Abstract
Competency for DNA replication is functionally coupled to the activation of histone gene expression at the onset of S phase to form chromatin. Human histone nuclear factor P (HiNF-P; gene symbol HINFP) bound to its cyclin E/cyclin-dependent kinase 2 (CDK2) responsive coactivator p220NPAT is a key regulator of multiple human histone H4 genes that encode a major subunit of the nucleosome. Induction of the histone H4 transcription factor (HINFP)/p220NPAT coactivation complex occurs in parallel with the CDK-dependent release of pRB from E2F at the restriction point. Here, we show that the downstream CDK-dependent cell cycle effector HINFP is genetically required and, in contrast to the CDK2/cyclin E complex, cannot be compensated. We constructed a mouse Hinfp-null mutation and found that heterozygous Hinfp mice survive, indicating that 1 allele suffices for embryogenesis. Homozygous loss-of-function causes embryonic lethality: No homozygous Hinfp-null mice are obtained at or beyond embryonic day (E) 6.5. In blastocyst cultures, Hinfp-null embryos exhibit a delay in hatching, abnormal growth, and loss of histone H4 gene expression. Our data indicate that the CDK2/cyclin E/p220NPAT/HINFP/histone gene signaling pathway at the G1/S phase transition is an essential, nonredundant cell cycle regulatory mechanism that is established early in embryogenesis.
Keywords: blastocyst, development, embryogenesis, p220NPAT, human embryonic stem cells
The cell cycle controlled activation of histone gene expression at the onset of S phase is essential for chromatin packaging of nascent DNA. Histones are highly conserved proteins (H1, H2A, H2B, H3, and H4) that form chromatin and carry epigenetic marks that control gene transcription (1, 2). Expression of histone genes in mammalian cells is both temporally and functionally coupled with DNA replication (1, 3–7) and provides a paradigm for understanding gene regulatory signaling mechanisms operative at the G1/S transition. Stringent cell cycle control of histone gene expression is evident in all somatic cell types examined and is already firmly programmed in human embryonic stem cells (8–10). Regulation of histone gene expression occurs at multiple levels including transcription, pre-mRNA processing and mRNA stability. However, cell cycle dependent modulation of histone gene transcription is a key rate-limiting step in the induction of histone protein synthesis.
The human histone gene transcription factor HINFP is the final and essential link in the cyclin E/CDK2/p220NPAT/HINFP pathway that is required for cell cycle-dependent activation of histone H4 genes at the G1/S phase transition (11–13). This pathway is activated at the restriction point (R-point) by cyclin E/CDK2, in parallel with the E2F/pRB pathway that controls transcription of genes involved in nucleotide metabolism and DNA synthesis (14). Histone H4 gene transcription is stimulated at the G1/S phase transition via a critical multipartite promoter element, Site II (15–17). HINFP interacts with Site II and integrates signals emanating from the cyclin E/CDK2 kinase cascade (11–13, 15, 18). HINFP recruits p220NPAT to multiple human histone H4 genes in the human diploid genome (13). p220NPAT is a cyclin E/CDK2 substrate (19–21) that functions as the primary coactivator of HINFP activity to mediate cell cycle responsiveness of histone H4 gene transcription (12). p220NPAT simultaneously interacts with multiple tandemly repeated genes encoding other core histone proteins (e.g., H2A, H2B, and H3) (20–22). The interaction of p220NPAT with HINFP stabilizes the resulting complex (12, 23), whereas HINFP can also bind other factors with functions in cell signaling, transcription, or RNA processing (24–26). HINFP together with p220NPAT supports the assembly of the histone gene expression machinery at histone locus bodies, specific subnuclear domains that are associated with human histone gene clusters (10, 12, 27). Although other cell cycle-regulated and CDK-dependent signaling pathways contribute to histone gene regulation (e.g., cyclin A/CDK1/pRB/CDP-cut and IRF proteins) (28–31), the cyclin E/CDK2/p220NPAT/ HINFP pathway is clearly the key regulator of histone H4 gene expression in cultured cells. The histone H4 gene-related transcriptional pathways that have been identified function independently from pathways involving transcription factor E2F. Hence, HINFP-dependent regulatory events reflect a fundamental cell cycle regulatory mechanism.
Recent studies on HINFP and histone gene regulation using cell culture models have defined molecular pathways and subnuclear parameters operative in control of histone gene expression in normal diploid somatic cells, cancer cells, or human embryonic stem cells (9, 10, 27, 32). However, defining biological relevance requires establishing developmental control of mammalian histone gene expression in vivo. Therefore, we have constructed a gene knockout of the mouse Hinfp gene and show that loss of Hinfp function causes early embryonic lethality. Our data indicate that, in contrast to cyclin-dependent kinases that can be compensated, HINFP is a nonredundant CDK2 dependent effector during early embryogenesis.
