Abstract
Makorin Ring Finger Protein 1 (MKRN1) is a transcriptional co-regulator and an E3 ligase. Here, we show that MKRN1 simultaneously functions as a differentially negative regulator of p53 and p21. In normal conditions, MKRN1 could destabilize both p53 and p21 through ubiquitination and proteasome-dependent degradation. As a result, depletion of MKRN1 induced growth arrest through activation of p53 and p21. Interestingly, MKRN1 used earlier unknown sites, K291 and K292, for p53 ubiquitination and subsequent degradation. Under severe stress conditions, however, MKRN1 primarily induced the efficient degradation of p21. This regulatory process contributed to the acceleration of DNA damage-induced apoptosis by eliminating p21. MKRN1 depletion diminished adriamycin or ultraviolet-induced cell death, whereas ectopic expression of MKRN1 facilitated apoptosis. Furthermore, MKRN1 stable cell lines that constantly produced low levels of p53 and p21 exhibited stabilization of p53, but not p21, with increased cell death on DNA damage. Our results indicate that MKRN1 exhibits dual functions of keeping cells alive by suppressing p53 under normal conditions and stimulating cell death by repressing p21 under stress conditions.
Keywords: Hdm2 (Mdm2), MKRN1, p21, p53, ubiquitination
Introduction
p53 has important functions for tumour suppression, which is accomplished by the induction of cell cycle arrest, DNA repair, and apoptosis in response to various cellular stresses, including DNA damage, oncogenic stress, telomere dysfunction, and hypoxia (Vogelstein et al, 2000; Bode and Dong, 2004; Lavin and Gueven, 2006; Vousden and Lane, 2007). On account of these cytotoxic effects, strict controls of its steady-state levels and activities exist in normal cells. One of the representative examples of cell protection from p53-mediated cytotoxicity is the ubiquitin-proteasome system, which removes p53 from the cell. Hdm2, the first identified E3 ubiquitin ligase for p53, is a major protein that facilitates ubiquitin-dependent degradation of p53 (Haupt et al, 1997; Honda et al, 1997; Kubbutat et al, 1997). Nevertheless, p53 is degraded in Mdm2-null tissues, although it not as efficient as in wild-type tissues (Ringshausen et al, 2006). Furthermore, other E3 ligases, such as pirh2, COP1, and ARF-BP1, stimulate p53 ubiquitination and degradation (Leng et al, 2003; Dornan et al, 2004; Chen et al, 2005). In addition, a variety of p53 cofactors, including ARF, MdmX, PML, ATM, and ribosomal proteins, are involved in the regulatory pathways affecting stabilization of p53 (Lavin and Gueven, 2006).
Although transcriptional activation of p53 is restricted by several E3 ligases and cofactors under normal conditions, p53 can escape from the numerous negative regulatory pathways under stress conditions (Brooks and Gu, 2006; Dornan et al, 2006). Post-translational modification of p53 accompanying phosphorylation and acetylation enables p53 to resist the attack of E3 ligases, resulting in the induction of cellular responses (Brooks and Gu, 2006; Dornan et al, 2006; Tang et al, 2008). The responses against p53-dependent stress signals result in the production of hundreds of p53 target genes, which introduce differentiated and graded responses against different stress levels. Indeed, many cofactors and specific modifiers of p53, such as HIPK2, p63, p73, JMY, and ASPP, differentially regulate the responses mediated by p53 activation (Oren, 2003; Rodier et al, 2007).
Among p53 target genes, p21 (also called WAF1 or CIP1) is considered to be one of the key factors differentiating cell cycle arrest and apoptosis. p21 was first recognized as a member of the cyclin-dependent kinase inhibitors (CKIs) (Gartel and Radhakrishnan, 2005). p21 overexpression induces G1, G2, or S phase arrest, whereas p21-deficient cells display a defect in DNA damage-induced G1 and G2 arrest (Gartel and Radhakrishnan, 2005). Furthermore, sustained activation of p21 can lead to permanent cellular senescence (Chang et al, 2000; Roninson, 2002). However, inactivation or depletion of p21 sensitizes cells to apoptosis under conditions of low DNA damage, which induces cell cycle arrest in p21-positive cells (Waldman et al, 1996; Zhang et al, 1999; Mahyar-Roemer and Roemer, 2001; Han et al, 2002; Javelaud and Besancon, 2002; Martinez et al, 2002). Similarly, p21 deficiency decreases tumourigenesis and increases sensitivity to apoptosis in MMTV-Ras mice and ATM-deficient mice (Wang et al, 1997; Bearss et al, 2002). In contrast, ectopically expressed p21 delays apoptosis induced by several kinds of DNA damage in various cell lines and in a xenograft mouse model (Gorospe et al, 1996, 1997). Clinical data also support the observation that tumours overexpressing p21 are more resistant to chemotherapeutic treatment (Aaltomaa et al, 1999; Baretton et al, 1999). Consistent with these findings, p21 accumulation is evident in several tumours (Erber et al, 1997; Fizazi et al, 2002). As p21 functions as a mediator of cell cycle arrest and an inhibitor of apoptosis, the levels and activities of p21 are thought be regulated by different levels of DNA damage. The steady-state levels of p21 decrease with lethal doses of adriamycin, whereas they increase in the presence of sub-lethal doses (Martinez et al, 2002). Recently, p21 transcription was shown to be repressed under lethal DNA damage by several transcriptional cofactors, such as Myc, Miz, Zbtb4, and Pdcd4 (Seoane et al, 2002; Bitomsky et al, 2008; Weber et al, 2008). Furthermore, p21 is inactivated through caspase-3-mediated cleavage or proteasome-dependent degradation to undergo efficient apoptosis (Zhang et al, 1999; Bloom et al, 2003; Jin et al, 2003; Nishitani et al, 2008).
Makorin Ring Finger Protein 1 (MKRN1) was first identified as an intron-containing source gene for the MKRN gene family, the majority of which has been identified as being intronless and imprinted (Gray et al, 2000). The MKRN family contains several C3H, C3HC4, and unique Cys-His motifs, all of which are highly conserved from invertebrate to vertebrate species (Gray et al, 2000). Furthermore, MKRN1 is constitutively expressed in various human and mouse tissues from fetus to adult (Gray et al, 2000). Recently, MKRN1 was shown to act as an E3 ligase for hTERT, and the transcriptional co-regulator of androgen receptor and retinoic acid receptor (Kim et al, 2005; Omwancha et al, 2006). However, the cellular and physiological function(s) of MKRN1 is yet to be defined.
Here, we report new roles of MKRN1 as an E3 ubiquitin ligase for both p53 and p21. Ablation of MKRN1 resulted in cellular growth defect with the stabilization of p53 and p21. Interestingly, MKRN1 was able to induce the ubiquitination and degradation of both p53 and p21. Although the negative regulatory effects on p53 by MKRN1 were disrupted under DNA-damaging stresses, MKRN1 was still able to mediate destabilization of p21. Accordingly, knockdown of MKRN1 attenuated adriamycin-induced apoptosis, which is due in part to up-regulated levels of p21. Taken together, our data indicate that MKRN1 is a novel modulator of p53 and p21, preferentially leading cells to p53-dependent apoptosis by suppressing p21.
Results
Depletion of MKRN1 induces cell cycle arrest by activating p53 and p21
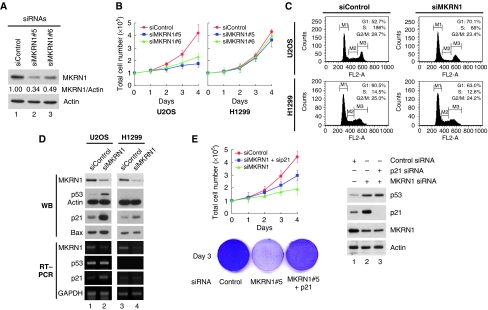
The activities of MKRN1 as an E3 ligase for hTERT (Kim et al, 2005) prompted us to test the effects of MKRN1 on the proliferation of tumour cells. The H1299 and U2OS tumour cells lines, which possess and lack telomerase activity, respectively, were used for this purpose. First, we tested four kinds of MKRN1 small interfering RNA (siRNA) purchased from Qiagen-Xeragon. Among the four sequences tested, siMKRN1#5 and siMKRN1#6 were able to effectively deplete endogenous MKRN1 up to 66% (Figure 1A). When these MKRN1 siRNAs were transfected into H1299 or U2OS cells, we unexpectedly observed that MKRN1-depleted cells had a severe defect in U2OS cell proliferation, although not displaying any noticeable effects with H1299 cells (Figure 1B; Supplementary Figure S1A). The growth defect shown in U2OS cells seemed not to be due to cell death, as MKRN1 depletion had little effect on U2OS cell death as determined by trypan blue staining (Supplementary Figure S1B). To establish whether ablation of MKRN1 resulted in cell cycle arrest, we analysed cell cycle profile of U2OS and H1299 cells treated with MKRN1 siRNA (siMKRN1#5) or control siRNA. As siMKRN1#5 was marginally more efficient in suppressing cell growth, we used this siRNA in subsequent experiments. Ablation of MKRN1 in U2OS cells resulted in increased cell fractions in G1 phase from 53 to 70%, and in decreased cell fractions in S and G2/M cells (Figure 1C, upper panel). However, MKRN1 knockdown had little effect on cell cycle distribution in H1299 cells (Figure 1C, lower panel).
