Abstract
Rationale: A cellular prooxidant state promotes cells to neoplastic growth, in part because of modification of proteins and their functions. Reactive nitrogen species formed from nitric oxide (NO) or its metabolites, can lead to protein tyrosine nitration, which is elevated in lung cancer. Objective: To determine the alteration in these NO derivatives and the role they may play in contributing to lung carcinogenesis. Methods: We analyzed levels of NO, nitrite (NO2−), nitrate (NO3−), and the location of the protein nitration and identified the proteins that are modified. Measurements and Main Results: Although exhaled NO and NO2− were increased, endothelial NO synthase or inducible NO synthase expression was similar in the tumor and tumor-free regions. However, immunohistochemistry showed that nitrotyrosine was increased in the tumor relative to non–tumor-bearing sections. We used proteomics to identify the modified proteins (two-dimensional polyacrylamide gel electrophoresis; mass spectrometry). Both the degree of nitration and the protein nitration profile were altered. We identified more than 25 nitrated proteins, including metabolic enzymes, structural proteins, and proteins involved in prevention of oxidative damage. Alterations of the biology of NO metabolites and nitration of proteins may contribute to the mutagenic processes and promote carcinogenesis. Conclusions: This study provides evidence in favor of a role for reactive nitrogen and oxygen species in lung cancer.
Keywords: lung cancer, nitric oxide, nitrotyrosine, protein nitration, proteomics
Nitric oxide (NO) plays a variety of regulatory roles in vivo, including control of vascular tone and host defense. However, recent data have suggested that NO metabolites, such as nitrite or NO-modified peptides or proteins, may also play important physiologic roles. Enzymatic generation of NO in mammals is by three isoforms of NO synthase (NOS), inducible NOS (iNOS), endothelial NOS (eNOS), and neuronal NOS (nNOS) (1). The function and regulation of these proteins reflect the role they play in normal physiology. Excessive or inappropriate production of endogenous or exogenous reactive oxygen species (ROS) and NO are implicated in the pathogenesis of cancer (2, 3). For example, cigarette smoke, a major source of exogenous oxidants, is associated with the development of lung cancer (2, 4). Furthermore, cigarette smoking leads to chronic airway inflammation with accumulation and activation of leukocytes, which produce high levels of ROS and NO. Both these processes lead to oxidative damage (5). Although it is generally accepted that the prooxidant state promotes cells to neoplastic growth through DNA damage or modification of proteins and their functions (3, 6–8), precisely how NO is involved in this process is unclear because it has been shown to have bipolar cellular effects, leading to either promoting or inhibiting tumor growth (9). There is considerable evidence supporting a role for NO in carcinogenesis (10–12). It is believed that high levels of NO may be cytostatic, or cytotoxic for tumor cells, whereas low levels may promote tumor growth (13, 14); however, the role of NO in tumor biology is still poorly understood.
NO and its metabolites interact with ROS to generate potent nitrating agents leading to 3-nitrotyrosine formation in proteins (15, 16), which is one of several chemical modifications that occur during oxidative/nitrosative stress (16, 17). Patients with lung cancer have significantly higher levels of nitrated proteins in serum, supporting the presence of oxidative and nitrosative stress (12, 18). We believe that protein nitration may be involved in pathogenesis of lung cancer because it alters enzymatic activity, structural properties, and signaling pathways by affecting tyrosine phosphorylation cascades (17, 19). Although increases in tyrosine nitration have been observed in cancers (18, 20), specific locations and targets have not been detailed, which are required to ultimately understand the molecular consequences.
Here, we have determined the alterations in NO and its metabolites in lung cancer and demonstrated that NO, nitrite, and nitrotyrosine are increased in patients with lung cancer. Increased nitrotyrosine immunostaining is limited to the tumor, suggesting a unique environment inside the tumor that may contribute to the disease process. Using proteomic and genomic approaches, we have identified the protein targets. The physiologic effects of abnormalities in NO metabolites and the contribution to the multistage carcinogenesis process are discussed. Some of the results of these studies have been previously reported in the form of an abstract (21).
METHODS
Redundant deidentified samples of lung tumors and adjacent normal lung tissue were obtained at the time of surgery of patients at the Cleveland Clinic Foundation between 1993 and 2001. Patients with lung cancer were evaluated for exhaled NO levels, and NO reaction products in exhaled breath condensate. Preoperative lung function indices and histologic or computed tomographic evidence of emphysema were recorded. Postoperative adjuvant or rescue treatments, time to tumor progression, and survival time were also described. Lung parenchymal tissues representing the tumor-involved and nearby tumor-free areas were stored immediately after resection in liquid nitrogen until subsequent processing. The study was approved by the Cleveland Clinic Institutional Review Board, and all individuals for exhaled breath analyses signed informed consent.
Exhaled NO levels were measured by an offline method according to established guidelines by American Thoracic Society using a chemiluminescent analyzer (NOA 280; Sievers, Boulder, CO) (22).
Nitrite and nitrate concentrations were determined using the ISO-NOP nitric oxide sensor (World Precision Instruments, Sarasota, FL), an amperometric sensor specific for NO (23).
Immunohistochemistry was done using paraffin-embedded tissue sections with a monoclonal antinitrotyrosine antibody.
Proteomic analysis was performed as described by Aulak and colleagues (24).
Proteins were identified either by matrix-assisted laser desorption ionization/time-of-flight mass spectrometric or by LCQ-Deca ion trap mass spectrometer system (Finnegan, La Jolla, CA), as described by Willard and colleagues (25). This methodology has limitations, as only approximately 10% of the proteome is visualized by this system.
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) activity was measured as described by Krebs and colleagues (26). Quantitation of GAPDH was determined by Western analysis using a mouse monoclonal GAPDH antibody (1:200,000; Research Diagnostics Inc., Concord, MA) and a standard curve of known amounts of GAPDH (8–50 ng). Density of the signal on autoradiographs was used to determine exact values.
Expression of eNOS and iNOS in tissue homogenates were detected by Western blot analysis.