Results
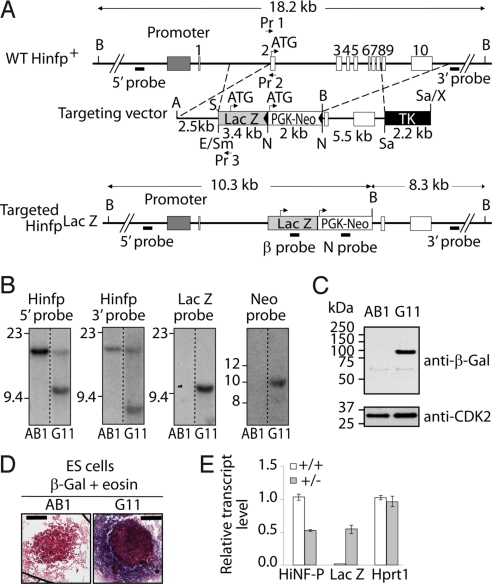
To examine the biological role of HINFP protein during mammalian development, we constructed a null mouse model. One Hinfp allele was inactivated in mouse embryonic stem cells by replacing the initial Hinfp-coding sequences with the β-galactosidase (LacZ) marker gene through homologous recombination to yield the HinfpLacZ-null allele (Fig. 1A). Southern blot analysis revealed that proper gene targeting was achieved via homologous recombination (Fig. 1B). Using immunoblot analysis and β-galactosidase staining, we confirmed that β-galactosidase is placed under transcriptional control of the endogenous Hinfp promoter in targeted embryonic stem cells (Fig. 1 C and D). After blastocyst injection, the resulting mouse chimeras exhibited germ-line transmission of the targeted allele. Mouse embryonic fibroblasts (MEFs) from offspring of wild-type female and heterozygous male HinfpLacZ/+ mice were examined for LacZ and Hinfp gene expression (Fig. 1E). Relative to wild-type MEFs, HinfpLacZ/+ MEFs express half the amount of Hinfp mRNA and robust levels of LacZ. Despite reduced Hinfp gene dosage, heterozygous HinfpLacZ/+ mice thrive, reproduce, and do not exhibit overt phenotypes that distinguish them from their wild-type littermates.
Fig. 1.
Generation of a Hinfp-null allele by homologous recombination. (A) Restriction enzyme maps of the Hinfp locus located on mouse chromosome 9 (Top), the targeting vector (Middle), and the recombined locus that generates a HinfpLacZ/+-null mutation (Bottom). The following restriction enzyme sites are indicated: BspHI, B; AatII, A; SacII, S; EcoRV, E; SmaI, Sm; NotI, N; SalI, Sa; and XhoI, X. For the targeting vector, a 2.5-kb 5′ fragment (left arm) ending at and excluding the ATG of the Hinfp locus, was inserted at the AatII and SacII sites. The left arm is followed by a 3.4-kb Smal-NotI fragment spanning β-gal-coding sequences containing a Kozak consensus sequence and ATG (inserted between EcoRV and NotI), and a 2.0-kb NotI-NotI fragment spanning a floxed neomycin gene cassette [LoxP-PGK-Neo-LoxP; black triangles are Lox P sites flanking the neomycin phosphotransferase (Neo) gene driven by the phosphoglycerate kinase promoter (PGK)] (inserted at a NotI site). The right arm is a 5.5-kb NotI-SalI 3′ fragment (inserted into NotI-SalI sites) that is followed by a 2.2-kb SalI-XhoI fragment containing a cassette with the PGK promoter driving the thymidine kinase (TK) gene that was inserted into SalI sites of the targeting vector. Arrow heads indicate the direction of transcription for the LacZ and Neo genes. Lines with double arrowheads in the maps for the wild-type and targeted alleles represent restriction enzyme fragments used for validation of homologous recombination by Southern blot analysis by using the indicated hybridization probes (short thick lines). (B) Southern blot analysis of mouse ES cell clones (AB1, wild type; G11, a representative recombined mES clone) that were digested with BspHI and hybridized to 2 external probes (5′ probe and 3′ probe) and 2 internal probes (β-gal and Neo) that are all indicated at the top of each autoradiogram. DNA from HinfpLacZ/+ clones hybridized to the 5′ and 3′ probes will each give rise to a 18.2-kb signal for the wild-type allele. The targeted allele will generate either 10.3-kb (5′ probe) or 8.3-kb (3′ probe) signals. Unique integration and recombination was further confirmed with 2 internal probes spanning the β-galactosidase and neomycin cassettes. As expected, hybridization of HinfpLacZ/+ clones with either the 760-bp β-gal probe or the 557-bp Neo probe yields only the 10.3-kb mutant signal and no signal for the wild-type fragment, whereas hybridization with a 542-bp 3′ probe yields a 18.2-kb wild-type signal and the 8.3-kb targeted allele. (C) Western blot analysis of β-gal expression driven by the endogenous promoter in wild-type (AB1) or recombinant (G11) ES cells. CDK2 protein levels were monitored to account for protein loading. (D) β-galactosidase staining of targeted ES cells (G11), but not wild-type (AB1) ES cells, shows in situ expression of LacZ under control of the native Hinfp gene promoter. (Scale bar, 500 μm.) (E) Genotype dependent reciprocal expression of Hinfp and LacZ mRNAs from mouse embryonic fibroblasts (MEFs) at E12.5 in wild-type and heterozygous HinfpLacZ/+ fetal pups. Homozygous HinfpLacZ/LacZ-null pups were not recovered at E12.5
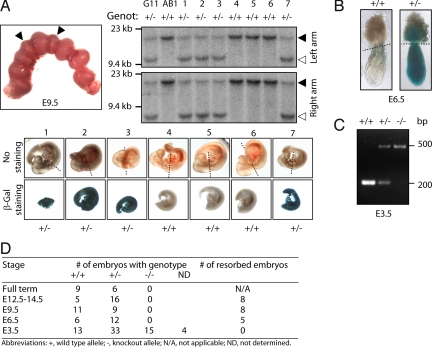
The Hinfp gene is actively expressed throughout murine development. HinfpLacZ/+ embryos show strong and ubiquitous β-galactosidase staining at E9.5 or E6.5 (Fig. 2 A and B). Genotyping of litters obtained from crosses between heterozygous HinfpLacZ/+ mice revealed that there are no live births of homozygous HinfpLacZ/LacZ mice (Fig. 2). HinfpLacZ/LacZ-null embryos are also not observed at earlier stages of fetal development, including E12.5–E14.5, E9.5, and E6.5, concurrent with an increased incidence of resorbed embryos. However, HinfpLacZ/LacZ-null blastocysts were recovered at E3.5 in ratios expected from Mendelian inheritance (Fig. 2D). Thus, the HinfpLacZ-null mutation induces embryonic lethality between E3.5 and E6.5.