Figure 1.
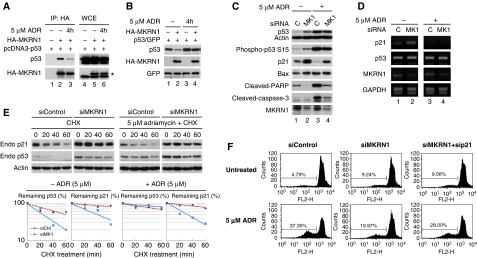
Ablation of MKRN1 induces cell growth arrest by up-regulating p53 and p21. (A) U2OS cells were transfected with 50 nM of control or MKRN1 siRNAs (siMKRN1#5, siMKRN1#6) for 48 h. Total cell lysates were immunoblotted using α-MKRN1 or α-actin antibodies. (B) U2OS and H1299 cells were transfected with the indicated siRNAs. Total cell number was counted at the indicated times using a hemacytometer. Three independent experiments were carried out, and the standard deviation (s.d.) is indicated by error bars. (C) Cell cycle analysis of MKRN1-knockdown cells. U2OS and H1299 cells were transfected with control siRNA (siControl) or MKRN1 siRNA (siMKRN1#5). At 72 h after transfection, cells were stained by propidium iodide and analysed by FACS. (D) Western blot and RT–PCR analysis of p53 and p21 in MKRN1-knockdown cells. U2OS and H1299 cells were transfected with the indicated siRNAs for 48 h. Cell lysates were immunoblotted with antibodies against MKRN1, p53, p21, Bax, and actin. For RT–PCR analysis, total RNAs in transfected cells were purified using Trizol reagent, reverse transcribed, and amplified using specific primers. (E) Proliferation of p21- and MKRN1-knockdown cells. U2OS cells were transfected with the indicated siRNAs (40 nM for MKRN1, 20 nM for p21). Total cells were counted and viable cells were stained with crystal violet. The protein levels of p53, p21, and MKRN1 were analysed by western blot analysis.
As MKRN1 seemed to induce cell cycle arrest in cells having wild-type p53, we sought to determine whether there were any changes in the level of p53. We also determined the level of p21 and Bax1, p53-dependent mediators of cell cycle arrest and apoptosis, respectively. Indeed, depletion of MKRN1 induced up-regulation of p53 protein levels, but not mRNA, in U2OS cells (Figure 1D, lanes 1 and 2). MKRN1 depletion also induced strong up-regulation of p21 and Bax proteins with increased levels of p21 mRNA in U2OS cells (Figure 1D). Interestingly, p21 protein levels in p53-null H1299 cells were also increased by MKRN1 knockdown (Figure 1D, lanes 3 and 4). Although these increases were not enough to induce cell cycle arrest, they suggest the possibility that p21 might be regulated by MKRN1 (Figure 1D). The mechanism by which MKRN1 directly regulates p21 protein will be discussed later. As growth retardation of U2OS cells by MKRN1 depletion seemed to be associated with p21 stabilization, we tested the effect of p21 depletion on MKRN1 knockdown. The data indicated that ablation of p21 could partially prevent cell growth retardation induced by MKRN1 knockdown (Figure 1E). These results suggest that the accumulation of p21 on depletion of MKRN1 was involved in inducing growth retardation. As the lack of p21 could not completely rescue cells from growth retardation by MKRN1 depletion in U2OS cells, we could not exclude the possibility of other factors being involved in this process as well.
Although the cell growth retardation of U2OS and H1299 cells seems to indicate a plausible connection between MKRN1, p53, and p21, we could not exclude the possibility of genome variation between two cell lines producing the results shown above. To convincingly show the effects of MKRN1 on the levels of p53 and p21 for cell growth, we used the isogenic cell lines, HCT116, HCT116 p53−/−, and HCT116 p21−/−. On depletion of MKRN1, the growth rate of HCT116 cells was retarded >60% with concomitant increases in the levels of p53 and its targets, p21and Bax (Supplementary Figure S2A, upper panel, and S2C, lanes 1 and 2). The cell cycle analyses using fluorescence-activated cell sorting (FACS) indicated that MKRN1 depletion also induced G1 cell cycle arrest of HCT116 cells from 63 to 75% (Supplementary Figure S2B). Cell death under the conditions of MKRN1 depletion was not significant for HCT116 cell line, the results of which were similar to U2OS cells (Supplementary Figure S2A, lower pannel). Contrary to HCT116 cells, MKRN1 knockdown in HCT116 p53−/− or HCT116 p21−/− cells had little effect on cell growth retardation and cell cycle arrest (Supplementary Figure S2A–C). Particularly, the results for HCT116 p21−/− cells showing that MKRN1 depletion-mediated stabilized p53 did not induce cell cycle arrest or growth retardation indicate that p21 might be directly involved in MKRN1 depletion-induced growth retardation.
Collectively, these data suggest that MKRN1 depletion can induce accumulation of p53 and its target, p21, resulting in the growth arrest of tumour cell lines. Moreover, the results suggest that MKRN1 might be involved in regulatory pathways affecting both p53 and p21 levels.
MKRN1 induces the ubiquitination and proteasomal degradation of p53
The observations that MKRN1 could affect the growth rate of cells by affecting p53 levels led us to test whether p53 and MKRN1 could interact with each other. Immunoprecipitation analyses showed that exogenous MKRN1 could bind to transiently overexpressed p53 (Supplementary Figure S3A and B). We then tested whether endogenous MKRN1 could form a complex with endogenous p53. Reciprocal immunoprecipitation analyses indicated that the endogenous complex between MKRN1 and p53 was detectable in U2OS cells (Supplementary Figure S3C and D). To identify the domain of p53 responsible for MKRN1 interaction, we first used several serial deletion mutants of p53, all of which interacted with MKRN1 (Supplementary Figure S4A). These data indicate that MKRN1 might have multiple-binding sites on p53. Using different fragments of p53, we concluded that there are two domains, 43–200 and 201–300, which are responsible for its interaction with MKRN1 (Supplementary Figure S4B). Next, we investigated the domain of MKRN1 responsible for binding to p53. Although ΔC1 mutant of MKRN1 (1–280) was unable to bind to p53, ΔC2 (1–369) and ΔN (264–482) deletion mutants could bind (Supplementary Figure S4C). As the ΔN mutant bound to p53 with a higher affinity than ΔC2, the region between residues 369 and 482 might be a major binding site for p53 (Supplementary Figure S4C). Finally, a direct interaction between MKRN1 and p53 was further confirmed using recombinant proteins. When GST-MKRN1 was incubated with His-tagged p53-core domain (94–292), we were able to identify the interaction between the two proteins through GST pull down analyses (Supplementary Figure S3E).
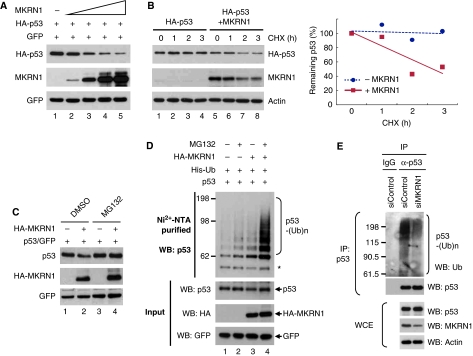
As depletion of MKRN1 stabilizes p53 and both proteins interact directly, we monitored the steady-state levels of p53 with increasing levels of exogenous MKRN1 in H1299 cells. As expected, the levels of HA-p53 decreased in the presence of exogenous MKRN1 (Figure 2A). Kinetic analyses of p53 degradation by MKRN1 in the presence of the protein synthesis inhibitor cycloheximide (CHX) indicated that although ectopically expressed p53 was relatively stable for 3 h, stability of p53 was drastically decreased up to 60% by exogenous MKRN1 after the same time (Figure 2B). MKRN1 was also destabilized by the CHX treatment, supporting the earlier study that MKRN1 is degraded by self-ubiquitination (Kim et al, 2005). As MKRN1 is an E3 ligase, we tested whether p53 degradation is dependent on polyubiquitination and the 26S proteasome. The results indicated that the proteasome inhibitor MG132 could reverse the MKRN1-mediated degradation process of p53 suggesting the necessity of 26S proteasome-dependent degradation (Figure 2C). Accordingly, ubiquitination analyses showed that MKRN1 was able to induce polyubiquitination of p53 when proteins were exogeneously introduced into cells (Figure 2D). To further analyse the ubiquitination process in a more relevant physiological context, the ubiquitination of endogenous p53 was analysed with endogenous MKRN1 depleted by MKRN1 siRNA. Endogenous levels of ubiquitinated p53 were reduced suggesting the necessity of endogenous MKRN1 in the ubiquitination process of p53 under normal conditions (Figure 2E).
Figure 2.
