Abstract
The structure and dynamic properties of an 80-residue fragment of Ste2p, the G-protein-coupled receptor for α-factor of Saccharomyces cerevisiae, was studied in LPPG micelles with the use of solution NMR spectroscopy. The fragment Ste2p(G31-T110) (TM1-TM2) consisted of 19 residues from the N-terminal domain, the first TM helix (TM1), the first cytoplasmic loop, the second TM helix (TM2), and seven residues from the first extracellular loop. Multidimensional NMR experiments on [15N], [15N, 13C], [15N, 13C, 2H]-labeled TM1-TM2 and on protein fragments selectively labeled at specific amino acid residues or protonated at selected methyl groups resulted in >95% assignment of backbone and side-chain nuclei. The NMR investigation revealed the secondary structure of specific residues of TM1-TM2. TALOS constraints and NOE connectivities were used to calculate a structure for TM1-TM2 that was highlighted by the presence of three α-helices encompassing residues 39–47, 49–72, and 80–103, with higher flexibility around the internal Arg58 site of TM1. RMSD values of individually superimposed helical segments 39–47, 49–72, and 80–103 were 0.25 ± 0.10 Å, 0.40 ± 0.13 Å, and 0.57 ± 0.19 Å, respectively. Several long-range interhelical connectivities supported the folding of TM1-TM2 into a tertiary structure typified by a crossed helix that splays apart toward the extracellular regions and contains considerable flexibility in the G56VRSG60 region. 15N-relaxation and hydrogen-deuterium exchange data support a stable fold for the TM parts of TM1-TM2, whereas the solvent-exposed segments are more flexible. The NMR structure is consistent with the results of biochemical experiments that identified the ligand-binding site within this region of the receptor.
Abbreviations: IMP, integral membrane protein; GPCR, G-protein coupled receptor; NOE, nuclear Overhauser effect; CD, circular dichroism; DPC, dodecylphosphocholine; LPPG, 1-palmitoyl-2-hydroxy-sn-glycero-3-[phospho-rac-(1-glycerol)]; IPTG, isopropyl-β-thiogalactopyranoside; HSQC, heteronuclear single quantum coherence; NOESY, nuclear Overhauser enhancement spectroscopy; 15N{1H}-NOE, heteronuclear Overhauser enhancement of 15N after saturation of 1H; 2D/3D, two-dimensional/three-dimensional; ppm, parts per million; NT, N-terminus; RMSD, root mean-square deviation; RP-HPLC, reverse-phase high-performance liquid chromatography; TM, transmembrane
Introduction
Relatively few high-resolution structures for membrane receptors and transporters have appeared in the Protein Data Bank, despite the fact that these IMPs have been estimated to constitute 25–30% of eukaryotic proteins (1,2). Among the IMPs, GPCRs represent a biomedically important superfamily of eukaryotic proteins. Eight hundred GPCRs have been identified by analysis of the human genome, and 30–50% of prescription drugs target these proteins (3). However, the high-resolution structures reported to date are limited to those of rhodopsin (4), the β-adrenergic receptors (5,6), and the human A2A adenosine receptor (7). To alleviate this underrepresentation, many laboratories are actively involved in developing new techniques to express, isolate, and crystallize membrane proteins (8,9) and to study them in membrane-like environments (10,11). Crystallization of membrane transporters and receptors has been aided by the use of chimeras, antibodies, and mutations to decrease the inherent flexibility of these IMPs. Although significant successes are being reported, crystallization of IMPs is still more art than science. Noteworthy progress has also been achieved in solid-state NMR studies (12–15), and most recently solution-state NMR experiments in detergent resulted in the complete backbone assignments of sensory rhodopsin II from Natronomonas pharaonis (16).
The use of peptides that represent individual regions of IMPs as surrogates for the partial structure of IMPs was predicated on a model of membrane protein assembly (17,18). In the first stage of the two-state model of IMP folding originally proposed by Popot and Engelman (17), upon partitioning into the membrane interface the peptide forms α-helical TMs, which then spontaneously insert into the bilayer core due to entropic driving forces. In stage two, these independent domains assemble into the 3D protein structure. TM-TM assembly likely involves van der Waals packing forces, a few polar or electrostatic interactions, and C-H—O=C hydrogen bonds, and often is influenced by GXXXG motifs and the presence of proline residues (19). The two-stage model was later extended to include additional interactions with membrane-lipid headgroups (18). Peptides corresponding to single TM domains of bacteriorhodopsin (20–25), rhodopsin (26–30), Ste2p, the α-factor receptor (28,31–36), and the adenosine A2 receptor (37,38) were shown to assume helices in membrane mimetic solvents, thereby providing evidence for the two-stage model.
Despite the extensive use of peptide fragments to elucidate the biophysical properties of regions of IMPs, significant skepticism remains concerning the biological significance of the information obtained from such investigations. Few studies have been conducted on peptides longer than a single TM. CD studies revealed that two TM fragments of the μ-opioid receptor and the CB2 cannabinoid receptor were highly helical in membrane mimetic solvents such as trifluoro-ethanol/water and a variety of detergents (39–41). Well-resolved 2D NMR spectra were measured and 80% of the peaks were assigned for the CB2 double TM fragment in DMSO solution. An NMR structure was reported for a two-TM fragment of the human glycine receptor in trifluoro-ethanol (42), and a series of biophysical studies from the CFTR protein provided insights into the influence of turn structures and residue effects on helical hairpin formation (43). Nevertheless, no high-resolution information is presently available regarding multitopic GPCR fragments in a lipid-like environment, and it is not clear whether two contiguous domains of these heptahelical receptors will pack to a stable tertiary structure in a detergent micelle.
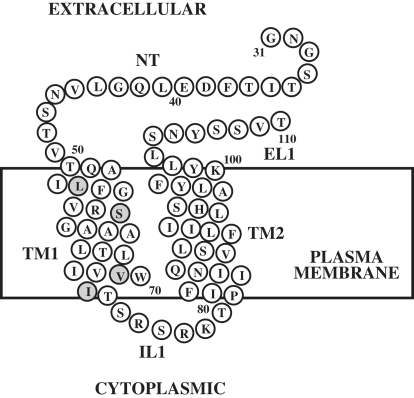
Herein we present a detailed high-resolution NMR study on an 80-residue fragment of the yeast α-factor receptor Ste2p(G31-T110) containing a short stretch of the N-terminus (NT), TM domain 1 (TM1), the first intracellular loop (IL1), TM domain 2 (TM2), and a short stretch of the first extracellular loop (EL1) (Fig. 1). This polypeptide was biosynthesized with [15N], [15N,13C], [15N,13C,2H] uniform isotope labeling. In addition, it was labeled at specific amino acids or at unique methyl protons in an otherwise perdeuterated background. With the use of triple-resonance NMR experiments, nearly complete assignment of backbone and side-chain resonances in LPPG micelles was accomplished. 3D NOESY spectra in combination with deuterated LPPG allowed assignment of a large number of medium-range NOEs that unambiguously established the secondary structure. Moreover, the use of a labeling pattern introduced by Tugarinov and Kay (44) allowed determination of several interhelical long-range NOEs among methyl groups, and these NOE-derived restraints were used to calculate a model of the structure of the 80-residue fragment. The structure represents the first high-resolution structure, to our knowledge, of a double TM domain fragment of a GPCR in lipid. The data help to explain biochemical cross-linking studies that revealed an interaction of the 13th residue of the α-factor tridecapeptide with residues 58 and 59 of the α-factor receptor.
