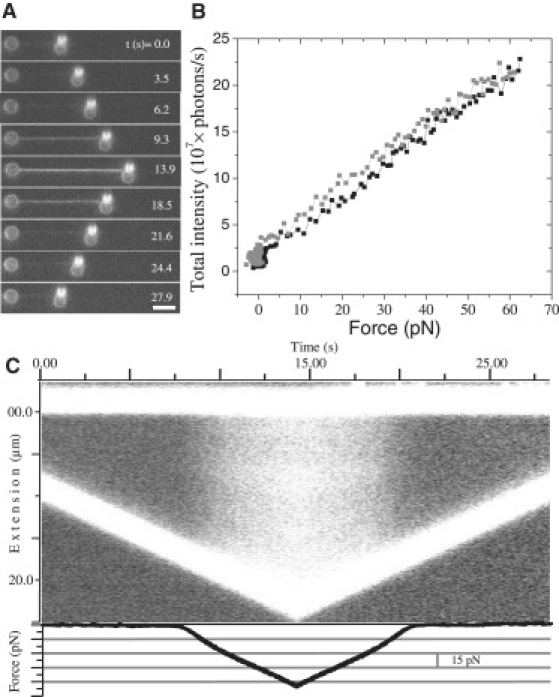
(A) Fluorescence images taken from a sequence recorded during the extension and relaxation of the DNA-YO complex in the presence of 100 nM YO (pulling speed = 1 μm/s, scale bar = 5 μm). (B) Total fluorescence intensity plotted as a function of force (black squares, extension; gray squares, relaxation). (C) Kymograph created from the sequence of images recorded simultaneously while carrying out force spectroscopy, where the horizontal axis represents time, and the vertical axis is extension. The two bright lines in the kymograph represent the streptavidin-coated polystyrene beads at either end of the DNA molecule. The force exerted on the complex is plotted as a function of time in the plot below the kymograph.
The fractional elongation was plotted against the fluorescence intensity (Fig. 7) of the DNA-YO complex at 100 nM. Here, two distinct regimes can be distinguished, that both can be fitted with a linear function. A fractional elongation of 0.14 that is the boundary between the two regimes corresponds with an extension of 18 μm, and this coincides with the extension at which the slope of the force extension curve decreases. The linear fit for fractional elongations below 0.14 corresponds to an emission of (23.2 ± 0.8) × 103 photons/s per YO molecule at 0.4 kW/cm2 power in the focal plane. This is about half the value of YOYO (Fig. 3), which may be expected from the chemical composition of the molecule. However, at an extension beyond 18 μm a reduced (14.5 ± 0.8) × 103 photons/s per YO molecule was observed, which may be attributed to a force-induced structural transition in the DNA-YO complex.
Figure 7.
Total fluorescence intensity (■) versus the fractional elongation obtained during the stretching of a DNA molecule in 100 nM YO. The number of YO (top x axis) molecules, calculated using Eq. 2. Data are fitted with two linear functions (gray lines), change in slope is presented by dotted gray line.
Constant extension experiments were carried out for DNA-YO (Fig. S6, A and B). After stretching to extensions of 18, 20, 22, and 24 μm at 3 μm/s no relaxation of the force was observed, corroborating the notion that the DNA-YO complex is in equilibrium. Assuming an exponential decay in force to reach the equilibrium an upper limit for the characteristic time constant of 8 ms was determined based on the time resolution of instrument.
Comparing interaction constants and kinetics of YOYO and YO with DNA
From this study it is evident that the interaction of DNA with the oxazole yellow dyes YO and YOYO are completely different. First, we compared the fluorescence intensities at forces below 5 pN. The intensity of the DNA-YOYO complex is ∼4 times larger than that of the DNA-YO complex, indicating that twice the amount of dye molecules are bound to DNA. This difference is also reflected in the contour length measured in the presence of various concentrations of either dye (Tables 1 and 3).