MKRN1 induces proteasome-dependent degradation and ubiquitination of p53 through its ring finger domain. (A) Degradation of p53 by MKRN1. H1299 cells, cultured in six-well plates, were transfected with the plasmid expressing HA-p53 (0.2 μg) and increasing amounts of MKRN1 (0.4, 0.8, 1.0, and 1.2 μg). Steady-state levels of p53 and MKRN1 were determined by western blotting. pEGFP-C2 expressing enhanced green fluorescent protein (EGFP) was cotransfected as a transfection control. (B) Destabilization of p53 by MKRN1. H1299 cells were treated with 100 μg/ml CHX for the indicated time after transfection with plasmids expressing HA-p53 (0.2 μg) and MKRN1 (1.2 μg), and analysed by western blotting. Relative amounts of p53 were calculated after normalizing to actin and are shown in the right panel. (C) Proteasomal-dependent degradation of p53 by MKRN1. H1299 cells, transfected with the indicated plasmids, were treated with 20 μM MG132 for 3 h, followed by western blot analysis. (D) MKRN1-mediated ubiquitination of p53. H1299 cells were transfected with plasmids expressing p53 (2 μg), EGFP (1 μg), His-Ub (4 μg), and MKRN1 (10 μg), and treated with MG132 for 3 h before harvest. After purification of ubiquitinated proteins using Ni2+-NTA beads, total extracts (Input) and His-purified proteins were detected by western blot analysis using α-p53 (DO-1) antibody. The asterisk (*) indicates non-specific bands. (E) Endogenous ubiquitination of p53. U2OS cells were transfected with control or MKRN1 siRNA and treated with MG132 for 3 h before harvest. Whole cell lysates were immunoprecipitated with α-p53 rabbit (FL393) antibodies and immunoblotted with α-Ub mouse antibodies (lanes 2 and 3). Normal rabbit IgG was used for negative control (lane 1).
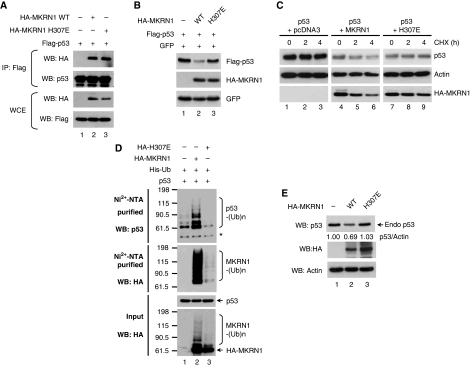
The MKRN1 histidine-to-aspartate point mutation involving histidine residue 307 within the ring finger domain (H307E) deprives MKRN1 of its E3 ligase activities (Kim et al, 2005). Although the MKRN1 H307E mutant was able to bind to p53, its ability to induce p53 degradation and destabilization was blocked (Figure 3A–C). The inability of MKRN1 H307E to induce ubiquitination was further confirmed (Figure 3D, upper panel, lane 3). In the lower panel of the same figure, the self-ubiquitinated form of MKRN1 was completely abolished compared with lane 2, indicating the defective E3 ligase activity of MKRN1 H307E. We then tested whether MKRN1 was indeed able to degrade endogenous p53. As expected, U2OS cells transiently transfected with HA-tagged MKRN1 showed decreased protein levels of p53 and p21 (Figure 3E), whereas HA-MKRN1 H307E could not affect p53 levels. In summary, MKRN1 leads to the polyubiquitination and proteasome-dependent degradation of p53 through direct interaction with the core domain of p53. The importance of the E3 ligase activities of MKRN1 in mediating degradation of p53 was clearly indicated by the observations that MKRN1 H307E was unable to direct either polyubiquitination or degradation of p53. These data indicate that MKRN1 is a novel E3 ligase of p53.
Figure 3.
The E3 ligase defective mutant form of MKRN1 is not able to mediate p53 ubiquitination. (A) Interaction between MKRN1 H307E and p53. The plasmids expressing Flag-p53 and HA-MKRN1 H307E mutant were transfected into 293T cells. Cell lysates were immunoprecipitated with α-Flag antibody, and were immunoblotted using the indicated antibodies. (B) Effects of the MKRN1 H307E mutant on p53 degradation. H1299 cells were transfected with plasmids expressing the indicated proteins. Western blotting was performed using α-HA and α-Flag antibody (C) Stability of p53 by MKRN1 H307E mutant. H1299 cells, transfected with the indicated plasmid, were analysed after treating with CHX as described in the legend of Figure 2B. (D) Ubiquitination of p53 by the MKRN1 H307E mutant. Ubiquitination assay using H1299 cells transfected with the indicated plasmid were performed as described in the legend of Figure 2D. The asterisk (*) indicates non-specific bands. (E) Degradation of endogenous p53 by MKRN1 wild type and H307E mutant. U2OS cells were transfected with pcDNA3-HA-MKRN1 wild type or H307E. At 24 h after transfection, cells were harvested, lysed, and immunoblotted with the indicated antibodies.
MKRN1 targets novel p53 lysine sites, K291, and K292 for mediation of ubiquitination
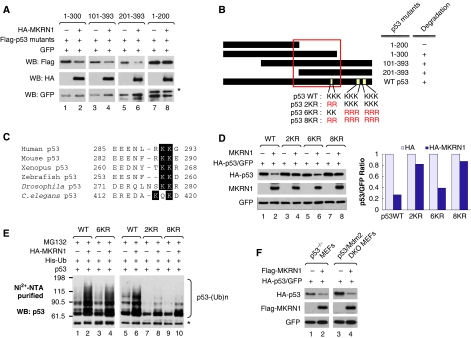
As MKRN1 was able to bind to two regions on the p53-core domain, 43–200 and 201–300, we tested whether the protein levels of p53 deletion mutants containing either of these MKRN1-binding regions could be destabilized by MKRN1. Indeed, we observed that 1–300, 101–393, and 201–393 were degraded by MKRN1, whereas the 1–200 domain was not affected (Figure 4A). When Hdm2 was tested for its ability to degrade these mutants as a control, we observed that it hardly affected their stabilities (Supplementary Figure S5A). The common motif among p53 mutants that are degraded by MKRN1 ranges from residues 200 to 300 (Figure 4B). Interestingly, there are two sequentially located lysine residues, K291 and K292 (Figure 4B), which are mutated in several different human cancers (Zhang et al, 1993; Zou et al, 1993; Miwa et al, 1995; Gao et al, 1997; Tseng et al, 1999; Wallerand et al, 2005; Hiraga et al, 2006) (http://p53.free.fr). These two lysine residues are also highly conserved from Caenorhabditis elegans to human beings (Figure 4C). To confirm whether these sequences are essential for MKRN1-dependent ubiquitination, we constructed a p53-2KR mutant mutated at sites 291 and 292 from lysine to arginine residues (Figure 4B). The point mutant 6KR, that is mutated at six conventionally known lysine sites for ubiquitination (Rodriguez et al, 2000), was used as a control. Moreover, we also constructed an 8KR mutant, which had both 2KR and 6KR residues mutated (Figure 4B). The degradation analyses showed that 2KR and 8KR were quite resistant to the MKRN1-dependent degradation with only 14–18% of both proteins reduced in the presence of MKRN1, whereas 6KR was degraded in a similar degree as p53 wild type with up to a 60–70% reduction (Figure 4D). These observations were further supported by ubiquitination assays showing that 2KR or 8KR were not polyubiquitinated by MKRN1, whereas wild type and 6KR were polyubiquitinated (Figure 4E). These results suggest that MKRN1 might be involved in the regulatory pathway of p53 independent of Hdm2, as Hdm2 is known to preferentially interact with N-terminus of p53 and ubiquitinate conventional six lysine residues (Rodriguez et al, 2000). To eliminate the involvement of Hdm2 in the MKRN1-p53 pathway, we used a p53 degradation assay in p53/Mdm2 double knockout (DKO) MEFs. MKRN1 was still able to induce p53 degradation in p53/Mdm2 DKO MEFs, as well as in p53 KO MEFs (Figure 4F). We further tested whether Hdm2 was involved in MKRN1-mediated p53 degradation by using Nutlin-3, which specifically inhibits the interaction between Hdm2 and p53 (Vassilev et al, 2004). Consistent with an earlier study (Vassilev et al, 2004), Hdm2-induced p53 degradation and ubiquitination was blocked by Nutlin-3 treatment, whereas MKRN1 was able to induce p53 degradation and ubiquitination (Supplementary Figure S5B and C), supporting the notion that MKRN1 can ubiquitinate and degrade p53 independently of Hdm2. Overall, our data show that MKRN1 mediates the degradation of p53 fragments containing the 201–300 motif. The two consecutively located lysine residues, 291 and 292, within this motif are required for MKRN1-mediated polyubiquitination and degradation of p53, whereas six lysine residues located on the C-terminal end of p53 are not necessary for MKRN1-dependent degradation. These observations reveal novel ubiquitination sites on p53 and suggest a possible existence of a whole new set of regulatory pathways for p53 stabilization that can be distinguished from the well-established Hdm2-mediated p53 degradation pathways.
Figure 4.