Figure 1.
Schematic representation of Ste2p(G31-T110) [TM1-TM2]. NT: 19 residues of the NT domain of Ste2p; TM1: first TM helix; IL1: first intracellular loop; TM2: second TM helix; EL1: first extracellular loop. The mutated methionines and cysteine are shaded. The numbering used follows that of the intact receptor.
Materials and Methods
Chemicals and solutions
Deuterated water, α-ketobutyric (13C4, 98%; 3,3-D2, 98%) and α-ketoisovaleric (1,2,3,4,-13C4, 99%; 3,4,4,4-D4, 98%) acids were bought from Cambridge Isotopes (Andover, MA) and (deuterated) lipids were purchased from Avanti Polar Lipids (Alabaster, AL). All other chemicals used were ordered from Sigma-Aldrich (St. Louis, MO) or Fluka (Buchs, Switzerland).
Cloning, expression, and purification of isotopically-labeled Ste2p(G31-T110)[TM1-TM2]
TM1-TM2 was successfully expressed as a TrpΔLE fusion protein, from which it was liberated by cyanogen bromide (CNBr) cleavage and purified by RP-HPLC. The cloning, expression, and isolation of Ste2p(G31-T110) labeled with 15N, 13C and 2H; [15N,13C,2H]-TM1-TM2 (G31NGST ITFDE L41QGLV NSTVT Q51AILF GVRSG A61AALT LIVVW I71TSRS RKTPI F81IINQ VSLFL I91ILHS ALYFK Y101LLSN YSSVT) were carried out using procedures described previously (45). This protein fragment contains four replacements of natural residues (three methionines were replaced with leucine, valine, and isoleucine, and Cys was replaced with Ser) to enable the CNBr cleavage and to stabilize the peptide against oxidation. The rationale for using these exact replacements was discussed previously (45). Cys59 can be replaced by Ser without any effect on the biological activity of Ste2p (46), and replacement of individual methionine residues with leucine, valine, or isoleucine has been shown to be biologically acceptable (47). TM1-TM2 protein fragments selectively labeled with [15N]-alanine, [15N]-isoleucine, [15N]-leucine, [15N]-valine, or [15N]-phenylalanine were prepared in defined minimal medium supplemented with all unlabeled amino acids and an excess of the [15N]-labeled amino acid as described by Cohen et al. (45).
A sample of TM1-TM2 that contained protonated methyl groups in an otherwise fully deuterated background was prepared as described by Tugarinov et al. (48). Briefly, BL21-AI cells containing pLC01 were streaked onto LBAmp plates and incubated overnight at 37°C. A 6 mL LBAmp culture was inoculated with one colony from the overnight growth and incubated at 37°C, 250 rpm, to OD600 0.7–0.8. These cells were pelleted and resuspended in M9 minimal media in H2O to OD600 ∼ 0.05–0.1 and then incubated at 37°C as above to OD600 0.6. The cells were once again pelleted and resuspended in 100 mL M9 minimal media in D2O containing 13C/2H-glucose and 15NH4Cl (M9/D2O), and cells were grown until OD600 ∼ 0.4–0.5 was reached. The cells were then diluted to 200 mL with M9/D2O, incubated as above to OD600 ∼ 0.4–0.5 and then diluted to 1L in M9 medium in D2O supplemented with 70 mg/L α-ketobutyric acid (13C4, 98%; 3,3-D2, 98%) and 120 mg/L α-ketoisovaleric acid (1,2,3,4-13C4, 99%; 3,4′,4′,4′-D4, 98%). These cells were incubated at 37°C, 250 rpm, to OD600 ∼ 0.3–0.4 and expression was induced with 0.5% L-arabinose. They were then grown at 37°C, 250 rpm, for 6–8 h and harvested by centrifugation. Further details regarding the purification are described in Cohen et al. (45).
NMR sample preparation
Partially, uniformly or selectively 15N/13C/2H/1H-labeled TM1-TM2 NMR samples were obtained by dissolving the protein (0.3–1 mg) and detergent LPPG (28.4 mg) in sodium phosphate buffer (200 μL, 20 mM, pH = 6.4, 1–2 min of shaking), followed by sonication (2 × 15 min) and incubation (30 min) at 37°C before transfer to a Shigemi NMR tube. The final concentrations used for NMR measurements were ∼0.1–0.4 mM and 200 mM for the protein and LPPG, respectively. The samples prepared using this method were sufficiently stable for measurement of NMR spectra at 320 K, and displayed degradation in the form of a white precipitate only after 2 weeks. 2D, 3D triple-resonance, and 15N-resolved NOESY spectra were recorded at 320 K using a Bruker AV700 spectrometer equipped with a triple-resonance cryoprobe. The 13C-edited HSQC and NOESY spectra centered on methyl (19 ppm), aliphatic (39 ppm), and aromatic (125 ppm) carbons were recorded at 320 K using 900 MHz spectrometers at the New York Center for Structural Biology and at the Swiss Federal Institute of Technology. All proton chemical shifts were referenced to the water line at 4.48 ppm at 47°C, from which the nitrogen and carbon scales were derived indirectly by using the conversion factors of 0.10132900 (15N) and 0.25144954 (13C). Chemical shifts were deposited in the BMRB database under accession code 15995.
NMR spectroscopy
Sequence-specific resonance assignment was accomplished based on a set of triple-resonance experiments as well as 15N- and 13C-resolved NOESY spectra. Backbone assignment was performed based on the set of HNCO/HN(CA)CO experiments (49) and HNCA/HN(CO)CA experiments (49). HNCACB and CBCA(CO)NH spectra (50) were evaluated to additionally support assignments made and to derive information on Cβ chemical shifts. Side-chain resonance assignment started with (H)CCH-TOCSY/COSY experiments (51,52). Finally, chemical shifts were obtained by picking peaks in a 13.3 ms constant-time (ct)-[13C,1H]-HSQC spectrum (53). Unfortunately, the signal/noise ratio in the (H)CCH data set was insufficient, so extensive use of 13C-resolved NOESYs had to be made. Methyl groups of Val(Hγ), Ile(Hδ1), and Leu(Hδ1/δ2) residues were assigned using a HMCMCBCANH experiment developed by Tugarinov and Kay (44). Peak positions were adjusted to the 13.3 ms ct-[13C,1H]-HSQC spectrum and cross-validated against the 13C-resolved NOESY experiments recorded on fully protonated and partially methyl-protonated samples. The aromatic ring systems of Tyr, His, and Trp residues were picked in an 8.8 ms ct-[13C,1H]-HSQC and correlated with β-carbons via the HBCBCGCDHD experiment (54) whenever possible or via NOEs from the 13C-NOESY centered on aromatic carbons. All chemical shifts were finally derived from peaks in the [15N,1H]-HSQC and the ct-[13C,1H]-HSQC spectra. All experiments employed pulsed-field gradients (55).
The extent of amide hydrogen exchange was probed by recording a [15N,1H]-HSQC experiment, in which low-power irradiation was applied on the water resonance during the relaxation delay. 15N-relaxation data were recorded using a proton-detected version of the 15N{1H}-steady-state NOE experiment (56,57) with a 2.7 s recycle delay.