Changes in gene expression were determined using Affymetrix Hu95A GeneChips (Affymetrix, Santa Clara, CA) containing 12,400 unique genes, with one GeneChip used for each human lung tissue (tumor-free or tumor). Detailed protocols for data extraction from Affymetrix oligonucleotide array and documentation on the sensitivity and quantitative aspects of the method have been described (27). Additional detail on all methods is provided in an online supplement.
Statistical Analysis
The demographic and clinical data are shown as mean ± SE unless otherwise stated in the text. All statistical comparisons between patients with lung cancer and healthy control subjects were performed using the Student's t test unless otherwise stated in the text. The level of significance for p value was chosen at 0.05. All data were analyzed with the SigmaStat Statistical Program (version 1.0; Systat, Inc., Evanston, IL).
RESULTS
Demographic and Clinical Data
There were 47 lung samples collected from 31 individuals between 1993 and 2001. Forty-two samples were taken as paired samples of tumor and tumor-free lung tissues from 21 individuals. Exhaled NO and its products were investigated in 11 patients with lung cancer and 35 healthy control subjects. Not all analyses could be performed on all samples because of the limited amounts of available surgical samples and inability of all patients to participate in all studies because of impaired health. The number of samples used in each experiment is stated in the text. The demography, tumor characteristics, preoperative lung function indices, and postoperative treatments are summarized in Table 1. Two-thirds of individuals with lung cancer were males, with a mean age of 66.7 ± 10.3 years (43–83 years). All except three patients were cigarette smokers with 49 ± 37 pack-years. The main tumor cell types were adenocarcinoma (38%) and squamous cell carcinoma (40%), predominantly poorly differentiated (69%). There were more than 40% of individuals with stage IA and IB diseases. The preoperative lung functions were preserved with FEV1 (71 ± 20.5% predicted) and FVC (82.6 ± 19.3% predicted).
TABLE 1.
Demographic and clinical data
| Clinical Parameters | |
|---|---|
| Sex | 14 females, 28 males |
| Age | 66.7 ± 10.3 |
| Smoking status | |
| Current smoker | 11 (26.2) |
| Ex-smoker | 28 (66.7) |
| Nonsmoker | 3 (7.1) |
| Smoking, pack-yr | 49 ± 37 |
| Histology | |
| Squamous cell | 17 (40.5) |
| Adenocarcinoma | 16 (38.1) |
| Poorly differentiated non–small cell | 9 (21.4) |
| Grade | |
| Well | 13 (31) |
| Poor | 29 (69) |
| Stage | |
| IA | 9 (21.4) |
| IB | 8 (19) |
| IIB | 9 (21.4) |
| IIIA | 5 (12) |
| IIIB | 6 (14.2) |
| IV | 5 (12) |
| FEV1, %predicted | 71.6 ± 20.5 |
| FVC, %predicted | 82.6 ± 19.3 |
| FEV1/FVC | 75.5 ± 16.7 |
All data are mean ± SD; values in brackets are the percentage of the total population studied.
Exhaled Breath NO and NO Reaction Products
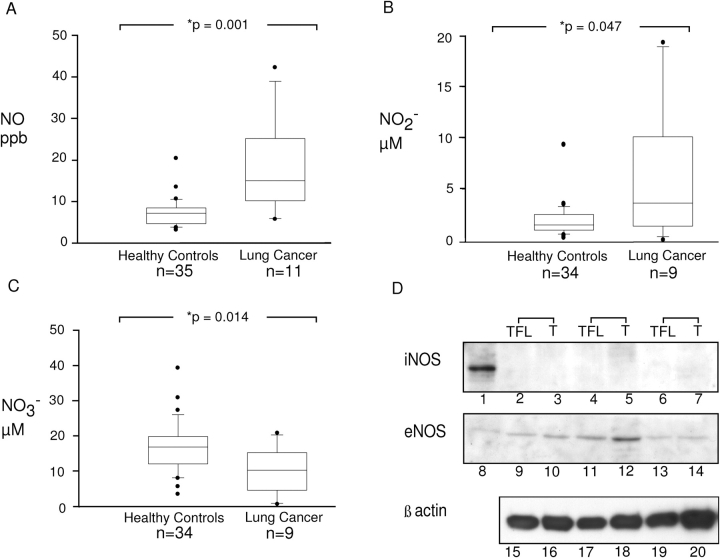
To investigate NO generation in patients, we analyzed exhaled NO and NO reaction products. Patients with lung cancer had higher levels of exhaled NO compared with control patients (control subjects, 7.4 ± 0.54, n = 35; patients with cancer, 18.4 ± 3.16 ppb, n = 11; p = 0.001; Figure 1A). The level of total nitrite in exhaled breath condensate was also elevated (control subjects, 1.93 ± 0.27, n = 34; patients with cancer, 5.83 ± 2.1, n = 9; p = 0.047; Figure 1B), but levels of nitrate were lower (control subjects, 16.75 ± 1.2, n = 34; patients with cancer, 10.01 ± 2.1 μM, n = 9; p = 0.014 Figure 1C).
Figure 1.
Levels of inducible nitric oxide synthase (iNOS), endothelial nitric oxide synthase (eNOS), nitric oxide (NO), and its metabolites in lung cancer. Increased levels of exhaled NO were observed in patients with cancer (n = 11) compared with healthy control subjects when measured directly by chemiluminescence (A). Nitrite (B) was increased in the breath condensates of patients with cancer, whereas the levels of nitrate (C) were decreased. Graphs show the percentiles and the median of the data. The ends of the boxes define the 25th and 75th percentiles, with a line at the median and error bars defining the 10th percentiles. Levels of iNOS were undetectable (D; lanes 2–7) and eNOS were similar (D; lanes 9–14) between lung tumor and adjacent tumor-free lung parenchyma. Representative Western blot analysis of tissue lysates (50 μg/lane) from paired tumor-free lung (TFL) and tumor (T) is shown. Each of the paired samples (TFL and T) was obtained from the same individual. β-actin was used as control for sample integrity and loading (D; lanes 15–20). Lane 1 shows positive control for iNOS, A549 cells stimulated for 3 days with IFN-γ, interleukin 1β, and tumor necrosis factor α. Lane 8 shows purified eNOS positive control.