From the force extension curves measured at various YO concentrations (Fig. 5 A), it was possible to derive the fractional elongation as a function of different concentrations at one particular force (Fig. S7 A). These data sets were curve fitted with a McGhee-von Hippel binding isotherm (18) that resulted in a binding constant and binding site size as a function of force (Fig. 8, A and B). The force extension relaxation curves of the DNA-YOYO complex (Fig. 1 A) show clearly that the complex is not in equilibrium during the stretching and relaxation cycle. Thermodynamic parameters such as binding constants can be determined only from a situation in which the system is in equilibrium. By stepwise increasing the extension of the DNA-YOYO complex, and waiting until the force had reached a constant value, it was possible to find the force extension curve of the DNA-YOYO complex as it would be in equilibrium all the time (Fig. S8). These equilibrium force extension curves were used to determine the force dependent binding constant and binding site size as described above for YO.
Figure 8.
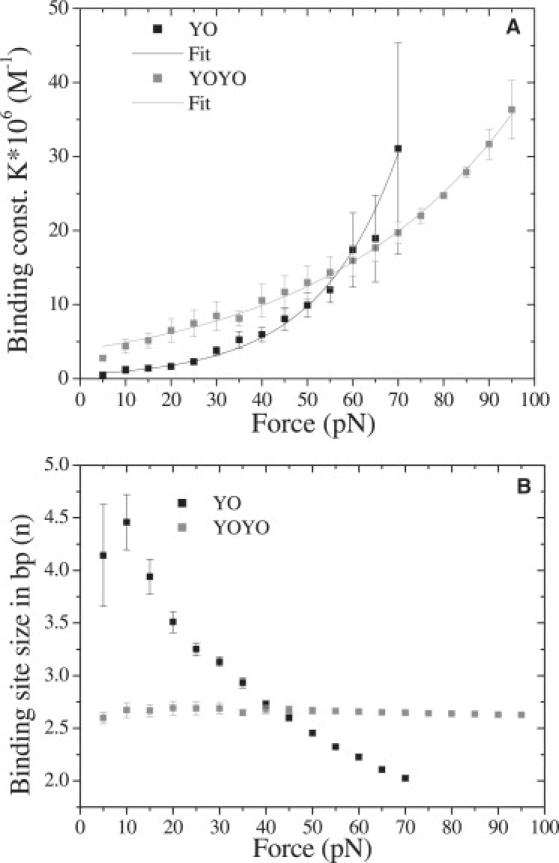
(A) Binding constant of YO (black squares) and YOYO (gray squares) as function of force. (B) Binding site size as function of force for YO (black squares) and YOYO (gray squares).
The binding constant of YO and YOYO increases as function of force (Fig. 8 A). By curve fitting the data points in Fig. 8 A with Eq. 3 as reported in Vladescu et al. (17) we can determine the zero force binding constant:
| (3) |
The zero force binding constant K0 for YO is (5.78 ± 0.80) × 105 M−1 and the distance over which the force is effectively performing work or extension per intercalator Δx is 0.233 ± 0.013 nm. These values are in good agreement with other intercalators such as ethidium (K0 = 4.5 × 105 M−1, Δx = 0.25 nm) (17). For YOYO the K0 value is (38.75 ± 1.28) × 105 M−1 that is much smaller than (1010–1012 M−1) as estimated in the literature (11), and the Δx value is 0.095 ± 0.002 nm that is three times smaller than that of YO, which is unexpected. YOYO is a bis-intercalator and the extension per intercalator was expected to be greater than that of YO. Additional research has to be formed to understand this discrepancy. Furthermore the binding site size as function of force decreases for YO where as it is almost constant in the case of YOYO, which further supports the difference in interaction of YOYO and YO molecules with double-stranded DNA.
The force extension curves of DNA in the presence of YOYO clearly shows hysteresis that was not observed in the presence of YO. This indicates a difference in binding and unbinding rates for the two intercalators. The hysteresis suggests that the complex is not in equilibrium during the stretching and relaxation experiments whereas the DNA-YO complex is. In the case of the bis-intercalator YOYO, typical time to restore equilibrium are on the order of seconds (see Fig. 4, Table 2). In the case of YO, relaxation was not observed at the present time resolution. This shows that for YO the characteristic time is <0.01 s, which is a difference of 2 orders of magnitude when compared to the bis-intercalator YOYO.