Processed data were transferred into the programs CARA (58) and XEASY (59) for data analysis. MOLMOL was used to calculate RMSD values and prepare structural representations (60).
Structure calculation
Distance restraints were obtained from 15N-resolved NOESY spectra recorded on [15N,13C]- and [15N,2H]-labeled TM1-TM2 samples with mixing times of 70 and 200 ms, respectively, and from 13C-resolved NOESY spectra recorded on [15N,13C]- and [15N,13C,2H(1H(methyl)-Ile, Leu, Val)]-labeled TM1-TM2 samples with mixing times of 100 and 200 ms, respectively. In addition, dihedral angle restraints derived from TALOS (61) using chemical shifts of 1Hα, 13Cα, 13Cβ, 13C′, and 15N nuclei were added. Structures were calculated with the program CYANA (62). The final CYANA calculation was performed with 100 randomized starting structures, and the 20 CYANA conformers with the lowest target function values were selected to represent the NMR ensemble and deposited in the Protein Data Bank under accession code 2k9p.
Results
Biosynthesis of selectively methyl protonated samples
Expression of selectively methyl protonated TM1-TM2 fusion protein in an otherwise deuterated background as described by Tugarinov and Kay (44) was performed using ketobutyric and ketoisovaleric acids that were isotopically labeled with 1H, 2H, and 13C as described by Tugarinov et al. (48). After the fusion protein was expressed, the cells were harvested, inclusion bodies were solubilized in 70% trifluoroacetic acid, and CNBr was used to remove the TrpΔLE peptide segment, immediately followed by purification by RP-HPLC in an acetonitrile/isopropanol/water gradient to >95% purity (see Fig. S1 in the Supporting Material) (45). The yield of purified protein fragment after lyophilization was 5.5 mg/L and the incorporation of 15N, 13C, and 2H was >95% (calculated MW = 9702.29, observed MW = 9660.42).
Backbone resonance assignment
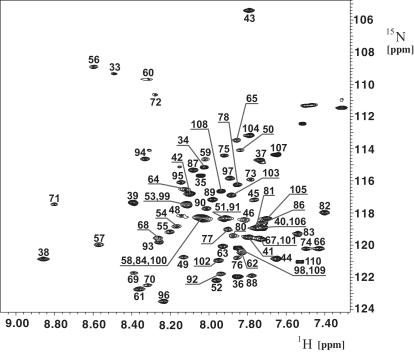
Resonance assignment of TM1-TM2 was accomplished with the use of 3D triple-resonance NMR experiments. Seventy-five crosspeaks were detected in the [15N,1H]-HSQC spectrum (Fig. 2), for which well-separated resonances in both HNCO and HNCA spectra were present; ∼30 of the crosspeaks in the [15N,1H]-HSQC spectrum had corresponding intra- and interresidual Cα and Cβ peaks in the [13C,1H] strips of the HNCACB and CBCA(CO)NH experiments. For ∼20 peaks in the [15N,1H]-HSQC spectrum, one or more resonances were missing in the corresponding strips from HNCACB or CBCA(CO)NH, and for the remaining ∼20 residues no peaks were observed in the corresponding strips (see Fig. S3). [15N,1H]-HSQC spectra measured on TM1-TM2 selectively labeled with 15N-amino acids (Ile, Leu, Phe, Ala, or Val) supported the assignments. Because of the large number of Ile and Leu residues in the TM1-TM2 sequence, we experienced significant difficulties with the backbone assignment for residues 63–68 and 87–94. We were able to resolve these difficulties by searching for sequential HN-HN, HN-H(aliphatic), and H(aliphatic)-H(aliphatic) crosspeaks in the 15N- and 13C-resolved NOESY spectra. Subsequently, these assignments were further cross-validated against (H)CCH-TOCSY and 13C-resolved NOESY data. Because of overlapping or missing peaks, we were initially not able to assign amide moieties of Gly31, Asn32, Ser47, and Gln85. However, in combination with knowledge of typical chemical shifts encountered for such residues, we were able to assign them using the combination of the ct-[13C,1H]-HSQC and 13C-resolved NOESY. The overall completeness of the backbone assignment for HN, N, Hα, Cα, and C′ was above 95%.
Figure 2.
[15N,1H]-HSQC spectrum of Ste2p(G31-T110). The spectrum was recorded at 700 MHz, 320K, on a 0.4 mM sample of the protein fragment in 200 mM LPPG at pH 6.4. The sequence-specific assignments are annotated.
Side-chain resonance assignment
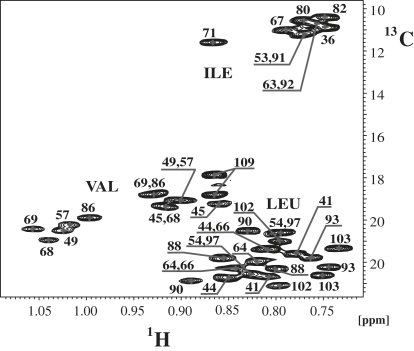
Based on the unique Hα and Cα resonance assignments derived from the backbone assignment process described above, the Hα/Cα crosspeaks were used as anchoring points for assignment of the aliphatic side chains. Because the signal/noise ratio in both the (H)CCH-TOCSY and (H)CCH-COSY spectra was often insufficient, extensive use of 13C-resolved NOESY and 15N-resolved NOESY spectra was required. Again, severe difficulties for side-chain assignments of Ile and Leu residues were encountered, especially in the methyl region that was partially covered by peaks from the nondeuterated LPPG. However, when partially deuterated d36-LPPG was used, the signals cleared up and NOEs could be used for assignment purposes (Fig. S4). Moreover, we observed significant line narrowing, most likely due to the reduced intermolecular dipolar broadening in the presence of the deuterated palmitoyl chains. To further facilitate assignments, a protein sample was prepared following the methodology developed by Tugarinov and Kay (44), whereby protonated methyl groups from Ile, Leu, and Val are introduced into an otherwise completely perdeuterated background. Assignment of methyl groups of Val(Hγ), Ile(Hδ1), and Leu(Hδ1/δ2) residues was accomplished by using an HMCMCBCANH experiment (44). However, the higher resolution in the 900 MHz 13C-resolved NOESY spectrum recorded on the selectively [15N,13C,2H(1H(methyl)-Ile, Leu, Val)]-labeled sample (Fig. 3) was required to unambiguously establish a number of methyl-methyl NOEs that were critically important for establishing interhelical contacts.
Figure 3.
Methyl region from the 900 MHz ct- [13C,1H]-HSQC of the selectively methyl-labeled Ste2p(G31-T110) sample in the d36-LPPG solution. Assignments of methyl groups corresponding to Ile, Leu, and Val residues are annotated.
The unique aromatic side-chain resonances of Trp70, His94, Tyr98, Tyr101, and Tyr106 were picked in the aromatic ct-[13C,1H]-HSQC and assigned from 2D (HB)CB(CGCD)HD experiments. Aromatic protons from Phe residues 38, 55, 81, 89, and 99 were assigned using the 13C-resolved aromatic NOESY experiment, but signal dispersion was so small that unambiguous assignments could only be established for Phe38. Ultimately, the percentage of the unambiguously assigned side-chain resonances was ∼95%, whereas assignments of ∼5% of the resonances, including aromatic spin systems of phenylalanine residues 55, 81, 89, and 99, and aliphatic spin systems of leucine residues 54, 64, and 97, were ambiguous.