Fig. 2.
A homozygous HinfpLacZ/LacZ null allele causes embryonic lethality between E3.5 and E6.5. (A) Characterization of embryos derived from heterozygous HinfpLacZ/+ crosses at E9.5. A representative uterus is shown that contained 7 embryos and 2 resorption sites (arrow heads) (Upper Left) and was genotyped by Southern blot analysis with 5′ and 3′ probes (Upper Right); DNA of wild-type AB1 (+/+) and heterozygous G11 (+/−) ES cells were used as controls genotyping. Genotyping results for each of the pups (below micrographs) reveal that β-galactosidase activity (blue) correlates with presence of 1 HinfpLacZ-null allele, but embryos with homozygous for the Hinfp-null allele (HinfpLacZ/LacZ) were not recovered, presumably because of resorption before E9.5. Tissue rostral to the dotted line was used for genotyping. (B) Embryos derived from heterozygous HinfpLacZ/+ crosses at E6.5 were stained with X-gal. Embryonic tissue below the dotted line was used for genotyping. Two representative embryos are shown. No homozygous HinfpLacZ/LacZ (−/−) genotypes were obtained. (C) PCR analysis was used to determine the genotypes of offspring from heterozygous HinfpLacZ/+-null mice. A representative ethidium bromide-stained gel of PCR products from blastocysts is shown with each of the 3 expected genotypes (wild type, heterozygous, and homozygous for Hinfp). Homozygous HinfpLacZ/LacZ-null mutants were not recovered after the blastocyst stage. The location of the PCR primers (Pr1, Pr2, and Pr3) used for the analyses are indicated in Fig. 1A. (D) The table shows the recovery of embryos with the indicated genotypes at different stages of development and the number of resorbed embryos (ND, not determined; N/A, not applicable).
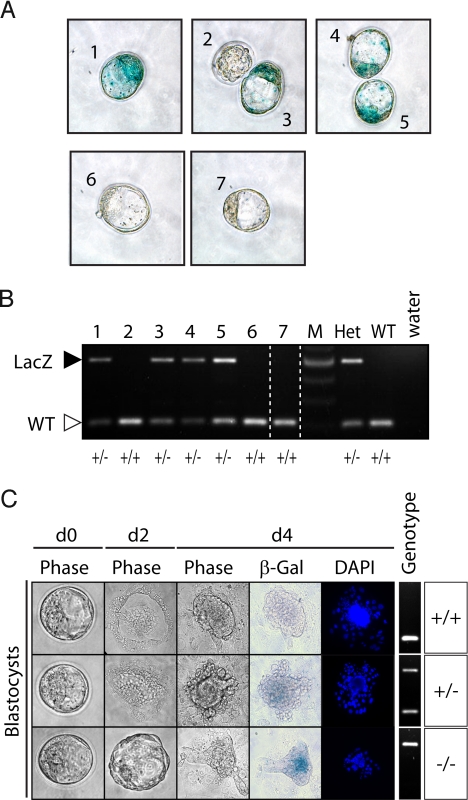
Zygotic expression of histone genes occurs after the 4- to 8-cell stage when maternal storage pools of histone mRNAs have been degraded (33). Because HINFP controls expression of multiple histone H4 genes in cultured cells (13, 34), we assessed whether the Hinfp gene is transcriptionally active in blastocysts. Clear β-galactosidase staining was only observed in HinfpLacZ/+ embryos (Fig. 3A), indicating that Hinfp is already expressed at the blastocyst stage. Recovery of homozygous HinfpLacZ/LacZ-null blastocysts (E3.5) (Fig. 2 C and D) indicates that zygotic Hinfp gene expression is dispensable for the earliest stages of embryogenesis.
Fig. 3.