K291 and K292 of p53 are required for MKRN1-mediated degradation and ubiquitination of p53. (A) Degradation of p53 deletion mutants by MKRN1. Plasmids expressing Flag-p53 deletion mutants were cotransfected with the plasmid expressing HA-MKRN1 into H1299 cells. The levels of p53 mutants were detected by western blot analysis. The asterisk (*) indicates the Flag-p53 1–200 fragment. (B) Schematic diagram of the motif responsible for the MKRN1-mediated p53 degradation and the putative p53 ubiquitination sites. The motif required for p53 degradation is shown by the square. (C) Alignment of p53 orthologues containing conserved lysine residues. Black shading indicates conserved lysine residues throughout species. (D) Degradation of p53 wild type, 2KR, and 6KR by MKRN1. H1299 cells were transfected with plasmids expressing HA-p53 (wild type, 2KR, and 6KR) and/or MKRN1. Cell lysates were immunoblotted as in Figure 2A. Relative amounts of p53 were quantified by densitometry, normalized to GFP, and are shown in the graph. (E) Ubiquitination of p53 2KR, 6KR, and 8KR mutants. Ubiquitination assays were performed as described in Figure 2D. (F) MKRN1-dependent p53 degradation in p53/Mdm2 DKO MEFs. p53 KO MEFs or p53/Mdm2 DKO MEFs were transfected with the indicated plasmid. Cell lysates were immunoblotted using α-HA and α-Flag antibodies.
Depletion of MKRN1 under DNA-damaging conditions stabilizes p21 and prevents cell death
Under DNA damage-related stress, p53, post-translationally modified by several kinases and acetylases, becomes stabilized and is prevented from interaction with Hdm2 (Bode and Dong, 2004; Kruse and Gu, 2008). Thus, we tested whether the interaction between MKRN1 and p53 could be affected under stress conditions. When 5 μM of adriamycin was applied to H1299 cells followed by MKRN1 and p53 immunoprecipitation, we observed that the levels of p53 bound to MKRN1 were attenuated up to 50% (Figure 5A, lanes 2 and 3). Under the same conditions, MKRN1 could not induce the degradation of p53 (Figure 5B, lanes 2 and 4), which might have resulted from the weakened interaction between p53 and MKRN1. Interaction between endogenous MKRN1 and endogenous p53 was also weakened on adriamycin treatment (Supplementary Figure S6A, lanes 2 and 3). As a result, when MKRN1 was depleted in U2OS cells by MKRN1 siRNA under the same DNA damage-mediated stress, there was no increase in endogenous p53 levels, whereas the levels of p53 increased under normal condition (Figure 5C). Furthermore, there was no increased p53 phosphorylation at the 15 serine residue under stress conditions. Unexpectedly, the depletion of MKRN1 under normal or stress conditions elicited the stabilization of p21 in the same cell extracts (Figure 5C). These observations pose a very interesting issue, as the increased levels of p21 do not seem to be the result of activated p53 under this stress condition. Under the treatment with 5 μM adriamycin, the condition that induces cell death rather than cell cycle arrest, there was no increase in p21 mRNA (Figure 5D, lanes 3 and 4) as reported earlier (Rinaldo et al, 2007). When kinetics of p21 and p53 stabilization were tested under CHX treatment, the stability of p21 protein was increased up to 80% by MKRN1 knockdown, regardless of the presence of adriamycin (Figure 5E upper panels, second and fourth graphs). On the other hand, MKRN1 knockdown induced stabilization of p53 only in the absence of adriamycin (Figure 5E, middle panels, first and third graphs). Similar to 5 μM adriamycin treatment, ultraviolet (UV) irradiation abrogated the interaction between endogenous MKRN1 and p53, resulting in the inhibition of MKRN1-dependent p53 degradation (Supplementary Figure S6A, lanes 2 and 4, and S6B).
Figure 5.
MKRN1 regulates p53 and p21 differently under stress conditions. (A) Interaction between p53 and MKRN1 under stress conditions. H1299 cells transfected with the indicated plasmid were treated with 5 μM adriamycin for 4 h. Whole cell extracts were immunoprecipitated with α-HA mouse antibody and immunoblotted with α-p53 or α-HA rabbit antibodies. The asterisk (*) indicates p53. (B) Degradation of p53 by MKRN1 in the presence of adriamycin. H1299 cells, transfected with the indicated, were treated with 5 μM adriamycin for 4 h, and analysed by western blotting. (C) Detection of p53, p21, and apoptotic markers under MKRN1 ablation in DNA-damaging conditions. U2OS cells were transfected with control siRNA and MKRN1 siRNA, treated with 5 μM adriamycin for 12 h, and analysed by western blotting. (D) RT–PCR analysis in U2OS cells transfected with the indicated siRNAs in the absence or presence of adriamycin. RT–PCR analysis was carried out as in Figure 1C. (E) Stability of p53 and p21 in MKRN1 knockdown cells on adriamycin treatment. U2OS cells pretreated with control or MKRN1 siRNA were treated with or without adriamycin for 2 h. Afterwards, the cells were treated with CHX for the indicated times. The percentage of remaining proteins were quantified by densitometry, normalized to actin, and are indicated in the graph. (F) Analysis of cell death affected by MKRN1 ablation in the presence of ADR. U2OS cells were transfected with the indicated siRNAs followed by the treatment of 5 μM adriamycin for 24 h. Subsequently, cells were stained with propidium iodide and analysed using FACS. The percentage of cells in subG1 is indicated in the figure.
It has been suggested that stabilization of p21 prevents apoptosis by inducing cell cycle arrest and by interacting with pro-caspase-3, caspase-8, or ASK-1 (Gartel and Tyner, 2002). Thus, down-regulation of p21 is thought to be required when cells undergo apoptosis (Han et al, 2002; Javelaud and Besancon, 2002; Martinez et al, 2002; Seoane et al, 2002; Bitomsky et al, 2008). Indeed, it has been shown that lethal stress suppresses p21 expression despite p53 activation, whereas a low level of stress induces p21 expression by p53 (Oda et al, 2000; Martinez et al, 2002; Rinaldo et al, 2007). Consistent with earlier data, the treatment of U2OS cells with 5 μM adriamycin or 70 J/m2 UV resulted in apoptosis detected by cleavage of caspase-3 and poly-ADP ribose polymerase (PARP), and a drastic decrease in p21 expression (Figure 5C, lanes 1 and 3; Supplementary Figure S6C, lanes 1 and 3). Cell death induced by adriamycin or UV treatment was further confirmed by determining subdiploid cells using FACS analyses (Figure 5F and Supplementary Figure S6D).
As mentioned above, depletion of MKRN1 resulted in p21 up-regulation in the cells treated with adriamycin or UV without altering p53 levels (Figure 5C, lanes 3 and 4; Supplementary Figure S6C, lanes 3 and 4). When MKRN1 siRNA was applied to U2OS cells treated with adriamycin or UV, the apoptotic process evident by the cleavage of caspase-3 and PARP was drastically attenuated (Figure 5C, lanes 3 and 4; Supplementary Figure S6C, lanes 3 and 4). Accordingly, MKRN1 depletion induced reduction in cell death (Figure 5F; Supplementary Figure S6D). To clarify whether stabilized p21 could be directly involved in the resistance to apoptosis in MKRN1-ablated cells on adriamycin or UV treatment, U2OS cells were simultaneously treated with p21 and MKRN1 siRNA. Resistance to apoptosis by MKRN1 ablation on adriamycin or UV treatment was relieved by p21 deficiency (Figure 5F; Supplementary Figure S6D). Collectively, our data indicated that MKRN1 was no longer able to negatively regulate p53 under stress conditions possibly because of putative post-translational modification of p53. Yet, depletion of MKRN1 was able to stabilize p21 either under stress or normal conditions. Furthermore, ablation of MKRN1 could induce prevention of adriamycin or UV-dependent apoptosis as shown from the detection of apoptotic marker or FACS analyses. Finally, although the lack of MKRN1 protected cells from stress-induced cell death, this process was reversed by p21 knockdown.
MKRN1 is an E3 ligase for p21, inducing its degradation under normal or stress conditions
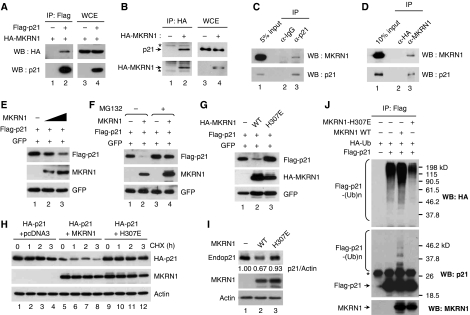
The stabilization of p21 on knockdown of MKRN1 supports a physical interaction between MKRN1 and p21. To confirm that the stabilization of p21 under depletion of MKRN1 was due to their interaction, we tested whether p21 and MKRN1 could bind to each other. Immunoprecipitation of exogenous p21 and MKRN1 showed this binding (Figure 6A). We also observed that ectopically expressed HA-MKRN1 could bind to endogenous p21 (Figure 6B). Reciprocal immunoprecipitation analyses of endogenous MKRN1 and p21 clearly indicate that both proteins could interact with each other (Figure 6C and D). Furthermore, exogenous MKRN1 was able to degrade p21 in a concentration-dependent manner (Figure 6E). MKRN1-facilitated degradation of p21 was prevented by MG132, indicating that degradation occurred through the proteasome-dependent pathway (Figure 6F). MKRN1 H307E, the E3 ligase defective mutant of MKRN1, was not able to decrease the steady-state levels of p21, as well as stability measured by CHX treatment, as expected (Figure 6G and H). Furthermore, protein levels of endogenous p21 were also affected by the MKRN1 wild –type, but not by the H307E mutant (Figure 6I). Finally, ubiquitination analysis showed that MKRN1 increased p21 polyubiquitination, whereas MKRN1 H307E did not (Figure 6J). The inability of MKRN1 H307E to induce p21 degradation and ubiquitination indicated that p21 might be degraded through an MKRN1-dependent ubiquitination process.