To investigate the mechanism of increased NO levels, we analyzed the expression of eNOS and iNOS in tissues. Using protein lysates from tumor-bearing samples and control regions from the same patient, iNOS and eNOS levels were not different (n = 5; Figure 1D). This does not exclude the possibility that elevation of NOS exist compared with control patients, because healthy nondiseased control tissue samples were unavailable for comparison. Levels of arginine, a substrate for NOS, and an endogenous inhibitor of NOS, asymmetric NG,NG-dimethyl-l-arginine were also investigated. However, serum levels of arginine and asymmetric NG,NG-dimethyl-l-arginine were similar among patients with cancer and healthy control subjects (data not shown; all p > 0.2).
Nitrotyrosine in Tissues
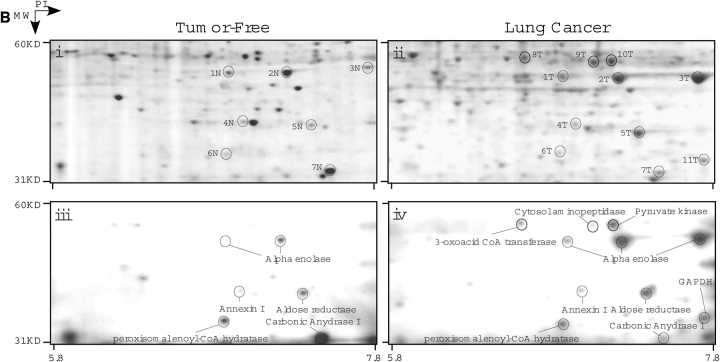
To investigate the location of nitrated proteins, immunohistochemistry with an antinitrotyrosine antibody was performed on tissue sections. Positive controls to test the specificity of the antibody included tissue sections treated with peroxynitrite, a potent nitrating agent, and negative controls were samples incubated with secondary antibody alone (data not shown). Six adenocarcinomas and six squamous cell carcinomas were evaluated (Figures 2Ai and 2Aiv). Ten patients were positive and two patients were negative for nitrotyrosine. Nitrotyrosine immunopositivity was observed in squamous cell carcinomas (Figure 2Aii) and adenocarcinomas (Figure 2Av). In both tumor types, regions that were free of the tumor contained little nitrotyrosine staining (Figures 2Aiii and 2Avi). Most of the reactivity was present inside the tumor itself (Figures 2Aii and 2Av). Quantitation of three different fields of each of those 10 positive patients indicates that nitrotyrosine staining was strongly positive in the tumor itself, but negative or weakly reactive in the adjacent tumor-free lung regions. Approximately 20 to 85% of tumor cells were stained positive (red) with antinitrotyrosine (tumor, 49.5 ± 5.5%; n = 10).
Figure 2.
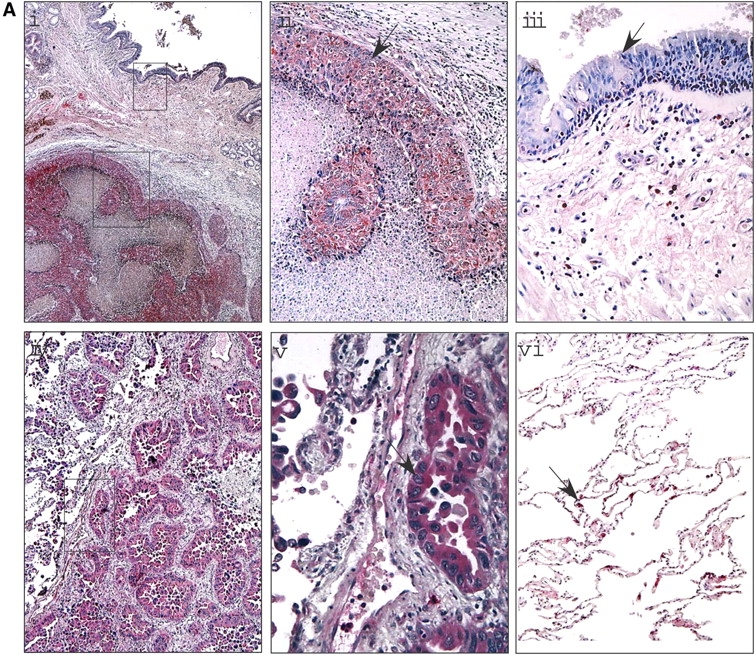
Nitrotyrosine location and targets in lung cancer. (A) Nitrotyrosine staining in lung cancer. Immunochemical staining using an antinitrotyrosine antibody demonstrated that the nitrotyrosine-modified proteins were localized mainly to the tumor and not to surrounding normal tissue. This was noted in squamous cell carcinoma (i–ii) as well as in the well differentiated adenocarcinoma (iv–v). Nitrotyrosine staining was mainly observed in the tumor itself (ii, v), but not in the adjacent tumor-free lung regions (iii), or weakly reactive in different regions of the lung from the same patients with cancer (vi). (B) Proteomic analyses of human lung tissue. Regions of tumor and tumor-free samples were subjected to proteomic analysis. Samples were solubilized in the denaturing buffer and 400 μg were subjected to two-dimensional electrophoresis. The gels were then partially transferred to polyvinylidene difluoride membrane and subjected to Western blot analysis using an antinitrotyrosine antibody. Coomassie blue–stained polyacrylamide gels of tumor-free (i) and tumor (ii) are shown with the corresponding Western blots (iii and iv, respectively). The protein spots that corresponded to the immunoreactive proteins observed in the Western blot were cut out, subjected to mass spectrometry, and identified. Some of the identified proteins are named on the Western blot and the corresponding spot on the Coomassie gel marked. Both samples show some degree of nitrotyrosine-containing proteins; however, more intense nitration is observed in tumor samples. Comparison of the Western blots and the respective Coomassie-stained gels demonstrate that selective proteins are modified and that these are not the most abundant proteins. Some proteins (2T, 3T, 5T) show increased specific 3-nitrotyrosine immunoreactivity, whereas others (8T, 9T, 10T) are nitrated only in the tumor. Corresponding spots from tumor-free tissue samples are suffixed with N.