Homozygous Hinfp-null blastocysts exhibit growth defects. Blastocysts (numbered 1–7) were flushed at E3.5 from wild-type females crossed with HinfpLacZ/+ males and directly stained for β-galactosidase. (A) Staining results show that β-gal expression, driven by the native Hinfp gene promoter, occurs in blastocysts. (B) Blastocysts shown in A were genotyped by PCR as visualized by ethidium bromide staining of PCR products (using Pr1, Pr2, and Pr3 primers indicated in Fig. 1A). Genotypes are indicated at the bottom of the gel. The last 3 lanes show DNA from heterozygous G11 HinfpLacZ/+ (+/−) and wild-type AB1 Hinfp+/+ (+/+) ES cells or distilled water as PCR controls. (C) Blastocysts isolated from heterozygous crosses (E3.5) reveal that Hinfp-null embryos exhibit a delay in hatching on ex vivo culture. On incubation in outgrowth culture for 96 h (d 4), cultured embryos with at least 1 HinfpLacZ-null allele (but not wild-type embryos) display β-galactosidase staining. Hatching from the zona pellucida by day 2 in culture is observed for wild-type and heterozygous embryos, but homozygous HinfpLacZ/LacZ-null embryos hatch later (≈day 3). Furthermore, the latter embryos do not spread to form an inner cell mass and do not develop normally. Phase contrast micrograph and fluorescence microscopy for DAPI staining are presented to show embryonic morphology. Genotype is shown at right.
To assess whether deletion of Hinfp causes early cell growth abnormalities, we harvested blastocysts with Hinfp+/+, HinfpLacZ/+, and HinfpLacZ/LacZ genotypes and cultured the embryos in vitro (Fig. 3 B and C). Wild-type and heterozygous embryos hatched from the zona pellucida at day 2 in culture and attached to the tissue culture plate. The embryos showed the typical morphology of cultured attached blastocysts: The trophectoderm expanded over the surface of the plate, whereas the inner cell mass formed an outgrowth at the center of the trophectoderm growth (Fig. 3C). In contrast, homozygous HinfpLacZ/LacZ-null embryos exhibited delayed hatching (∼day 3) and failed to expand (i.e., ≈1- to 1.5-day delay in development) (Fig. 3C). These results indicate that Hinfp function is essential for early embryonic development.
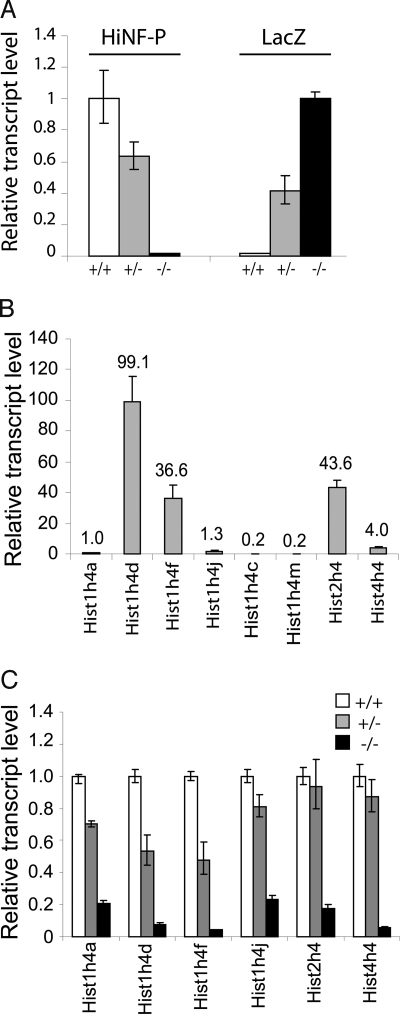
The molecular consequences of Hinfp deficiency were investigated by analyzing the expression of histone H4 genes, the principal gene regulatory targets of HINFP protein. Our analysis includes the Hist2h4 gene, which is the mouse equivalent of the human HIST2H4 gene (previously referred to as H4/n or pFO108) that provided the model for identification of the key role of HINFP protein in controlling human histone H4 gene expression (13, 35). In blastocysts, expression of Hinfp and the LacZ marker is directly linked to the number of wild-type or HinfpLacZ-null alleles (Fig. 4A). Wild-type mouse blastocysts cultured ex vivo express multiple histone H4 genes with expression of 3 representative genes (i.e., Hist1h4d, Hist1h4f, and Hist2h4) being more prominent than that of others (Fig. 4B). Ex vivo cultures of heterozygous HinfpLacZ/+ blastocysts express this same set of histone H4 genes at levels comparable to wild type (Fig. 4C). In contrast, homozygous HinfpLacZ/LacZ-null embryos cultured ex vivo exhibit a dramatic reduction (>10-fold) in the expression of multiple distinct histone H4 gene copies. These data indicate that HINFP is the master regulator of mammalian histone H4 genes in vivo. Moreover, the results indicates that loss of Hinfp and reduced expression of histone H4 genes results in a cell growth phenotype and induces embryonic lethality at the peri-implantation stage of embryonic development.
Fig. 4.
Histone H4 gene expression is perturbed in homozygous Hinfp-null blastocysts. Expression of multiple histone H4 genes is significantly decreased in homozygous HinfpLacZ/LacZ-null blastocysts (E3.5) that were cultured ex vivo for 4 days. (A) Hinfp and LacZ gene expression were detected by qRT-PCR in wild-type Hinfp+/+ (+/+), heterozygous HinfpLacZ/+ (+/−), and homozygous HinfpLacZ/LacZ (−/−) mouse blastocysts on ex vivo culture. (B) The expression profile of a representative set of mouse histone H4 genes was determined in wild-type mouse blastocysts (E3.5) that were maintained in ex vivo outgrowth cultures. Each gene transcript level is compared with the Hist1h4a mRNA level that was arbitrarily set as 1. (C) Same as B, but expression of histone H4 was examined as a function of genotype in blastocysts cultured ex vivo. Expression of histone H4 genes is strongly decreased in homozygous HINFP-null embryos compared with outgrowth cultures of wild-type and heterozygous blastocysts. All transcript levels were normalized to GAPDH mRNA and represent the mean ± SD (n = 2). For each histone H4 gene, expression levels were numerically adjusted relative to expression observed in wild-type embryos (arbitrarily set as 1).