Backbone dynamics of TM1-TM2 in LPPG
Values of the heteronuclear 15N{1H}-NOE (H-NOE) have been used to discriminate well-structured regions from those that display increased flexibility (57). In the 2-TM protein, most residues in segments 37–44, 50–72, and 80–101 had H-NOE values of >0.75 (Fig. S5). The H-NOE values for many residues in segments 45–49, 58–62, and 74–81 were reduced to ∼0.6. The predicted TM helices TM1 and TM2 are characterized by comparably large H-NOEs, whereas most values for residues S72–P79, which are predicted to constitute the first intracellular loop, are lower. From residue Ser107 to the C-terminus, and from Ile36 to the NT, the H-NOE values steadily decreased, indicating that both termini are rather flexible, which is supported by the absence of medium-range NOEs in these regions (vide infra).
Structure calculation
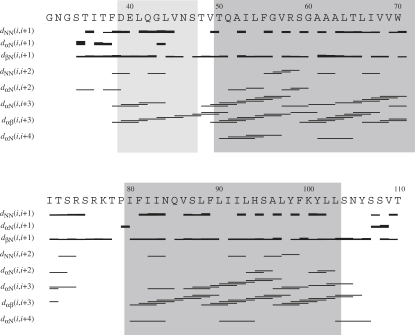
Once the chemical shifts were available, structure calculations were performed with the program CYANA and the internally implemented algorithm for automatic NOE assignment (62). Distance restraints were obtained from 15N-resolved NOESY and 13C-resolved NOESY spectra. In addition, 42 dihedral angle restraints were derived from 13C chemical shifts of 13Cα, 13Cβ, 13C′and 15N atoms using the program TALOS (61). Initially, hydrogen-bond restraints were applied in the regions of the putative helices to facilitate automatic assignment of medium-range NOEs. After the hydrogen-bond restraints were removed, almost all medium-range contacts remained, and these were manually checked in the 3D spectra. As shown in Fig. 4, almost complete sets of α,β(i,i+3) and numerous α,N(i,i+3) and several α,N(i,i+4) contacts for the segments comprising residues 49–72 and 80–103, which correspond to the predicted TM regions (TM1 50–72, TM2 79–103), were observed throughout the helices. Moreover, we observed characteristic α,β (i,i+3) contacts for residues 39–47 of the NT domain. This region of the protein fragment also appeared to form an α-helix that is most probably surface-associated (vide infra).
Figure 4.
Sequence plot displaying characteristic upper distance restraints along the Ste2p(G31-T110) sequence derived from NOEs. Regions of the predicted TM helices and the extracellular helix are shaded in gray.
Using the above restraints (Fig. 4) and several long-range restraints measured on selectively methyl protonated samples, we calculated the ensemble of the 20 lowest-energy NMR conformations depicted in Fig. 5. The superposition of NMR-derived conformers reveals the presence of two TM helices corresponding to the predicted TM1 and TM2 segments, preceded by an additional helical region encompassing residues 39–47. In 11 of the 20 conformers, the first α-helix starts at residue 38 according to the criteria of Kabsch and Sander (63). The TM1 and TM2 α-helices extend from residues 49–72 and 80–103 in all conformers. When backbone atoms from the TM1 and TM2 helices are individually superimposed, the RMSD values are 0.40 ± 0.13 Å and 0.57 ± 0.19 Å for backbone atoms of residues 49–72 and 80–103, respectively, whereas the RMSD is 2.36 ± 0.97 Å when superimposing backbone atoms of residues 49–103. For the amphiphilic α-helix of the NT domain, the RMSD value is 0.25 ± 0.10 Å. Considering that residues 55–60 display lower H-NOE values (Fig. S5) and fewer correlations in the corresponding 3D NOESY spectrum (Fig. 4), we suspect that the segment comprising residues 55–60 around the internal Arg58 undergoes slow conformational exchange corresponding to a kink motion. A similar behavior was observed for residues 80–88, indicating that the entire NT half of the second TM helix is destabilized. The segments encompassing residues 31–38, 71–79, and 104–110 display no contacts characteristic of helices and have decreased H-NOE values, and therefore are more flexible. At present, the number of tertiary contacts between the TM helices remains insufficient to unambiguously establish their relative orientation. However, we observed unambiguous long-range NOE contacts between methyl groups of the residues Leu66–Val86, Ala63–Val86, Ala63–Leu90, and Val69–Ser75 that help to partially restrain the tertiary structure in the helical regions adjacent to the loop (Fig. 5). No unambiguous long-range contacts involving the π-systems of the aromatic residues were detected.
Figure 5.
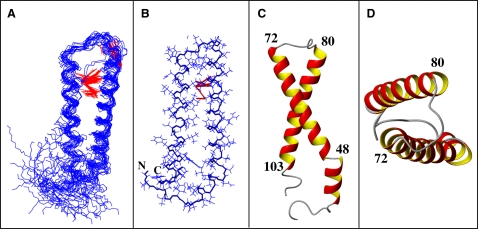
(A) Backbone representation of the ensemble of the 20 lowest-energy conformers of Ste2p(G31-T110) superimposed over backbone atoms in the region comprising residues 39–103. Observed long-range NOE contacts are highlighted in red. (B) A single conformer from the ensemble additionally displaying the side chains. (C) Structure of a single conformer—view from the side of membrane interior. (D) The same as C but viewed from the cytoplasmic side.
We have superimposed the experimental structure determined in this work with the corresponding segment from the rhodopsin-based model of the Ste2p receptor published by Eilers et al. (64) (see Fig. S6). Of interest, the overall features of the helical hairpin are very similar in both structures (RMSD of 1.8 Å for the superimposition of the backbone atoms of TM1 and TM2). The angle between the two helices is larger in the experimental structure, and the helices are slightly more closely packed in the structure derived from homology modeling.
Amide proton exchange
To investigate the extent to which amide protons are protected from solvent exchange, we measured reductions due to saturation transfer in a [15N,1H]-HSQC spectrum (Fig. S7). Significantly reduced saturation transfer (>80% remaining peak intensity) was observed in segments 37–48, 52–64, 66–72, and 80–107, whereas a more rapid exchange was measured in the NT region (residues 32–36) of the protein fragment and in the loop Thr72–Thr78. The observation that residues 37–48 from the NT of the protein were protected from solvent exchange is in agreement with the occurrence of a nascent, probably surface-associated helix in that part of the polypeptide chain.
Discussion
Despite intense efforts, no high-resolution NMR structure of an entire GPCR has been reported to date. Very recently, the nearly complete backbone assignment of sensory rhodopsin in DHPC was published (16). Moreover, although many biophysical studies on peptide surrogates representing regions of IMPs have been conducted, only a few detailed NMR studies on membrane-spanning peptide fragments of GPCRs in detergent micelles appear in the literature. We recently described a solution-state NMR analysis on a fragment comprising the seventh TM domain plus 40 residues from the cytosolic region of Ste2p in DPC micelles (35). A peptide corresponding to the sixth TM domain of Ste2p was also analyzed in lipid bilayers by solid-state NMR and was found to be very similar in structure to that observed in TFE/water (36).