All positive immunostained regions were scored semiquantitatively on a range of 0 to 4+. Among the 10 patients, 2 were scored as 4+, four were scored as 3+, three were 2+, and one was 1+. Thus, higher levels of nitrotyrosine were observed in the tumor compared with the surrounding tumor-free region.
Identification of Nitrotyrosine-containing Proteins
A proteomic approach was used to identify and analyze for changes in expression of nitrotyrosine-containing proteins. Protein lysates from tumor and tumor-free regions (n = 6) were run on two-dimensional gels using isoelectrofocusing in the first dimension and sodium dodecyl sulfate–polyacrylamide gel electrophoresis in the second dimension (Figures 2Bi and 2Bii). Gels were partially transferred onto polyvinylidene difluoride membrane and probed with an antinitrotyrosine antibody (Figures 2Biii and 2Biv). Immunopositive proteins were isolated from the original polyacrylamide gels and analyzed by mass spectrometry (Table 2). Identified proteins include proteins involved in glycolysis, cell structure and intracellular transport, antioxidant defense, and RNA binding. Notably, nitration of succinyl-CoA: 3-oxoacid CoA-transferase (SCOT), cytosolic aminopeptidase, GAPDH, and pyruvate kinase were seen in the tumor but not in tumor-free samples, whereas nitration of aldose reductase and α-enolase were present in tumor-free samples but increased in the tumor samples. Peroxisomal enoyl-CoA hydratase and carbonic anhydrase I nitration were reduced in tumor samples.
TABLE 2.
Nitrated proteins in lung cancer
| Proteins | pI | Mol. Wt. (kD) | Accession No. | Function |
|---|---|---|---|---|
| Antioxidant proteins | ||||
| Mn superoxide dismutase | 8.4 | 24.9 | 1070454 | Dismutation of superoxide radicals generating H2O2 |
| Antioxidant protein 2 | 6.0 | 25.1 | 4758638 | H2O2 breakdown and phospholipid turnover |
| Carbonic andydrase I* | 6.5 | 29.1 | 4502517 | Catalyze the reversible hydration of CO2 |
| Selenium binding protein | 5.8 | 53.1 | 16306550 | Participates in intra-golgi protein transport |
| Aldose reductase | 6.56 | 35.85 | 113596 | Reduction of carbonyl-containing compounds |
| Metabolic enzymes | ||||
| Aldolase A* | 8.4 | 39.28 | 229674 | Glycolytic enzyme |
| Triosephosphate isomerase* | 6.4 | 26.7 | 4507645 | Glycolytic enzyme |
| GAPDH | 8.57 | 36.2 | 62503 | Glycolytic enzyme |
| Phosphoglycerate mutase-1 | 6.7 | 28.8 | 4505753 | Glycolytic enzyme |
| Pyruvate kinase | 7.57 | 58.3 | 20178296 | Glycolytic enzyme |
| α-Enolase* | 6.9 | 47.3 | 119339 | Glycolytic enzyme |
| 3-oxoacid CoA transferase | 7.2 | 56.6 | 14290472 | Key enzyme in ketone body metabolism |
| Mit. aldehyde dehydrogenase-2 | 6.6 | 56.4 | 25777732 | Involved in proline degradation pathway |
| Cytosol aminopeptidase | 6.3 | 53.1 | 12643394 | Processing and turnover of intracellular proteins |
| Lipid metabolism | ||||
| Peroxisomal enoyl-CoA hydratase* | 6.6 | 36.3 | 2135896 | Key protein in fatty acid metabolism |
| Apolipoprotein A | 5.6 | 30.7 | 4557321 | Lipid metabolism |
| Apoptosis | ||||
| Annexin I* | 6.6 | 38.9 | 71756 | Regulates activity of Ca2+-phospholipid |
| Annexin III | 5.6 | 36.4 | 4826643 | Regulate cellular growth and signal transduction |
| Annexin VII | 6.3 | 50.4 | 4502111 | Ca2+- and phospholipid-binding protein |
| Structure proteins | ||||
| Vimentin | 4.9 | 53.3 | 13629179 | Structural protein |
| α1-Actin* | 5.2 | 42.4 | 4501883 | Structural protein |
| β-Tubulin* | 4.7 | 48.9 | 2119276 | Structural protein |
| α-Tubulin | 4.9 | 49.9 | 32015 | Structural protein |
| Tubulin α-β dimer | 5.2 | 47.9 | 3745822 | Structural protein |
| Others | ||||
| Transferrin | 6.8 | 77.1 | 4557871 | Plasma protein and carrier for iron |
| Guanine nucleotide binding protein | 7.6 | 35.0 | 1321585 | Activation of signal transduction |
| Fibrinogen γ–α chain* | 5.7 | 49.5 | 71827 | Involved in clot formation |
| β-Fibrinogen | 8.3 | 54.8 | 182430 | Involved in clot formation |
| hnRNPK | 5.4 | 50.9 | 241478 | Pre-mRNA processing, mRNA transport and transcription factor |
Definition of abbreviations: GAPDH = glyceraldehyde 3-phosphate dehydrogenase; hnRNPK = heterogenous nuclear ribonucleoprotein K.
Nitrated proteins in tumor and tumor-free tissue.