Figure 6.
MKRN1 binds to, degrades, and ubiquitinates p21. (A) Interaction between exogenous HA-MKRN1 and Flag-p21. H1299 cells were transfected with the plasmids expressing HA-MKRN1 and Flag-p21. Whole cell extracts and immunoprecipitates were analysed by western blotting using α-p21 and α-HA antibodies. (B) Interaction between HA-MKRN1 and endogenous p21. H1299 cells were transfected with the plasmid expressing HA-MKRN1 or empty vector. WCE and IP were analysed by western blotting using α-p21 and α-HA antibodies. The asterisk (*) indicates heavy (lower panel) and light (upper panel) chains of IgG. (C, D) Reciprocal immunoprecipitation analysis between endogenous MKRN1 and endogenous p21. Lysates of U2OS cells were immunoprecipitated with α-p21 mouse (C) or α-MKRN1 rabbit (D) antibodies and immunoblotted with α-p21 mouse or α-MKRN1 rabbit antibodies. (E) Degradation of p21 by MKRN1. H1299 cells were transfected with plasmids expressing Flag-p21 and GFP with or without MKRN1. Levels of p21 were detected by western blot using α-p21 antibody. (F) Proteasomal-dependent degradation of p21 by MKRN1. H1299 cells, transfected with the indicated plasmids, were treated with MG132 for 3 h. Western blot analysis was performed as described above. (G, H) Effect of MKRN1 H307E on the stead-state levels and stability of p21. H1299 cells were transfected with the indicated plasmids. For determination of p21 stability, cells were treated with 100 μg/ml CHX and harvested at the indicated time. Cell lysates were analysed by western blotting. (I) Endogenous p21 degradation by MKRN1. H1299 cells were transfected with the plasmid expressing MKRN1 wild type or H307E. Cells were lysed and immunoblotted using α-p21 antibody. (J) Ubiquitination of p21 by MKRN1. H1299 cells, transfected with the indicated plasmids, were treated with MG132 for 3 h and then harvested. Ubiquitinated p21 was detected by immunoprecipitation assay using α-Flag mouse antibody, followed by western blot using α-HA rabbit or α-p21 rabbit antibody. The asterisk (*) indicates non-specific bands.
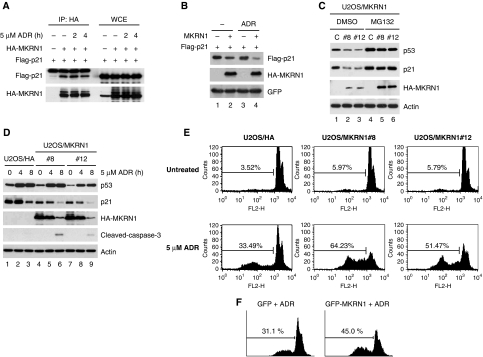
As p21 stabilization on MKRN1 knockdown was not affected by adriamycin or UV treatment (Figure 5C; Supplementary Figure S6C), we hypothesized that p21 would still be degraded by MKRN1 under the same conditions. As anticipated, MKRN1 was able to bind to and degrade p21 in the presence or absence of adriamycin (Figure 7A and B). MKRN1 also induced degradation of p21 with UV treatment (Supplementary Figure S7A). In addition, we were able to observe that the interaction between endogenous MKRN1 and p21 were not changed under stress conditions (Supplementary Figure S7B). As p21 was rapidly degraded on adriamycin and UV treatment, cells were treated with MG132 before immunoprecipitation. To further elucidate the roles of MKRN1 on the endogenous p53 and p21, we established two U2OS cell lines, U2OS/MKRN1#8 and #12, which stably express HA-MKRN1. As expected, these MKRN1 stable cell lines displayed relatively lower levels of p53 and p21 compared with the control, which could be stabilized by the MG132 proteasome inhibitor indicating that p53 and p21 were degraded by an MKRN1-dependent proteasome pathway (Figure 7C). When 5 μM of adriamycin was applied to these cell lines, p53 was stabilized within 4 h as expected (Figure 7D, lanes 2, 3, 5, 6, 8, and 9). On the other hand, the steady-state levels of p21 were kept at reduced levels in the U2OS/MKRN1#8 and #12 stable cell lines (Figure 7D, lanes 4 and 7). Of note, the steady-state levels of p21 were rapidly decreased between 4 and 8 h after stress in control cells, indicating p53-mediated preferential stimulation of other apoptotic target promoters compared with p21 under lethal apoptotic stimuli in the early phase of stress (Figure 7D) as reported before (Oda et al, 2000; Martinez et al, 2002). In addition to the down-regulation of p21, both stable cell lines displayed increased levels of cleaved-caspase-3 under stress conditions suggesting that the decrease of p21 levels at the early phase of apoptosis could accelerate the cell death processes of the stable cell lines (Figure 7D, lanes 6 and 9). Moreover, we observed that U2OS/MKRN1#8 and #12 stable cells were highly vulnerable to adriamycin-induced cell death compared with control cells (Figure 7E, lower panels of graphs), concurrent with the observations of reduced p21 levels in stable cell lines. We also observed that stable expression of MKRN1 sensitized the cells to UV-induced cell death by accelerating p21 degradation (Supplementary Figures S7C and D). Finally, we also observed that U2OS cells transiently expressing GFP-MKRN1 were more sensitive to adriamycin-induced cell death (Figure 7F). Taken together, our results indicate that MKRN1 binds to and functions as an E3 ligase of p21. The MKRN1-mediated degradation of p21 was not influenced by apoptotic stresses. As a result, anti-apoptotic activities of p21 were abrogated in the presence of MKRN1.
Figure 7.
MKRN1 degrades p21 under adriamycin treatment and stimulates adriamycin-induced cell death. (A) Binding of MKRN1 and p21 under stress conditions. H1299 cells, transfected with the indicated plasmids, were treated with 5 μM of adriamycin for the indicated times. WCE were immunoprecipitated with α-HA antibody, and immunoblotted with α-Flag antibody. (B) Degradation of p21 by MKRN1 under stress conditions. H1299 cells, transfected with plasmids expressing the indicated proteins, were treated with 5 μM of adriamycin for 4 h. Cell lysates were analysed by western blotting. (C) U2OS cells stably expressing HA or HA-MKRN1 were treated with MG132 for 3 h. The protein levels of p53 and p21 were detected by western blotting. (D) Western blot analysis of U2OS cells stably expressing HA-MKRN1 with or without adriamycin. U2OS stable cell lines (MK1#8 and #12) were treated with 5 μM of adriamycin for 4 and 8 h. Levels of p53, p21, and MKRN1 on adriamycin treatment were monitored using western blot analysis. Activation of caspase-3 was also shown by α-cleaved-caspase-3 antibody. (E) FACS analysis of cell death induced by adriamycin in MKRN1 stable cell lines. Cells were treated with adriamycin for 24 h, stained with propidium iodide, and analysed using FACS. (F) FACS analysis of U2OS cells overexpressing GFP or GFP-MKRN1 in the presence of adriamycin. U2OS cells, transfected with GFP or GFP-MKRN1, were stained with propidium iodide and analysed by FACS after gating with GFP signal. The percentage of cells in subG1 is shown in the figure.
Discussion
To repress p53 activities as a tumour suppressor, polyubiquitination-mediated degradation of p53 is usually used as a critical event. Hdm2 (human Mdm2), an E3 ubiquitin ligase containing a ring finger domain, is a major negative mediator for p53 ubiquitination and degradation (Bode and Dong, 2004; Brooks and Gu, 2006; Lee et al, 2006). Hdm2 was initially reported to target six lysine residues located at the C-terminus of p53 for ubiquitination (Rodriguez et al, 2000). However, numerous studies have suggested that the six C-terminal lysines might not be the only sites for p53 ubiquitination and degradation. Indeed, the p53 6KR mutant, which has its six lysine residues substituted with arginine, can still be ubiquitinated and degraded in cells, although less effectively (Camus et al, 2003; Tang et al, 2008). Furthermore, emerging evidence shows that there might exist various p53's E3 ligases, such as pirh2, COP1, and ARF-BP1 (Brooks and Gu, 2006). Indeed, E4F1 and MSL2 ubiquitinate p53 at the sites different from six conventional lysine residues, although these ubiquitination does not lead to the degradation process (Le Cam et al, 2006; Kruse and Gu, 2009). Together, p53 can be ubiquitinated at the multiple sites by the numerous E3 ligases to be controlled elaborately.
Here, we have identified MKRN1 as another E3 ubiquitin ligase for p53. Interestingly, MKRN1 could induce degradation and ubiquitination of the p53 6KR mutant (Figure 4D and E). Observations that MKRN1 could degrade p53 without its six lysines at the C-terminus led us to identify two novel ubiquitination sites, K291 and K292, on p53 (Figure 4B). These two lysines are highly conserved in most species that express p53 (Figure 4C), and are also mutated in human cancers, implicating possibly yet unknown critical roles for K291 and K292 in p53 function (http://p53.free.fr). As lysine residues also can be post-translationally modified by acetylation, methylation, sumoylation, and neddylation, it is plausible that these modifications compete with MKRN1-dependent ubiquitination for the control of p53 activities.