Although important insights into the biophysical properties of peptides corresponding to single TM regions of IMPs have been derived, it remains uncertain whether such peptides are good surrogates to learn about the actual structure of these regions in the entire GPCR. Even though peptides corresponding to single TMs of GPCRs did assume helices in the presence of detergents and in organic aqueous media (28,65,66), long-range stabilizing interactions between the individual TM helices may be required for the GPCR fragment to fold into the biologically relevant conformation. Such long-range interactions are missing in a single TM fragment of the heptahelical GPCR. Our goal in this investigation was to extend the high-resolution analysis on membrane protein fragments to a region of a model GPCR that consists of two TMs and the intervening loop. Previous studies on the MerF protein, the human Gly transporter, and subunit C of the F1F0 ATP synthase provided precedents for the notion that peptides, including those corresponding to IMP fragments consisting of two TMs, can fold into a defined tertiary structure in both detergent micelles (67) and organic solvents (42,68). The only previous NMR study on a double TM fragment of a GPCR was conducted in DMSO on the TM1-TM2 fragment of the cannabinoid receptor (40). That study resulted in nearly complete assignments of the backbone atoms and indicated that this region of the receptor is highly helical, but it lacked insight into the tertiary structure of the fragment. Most recently, a detailed NMR analysis of the TM3-TM4 hairpin of the CFTR receptor in perfluorooctanoate micelles led to nearly complete assignments of the backbone resonances and, with the help of a number of specific mutations, provided insights into the structure of the helical hairpin (43).
Here, using a variety of isotope labeling patterns and NMR experiments, we achieved a nearly complete assignment of the backbone and side-chain nuclei in detergent micelles for an 80-residue protein fragment corresponding to the first two TM domains of Ste2p, the yeast α-factor GPCR. During the course of this project, we noted that the use of deuterated LPPG significantly improved spectral quality by both eliminating interfering micellar resonances and decreasing intermolecular relaxation pathways that likely result in broadening of protein resonances. In addition, only when we used proteins specifically labeled at certain methyl groups (Fig. 3) and NMR experiments suggested by the Kay laboratory (69) were we able to resolve individual methyl resonances. The combination of deuterated LPPG and methyl-labeled protein fragments allowed us to identify many medium-range NOEs (Fig. 4) that define secondary structure in the TM1 and TM2 helices and, most importantly, to discern long-range NOEs between residues on TM1 and TM2. These latter interactions provide support for the conclusion that in LPPG this fragment folds to a helical hairpin-like structure (Fig. 5).
In addition to defining the secondary structure of the TM regions and identifying the helical hairpin, our investigation revealed the presence of a helix in the amino-terminal portion of TM1-TM2 that according to its amphiphilic nature is most likely surface-associated. The identification of a helical element in the amine terminal tail of TM1-TM2 may be a biologically significant finding, as the NT of GPCRs plays essential roles in their biology. Although studies on chimeric Ste2p from Saccharomyces cerevisiae and Saccharomyces kluyveri provided evidence that the first 45 residues in the NT of Ste2p from S. cerevisiae were not critical for ligand-binding specificity or signaling (70), it was observed that substitutions in residues 47–49 affected binding specificity but not signaling (71). Moreover, deletion of the first 30 residues of the 51 residue NT of Ste2p led to MATα cells that signaled but could not mate (72). The helical region (residues 39–47) of the NT of TM1-TM2 that our NMR studies discerned is likely an amphiphilic helix that interacts with the phospholipid headgroups of the LPPG micelles. If similar interactions occur in the native environment, they may help to define the biologically active structure of the pheromone receptor. Previous crystal structures published on GPCRs either do not define their NTs or they reveal some tendency to β-sheet conformations in the termini of rhodopsin family receptors (73). An NMR study on a synthetic protein fragment corresponding to the first 110 residues of the N-Y4 receptor also found a short helical structure in the extracellular region of this NT GPCR fragment (74,75). Thus, it is conceivable that the extracellular NT domain in GPCRs contain specific secondary structures or incipient structures that can be stabilized on interaction with either ligands or other accessory proteins involved in signal transduction pathways.
The conformation of the segments corresponding to the putative TM helices 1 and 2 had reasonably low RMSD values when the individual helices were superimposed. The α-helical character is very well supported by numerous medium-range NOEs. The observation of these medium-range NOEs indicates that the helices in the two-TM construct are much more stable than the single TM helix we previously observed in the seventh TM of the same receptor in DPC micelles (35). The TM1 helix is destabilized in the G56–G60 region, and the helices cross each other and splay apart near the putative extracellular surface of this GPCR domain. At least four independent contacts between residues adjacent to the first intracellular loop were established. No such contacts were observed between residues near the center or the extracellular face of the helical hairpin. This finding could reflect the fact that contacts with additional TMs are necessary to stabilize the structure of this region of Ste2p. Although the spatial relationship between the two TMs is presently underdetermined, it is important that the polypeptide appears to take a turn in the micellar environment rather than assuming a large distribution of structures. Because of the inverse sixth-power dependence of the nuclear Overhauser interaction, it is possible that the interhelical contacts may not be persistent, resulting in two TM segments whose relative orientation is not defined for short periods of time. In addition, the fact that not all of the individual helical hairpins superimpose well is consistent with the view that the relative orientation of the two helices may not be unique over time or over the ensemble. However, the presence of a number of long-range interhelical contacts indicates that hairpin-like structures make a significant contribution to the conformational distribution of TM1-TM2 in LPPG micelles. At present we are evaluating experimental approaches to gain additional restraints that will help to more accurately define the relative orientation of TM1 and TM2.
There are several aspects of the NMR structure of TM1-TM2 that should be considered in the context of biological and biochemical information about the function of this protein fragment. Previous analyses of Ste2p biology show that the first extracellular loop and the extracellular end of TM1 are involved in both the binding of pheromone and the signal transduction pathway (70,76–78). Cross-linking studies have revealed that Tyr13 of α-factor contacts residues 55–59 of TM1 (79), and we recently determined that the contact points are Arg58 and Cys59 by photochemical and oxidative cross-linking approaches (J. M. Becker and F. Naider, unpublished). Thus, it seems clear that the carboxyl terminus of the α-factor must penetrate into the TM interior of Ste2p upon binding to this receptor. If this is correct, it would not be possible for TM1 to be tightly packed against TM2, and our finding of destabilization in the G56–G60 region and splaying apart of the two TMs would be consistent with the cross-linking results. Part of the driving force for the destabilization most likely stems from the occurrence of polar and, in particular, charged residues within the membrane. For example, transferring an Arg residue into the membrane interior requires 1.8 kcal mol−1 (80). Even if the charges from these residues are partially compensated for by polar residues placed in other TM helices, they may still introduce some conformational instability that may be important for the creation of the pheromone-binding pocket.
In conclusion, we report here an NMR structure for Ste2p(G31-T110) of the GPCR mating receptor from S. cerevisiae in LPPG micelles. Almost complete resonance assignments were accomplished for the 80-residue fragment, representing more than 25% of the residues from the core of this receptor. The conformation was determined without introducing any artificial restraints, and its secondary structure was well defined. A few interhelical contacts demonstrate that the protein is folded in micelles into a helical hairpin that splays apart at the termini. A region of the receptor predicted to be in the NT receptor tail formed a helix that likely interacts with the surface of the micelles. To our knowledge, this structure is the first reported for a double TM containing a fragment of a GPCR in lipid. Its tendency to assume a specific tertiary structure supports the use of GPCR fragments as models to discern the structure of the intact receptor.