Alteration in Gene and Protein Expression
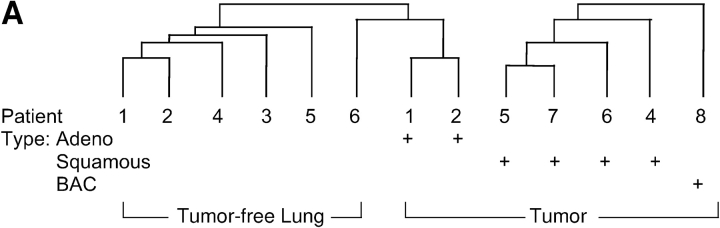
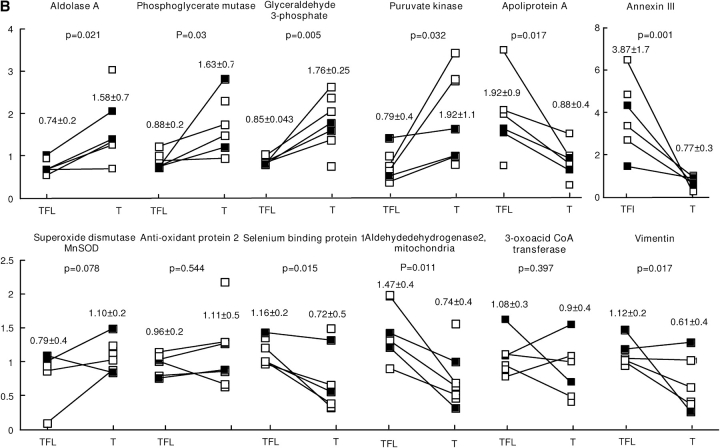
We investigated whether the observed increase in nitrated proteins was due to increases in the total expression of specific proteins or if a higher degree of nitration of the protein occurred. We used data obtained from gene chip analysis on tumors from adenocarcinoma (n = 3), squamous cell carcinoma (n = 4), and bronchial alveolar carcinoma (n = 1). Hierarchic cluster of gene expression profiles verified that the expression pattern of tumor-free versus tumor was different, even when tumor and tumor-free profiles were derived from the same person (Figure 3A). We analyzed individual genes to identify changes in RNA expression. As in the Western blot analysis, no change in eNOS or iNOS gene expression was noted. Some of the proteins that had higher nitration in tumors, such as selenium binding protein and SCOT, were not increased at the mRNA level in tumors compared with tumor-free samples (Figure 3B). However, in some, increased mRNA levels were detected, such as antioxidant protein 2 and Mn superoxide dismutase (Figure 3B).
Figure 3.
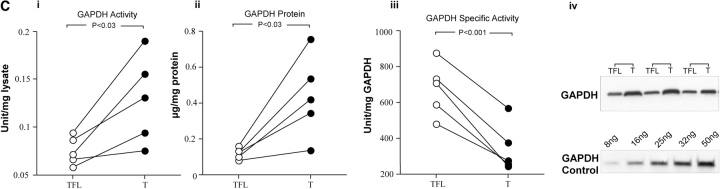
Alteration in RNA expression of nitrated proteins and decrease glyceraldehyde 3-phosphate dehydrogenase (GAPDH) activity in tumor samples. (A) Patterns of gene expression correspond to the major morphologic classes. Three main groups were identified by hierarchic cluster of gene-expression profiles using the 8,953 expressed genes. Tumor 3 is absent from the dendrogram because of lack of tumor-free pair for this patient. (B) GeneChip (Affymatrix, La Jolla, CA) analysis of genes for nitrated proteins in lung cancer. The genes for the targets of protein nitration are both upregulated and downregulated. Most genes from the glycolytic pathways are upregulated. Many of the other genes are either similar or downregulated in the tumor, suggesting that targeting is not from increased expression of the protein. (C) GAPDH activity in tumor. (i) GAPDH activities were higher in the tumor (T) than tumor-free lung (TFL; TFL: 0.08 ± 0.01, T: 0.13 ± 0.02 U/mg protein; n = 5). GAPDH expression was determined by Western analysis and a standard curve of known amount of GAPDH (8–50 ng). GAPDH protein expression was also higher in tumor (ii, iv; TFL: 0.12 ± 0.01, T: 0.44 ± 0.1 μg/mg protein; n = 5). However, specific activity of GAPDH was significantly lower in tumors (iii; TFL: 674.6 ± 149.7, T: 341.7 ± 135.45 U/GAPDH protein; n = 5).
GAPDH Activity
We investigated possible effects of protein nitration on one important glycolytic enzyme, GAPDH. GAPDH activities were higher in the tumor than in the tumor-free lung (tumor-free lung: 0.08 ± 0.01; tumor: 0.13 ± 0.02 U/mg protein; n = 5; p = 0.03; Figure 3Ci). However, GAPDH protein expression was also higher in tumor (tumor-free lung: 0.12 ± 0.01; tumor: 0.44 ± 0.1 μg/mg protein; n = 5; p = 0.03; Figures 3Cii and 3Civ). Specific activity of GAPDH (GAPDH activity/mg GAPDH protein) was significantly lower in tumors, indicating loss of GAPDH protein activity (tumor-free lung: 674.6 ± 149.7; tumor: 341.7 ± 135.45 U/mg GAPDH protein; n = 5; p = 0.001; Figure 3Ciii).