Discussion
HINFP is a ubiquitous regulator of histone H4 gene expression in all proliferating cell types examined to date, and the protein is down-regulated in post-proliferative differentiated cells (11, 13, 15, 36). HINFP transduces cyclin E/CDK2 signals into transcriptional responses through interactions with its obligatory coactivator p220NPAT (12, 19–21, 37, 38). Cyclin E/CDK2-dependent mechanisms are frequently deregulated in a broad spectrum of cancers (39, 40), and thus a subset of the resulting pathological consequences may be mediated through modifications in the activity of HINFP. To understand how HINFP may control cell cycle progression during normal development, we have constructed a knockout of the mouse Hinfp gene. Our results show that HINFP is essential for histone H4 gene expression in vivo and for embryonic survival beyond the blastocyst stage.
Mouse embryos implant at E4.5, but Hinfp-null embryos are resorbed before E6.5. This finding indicates that HINFP is rate-limiting for fetal development around and beyond the blastocyst stage, perhaps because alterations in the rate of cell proliferation may change how cells, tissues and morphogens interact at successive developmental stages. Hinfp-null blastocysts are delayed in hatching and exhibit severely reduced expression of histone H4 genes. Thus, consistent with cell culture studies, absence of HINFP is directly related to a molecular defect in histone H4 gene expression. Lack of histone H4 protein biosynthesis is predicted to prevent deposition of histone octamers onto newly replicated DNA. The absence of nucleosomes may cause DNA replication defects during S phase, chromosomal segregation defects during mitosis, and/or epigenetic alterations in progeny cells.
Zygotic expression of histone genes is first detected at the 4–8 cell stage (33). However, HINFP function appears to be dispensible for the early cleavage stages of embryogenesis, perhaps because of the presence of maternal mRNA pools for HINFP or histone H4. Alternatively, HINFP independent (and perhaps less efficient) transcriptional mechanisms may compensate until the blastocyst stage.
The human HINFP gene is located together with the gene for its coactivator p220 NPAT in the chromosomal interval 11q22-q23. Genes within this genomic region, which encompasses fragile sites and putative tumor suppressor genes, are frequently deleted and/or amplified in many different types of cancer (41, 42). Homozygous null mutations of both Hinfp (this work) and p220NPAT (43) cause embryonic lethality in mice, suggesting that there should be positive selection for cells that retain at least 1 of each allele during tumorigenesis. Because human 11q22-q23 contains putative tumor suppressors, the requirement to retain wild-type HINFP and p220NPAT alleles may oppose the growth advantage of homozygous deletions within this highly recombinogenic interval. Thus, the spectrum of permissible genetic alterations in 11q22-q23 in cancer cells is limited by the genetic necessity of the human HINFP gene for efficient cell proliferation.
Deregulation of cyclin E/CDK2 activity contributes to the molecular etiology of cancer. However, loss-of-function mutations that ablate cyclin E/CDK2 kinase activity are not embryonic lethal (44–47). The HINFP coactivator p220NPAT was identified as a key substrate of cyclin E/CDK2 and interacts with cyclin E (12, 19–21). Thus, both p220NPAT and HINFP have a shared molecular function in the activation of histone H4 gene expression (12). The early embryonic lethality of a nontargeted null mutation in p220NPAT (43) and the genetic demonstration that a defined Hinfp-null mutation also causes early embryonic lethality are consistent with the concept that these proteins are functionally and biochemically linked. The findings that these 2 specific downstream effectors of CDK2/cyclin E kinase are genetically indispensible, whereas CDK2 kinase activity is not, suggest that CDK2 effectors may be more suitable targets for molecular strategies to selectively treat proliferative diseases that include cancer.
Activation of histone H4 gene transcription by HINFP operates independently of the cell cycle regulatory events controlled by the E2F/pRB pathway. Although E2F proteins and retinoblastoma related factors (the pocket proteins pRB, p107, and p130) exhibit considerable genetic redundancy, our findings establish HINFP as a unique and irreplaceable transcription factor that functions as part of a CDK2 responsive gene regulatory mechanism that controls progression beyond the G1/S phase transition.
Materials and Methods
Construction of Mice with a HinfpLacZ-Null Allele.
We targeted the mouse Hinfp locus by homologous recombination over a DNA fragment upstream of the ATG and another spanning intron 9 to intron 11 of the Hinfp gene (see Fig. 1). Both DNA fragments were generated from mouse AB2.2 genomic DNA by using PCR primers (Table S1) and inserted into a targeting vector containing gene cassettes for LacZ, neomycin, and thymidine kinase (kindly provided by the Transgenic Animal Modeling Core Facility of the University of Massachusetts Medical School). The final targeting construct was subjected to DNA sequencing to confirm absence of random mutations and linearized for electroporation into AB1 (129S5/SvEvBrd) ES cells. Proper homologous recombination of the HinfpLacZ allele in a representative neomycin resistant clone (G11) was established by Southern blot analysis using restriction sites and probes external to the targeting vector.