Supporting Material
Six figures and two tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(09)00474-3.
Supporting Material
Acknowledgments
Fred Naider is the Leonard and Esther Kurtz Term Professor at the College of Staten Island, City University of New York. This work was supported by research grants GM22086 (F.N.) and GM22087 (J.M.B.) from the National Institutes of Health, and a grant from the Alfred Werner Legat (O.Z.). Professor Naider is a member of the New York Structural Biology Center, which is a STAR center supported by the New York State Office of Science, Technology, and Academic Research. The 900 MHz spectrometer at the New York Structural Biology Center was purchased with funds from the National Institutes of Health, USA; the Keck Foundation, New York State; and the New York City Economic Development Corp.
Contributor Information
Oliver Zerbe, Email: oliver.zerbe@oci.uzh.ch.
Fred Naider, Email: naider@mail.csi.cuny.edu.
References
- 1.Boyd D., Schierle C., Beckwith J. How many membrane proteins are there? Protein Sci. 1998;7:201–205. doi: 10.1002/pro.5560070121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stevens T.J., Arkin I.T. Do more complex organisms have a greater proportion of membrane proteins in their genomes? Proteins. 2000;39:417–420. doi: 10.1002/(sici)1097-0134(20000601)39:4<417::aid-prot140>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 3.Hopkins A.L., Groom C.R. The druggable genome. Nat. Rev. Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 4.Palczewski K., Kumasaka T., Hori T., Behnke C.A., Motoshima H. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 5.Rasmussen S.G.F., Choi H.J., Rosenbaum D.M., Kobilka T.S., Thian F.S. Crystal structure of the human β(2) adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 6.Warne T., Serrano-Vega M.J., Baker J.G., Moukhametzianov R., Edwards P.C. Structure of a β1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaakola V.P., Griffith M.T., Hanson M.A., Cherezov V., Chien E.Y. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarramegn V., Muller I., Milon A., Talmont F. Recombinant G protein-coupled receptors from expression to renaturation: a challenge towards structure. Cell. Mol. Life Sci. 2006;63:1149–1164. doi: 10.1007/s00018-005-5557-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grisshammer R., White J.F., Trinh L.B., Shiloach J. Large-scale expression and purification of a G-protein-coupled receptor for structure determination—an overview. J. Struct. Funct. Genomics. 2005;6:159–163. doi: 10.1007/s10969-005-1917-6. [DOI] [PubMed] [Google Scholar]
- 10.Page R.C., Moore J.D., Nguyen H.B., Sharma M., Chase R. Comprehensive evaluation of solution nuclear magnetic resonance spectroscopy sample preparation for helical integral membrane proteins. J. Struct. Funct. Genomics. 2006;7:51–64. doi: 10.1007/s10969-006-9009-9. [DOI] [PubMed] [Google Scholar]
- 11.Sanders C.R., Sönnichsen F. Solution NMR of membrane proteins: practice and challenges. Magn. Reson. Chem. 2006;44:S24–S40. doi: 10.1002/mrc.1816. [DOI] [PubMed] [Google Scholar]
- 12.Hu J., Qin H., Li C., Sharma M., Cross T.A. Structural biology of transmembrane domains: efficient production and characterization of transmembrane peptides by NMR. Protein Sci. 2007;16:2153–2165. doi: 10.1110/ps.072996707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williamson P.T., Verhoeven A., Miller K.W., Meier B.H., Watts A. The conformation of acetylcholine at its target site in the membrane-embedded nicotinic acetylcholine receptor. Proc. Natl. Acad. Sci. USA. 2007;104:18031–18036. doi: 10.1073/pnas.0704785104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vosegaard T., Kamihira-Ishijima M., Watts A., Nielsen N.C. Helix conformations in 7TM membrane proteins determined using oriented-sample solid-state NMR with multiple residue-specific 15N labeling. Biophys. J. 2008;94:241–250. doi: 10.1529/biophysj.107.116004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luca S., White J.F., Sohal A.K., Filippov D.V., van Boom J.H. The conformation of neurotensin bound to its G protein-coupled receptor. Proc. Natl. Acad. Sci. USA. 2003;100:10706–10711. doi: 10.1073/pnas.1834523100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gautier A., Kirkpatrick J.P., Nietlispach D. Solution-state NMR spectroscopy of a seven-helix transmembrane protein receptor: backbone assignment, secondary structure, and dynamics. Angew. Chem. Int. Ed. Engl. 2008;47:7297–7300. doi: 10.1002/anie.200802783. [DOI] [PubMed] [Google Scholar]
- 17.Popot J.L., Engelman D.M. Helical membrane protein folding, stability, and evolution. Annu. Rev. Biochem. 2000;69:881–922. doi: 10.1146/annurev.biochem.69.1.881. [DOI] [PubMed] [Google Scholar]
- 18.White S.H., Wimley W.C. Membrane protein folding and stability: physical principles. Annu. Rev. Biophys. Biomol. Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 19.Senes A., Engel D.E., DeGrado W.F. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr. Opin. Struct. Biol. 2004;14:465–479. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Barsukov I.L., Abdulaeva G.V., Arseniev A.S., Bystrov V.F. Sequence-specific 1H-NMR assignment and conformation of proteolytic fragment 163–231 of bacterioopsin. Eur. J. Biochem. 1990;192:321–327. doi: 10.1111/j.1432-1033.1990.tb19230.x. [DOI] [PubMed] [Google Scholar]
- 21.Grabchuk I.A., Orekhov V.Y., Arseniev A.S. 1H-15N backbone resonance assignments of bacteriorhodopsin. Pharm. Acta Helv. 1996;71:97–102. doi: 10.1016/0031-6865(95)00055-0. [DOI] [PubMed] [Google Scholar]
- 22.Lomize A.L., Pervushin K.V., Arseniev A.S. Spatial structure of (34–65)bacterioopsin polypeptide in SDS micelles determined from nuclear magnetic resonance data. J. Biomol. NMR. 1992;2:361–372. doi: 10.1007/BF01874814. [DOI] [PubMed] [Google Scholar]
- 23.Pervushin K.V., Arseniev A.S. Three-dimensional structure of (1–36)bacterioopsin in methanol-chloroform mixture and SDS micelles determined by 2D 1H-NMR spectroscopy. FEBS Lett. 1992;308:190–196. doi: 10.1016/0014-5793(92)81272-n. [DOI] [PubMed] [Google Scholar]
- 24.Pervushin K.V., Arseniev A.S., Kozhich A.T., Ivanov V.T. Two-dimensional NMR study of the conformation of (34–65)bacterioopsin polypeptide in SDS micelles. J. Biomol. NMR. 1991;1:313–322. doi: 10.1007/BF02192857. [DOI] [PubMed] [Google Scholar]
- 25.Pervushin K.V., Orekhov V.Y., Popov A.I., Musina L.Y., Arseniev A.S. Three-dimensional structure of (1–71)bacterioopsin solubilized in methanol/chloroform and SDS micelles determined by 15N-1H heteronuclear NMR spectroscopy. Eur. J. Biochem. 1994;219:571–583. doi: 10.1111/j.1432-1033.1994.tb19973.x. [DOI] [PubMed] [Google Scholar]