DISCUSSION
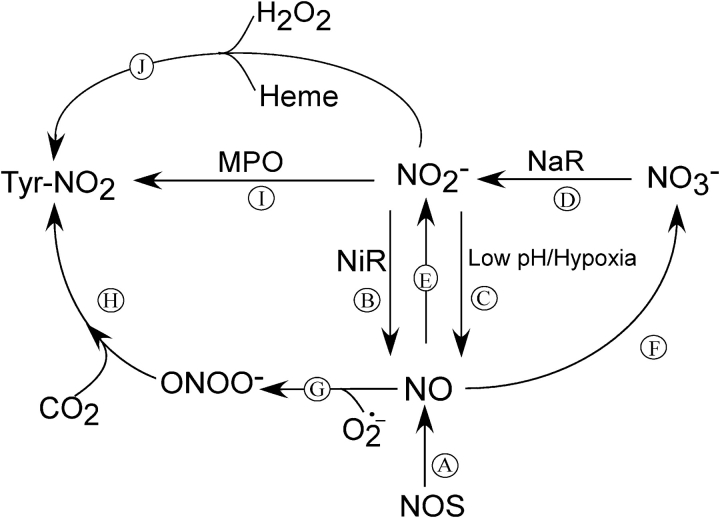
In this report, we have used in vivo measurements of NO and its reaction products to determine the role NO may play in lung cancer. We show that patients with lung cancer have increased exhaled NO and nitrite compared with healthy control subjects. This is in contrast to asthma, in which an increase in NO also occurs but levels of nitrate are higher and nitrite is unchanged compared with control subjects (28). This suggests that differences in NO metabolism occur in cancer that cannot simply be accounted for by elevated NO. Normally, NO2− is a good indicator of NOS activity. However, we detected no expression of iNOS in the tumor, and eNOS was not significantly increased in tumor-bearing lung samples compared with healthy regions of the same patient, using both Western blotting for the protein and gene chip analysis for mRNA. Because NO is increased, then either a systemic increase in NOS expression occurs, increasing NOS coupling (29), or nonenzymatic sources lead to higher release of NO in the lung. Liu and colleagues (30) also observed elevated exhaled NO in patients with cancer with increases in both the tumor-free and tumor-containing sides of the lung. However, in contrast to our finding with lung tissues lysates, they found increased iNOS expression in alveolar and tumor-associated macrophages. To understand the pathophysiology of NO and its related products, we proposed a scheme to explain our findings in light of current understanding of NO reactions (Figure 4). NO can be produced by enzymatic and nonenzymatic mechanisms (6, 31, 32). Enzymatically, NO can be generated either from l-arginine in the presence of NOS (reaction A, Figure 4) (1, 33) or by reduction of nitrite to NO in the presence of nitrite reductase (reaction B, Figure 4) (34, 35). The former reaction can occur in mammals, but humans lack nitrite reductases to carry out the latter reaction. The source of the latter enzyme may be from respiratory pathogens such as Haemophilus influenzae and Streptococcus pneumoniae (36) that chronically colonize patients with chronic obstructive pulmonary disease and lung cancer. These pathogens may generate NO by reducing nitrate to nitrite by nitrate reductase (reaction D, Figure 4) and then nitrite to NO by nitrite reductase (reaction B, Figure 4) (35, 37). These mechanisms have been demonstrated to occur in the normal oral cavity, where nitrate may be converted to nitrite by the bacteria colonization in the mouth (38). We speculate that these pathways lead to higher levels of NO and nitrite, and reduced levels of nitrate in the lung (39, 40). The local environment of the tumor may also increase NO synthesis. The altered metabolic states of tumors involve increased dependence on glycolysis that causes acidosis by the production of lactic acid (39, 41) and by the hydrolysis of adenosine triphosphate, which yields two protons per cycle of anaerobic glycolysis (39). This acidosis, together with the highly reducing environment of the tumor, may aid local generation of NO from nitrite (reaction C, Figure 4). In addition, further elevation of NO may be a result of nitrite reacting with deoxygenated heme (reaction E, Figure 4) (37, 42) in the hypoxic tumor environment. Studies with stable isotopes are needed to address these possibilities.
Figure 4.
Proposed scheme describing NO and its related products reactions in lung cancer. NO can be generated by enzymatic (A) and nonenzymatic mechanisms (B, C). NO reaction with superoxide produces peroxynitrite (ONOO−; G) with breakdown to nitrate (not shown). NO can be oxidized to nitrite (E), or nitrate in the presence of heme (F). In conditions in which the lung is colonized by bacteria, nitrate can be converted to nitrite by bacterial nitrate reductase (NaR; D), which can be further reduced to NO by bacterial nitrite reductase (NiR; B). In addition, nitrite may be reduced to NO at low pH and under hypoxic conditions (C). Another factor that may increase NO levels is myeloperoxidase (MPO) acting as a catalytic sink removing NO from autoinhibition of NOS (29). These factors contribute to both NO and nitrite being elevated and the levels of nitrate being decreased in patients with cancer. Altered nitration profiles may result from different mechanisms of protein nitration: either mediated via the generation of ONOO− (G, H) or via a peroxidase-mediated mechanism (I). In addition, NO2 may also be generated by the Fenton reaction. In the presence of H2O2, the heme is converted to ferryl π-cation radical species, which subsequently converts nitrite to ·NO2 to increase nitration (J).
NO by itself is relatively nonreactive (43). Many of its directly damaging properties may be derived by secondary reaction that may modify protein function (44–46) or may cause nucleotide modifications in DNA (47, 48). Increase in NO can lead to protein tyrosine nitration or nitrosylation. These types of modification are dependent on the flux of NO, concentration of potential targets, the microenvironment, and presence of cofactors. Nitrosylation may also lead to altered activity of target proteins, similar to nitration (49). Using immunohistochemistry, we demonstrated that protein nitration is increased in the tumor relative to the tumor-free region. The presence of nitrotyrosine appears tightly defined to the boundaries of the tumors. The mechanism of protein nitrotyrosine formation is unclear and may involve multiple pathways. These include the reaction of superoxide and NO to generate peroxynitrite, which may directly nitrate tyrosine residues or react with carbon dioxide to form the more potent nitrating agent ONOOCO2− (reactions G and H, Figure 4) (50). In addition, nitrite and hydrogen peroxide may be used by peroxidases, such as myeloperoxidase, to generate nitrogen dioxide (51), another potent nitrating agent (reaction I, Figure 4) (32). However, we did not detect alteration of the peroxidase-mediated oxidative markers chlorotyrosine, bromotyrosine, or ortho-tyrosine between tumor and tumor-free samples (data not shown). This suggests that peroxidase-related nitration is not a primary mechanism in cancer. Another possible mechanism involves the Fenton reaction. Increase in NO leads to release of heme from proteins that can then serve as catalysts for nitration of adjacent proteins (52). In the presence of H2O2, the heme is converted to ferryl π-cation radical species, which subsequently converts nitrite to ·NO2 to increase nitration (reaction J, Figure 4). The lack of increased NOS expression between the tumor-bearing and tumor-free regions suggests that non–NOS-mediated mechanisms probably lead to the increased localized nitration. This may be due to increased NO generation by nonenzymatic mechanisms in the hypoxic environment of the tumor or to localized increases in nitrite and ROS.