MKRN1 also seems to have an ubiquitination-independent role for suppressing p53 activities similarly to Hdm2 (Minsky and Oren, 2004; Ohkubo et al, 2006; Tang et al, 2008). Transcriptional activity of the p53 2KR mutant that is considerably resistant to MKRN1-mediated ubiquitination and degradation was suppressed by MKRN1 (Supplementary Figure S8A and B). In addition, the E3 ligase-deficient mutant MKRN1 was also able to suppress activation of a p53-dependent reporter promoter (Supplementary Figure S8C). These data imply that MKRN1, similar to Hdm2, could negatively regulate transcriptional activities of p53 through direct interaction (Minsky and Oren, 2004; Ohkubo et al, 2006; Tang et al, 2008). As a result, the ubiquitination-dependent and -independent roles of MKRN1 on the negative regulation of p53 prevent improper activation of p53-dependent cell cycle arrest and apoptosis in unstressed cells. On the other hand, ablation of MKRN1 resulted in p53 and p21 up-regulation, and led to growth arrest in p53-positive cells.
Most E3 ubiquitin ligases for p53, such as Hdm2, Pirh2, and COP1, have also been reported to be transcriptionally regulated by p53, and thus, forming a negative regulatory feedback loop (Brooks and Gu, 2006). However, there is no difference in expression of MKRN1 in p53-positive and -negative HCT116 pair cell lines (Supplementary Figure S2C). Similarly, H1299 cells, which lack p53 express a considerable amount of MKRN1 (Figure 1D). Furthermore, the MKRN1 promoter also has no putative p53-binding site suggesting that MKRN1 might not to be a target of p53.
In this report, we have also identified p21 as a target of MKRN1. MKRN1 binds to and degrades p21 by inducing its polyubiquitination. Even though p21 is truly ubiquitinated in cells, it is still controversial whether ubiquitination is required for p21 degradation (Sheaff et al, 2000; Bloom et al, 2003; Jin et al, 2003, 2008; Nishitani et al, 2008). As a p21 mutant with all lysine residues mutated to arginine is still degraded, ubiquitination of p21 seems to be dispensable for certain degradation pathways (Jin et al, 2003). Supporting this idea, it has been observed that no E3 ligase activities of Mdm2 and Mdmx are required for the degradation of p21 (Jin et al, 2003). On the other hand, several reports have suggested the necessity of p21 ubiquitination for its degradation through other E3 ligases (Maki and Howley, 1997; Fukuchi et al, 1999; Nakanishi et al, 2000; Nishitani et al, 2008). Our results indicate that MKRN1 mediates ubiquitination-dependent degradation of p21 as MKRN1 defective in E3 ligase activity is unable to induce p21 degradation as well as its ubiquitination.
Down-regulation of p21 under DNA-damaging stress is considered as an important step for facilitating efficient apoptosis, as the existence of p21 can stimulate a cells' responses towards cell cycle arrest through the suppression of apoptosis (Zhang et al, 1999; Gartel and Tyner, 2002; Han et al, 2002; Javelaud and Besancon, 2002; Martinez et al, 2002; Hill et al, 2008). Although inhibition of p21 activity through the transcriptional inhibition by Myc, Miz1, Ztbt4, Pdcd4, or the cleavage of p21 by caspase-3 are known to take place under apoptotic stimuli (Zhang et al, 1999; Seoane et al, 2002; Bitomsky et al, 2008; Weber et al, 2008), our data indicate that MKRN1-mediated p21 ubiquitination and degradation are also important processes for inducing apoptosis. The importance of MKRN1-dependent p21 degradation is highlighted by the observations that MKRN1 knockdown resulted in p21 stabilization and prevention of DNA damage-induced apoptosis (Figure 5).
Activity of MKRN1 also seems to be regulated in multiple ways. DNA-damaging stress impaired the function of MKRN1 related to p53 degradation (Figure 5A–C). However, MKRN1 was able to degrade p21 under stress conditions (Figures 5E and 7B; Supplementary Figure S7A), indicating that there might be separate regulatory pathways that control the function of MKRN1 towards p53 and p21 under stress conditions. Furthermore, we constantly observed a rapid decrease in the levels of MKRN1 at the later stage of stress, suggesting possible destabilizing mechanisms of MKRN1 (Figure 7D).
MKRN1 is highly conserved in vertebrates. Human MKRN1 is 92% identical (94% similar) and 73% identical (80% similar) at the amino-acid level to mouse MKRN1 and chicken MKRN1, respectively (Gray et al, 2000). Drosophila and C. elegans also express MKRN1 orthologues. Although MKRN1 in these species is only 27 and 25% identical (40 and 38% similar) at the amino-acid level, respectively, to human MKRN1, these MKRN1 orthologues have six zinc-finger motifs that are >70% similar to those of human MKRN1. As explained above, two lysine residues of p53, which are regulated by MKRN1 in human beings, were also conserved from C. elegans to human beings (Figure 4C). Thus, it should be investigated whether MKRN1 in lower species is also involved in the control of p53 activity through the regulation of these two conserved lysines. It is also interesting to note that among various p53 E3 ligases, only MKRN1 and ARF-BP1 are conserved from C. elegans to human beings. As Hdm2 and COP1 have no orthologue in Drosophila and C. elegans, and Pirh2 can be found in Drosophila, MKRN1 and ARF-BP1 might be ancient regulators of p53 (Brooks and Gu, 2006). Similar to ARF-BP1, MKRN1 is not regulated by p53, whereas Mdm2, COP1, and pirh2 display a p53-dependent feedback loop (Brooks and Gu, 2006). However, we cannot exclude the possibility that MKRN1 might be indirectly regulated by p53 target proteins under stress conditions. These observations indicate how various p53 E3 ligases are functionally evolved from different species.
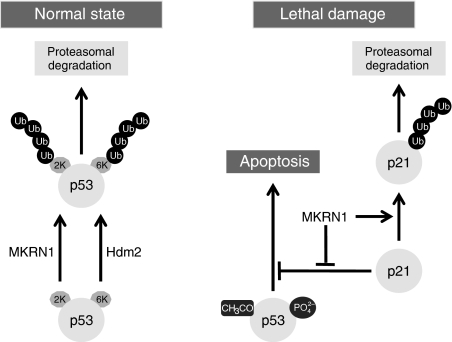
Overall, the comprehensive function of MKRN1 can be described as follows. Under normal conditions, MKRN1 is able to suppress p53 and p21 by ubiquitin-dependent degradation. Specifically, MKRN1 targets lysines on residues 291 and 292 of p53 for polyubiquitination (Figure 8, left panel). Furthermore, its binding to p53 prevents the transcriptional activities of p53 (Supplementary Figure S8). This inhibition ensures the regular growth of normal cells under unstressed conditions. Under stress conditions, MKRN1 ceases to have a negative effect on p53, allowing p53 to proceed with its cytotoxic effects on cells. However, the regulatory effect of MKRN1 on p21 is still active, preventing an excessive accumulation of p21 and, thus, stimulating the apoptotic process if necessary (Figure 8, right panel). The continuous destabilization of p21 by MKRN1 seems to be required to help cells become more susceptible to apoptotic stimuli.
Figure 8.
A schematic model for the p53/p21 regulatory pathway mediated by MKRN1.
MKRN1 is expressed in most human tissues, such as heart, brain, lung, liver, skeletal muscle, kidney, and pancreas (Gray et al, 2000). It has yet to be identified whether these endogenous MKRN1 have different roles in determining the fate of cells other than the ones suggested here. As MKRN1 seems to be tightly related to the maintenance of homeostasis for p53 and p21 as well as hTERT (Kim et al, 2005), future studies need to be extended further to investigate its role in cellular ageing and death process in the primary cells as well as in stem cells in various organs.
Our results identify the evolutionary conserved MKRN1 as a novel regulator of p53 and p21, which mediates p53-dependent cell cycle arrest and apoptosis. It seems that MKRN1 differently regulates p53 and p21 under different stresses, thus having an important function in facilitating p53-dependent apoptotic pathways. Eventually, these observations are expected to provide novel therapeutic targets for tumourigenesis.
Materials and methods
Plasmids
MKRN1 cDNA and p21 cDNA were purchased from the Frontier Human Gene Bank (Seoul, Korea). MKRN1 cDNA was also kindly provided by IK Chung (Yonsei University). MKRN1 cDNA was subcloned into pcDNA3.1, pcDNA3-HA, pcDNA3-Flag, pGEX-4T-1, and pEGFP-C2. MKRN1 H307E was generated by site-directed mutagenesis, and MKRN1 deletion mutants were generated by polymerase chain reaction (PCR). p21 cDNA was subcloned into pcDNA3-HA and pcDNA3-Flag vectors. pSV-p53, pcDNA3-p53 6KR, and pcDNA3-His-Ub were described earlier (Camus et al, 2003). pcDNA3-HA-p53 has been described earlier (Oh et al, 2006). p53 deletion mutants and the lysine-to-arginine mutant were also generated by PCR, and then subcloned into pcDNA3.1, pcDNA3-HA, and pcDNA3-Flag vectors. pHM6-HA-Ub has been described earlier (Yang et al, 2008). The pEGFP-C2 vector (Clontech, San Diego, CA) was used as a transfection control.