- 26.Chopra A., Yeagle P.L., Alderfer J.A., Albert A.D. Solution structure of the sixth transmembrane helix of the G-protein-coupled receptor, rhodopsin. Biochim. Biophys. Acta. 2000;1463:1–5. doi: 10.1016/s0005-2736(99)00212-6. [DOI] [PubMed] [Google Scholar]
- 27.Katragadda M., Chopra A., Bennett M., Alderfer J.L., Yeagle P.L. Structures of the transmembrane helices of the G-protein coupled receptor, rhodopsin. J. Pept. Res. 2001;58:79–89. doi: 10.1034/j.1399-3011.2001.00904.x. [DOI] [PubMed] [Google Scholar]
- 28.Arshava B., Taran I., Xie H., Becker J.M., Naider F. High resolution NMR analysis of the seven transmembrane domains of a heptahelical receptor in organic-aqueous medium. Biopolymers. 2002;64:161–176. doi: 10.1002/bip.10157. [DOI] [PubMed] [Google Scholar]
- 29.Xie H., Ding F.X., Schreiber D., Eng G., Liu S.F. Synthesis and biophysical analysis of transmembrane domains of a Saccharomyces cerevisiae G protein-coupled receptor. Biochemistry. 2000;39:15462–15474. doi: 10.1021/bi001432p. [DOI] [PubMed] [Google Scholar]
- 30.Reddy A.P., Tallon M.A., Becker J.M., Naider F. Biophysical studies on fragments of the α-factor receptor protein. Biopolymers. 1994;34:679–689. doi: 10.1002/bip.360340510. [DOI] [PubMed] [Google Scholar]
- 31.Arshava B., Liu S.F., Jiang H., Breslav M., Becker J.M. Structure of segments of a G protein-coupled receptor: CD and NMR analysis of the Saccharomyces cerevisiae tridecapeptide pheromone receptor. Biopolymers. 1998;46:343–357. doi: 10.1002/(SICI)1097-0282(199811)46:6<343::AID-BIP1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 32.Estephan R., Englander J., Arshava B., Samples K.L., Becker J.M. Biosynthesis and NMR analysis of a 73-residue domain of a Saccharomyces cerevisiae G protein-coupled receptor. Biochemistry. 2005;44:11795–11810. doi: 10.1021/bi0507231. [DOI] [PubMed] [Google Scholar]
- 33.Naider F., Arshava B., Ding F.X., Arevalo E., Becker J.M. Peptide fragments as models to study the structure of a G-protein coupled receptor: the α-factor receptor of Saccharomyces cerevisiae. Biopolymers. 2001;60:334–350. doi: 10.1002/1097-0282(2001)60:5<334::AID-BIP10175>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 34.Naider F., Khare S., Arshava B., Severino B., Russo J. Synthetic peptides as probes for conformational preferences of domains of membrane receptors. Biopolymers. 2005;80:199–213. doi: 10.1002/bip.20183. [DOI] [PubMed] [Google Scholar]
- 35.Neumoin A., Arshava B., Becker J., Zerbe O., Naider F. NMR studies in dodecylphosphocholine of a fragment containing the seventh transmembrane helix of a G-protein-coupled receptor from Saccharomyces cerevisiae. Biophys. J. 2007;93:467–482. doi: 10.1529/biophysj.106.103770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valentine K.G., Liu S.F., Marassi F.M., Veglia G., Opella S.J. Structure and topology of a peptide segment of the 6th transmembrane domain of the Saccharomyces cerevisae α-factor receptor in phospholipid bilayers. Biopolymers. 2001;59:243–256. doi: 10.1002/1097-0282(20011005)59:4<243::AID-BIP1021>3.0.CO;2-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thévenin D., Roberts M.F., Lazarova T., Robinson C.R. Identifying interactions between transmembrane helices from the adenosine A2A receptor. Biochemistry. 2005;44:16239–16245. doi: 10.1021/bi051422u. [DOI] [PubMed] [Google Scholar]
- 38.Thevenin D., Lazarova T. Stable interactions between the transmembrane domains of the adenosine A2A receptor. Protein Sci. 2008;17:1188–1199. doi: 10.1110/ps.034843.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kerman A., Ananthanarayanan V.S. Expression and spectroscopic characterization of a large fragment of the mu-opioid receptor. Biochim. Biophys. Acta. 2005;1747:133–140. doi: 10.1016/j.bbapap.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 40.Zheng H., Zhao J., Sheng W., Xie X.Q. A transmembrane helix-bundle from G-protein coupled receptor CB2: biosynthesis, purification, and NMR characterization. Biopolymers. 2006;83:46–61. doi: 10.1002/bip.20526. [DOI] [PubMed] [Google Scholar]
- 41.Kerman A., Ananthanarayanan V.S. Conformation of a double-membrane-spanning fragment of a G protein-coupled receptor: effects of hydrophobic environment and pH. Biochim. Biophys. Acta. 2007;1768:1199–1210. doi: 10.1016/j.bbamem.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 42.Ma D., Liu Z., Li L., Tang P., Xu Y. Structure and dynamics of the second and third transmembrane domains of human glycine receptor. Biochemistry. 2005;44:8790–8800. doi: 10.1021/bi050256n. [DOI] [PubMed] [Google Scholar]
- 43.Wehbi H., Gasmi-Seabrook G., Choi M.Y., Deber C.M. Positional dependence of non-native polar mutations on folding of CFTR helical hairpins. Biochim. Biophys. Acta. 2008;1778:79–87. doi: 10.1016/j.bbamem.2007.08.036. [DOI] [PubMed] [Google Scholar]
- 44.Tugarinov V., Kay L.E. Ile, Leu, and Val methyl assignments of the 723-residue malate synthase G using a new labeling strategy and novel NMR methods. J. Am. Chem. Soc. 2003;125:13868–13878. doi: 10.1021/ja030345s. [DOI] [PubMed] [Google Scholar]
- 45.Cohen L.S., Arshava B., Estephan R., Englander J., Kim H. Expression and biophysical analysis of two double-transmembrane domain-containing fragments from a yeast G protein-coupled receptor. Biopolymers. 2008;90:117–130. doi: 10.1002/bip.20950. [DOI] [PubMed] [Google Scholar]
- 46.Hauser M., Kauffman S., Lee B.K., Naider F., Becker J.M. The first extracellular loop of the Saccharomyces cerevisiae G protein-coupled receptor Ste2p undergoes a conformational change upon ligand binding. J. Biol. Chem. 2007;282:10387–10397. doi: 10.1074/jbc.M608903200. [DOI] [PubMed] [Google Scholar]
- 47.Martin N.P., Celic A., Dumont M.E. Mutagenic mapping of helical structures in the transmembrane segments of the yeast α-factor receptor. J. Mol. Biol. 2002;317:765–788. doi: 10.1006/jmbi.2002.5444. [DOI] [PubMed] [Google Scholar]
- 48.Tugarinov V., Kanelis V., Kay L.E. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat. Protocols. 2006;1:749–754. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- 49.Yamazaki T., Lee W., Arrowsmith C.H., Muhandiram D.R., Kay L.E. A suite of triple-resonance NMR experiments for the backbone assignment of N-15,C-13,H2- labeled proteins with high sensitivity. J. Am. Chem. Soc. 1994;116:11655–11666. [Google Scholar]