Protein nitration is increased in numerous diseases (53–56) and is typically considered to be a marker of oxidative damage rather than an active agent responsible for pathogenic effects. We have demonstrated that protein nitration is a specific process affecting select proteins in cells and tissues (24, 53, 54, 56). Recently, we showed that protein nitration and denitration is a tightly regulated dynamic process in cells and tissues (19). Here, we demonstrated that tyrosine nitrated proteins are present in lung tissues of both tumor-free and tumor-bearing samples. Comparisons show that patterns of protein nitration are different, with more nitrated proteins in the tumor-bearing samples. Although the intensity is generally increased in the tumor, some specific nitration is more intense in the tumor-free region. Most of the nitrated proteins fall into four categories: oxidant defense, energy production, structure, and those involved in apoptosis. A major group of proteins that are targets of tyrosine nitration is involved in oxidative stress. This includes MnSOD, antioxidant protein 2, aldose reductase, carbonic anhydrase, and selenium-binding protein. Accumulating evidence indicates that dysregulation of ROS promotes neoplastic growth (6, 57) with reactive nitrogen species and ROS often serving in signal transduction and having important biologic regulative activities. Smoking and chronic inflammation cause elevation of NO and ROS, affecting the cellular redox balance, both by providing increased oxidants and deactivating antioxidant mechanisms. With nitration and so inhibition of the antioxidant enzymes, a loss of antioxidant capacity would occur and so further promote injury and inflammation.
A second major group of proteins affected is involved in energy production. Tumors have an altered energy production pathway to keep up with the demand for increased growth in a hypoxic environment. Warburg, more than 70 years ago, introduced the concept that cancer cells relied on the glycolytic pathway for the majority of their energy production (58) and this is related to the hypoxic nature of the tumor. In this study, we found that many glycolytic enzymes are targets of protein nitration. Indeed, SCOT is the only other energy production pathway enzyme targeted for nitration that is not involved in glycolysis. Because protein nitration often leads to enzyme inhibition—for example, aldolase A (19) and GAPDH (59)—it is surprising to find increased glycolysis in tumor cells. However, here we show that mRNA from aldolase A, phosphoglycerate mutase, pyruvate kinase, and GAPDH are increased (Figure 3B). These observations are consistent with the finding of Unwin and colleagues (60), who demonstrated protein expression of six enzymes of the glycolysis pathway were increased in tumor cells. Increased expression of glycolytic enzymes appears to compensate for inhibition caused by protein nitration. For example, we demonstrate that activity of GAPDH is increased in the tumor compared with the tumor-free samples. However, specific activity of GAPDH is lower in the tumor samples, indicating protein modification inhibits the enzyme. Unwin and colleagues also demonstrated decreased expression of several mitochondrial proteins. We have previously shown that many of the targets of protein nitration in tissues are mitochondrial proteins (24). Their lack of nitration in tumors may reflect the reduced reliance on mitochondrial respiration and lower mitochondrial enzyme expression. In addition, under conditions of reduced oxygen levels, we demonstrated that net protein denitration in mitochondria occurs (19). Given the hypoxic environment in central tumor masses, denitration of mitochondrial protein targets may be preferred.
Here, we observe differences in protein nitration between tumor and nontumor tissue. It has been suggested that NO and protein nitration may have combined effects on tumorigenesis. NO consumptive mechanisms require oxygen. In the hypoxic environment of the tumor, NO consumption may decrease leading to local increases of NO. Thomas and colleagues have shown that levels of NO determine its biologic effects, including angiogenesis, erythropoiesis, and glycolysis (52). At low levels of NO, activation of hypoxia-inducible factor–1α occurs, but higher levels of NO inactivate p53, a tumor suppressor that regulates cell cycle genes such as p21waf/cip1 (61, 62). More than 90% of lung tumors are defective in p53. Other proteins previously associated with lung cancer, such as hnRNPK, were also nitrated and may significantly enhance cell proliferation and anchorage-independent growth (63). Likewise, nitration of annexin III may affect cell growth and signaling pathways (64). With broad effects on angiogenesis, glycolysis, p53 activity, antioxidant potential in the lung, and alterations in cell growth pathways, NO may create a microenvironment that can initiate tumorigenesis or promote tumor heterogeneity, leading to metastasis.
Overall, the observation of increased levels of NO, nitrite, and tumor-restricted tyrosine nitration in patients with cancer without increased NOS expression strongly supports an alteration of NO metabolism in the lungs of patients with cancer. Collectively, these studies and previous work provide evidence in favor of a role for reactive nitrogen and oxygen species in tumorigenesis and specifically in lung cancer.
Supplementary Material
Acknowledgments
The authors thank Daniel Laskowski, Roberto Machado, and Mohammad Taher for exhaled NO and clinical information. The authors also thank Carol Farver for tissue slides, Kavita Iyengar for RNA extraction, John Humes at Merck for anti-iNOS antibody, Stan Hazen with help with arginine and ADMA analyses, and Jim Lang for artwork.