Cell culture and transfection
The H1299 human lung carcinoma cell line and 293T kidney cell line were maintained in Dulbecco's modified Eagle's Medium supplemented with 10% fetal bovine serum (FBS) (GIBCO-BRL, Rochester, NY) and 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA) in 5% CO2 at 37°C. HCT116 pair cell lines were kindly provided by Dr B Vogelstein (Johns Hopkins University, Baltimore, MD). HCT116 cells were maintained in McCoy's 5A medium supplemented with 10% FBS and antibiotics. U2OS/HA-MKRN1 stable cell lines were selected and maintained in 800 μg/ml G418 (Sigma-Aldrich, St Louis, MO). Multiple independent single colonies were subcloned and checked for protein expression by western blotting. The transient transfections were performed using Lipofectamine 2000 (Invitrogen) or WelfectEX (Welgene Biotech, Daegu, Korea) according to the instructions of the manufacturer.
siRNA-mediated ablation of MKRN1
Four kinds of MKRN1 siRNA were purchased from Qiagen-Xeragon (Valencia, CA). MKRN1-5HP siRNA (siMKRN1#5) (5′-CAGGCGAAGCTGAGTCAAGAA-3′) and MKRN1-6HP siRNA (siMKRN1#6) (5′-CGGGATCCTCTCCAACTGCAA-3′) were resynthesized from Qiagen-Xeragon. Control siRNA was also purchased from Qiagen-Xeragon. Lipofectamin RNAiMax (Invitrogen) was used for siRNA transfection.
Growth curves and crystal violet staining
Cells were transfected with 50 nM of siRNAs by reverse transfection method using Lipofectamine RNAiMax. For determining cell growth, cells were seeded in wells of 12-well plates. Viable and dead cells were counted at the indicated times using trypan blue staining. For crystal violet staining, cells were plated into 60 mm-diameter culture dishes and stained with crystal violet 2 and 3 days after transfection.
FACS analysis
Cells were fixed in 70% ethanol and incubated in 40 μg/ml of RNase A for 30 min, followed by incubation in 100 μg/ml of propidium iodide for 30 min. Cell cycle was analysed by use of an FACScan apparatus (BD Biosciences, Franklin Lakes, NJ).
Information on antibodies, chemicals, and biochemical analyses are provided in the Supplementary data.
Supplementary Material
Supplementary Figures S1–S8
Supplementary Data
Review Process File
Acknowledgments
We thank Professor Jung In-Kwon for his generous and kind discussion. This study was supported by a Korea Science and Engineering Foundation (KOSEF) grant funded by the Korean government (MOST) (R01-2007-000-20327-0), and a grant from the National R&D Program for Cancer Control, Ministry of Health & Welfare, Republic of Korea (0820110).
Footnotes
The authors declare that they have no conflict of interest.
References
- Aaltomaa S, Lipponen P, Eskelinen M, Ala-Opas M, Kosma VM (1999) Prognostic value and expression of p21(waf1/cip1) protein in prostate cancer. Prostate 39: 8–15 [DOI] [PubMed] [Google Scholar]
- Baretton GB, Klenk U, Diebold J, Schmeller N, Lohrs U (1999) Proliferation- and apoptosis-associated factors in advanced prostatic carcinomas before and after androgen deprivation therapy: prognostic significance of p21/WAF1/CIP1 expression. B J Cancer 80: 546–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearss DJ, Lee RJ, Troyer DA, Pestell RG, Windle JJ (2002) Differential effects of p21(WAF1/CIP1) deficiency on MMTV-ras and MMTV-myc mammary tumor properties. Cancer Res 62: 2077–2084 [PubMed] [Google Scholar]
- Bitomsky N, Wethkamp N, Marikkannu R, Klempnauer KH (2008) siRNA-mediated knockdown of Pdcd4 expression causes upregulation of p21(Waf1/Cip1) expression. Oncogene 27: 4820–4829 [DOI] [PubMed] [Google Scholar]
- Bloom J, Amador V, Bartolini F, DeMartino G, Pagano M (2003) Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell 115: 71–82 [DOI] [PubMed] [Google Scholar]
- Bode AM, Dong Z (2004) Post-translational modification of p53 in tumorigenesis. Nat Rev 4: 793–805 [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W (2006) p53 ubiquitination: Mdm2 and beyond. Mol Cell 21: 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus S, Higgins M, Lane DP, Lain S (2003) Differences in the ubiquitination of p53 by Mdm2 and the HPV protein E6. FEBS Lett 536: 220–224 [DOI] [PubMed] [Google Scholar]
- Chang BD, Watanabe K, Broude EV, Fang J, Poole JC, Kalinichenko TV, Roninson IB (2000) Effects of p21Waf1/Cip1/Sdi1 on cellular gene expression: implications for carcinogenesis, senescence, and age-related diseases. Proc Natl Acad Sci USA 97: 4291–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Kon N, Li M, Zhang W, Qin J, Gu W (2005) ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell 121: 1071–1083 [DOI] [PubMed] [Google Scholar]
- Dornan D, Shimizu H, Mah A, Dudhela T, Eby M, O'Rourke K, Seshagiri S, Dixit VM (2006) ATM engages autodegradation of the E3 ubiquitin ligase COP1 after DNA damage. Science (New York, NY) 313: 1122–1126 [DOI] [PubMed] [Google Scholar]
- Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, O'Rourke K, Koeppen H, Dixit VM (2004) The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 429: 86–92 [DOI] [PubMed] [Google Scholar]
- Erber R, Klein W, Andl T, Enders C, Born AI, Conradt C, Bartek J, Bosch FX (1997) Aberrant p21(CIP1/WAF1) protein accumulation in head-and-neck cancer. Inter J Cancer 74: 383–389 [DOI] [PubMed] [Google Scholar]
- Fizazi K, Martinez LA, Sikes CR, Johnston DA, Stephens LC, McDonnell TJ, Logothetis CJ, Trapman J, Pisters LL, Ordonez NG, Troncoso P, Navone NM (2002) The association of p21((WAF-1/CIP1)) with progression to androgen-independent prostate cancer. Clin Cancer Res 8: 775–781 [PubMed] [Google Scholar]
- Fukuchi K, Maruyama H, Takagi Y, Gomi K (1999) Direct proteasome inhibition by clasto-lactacystin beta-lactone permits the detection of ubiquitinated p21(waf1) in ML-1 cells. Biochim Biophys Acta 1451: 206–210 [DOI] [PubMed] [Google Scholar]
- Gao HG, Chen JK, Stewart J, Song B, Rayappa C, Whong WZ, Ong T (1997) Distribution of p53 and K-ras mutations in human lung cancer tissues. Carcinogenesis 18: 473–478 [DOI] [PubMed] [Google Scholar]
- Gartel AL, Radhakrishnan SK (2005) Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res 65: 3980–3985 [DOI] [PubMed] [Google Scholar]
- Gartel AL, Tyner AL (2002) The role of the cyclin-dependent kinase inhibitor p21 in apoptosis. Mol Cancer Ther 1: 639–649 [PubMed] [Google Scholar]
- Gorospe M, Cirielli C, Wang X, Seth P, Capogrossi MC, Holbrook NJ (1997) p21(Waf1/Cip1) protects against p53-mediated apoptosis of human melanoma cells. Oncogene 14: 929–935 [DOI] [PubMed] [Google Scholar]
- Gorospe M, Wang X, Guyton KZ, Holbrook NJ (1996) Protective role of p21(Waf1/Cip1) against prostaglandin A2-mediated apoptosis of human colorectal carcinoma cells. Mol Cell Biol 16: 6654–6660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray TA, Hernandez L, Carey AH, Schaldach MA, Smithwick MJ, Rus K, Marshall Graves JA, Stewart CL, Nicholls RD (2000) The ancient source of a distinct gene family encoding proteins featuring RING and C(3)H zinc-finger motifs with abundant expression in developing brain and nervous system. Genomics 66: 76–86 [DOI] [PubMed] [Google Scholar]
- Han Z, Wei W, Dunaway S, Darnowski JW, Calabresi P, Sedivy J, Hendrickson EA, Balan KV, Pantazis P, Wyche JH (2002) Role of p21 in apoptosis and senescence of human colon cancer cells treated with camptothecin. J Biol Chem 277: 17154–17160 [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M (1997) Mdm2 promotes the rapid degradation of p53. Nature 387: 296–299 [DOI] [PubMed] [Google Scholar]
- Hill R, Bodzak E, Blough MD, Lee PW (2008) p53 binding to the p21 promoter is dependent on the nature of DNA damage. Cell Cycle 7: 2535–2543 [DOI] [PubMed] [Google Scholar]
- Hiraga J, Kinoshita T, Ohno T, Mori N, Ohashi H, Fukami S, Noda A, Ichikawa A, Naoe T (2006) Promoter hypermethylation of the DNA-repair gene O6-methylguanine-DNA methyltransferase and p53 mutation in diffuse large B-cell lymphoma. Inter J Hematol 84: 248–255 [DOI] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H (1997) Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett 420: 25–27 [DOI] [PubMed] [Google Scholar]
- Javelaud D, Besancon F (2002) Inactivation of p21WAF1 sensitizes cells to apoptosis via an increase of both p14ARF and p53 levels and an alteration of the Bax/Bcl-2 ratio. J Biol Chem 277: 37949–37954 [DOI] [PubMed] [Google Scholar]