Expression of β-galactosidase driven by the endogenous Hinfp promoter was confirmed by 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal) staining. Wild-type AB1 ES cells and G11 Hinfp+/LacZ ES cells were fixed in 0.5% glutaraldehyde and stained with a solution containing 1 mg/mL X-gal, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, and 2 mM MgCl2. LacZ gene expression was also confirmed by western blot analysis with antibodies for β-galactosidase (1:2,000 dilution; GenWay Biotech, Inc.) and Cdk2 (1:2,500 dilution; Santa Cruz Biotechnology, Inc.). Proteins were visualized by using a horseradish peroxidase-conjugated secondary antibody (1:8,000 dilution; Santa Cruz Biotechnology, Inc.) and enhanced chemiluminescence (Plus-ECL kit; PerkinElmer Life and Analytical Sciences).
Clone G11 was microinjected into C57BL/6 blastocysts to produce chimeric mice, and germ line transmission of the mutant allele was determined by genotyping of tail DNA from offspring by Southern blot analysis (as described above) and confirmed by PCR using genotyping primers (Table S1). HinfpLacZ/+ mice were crossed to generate HinfpLacZ/LacZ-null mice, and offspring were subjected to genotyping by PCR and Southern blot analysis as described above. Animals were treated in accordance with Institutional Animal Care and Use Committee guidelines.
Characterization of Early Embryos.
Blastocysts (E3.5) and implanted embryos (E6.5 and E9.5) from crosses between HinfpLacZ/+ males and wild-type female C57BL/6 mice were harvested in DMEM (Invitrogen-Gibco) plus 10% FBS (FBS), 20 mM Hepes (pH 7.5), and 1× penicillin-streptomycin using a dissecting microscope (Leica MZ16F, Leica Microsystems Inc.). Embryos were examined for expression of the LacZ reporter gene under control of the endogenous Hinfp promoter by using β-galactosidase staining and subjected to genotyping as described above.
Blastocysts harvested at E3.5 from heterozygous crosses between HinfpLacZ/+ mice were cultured in M15 media (Dulbecco's MEM, 15% FCS, 100 μM β-mercaptoethanol, 2 mM glutamine, and 1× penicillin-streptomycin) for up to 96 h. Embryo outgrowths were tested for β-galactosidase expression and genotyped retrospectively.
Preparation of Mouse Embryonic Fibroblasts.
Mouse embryonic fibroblasts (MEFs) were isolated from litters harvested at E12.5. Cell suspensions from embryos were prepared by repeated shearing using an 18 gauge needle in 1 mL 0.25% trypsin/1 mM EDTA (Invitrogen-Gibco). Trypsin was inactivated by addition of DMEM (Invitrogen-Gibco) containing 15% FBS and cells were cultured for 24 h to select for adherent cells. MEFs were expanded by passing preconfluent cultures at a 1:5 ratio.
Isolation and Analysis of RNA.
Total RNA was isolated from MEFs and cultured blastocysts (E3.5) that were obtained from crosses between HinfpLacZ/+ mice. Purified RNA was reverse transcribed using random hexamer primers and gene expression was assessed by quantitative reverse transcriptase PCR (qRT-PCR) using optimized qRT-PCR primer sets (Table S1), except for GAPDH primers (Applied Biosystems). Automated qRT-PCR were performed by using SYBR Green 2× master mixture (Applied Biosciences) and a 2-step cycling protocol (anneal and elongate at 60 °C, denature at 94 °C). Specificity of primers was verified by dissociation of amplicons by using SYBR Green as a detector. All transcript levels were normalized to GAPDH mRNA. Additional details are described in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank the members of our laboratories including Kaleem Zaidi, Tripti Gaur, Jitesh Pratap, Klaus Becker, Shirwin Pockwinse, Kathleen Hoover, and Jiufeng Cai for stimulating discussions. We also thank Judy Rask for assistance with manuscript preparation. This work was supported by National Institutes of Health Grants R01 GM032010 and DK32520.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905651106/DCSupplemental.