Kinetic model
A kinetic model is presented to explain the differences observed in the interaction kinetics of the oxazole yellow mono and bis-intercalator with DNA. The long range interaction between positively charged oxazole yellow dyes and negatively charged DNA is Coulombic interaction. In Fig. 9, A and B, the on and off rates of this interaction are described with k1 and k−1. This step is assumed to be diffusion-controlled and a rate of 1.6 × 109 M−1s−1 can be calculated for YOYO-1 using the Smoluchowski equation, indicating that this step is extremely fast with respect to the rate constants found in the results. For YO this rate is even higher. In the case of YO the next step in the intercalation process of YO to DNA is the actual intercalation, described by the rate constants k2 and k−2. (Fig. 9 A). From the force relaxation experiments the typical relaxation time for reaching equilibrium for YO was found to be <0.01 s. Considering the fact that k1 is so high, this measured rate is effectively k2. The YOYO molecule is a bis-intercalating agent that consists of two identical YO moieties each bearing +2e charge. The proposed model assumes that actual intercalation of YOYO occurs in two steps. In the first step, one of the YO moieties intercalate in the DNA, and in a second step the second YO moiety intercalates. The binding rate k2 of the first moiety is assumed to be similar to that of YO to DNA. With the first YO moiety of YOYO bound to DNA, the second YO moiety resides close to the DNA molecule and a possible available binding site (Fig. 9 B, iii). This leads to an increased effective concentration of the YO moiety locally and would result in a binding rate that is even higher than that for binding the first moiety (k2). This, however, is not observed. Both the force spectroscopy data as well as the force relaxation measurements show that YOYO binding to DNA is much slower than that of YO binding. To account for this observation, an intermediate state is introduced within the model that is relatively stable, and that effectively reduces the intercalation rate for the second YO moiety of a YOYO (Fig. 9 B, iii). The rate converting the intermediate state to the final state k3 has to be much lower than k2, which results in an overall reduction of the binding rate from the unintercalated to the bis-intercalated state for YOYO. For YOYO it means that k3 is the rate limiting step (Fig. 9 B, from iii to iv state).
Figure 9.
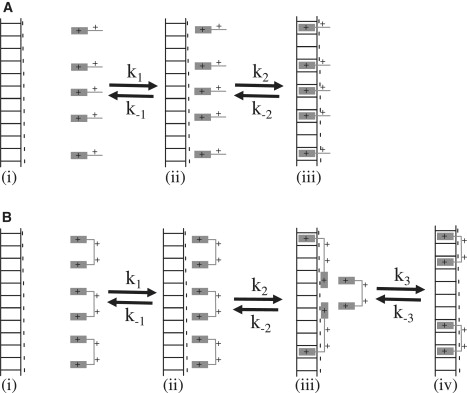
Schematic illustration of different steps in the interaction of (A) YO and (B) YOYO molecules with DNA.
Although the results presented in this study do not show the structure of the intermediate state, we assume that the intermediate state is one in which one YO moiety is intercalated and the second one is bound to DNA in another mode (Fig. 9 B, iii). It has been proposed that the nonintercalating moiety can strongly interact with the backbone of a DNA molecule (27). It is also possible that the nonintercalated YO moiety temporarily blocks some of the neighboring binding sites effectively reducing the number of available binding sites, and slowing down the kinetic on rate. The length of the linker that connects the two YO moieties is ∼1.14 nm (11) and combined with the size of a YO moiety it is possible to cover ∼5–6 bp. In the literature multiple force spectroscopy studies of single DNA molecules in the presence of mono-intercalating and bis-intercalating molecules have been presented, and in most cases the bis-intercalating variant shows more hysteresis than its mono-intercalating counterpart (14). We therefore think that the model proposed here is generally applicable to describe the binding of bis-intercalating agents to DNA.