- 50.Shan X., Gardner K.H., Muhandiram D.R., Rao N.S., Arrowsmith C.H. Assignment of N-15, C-13(α), C-13(β), and HN resonances in an N-15, C-13, H-2 labeled 64 kDa trp repressor-operator complex using triple-resonance NMR spectroscopy and H-2-decoupling. J. Am. Chem. Soc. 1996;118:6570–6579. [Google Scholar]
- 51.Kay L., Xu G.Y., Singer A.U., Muhandiram D.R., Forman-Kay J.D. A gradient-enhanced HCCH-TOCSY experiment for recording side-chan 1H and 13C correlations in H2O samples of proteins. J. Magn. Reson. B. 1993;101:333–337. [Google Scholar]
- 52.Olejniczak E.T., Xu R.X., Fesik S.W. A 4D HCCH-TOCSY experiment for assigning the side chain 1H and 13C resonances of proteins. J. Biomol. NMR. 1992;2:655–659. doi: 10.1007/BF02192854. [DOI] [PubMed] [Google Scholar]
- 53.Vuister G.W., Bax A. Resolution enhancement and spectral editing of uniformly 13C-enriched proteins by homonuclear broadband 13C decoupling. J. Magn. Reson. 1992;98:428. [Google Scholar]
- 54.Yamazaki T., Forman-Kay J.D., Kay L.E. 2-Dimensional NMR experiments for correlating C-13-β and H-1-δ/epsilon chemical-shifts of aromatic residues in C-13-labeled proteins via scalar couplings. J. Am. Chem. Soc. 1993;115:11054–11055. [Google Scholar]
- 55.Keeler J., Clowes R.T., Davis A.L., Laue E.D. Pulsed-field gradients: theory and practice. Methods Enzymol. 1994;239:145–207. doi: 10.1016/s0076-6879(94)39006-1. [DOI] [PubMed] [Google Scholar]
- 56.Noggle J.H., Schirmer R.E. Academic Press; New York: 1971. The Nuclear Overhauser Effect—Chemical Applications. [Google Scholar]
- 57.Palmer A.G. NMR characterization of the dynamics of biomacromolecules. Chem. Rev. 2004;104:3623–3640. doi: 10.1021/cr030413t. [DOI] [PubMed] [Google Scholar]
- 58.Keller, R. 2004. The Computer Aided Resonance Assignment. Cantina Verlag, Goldau, Switzerland.
- 59.Bartels C., Xia T.H., Billeter M., Güntert P., Wüthrich K. The program XEASY for computer-supported spectral analysis of biological macromolecules. J. Biomol. NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 60.Koradi R., Billeter M., Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 61.Cornilescu G., Delaglio F., Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 62.Güntert P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 63.Kabsch W., Sander C. Dictionary of protein secondary strucutre: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 64.Eilers M., Hornak V., Smith S.O., Konopka J.B. Comparison of class A and D G protein-coupled receptors: common features in structure and activation. Biochemistry. 2005;44:8959–8975. doi: 10.1021/bi047316u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hunt J.F., Rath P., Rothschild K.J., Engelman D.M. Spontaneous, pH-dependent membrane insertion of a transbilayer α-helix. Biochemistry. 1997;36:15177–15192. doi: 10.1021/bi970147b. [DOI] [PubMed] [Google Scholar]
- 66.Katragadda M., Alderfer J.L., Yeagle P.L. Assembly of a polytopic membrane protein structure from the solution structures of overlapping peptide fragments of bacteriorhodopsin. Biophys. J. 2001;81:1029–1036. doi: 10.1016/S0006-3495(01)75760-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Howell S.C., Mesleh M.F., Opella S.J. NMR structure determination of a membrane protein with two transmembrane helices in micelles: MerF of the bacterial mercury detoxification system. Biochemistry. 2005;44:5196–5206. doi: 10.1021/bi048095v. [DOI] [PubMed] [Google Scholar]
- 68.Girvin M.E., Rastogi V.K., Abildgaard F., Markley J.L., Fillingame R.H. Solution structure of the transmembrane H+-transporting subunit c of the F1F0 ATP synthase. Biochemistry. 1998;37:8817–8824. doi: 10.1021/bi980511m. [DOI] [PubMed] [Google Scholar]
- 69.Tugarinov V., Kay L.E. Methyl groups as probes of structure and dynamics in NMR studies of high-molecular-weight proteins. ChemBioChem. 2005;6:1567–1577. doi: 10.1002/cbic.200500110. [DOI] [PubMed] [Google Scholar]
- 70.Sen M., Marsh L. Noncontiguous domains of the α-factor receptor of yeasts confer ligand specificity. J. Biol. Chem. 1994;269:968–973. [PubMed] [Google Scholar]
- 71.Sen M., Shah A., Marsh L. Two types of α-factor receptor determinants for pheromone specificity in the mating-incompatible yeasts S. cerevisiae and S. kluyveri. Curr. Genet. 1997;31:235–240. doi: 10.1007/s002940050200. [DOI] [PubMed] [Google Scholar]
- 72.Shi C., Kaminskyj S., Caldwell S., Loewen M.C. A role for a complex between activated G protein-coupled receptors in yeast cellular mating. Proc. Natl. Acad. Sci. USA. 2007;104:5395–5400. doi: 10.1073/pnas.0608219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Park J.H., Scheerer P., Hofmann K.P., Choe H.W., Ernst O.P. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 74.Zou C., Kumaran S., Markovic S., Walser R., Zerbe O. Studies of the structure of the N-terminal domain from the Y4 receptor, a G-protein coupled receptor, and its interaction with hormones from the NPY family. ChemBioChem. 2008;9:2276–2284. doi: 10.1002/cbic.200800221. [DOI] [PubMed] [Google Scholar]
- 75.Zou C., Naider F., Zerbe O. Biosynthesis and NMR-studies of a double transmembrane domain from the Y4 receptor, a human GPCR. J. Biomol. NMR. 2008;42:257–269. doi: 10.1007/s10858-008-9281-z. [DOI] [PubMed] [Google Scholar]
- 76.Lee B.K., Khare S., Naider F., Becker J.M. Identification of residues of the Saccharomyces cerevisiae G protein-coupled receptor contributing to α-factor pheromone binding. J. Biol. Chem. 2001;276:37950–37961. doi: 10.1074/jbc.M103579200. [DOI] [PubMed] [Google Scholar]
- 77.Akal-Strader A., Khare S., Xu D., Naider F., Becker J.M. Residues in the first extracellular loop of a G protein-coupled receptor play a role in signal transduction. J. Biol. Chem. 2002;277:30581–30590. doi: 10.1074/jbc.M204089200. [DOI] [PubMed] [Google Scholar]
- 78.Lin J.C., Duell K., Konopka J.B. A microdomain formed by the extracellular ends of the transmembrane domains promotes activation of the G protein-coupled α-factor receptor. Mol. Cell. Biol. 2004;24:2041–2051. doi: 10.1128/MCB.24.5.2041-2051.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Son C.D., Sargsyan H., Naider F., Becker J.M. Identification of ligand binding regions of the Saccharomyces cerevisiae α-factor pheromone receptor by photoaffinity cross-linking. Biochemistry. 2004;43:13193–13203. doi: 10.1021/bi0496889. [DOI] [PubMed] [Google Scholar]
- 80.White S.H., Wimley W.C. Hydrophobic interactions of peptides with membrane interfaces. Biochim. Biophys. Acta. 1998;1376:339–352. doi: 10.1016/s0304-4157(98)00021-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.