References
- 1.Osley MA. The regulation of histone synthesis in the cell cycle. Annu Rev Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- 2.Kingston RE, Bunker CA, Imbalzano AN. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 3.Prescott DM. The syntheses of total macronuclear protein, histone, and DNA during the cell cycle in Euplotes eurystomus. J Cell Biol. 1966;31:1–9. doi: 10.1083/jcb.31.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stein GS, Stein JL, van Wijnen AJ, Lian JB. Transcriptional control of cell cycle progression: The histone gene is a paradigm for the G1/S phase and proliferation/differentiation transitions. Cell Biol Int. 1996;20:41–49. doi: 10.1006/cbir.1996.0007. [DOI] [PubMed] [Google Scholar]
- 5.Stein G, Park W, Thrall C, Mans R, Stein J. Regulation of cell cycle stage-specific transcription of histone genes from chromatin by non-histone chromosomal proteins. Nature. 1975;257:764–767. doi: 10.1038/257764a0. [DOI] [PubMed] [Google Scholar]
- 6.Stein GS, et al. An architectural perspective of cell cycle control at the G1/S phase cell cycle transition. J Cell Physiol. 2006;209:106–110. doi: 10.1002/jcp.20843. [DOI] [PubMed] [Google Scholar]
- 7.Marzluff WF. Histone 3′ ends: Essential and regulatory functions. Gene Expr. 1992;2:93–97. [PMC free article] [PubMed] [Google Scholar]
- 8.Becker KA, et al. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J Cell Physiol. 2006;209:883–893. doi: 10.1002/jcp.20776. [DOI] [PubMed] [Google Scholar]
- 9.Becker KA, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Establishment of histone gene regulation and cell cycle checkpoint control in human embryonic stem cells. J Cell Physiol. 2007;210:517–526. doi: 10.1002/jcp.20903. [DOI] [PubMed] [Google Scholar]
- 10.Ghule PN, et al. Cell cycle dependent phosphorylation and subnuclear organization of the histone gene regulator p220NPAT in human embryonic stem cells. J Cell Physiol. 2007;213:9–17. doi: 10.1002/jcp.21119. [DOI] [PubMed] [Google Scholar]
- 11.Mitra P, et al. Identification of HiNF-P, a key activator of cell cycle controlled histone H4 genes at the onset of S phase. Mol Cell Biol. 2003;23:8110–8123. doi: 10.1128/MCB.23.22.8110-8123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miele A, et al. HiNF-P directly links the cyclin E/CDK1/p220NPAT pathway to histone H4 gene regulation at the G1/S phase cell cycle transition. Mol Cell Biol. 2005;25:6140–6153. doi: 10.1128/MCB.25.14.6140-6153.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holmes WF, et al. Coordinate control and selective expression of the full complement of replication-dependent histone H4 genes in normal and cancer cells. J Biol Chem. 2005;280:37400–37407. doi: 10.1074/jbc.M506995200. [DOI] [PubMed] [Google Scholar]
- 14.Grana X, Garriga J, Mayol X. Role of the retinoblastoma protein family, pRB, p107, and p130 in the negative control of cell growth. Oncogene. 1998;17:3365–3383. doi: 10.1038/sj.onc.1202575. [DOI] [PubMed] [Google Scholar]
- 15.van Wijnen AJ, van den Ent FM, Lian JB, Stein JL, Stein GS. Overlapping and CpG methylation-sensitive protein-DNA interactions at the histone H4 transcriptional cell cycle domain: Distinctions between two human H4 gene promoters. Mol Cell Biol. 1992;12:3273–3287. doi: 10.1128/mcb.12.7.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Wijnen AJ, Wright KL, Lian JB, Stein JL, Stein GS. Human H4 histone gene transcription requires the proliferation-specific nuclear factor HiNF-D. Auxiliary roles for HiNF-C (Sp1- like) and HiNF-A (high mobility group-like) J Biol Chem. 1989;264:15034–15042. [PubMed] [Google Scholar]
- 17.Ramsey-Ewing A, van Wijnen AJ, Stein GS, Stein JL. Delineation of a human histone H4 cell cycle element in vivo: The master switch for H4 gene transcription. Proc Natl Acad Sci USA. 1994;91:4475–4479. doi: 10.1073/pnas.91.10.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pauli U, Chrysogelos S, Stein G, Stein J, Nick H. Protein-DNA interactions in vivo upstream of a cell cycle- regulated human H4 histone gene. Science. 1987;236:1308–1311. doi: 10.1126/science.3035717. [DOI] [PubMed] [Google Scholar]
- 19.Zhao J, Dynlacht B, Imai T, Hori T, Harlow E. Expression of NPAT, a novel substrate of cyclin E-CDK2, promotes S-phase entry. Genes Dev. 1998;12:456–461. doi: 10.1101/gad.12.4.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao J, et al. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 2000;14:2283–2297. [PMC free article] [PubMed] [Google Scholar]
- 21.Ma T, et al. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 2000;14:2298–2313. doi: 10.1101/gad.829500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su C, et al. DNA damage induces down-regulation of histone gene expression through the G1 checkpoint pathway. EMBO J. 2004;23:1133–1143. doi: 10.1038/sj.emboj.7600120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Medina R, van Wijnen AJ, Stein GS, Stein JL. The histone gene transcription factor HiNF-P stabilizes its cell cycle regulatory co-activator p220NPAT. Biochemistry. 2006;45:15915–15920. doi: 10.1021/bi061425m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitra P, et al. HiNF-P is a bifunctional regulator of cell cycle controlled histone H4 gene transcription. J Cell Biochem. 2007;101:181–191. doi: 10.1002/jcb.21157. [DOI] [PubMed] [Google Scholar]