Conclusions
Using a combined optical tweezers and fluorescence microscope we have studied the interaction of the mono-intercalator YO and bis-intercalator YOYO with a double-stranded DNA molecule as a function of force. The contour length of the DNA in the presence of YO and YOYO was larger, which is a direct result of intercalation of the dye between the basepairs of DNA. The number of molecules determined from the increase in contour length was correlated with the fluorescence intensity. The binding constants of YO and YOYO were found to increase as the force increases. Furthermore, from the hysteresis observed in the force extension curve and the results of the force relaxation experiments, it was concluded that the DNA-YOYO complex, as opposed to the DNA-YO complex was not in equilibrium with the surrounding medium. From the force relaxation experiments at constant extension, the typical relaxation time was determined for the DNA-dye complex at different levels of initial loading. In the case of YOYO this characteristic time increased as more YOYO molecules were initially on the DNA. We determine the zero force binding constant for YO (5.78 × 105 M−1). In the case of YO no hysteresis was observed, suggesting that the characteristic relaxation time is <0.01 s. To explain this unexpected behavior in the kinetics, we propose a model in which the YOYO binding occurs via a relatively stable intermediate step, in which the second intercalative step is the limiting step. This model is thought to be more generally applicable for bis-intercalating molecules.
Supporting Material
Eight figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(09)01029-7.
Supporting Material
Acknowledgments
This work was supported by the Stichting voor Fundamenteel Onderzoek der Materie (FOM) through the program “Physical Biology II”.
References
- 1.Lerman L.S. The structure of the DNA acridine complex. Proc. Natl. Acad. Sci. USA. 1963;49:94–102. doi: 10.1073/pnas.49.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu C., Losytskyy M.Y., Kovalska V.B., Kryvorotenko D.V., Yarmoluk S.M. Novel, monomeric cyanine dyes as reporters for DNA helicase activity. J. Fluoresc. 2007;17:671–685. doi: 10.1007/s10895-007-0215-z. [DOI] [PubMed] [Google Scholar]
- 3.Hurley L.H. DNA and its associated processes as targets for cancer therapy. Nat. Rev. Cancer. 2002;2:188–200. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 4.Leng F., Priebe W., Chaires J.B. Ultratight DNA binding of a new bisintercalating anthracycline antibiotic. Biochemistry. 1998;37:1743–1753. doi: 10.1021/bi9720742. [DOI] [PubMed] [Google Scholar]
- 5.Rye H.S., Yue S., Wemmer D.E., Quesada M.A., Haugland R.P. Stable fluorescent complex of double-stranded DNA with bis-intercalating asymmetric cyanine dyes: properties and application. Nucleic Acids Res. 1992;20:2803–2812. doi: 10.1093/nar/20.11.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glazer A.N., Rye H.S. Stable dye-DNA intercalation complex as reagents for high-sensitivity fluorescence detection. Nature. 1992;359:859–861. doi: 10.1038/359859a0. [DOI] [PubMed] [Google Scholar]
- 7.Calsson C., Larsson A., Jonsson M., Albinsson B., Norden B. Optical and photophysical properties of the oxazole yellow DNA probes YO and YOYO. J. Phys. Chem. 1994;98:10313–10321. [Google Scholar]
- 8.Bianco P.R., Brewer L.R., Corzett M., Balhorn R., Yeh Y. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature. 2001;409:374–378. doi: 10.1038/35053131. [DOI] [PubMed] [Google Scholar]
- 9.Ladoux B., Quivy J., Doyle P., Roure O., Almouzni G. Fast kinetics of chromatin assembly revealed by single molecule videomicroscopy and scanning force microscopy. Proc. Natl. Acad. Sci. USA. 2000;97:14251–14256. doi: 10.1073/pnas.250471597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johansen F., Jacobsen J.P. 1H NMR studies of the bis-intercalation of a homodimeric oxazole yellow dye in DNA oligonucleotides. J. Biomol. Struct. Dyn. 1998;16:205–222. doi: 10.1080/07391102.1998.10508240. [DOI] [PubMed] [Google Scholar]