- Jin Y, Lee H, Zeng SX, Dai MS, Lu H (2003) MDM2 promotes p21waf1/cip1 proteasomal turnover independently of ubiquitylation. EMBO J 22: 6365–6377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Zeng SX, Sun XX, Lee H, Blattner C, Xiao Z, Lu H (2008) MDMX promotes proteasomal turnover of p21 at G1 and early S phases independently of, but in cooperation with, MDM2. Mol Cell Biol 28: 1218–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Park SM, Kang MR, Oh SY, Lee TH, Muller MT, Chung IK (2005) Ubiquitin ligase MKRN1 modulates telomere length homeostasis through a proteolysis of hTERT. Genes Dev 19: 776–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse JP, Gu W (2008) SnapShot: p53 posttranslational modifications. Cell 133: 930–930.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse JP, Gu W (2009) MSL2 promotes MDM2 independent cytoplasmic localization of p53. J Biol Chem 284: 3250–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH (1997) Regulation of p53 stability by Mdm2. Nature 387: 299–303 [DOI] [PubMed] [Google Scholar]
- Lavin MF, Gueven N (2006) The complexity of p53 stabilization and activation. Cell Death Differ 13: 941–950 [DOI] [PubMed] [Google Scholar]
- Le Cam L, Linares LK, Paul C, Julien E, Lacroix M, Hatchi E, Triboulet R, Bossis G, Shmueli A, Rodriguez MS, Coux O, Sardet C (2006) E4F1 is an atypical ubiquitin ligase that modulates p53 effector functions independently of degradation. Cell 127: 775–788 [DOI] [PubMed] [Google Scholar]
- Lee EW, Oh W, Song J (2006) Jab1 as a mediator of nuclear export and cytoplasmic degradation of p53. Mol Cells 22: 133–140 [PubMed] [Google Scholar]
- Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, Parant JM, Lozano G, Hakem R, Benchimol S (2003) Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 112: 779–791 [DOI] [PubMed] [Google Scholar]
- Mahyar-Roemer M, Roemer K (2001) p21 Waf1/Cip1 can protect human colon carcinoma cells against p53-dependent and p53-independent apoptosis induced by natural chemopreventive and therapeutic agents. Oncogene 20: 3387–3398 [DOI] [PubMed] [Google Scholar]
- Maki CG, Howley PM (1997) Ubiquitination of p53 and p21 is differentially affected by ionizing and UV radiation. Mol Cell Biol 17: 355–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez LA, Yang J, Vazquez ES, Rodriguez-Vargas Mdel C, Olive M, Hsieh JT, Logothetis CJ, Navone NM (2002) p21 modulates threshold of apoptosis induced by DNA-damage and growth factor withdrawal in prostate cancer cells. Carcinogenesis 23: 1289–1296 [DOI] [PubMed] [Google Scholar]
- Minsky N, Oren M (2004) The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol Cell 16: 631–639 [DOI] [PubMed] [Google Scholar]
- Miwa K, Miyamoto S, Kato H, Imamura T, Nishida M, Yoshikawa Y, Nagata Y, Wake N (1995) The role of p53 inactivation in human cervical cell carcinoma development. Br J Cancer 71: 219–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi Y, Pei XH, Takayama K, Bai F, Izumi M, Kimotsuki K, Inoue K, Minami T, Wataya H, Hara N (2000) Polycyclic aromatic hydrocarbon carcinogens increase ubiquitination of p21 protein after the stabilization of p53 and the expression of p21. Am J Respir Cell Mol Biol 22: 747–754 [DOI] [PubMed] [Google Scholar]
- Nishitani H, Shiomi Y, Iida H, Michishita M, Takami T, Tsurimoto T (2008) CDK inhibitor p21 is degraded by a PCNA coupled Cul4-DDB1Cdt2 pathway during S phase and after UV irradiation. J Biol Chem 283: 29045–29052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, Nakamura Y, Taya Y (2000) p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 102: 849–862 [DOI] [PubMed] [Google Scholar]
- Oh W, Lee EW, Sung YH, Yang MR, Ghim J, Lee HW, Song J (2006) Jab1 induces the cytoplasmic localization and degradation of p53 in coordination with Hdm2. J Biol Chem 281: 17457–17465 [DOI] [PubMed] [Google Scholar]
- Ohkubo S, Tanaka T, Taya Y, Kitazato K, Prives C (2006) Excess HDM2 impacts cell cycle and apoptosis and has a selective effect on p53-dependent transcription. J Biol Chem 281: 16943–16950 [DOI] [PubMed] [Google Scholar]
- Omwancha J, Zhou XF, Chen SY, Baslan T, Fisher CJ, Zheng Z, Cai C, Shemshedini L (2006) Makorin RING finger protein 1 (MKRN1) has negative and positive effects on RNA polymerase II-dependent transcription. Endocrine 29: 363–373 [DOI] [PubMed] [Google Scholar]
- Oren M (2003) Decision making by p53: life, death and cancer. Cell Death Differ 10: 431–442 [DOI] [PubMed] [Google Scholar]
- Rinaldo C, Prodosmo A, Mancini F, Iacovelli S, Sacchi A, Moretti F, Soddu S (2007) MDM2-regulated degradation of HIPK2 prevents p53Ser46 phosphorylation and DNA damage-induced apoptosis. Mol Cell 25: 739–750 [DOI] [PubMed] [Google Scholar]
- Ringshausen I, O'Shea CC, Finch AJ, Swigart LB, Evan GI (2006) Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer Cell 10: 501–514 [DOI] [PubMed] [Google Scholar]
- Rodier F, Campisi J, Bhaumik D (2007) Two faces of p53: aging and tumor suppression. Nucleic Acids Res 35: 7475–7484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MS, Desterro JM, Lain S, Lane DP, Hay RT (2000) Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol Cell Biol 20: 8458–8467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roninson IB (2002) Oncogenic functions of tumour suppressor p21(Waf1/Cip1/Sdi1): association with cell senescence and tumour-promoting activities of stromal fibroblasts. Cancer Lett 179: 1–14 [DOI] [PubMed] [Google Scholar]
- Seoane J, Le HV, Massague J (2002) Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature 419: 729–734 [DOI] [PubMed] [Google Scholar]
- Sheaff RJ, Singer JD, Swanger J, Smitherman M, Roberts JM, Clurman BE (2000) Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol Cell 5: 403–410 [DOI] [PubMed] [Google Scholar]
- Tang Y, Zhao W, Chen Y, Zhao Y, Gu W (2008) Acetylation is indispensable for p53 activation. Cell 133: 612–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng JE, Rodriguez M, Ro J, Liu D, Hong WK, Mao L (1999) Gender differences in p53 mutational status in small cell lung cancer. Cancer Res 59: 5666–5670 [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA (2004) In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science (New York, NY) 303: 844–848 [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ (2000) Surfing the p53 network. Nature 408: 307–310 [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lane DP (2007) p53 in health and disease. Nat Rev 8: 275–283 [DOI] [PubMed] [Google Scholar]
- Waldman T, Lengauer C, Kinzler KW, Vogelstein B (1996) Uncoupling of S phase and mitosis induced by anticancer agents in cells lacking p21. Nature 381: 713–716 [DOI] [PubMed] [Google Scholar]
- Wallerand H, Bakkar AA, de Medina SG, Pairon JC, Yang YC, Vordos D, Bittard H, Fauconnet S, Kouyoumdjian JC, Jaurand MC, Zhang ZF, Radvanyi F, Thiery JP, Chopin DK (2005) Mutations in TP53, but not FGFR3, in urothelial cell carcinoma of the bladder are influenced by smoking: contribution of exogenous versus endogenous carcinogens. Carcinogenesis 26: 177–184 [DOI] [PubMed] [Google Scholar]
- Wang YA, Elson A, Leder P (1997) Loss of p21 increases sensitivity to ionizing radiation and delays the onset of lymphoma in atm-deficient mice. Proc Natl Acad Sci USA 94: 14590–14595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A, Marquardt J, Elzi D, Forster N, Starke S, Glaum A, Yamada D, Defossez PA, Delrow J, Eisenman RN, Christiansen H, Eilers M (2008) Zbtb4 represses transcription of P21CIP1 and controls the cellular response to p53 activation. EMBO J 27: 1563–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MR, Lee SR, Oh W, Lee EW, Yeh JY, Nah JJ, Joo YS, Shin J, Lee HW, Pyo S, Song J (2008) West Nile virus capsid protein induces p53-mediated apoptosis via the sequestration of HDM2 to the nucleolus. Cell Microbiol 10: 165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Feng X, Koga H, Ichikawa T, Abe S, Kumanishi T (1993) p53 gene mutations in pontine gliomas of juvenile onset. Biochem Biophys Res Commun 196: 851–857 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Fujita N, Tsuruo T (1999) Caspase-mediated cleavage of p21Waf1/Cip1 converts cancer cells from growth arrest to undergoing apoptosis. Oncogene 18: 1131–1138 [DOI] [PubMed] [Google Scholar]
- Zou M, Shi Y, Farid NR (1993) p53 mutations in all stages of thyroid carcinomas. JClin Endocrinol Metab 77: 1054–1058 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–S8
Supplementary Data
Review Process File