- 25.Miele A, Medina R, van Wijnen AJ, Stein GS, Stein JL. The interactome of the histone gene regulatory factor HiNF-P suggests novel cell cycle related roles in transcriptional control and RNA processing. J Cell Biochem. 2007;102:136–148. doi: 10.1002/jcb.21284. [DOI] [PubMed] [Google Scholar]
- 26.Mitra P, et al. CDK inhibitors selectively diminish cell cycle controlled activation of the histone H4 gene promoter by p220(NPAT) and HiNF-P. J Cell Physiol. 2009;219:438–448. doi: 10.1002/jcp.21687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghule PN, et al. The subnuclear organization of histone gene regulatory proteins and 3′ end processing factors of normal somatic and embryonic stem cells is compromised in selected human cancer cell types. J Cell Physiol. 2009;220:129–135. doi: 10.1002/jcp.21740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaughan PS, et al. Activation of a cell-cycle-regulated histone gene by the oncogenic transcription factor IRF-2. Nature. 1995;377:362–365. doi: 10.1038/377362a0. [DOI] [PubMed] [Google Scholar]
- 29.van Wijnen AJ, et al. CDP/cut is the DNA-binding subunit of histone gene transcription factor HiNF-D: A mechanism for gene regulation at the G1/S phase cell cycle transition point independent of transcription factor E2F. Proc Natl Acad Sci USA. 1996;93:11516–11521. doi: 10.1073/pnas.93.21.11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie R, et al. The cell cycle control element of histone H4 gene transcription is maximally responsive to interferon regulatory factor pairs IRF-1/IRF-3 and IRF-1/IRF-7. J Biol Chem. 2001;276:18624–18632. doi: 10.1074/jbc.M010391200. [DOI] [PubMed] [Google Scholar]
- 31.Xie RL, van Wijnen AJ, van der Meijden CM, Stein JL, Stein GS. Forced expression of the interferon regulatory factor 2 oncoprotein causes polyploidy and cell death in FDC-P1 myeloid hematopoietic progenitor cells. Cancer Res. 2002;62:2510–2515. [PubMed] [Google Scholar]
- 32.Ghule PN, et al. Staged assembly of histone gene expression machinery at subnuclear foci in the abbreviated cell cycle of human embryonic stem cells. Proc Natl Acad Sci USA. 2008;105:16964–16969. doi: 10.1073/pnas.0809273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clarke HJ, Oblin C, Bustin M. Developmental regulation of chromatin composition during mouse embryogenesis: Somatic histone H1 is first detectable at the 4-cell stage. Development. 1992;115:791–799. doi: 10.1242/dev.115.3.791. [DOI] [PubMed] [Google Scholar]
- 34.Lichtler AC, et al. Multiple H4 histone mRNAs of HeLa cells are encoded in different genes. Nature. 1982;298:195–198. doi: 10.1038/298195a0. [DOI] [PubMed] [Google Scholar]
- 35.Medina R, et al. The HiNF-P/p220NPAT cell cycle signaling pathway controls non-histone target genes. Cancer Res. 2007;67:10334–10342. doi: 10.1158/0008-5472.CAN-07-1560. [DOI] [PubMed] [Google Scholar]
- 36.Hovhannisyan H, et al. Maintenance of open chromatin and selective genomic occupancy at the cell-cycle-regulated histone H4 promoter during differentiation of HL-60 promyelocytic leukemia cells. Mol Cell Biol. 2003;23:1460–1469. doi: 10.1128/MCB.23.4.1460-1469.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei Y, Jin J, Harper JW. The cyclin E/Cdk2 substrate and Cajal body component p220(NPAT) activates histone transcription through a novel LisH-like domain. Mol Cell Biol. 2003;23:3669–3680. doi: 10.1128/MCB.23.10.3669-3680.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye X, Wei Y, Nalepa G, Harper JW. The cyclin E/Cdk2 substrate p220(NPAT) is required for S-phase entry, histone gene expression, and Cajal body maintenance in human somatic cells. Mol Cell Biol. 2003;23:8586–8600. doi: 10.1128/MCB.23.23.8586-8600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loeb KR, et al. A mouse model for cyclin E-dependent genetic instability and tumorigenesis. Cancer Cell. 2005;8:35–47. doi: 10.1016/j.ccr.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 40.Hunt KK, Keyomarsi K. Cyclin E as a prognostic and predictive marker in breast cancer. Semin Cancer Biol. 2005;15:319–326. doi: 10.1016/j.semcancer.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 41.Harper DP, Aplan PD. Chromosomal rearrangements leading to MLL gene fusions: Clinical and biological aspects. Cancer Res. 2008;68:10024–10027. doi: 10.1158/0008-5472.CAN-08-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baysal BE, et al. A high-resolution integrated map spanning the SDHD gene at 11q23: A 1.1-Mb BAC contig, a partial transcript map and 15 new repeat polymorphisms in a tumour-suppressor region. Eur J Hum Genet. 2001;9:121–129. doi: 10.1038/sj.ejhg.5200585. [DOI] [PubMed] [Google Scholar]
- 43.Di Fruscio M, et al. Proviral inactivation of the Npat gene of Mpv 20 mice results in early embryonic arrest. Mol Cell Biol. 1997;17:4080–4086. doi: 10.1128/mcb.17.7.4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geng Y, et al. Cyclin E ablation in the mouse. Cell. 2003;114:431–443. doi: 10.1016/s0092-8674(03)00645-7. [DOI] [PubMed] [Google Scholar]
- 45.Parisi T, et al. Cyclins E1 and E2 are required for endoreplication in placental trophoblast giant cells. EMBO J. 2003;22:4794–4803. doi: 10.1093/emboj/cdg482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berthet C, Aleem E, Coppola V, Tessarollo L, Kaldis P. Cdk2 knockout mice are viable. Curr Biol. 2003;13:1775–1785. doi: 10.1016/j.cub.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 47.Ortega S, et al. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat Genet. 2003;35:25–31. doi: 10.1038/ng1232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.