- 11.Larsson A., Carlsson C., Jonssson M., Albinsson B. Characterization of the binding of the fluorescent dyes YO and YOYO to DNA by polarized light spectroscopy. J. Am. Chem. Soc. 1994;116:8459–8465. [Google Scholar]
- 12.Eckel R., Ros R., Ros A., Wilking S.D., Sewald N. Identification of binding mechanisms in single molecule-DNA complex. Biophys. J. 2003;85:1968–1973. doi: 10.1016/S0006-3495(03)74624-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krautbauer R., Fischerlander S., Allen S., Gaub H.E. Mechanical fingerprints of DNA drug complex. Single. Mol. 2002;3:97–103. [Google Scholar]
- 14.Sischka A., Toensing K., Eckel R., Wilking S.D., Sewald N. Molecular mechanisms and kinetics between DNA and DNA binding ligands. Biophys. J. 2005;88:404–411. doi: 10.1529/biophysj.103.036293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennink M.J., Scharer O.D., Kanaar R., Sakata-Sogawa K., Schins J.M. Single-molecule manipulation of double-stranded DNA using optical tweezers: interaction studies of DNA with RecA and YOYO-1. Cytometry. 1999;36:200–208. doi: 10.1002/(sici)1097-0320(19990701)36:3<200::aid-cyto9>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 16.Vladescu I.D., McCauley M.J., Rouzina I., Williams M.C. Mapping the phase diagram of single DNA molecule force-induced melting in the presence of ethidium. Phys. Rev. Lett. 2005;95:158102. doi: 10.1103/PhysRevLett.95.158102. [DOI] [PubMed] [Google Scholar]
- 17.Vladescu I.D., McCauley M.J., Nunez M.E., Rouzina I., Williams M.C. Quantifying force-dependent and zero-force DNA intercalation by single-molecule stretching. Nat. Methods. 2007;4:517–522. doi: 10.1038/nmeth1044. [DOI] [PubMed] [Google Scholar]
- 18.McGhee J.D., von Hippel P.H. Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974;86:469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- 19.Fallman E., Axner O. Design for fully steerable dual-trap optical tweezers. Appl. Opt. 1997;36:2107–2113. doi: 10.1364/ao.36.002107. [DOI] [PubMed] [Google Scholar]
- 20.Gurrieri S., Wells K.S., Johnson I.D., Bustamante C. Direct visualization of individual DNA molecules by fluorescence microscopy: characterization of the factors affecting signal/background and optimization of imaging conditions using YOYO. Anal. Biochem. 1997;249:44–53. doi: 10.1006/abio.1997.2102. [DOI] [PubMed] [Google Scholar]
- 21.Akerman B., Tuite E. Single and double strand photocleavage of DNA by YO, YOYO and TOTO. Nucleic Acids Res. 1996;24:1080–1090. doi: 10.1093/nar/24.6.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waterman-Storer C.M., Desai A., Bulinski J.C., Salmon E.D. Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr. Biol. 1998;8:1227–1230. doi: 10.1016/s0960-9822(07)00515-5. [DOI] [PubMed] [Google Scholar]
- 23.Marko J.F., Siggia E.D. Stretching DNA. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- 24.Smith S.B., Cui Y., Bustamante C. Overstretching B-DNA The elastic response of individual double stranded and single stranded DNA molecule. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 25.Cluzel P., Lebrun A., Heller C., Lavery R., Viovy J. DNA an extensible molecule. Science. 1996;271:792–794. doi: 10.1126/science.271.5250.792. [DOI] [PubMed] [Google Scholar]
- 26.Larsson A., Carisson C., Jonsoon M. Characterization of the binding of YO to [poly(dA-dT)]2 and [poly(dG-dC)]2, and of the fluorescence properties of YO and YOYO complexed with the polynucleotides and double-stranded DNA. Biopolymers. 1995;36:153–167. doi: 10.1002/bip.360360205. [DOI] [PubMed] [Google Scholar]
- 27.Pecq J.B., Bret M., Barbet J., Roques B. DNA polyintercalating drugs DNA binding of diacridine derivatives. Proc. Natl. Acad. Sci. USA. 1975;72:2915–2919. doi: 10.1073/pnas.72.8.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