Supported in part by grants HL60917, AI70649, HL076491, CA53914, and HL04265 from the National Institutes of Health; Debartolo Endowed Funds; American Heart Association 0325313B; a Betsy DeWindt American Cancer Society fellowship, GCRC M01RR018390; and Cafaro Endowed Funds.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Conflict of Interest Statement: None of the authors have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Knowles RG, Moncada S. Nitric oxide synthases in mammals. Biochem J 1994;298:249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kiyosawa H, Suko M, Okudaira H, Murata K, Miyamoto T, Chung MH, Kasai H, Nishimura S. Cigarette smoking induces formation of 8-hydroxydeoxyguanosine, one of the oxidative DNA damages in human peripheral leukocytes. Free Radic Res Commun 1990;11:23–27. [DOI] [PubMed] [Google Scholar]
- 3.Wiseman H, Halliwell B. Damage to DNA by reactive oxygen and nitrogen species: role in inflammatory disease and progression to cancer. Biochem J 1996;313:17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hurt RD, Ebbert JO. Preventing lung cancer by stopping smoking. Clin Chest Med 2002;23:27–36. [DOI] [PubMed] [Google Scholar]
- 5.Hoidal JR, Niewoehner DE. Lung phagocyte recruitment and metabolic alterations induced by cigarette smoke in humans and in hamsters. Am Rev Respir Dis 1982;126:548–552. [DOI] [PubMed] [Google Scholar]
- 6.Ara J, Przedborski S, Naini AB, Jackson-Lewis V, Trifiletti RR, Horwitz J, Ischiropoulos H. Inactivation of tyrosine hydroxylase by nitration following exposure to peroxynitrite and 1-methyl-4-phenyl-1, 2,3,6-tetrahydropyridine (MPTP). Proc Natl Acad Sci USA 1998;95:7659–7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohshima H, Tatemichi M, Sawa T. Chemical basis of inflammation-induced carcinogenesis. Arch Biochem Biophys 2003;417:3–11. [DOI] [PubMed] [Google Scholar]
- 8.Cerutti PA. Prooxidant states and tumor promotion. Science 1985;227:375–381. [DOI] [PubMed] [Google Scholar]
- 9.Wink DA, Mitchell JB. Nitric oxide and cancer: an introduction. Free Radic Biol Med 2003;34:951–954. [DOI] [PubMed] [Google Scholar]
- 10.Chhatwal VJ, Moochhala SM, Chan ST, Ngoi SS. Nitric oxide and cancer. Med Hypotheses 1996;46:21–24. [DOI] [PubMed] [Google Scholar]
- 11.Wink DA, Mitchell JB. Chemical biology of nitric oxide: insights into regulatory, cytotoxic, and cytoprotective mechanisms of nitric oxide. Free Radic Biol Med 1998;25:434–456. [DOI] [PubMed] [Google Scholar]
- 12.Gole MD, Souza JM, Choi I, Hertkorn C, Malcolm S, Foust RF III, Finkel B, Lanken PN, Ischiropoulos H. Plasma proteins modified by tyrosine nitration in acute respiratory distress syndrome. Am J Physiol Lung Cell Mol Physiol 2000;278:L961–L967. [DOI] [PubMed] [Google Scholar]
- 13.Jenkins DC, Charles IG, Thomsen LL, Moss DW, Holmes LS, Baylis SA, Rhodes P, Westmore K, Emson PC, Moncada S. Roles of nitric oxide in tumor growth. Proc Natl Acad Sci U S A 1995;92:4392–4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie K, Huang S. Contribution of nitric oxide-mediated apoptosis to cancer metastasis inefficiency. Free Radic Biol Med 2003;34:969–986. [DOI] [PubMed] [Google Scholar]
- 15.Beckman JS, Ischiropoulos H, Zhu L, van der Woerd M, Smith C, Chen J, Harrison J, Martin JC, Tsai M. Kinetics of superoxide dismutase- and iron-catalyzed nitration of phenolics by peroxynitrite. Arch Biochem Biophys 1992;298:438–445. [DOI] [PubMed] [Google Scholar]
- 16.Haddad IY, Pataki G, Hu P, Galliani C, Beckman JS, Matalon S. Quantitation of nitrotyrosine levels in lung sections of patients and animals with acute lung injury. J Clin Invest 1994;94:2407–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacMillan-Crow LA, Crow JP, Kerby JD, Beckman JS, Thompson JA. Nitration and inactivation of manganese superoxide dismutase in chronic rejection of human renal allografts. Proc Natl Acad Sci U S A 1996;93:11853–11858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pignatelli B, Li CQ, Boffetta P, Chen Q, Ahrens W, Nyberg F, Mukeria A, Bruske-Hohlfeld I, Fortes C, Constantinescu V, et al. Nitrated and oxidized plasma proteins in smokers and lung cancer patients. Cancer Res 2001;61:778–784. [PubMed] [Google Scholar]
- 19.Aulak KS, Koeck T, Crabb JW, Stuehr DJ. Dynamics of protein nitration in cells and mitochondria. Am J Physiol Heart Circ Physiol 2004;286:H30–H38. [DOI] [PubMed] [Google Scholar]
- 20.Maeda H, Akaike T. Nitric oxide and oxygen radicals in infection, inflammation, and cancer. Biochemistry (Mosc) 1998;63:854–865. [PubMed] [Google Scholar]
- 21.Masri F, Comhair SAA, Stuehr DJ, Erzurum SC, Aulak KS. Nitrosative modification of proteins in lung cancer. Free Radic Biol Med 2003;35:S170. [Google Scholar]
- 22.Dweik RA, Comhair SA, Gaston B, Thunnissen FB, Farver C, Thomassen MJ, Kavuru M, Hammel J, Abu-Soud HM, Erzurum SC. NO chemical events in the human airway during the immediate and late antigen-induced asthmatic response. Proc Natl Acad Sci USA 2001;98:2622–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berkels R, Purol-Schnabel S, Roesen R. A new method to measure nitrate/nitrite with a NO-sensitive electrode. J Appl Physiol 2001;90:317–320. [DOI] [PubMed] [Google Scholar]
- 24.Aulak KS, Miyagi M, Yan L, West KA, Massillon D, Crabb JW, Stuehr DJ. Proteomic method identifies proteins nitrated in vivo during inflammatory challenge. Proc Natl Acad Sci USA 2001;98:12056–12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willard BB, Ruse CI, Keightley JA, Bond M, Kinter M. Site-specific quantitation of protein nitration using liquid chromatography/tandem mass spectrometry. Anal Chem 2003;75:2370–2376. [DOI] [PubMed] [Google Scholar]
- 26.Krebs EG. Yeast glyceraldehyde-3-phosphate dehydrogenase: I: electrophoresis of fractions precipitated by nucleic acid. J Biol Chem 1953;200:471–478. [PubMed] [Google Scholar]
- 27.Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat Genet 1999;21:20–24. [DOI] [PubMed] [Google Scholar]
- 28.Dweik RA, Laskowski D, Ozkan M, Farver C, Erzurum SC. High levels of exhaled nitric oxide (NO) and NO synthase III expression in lesional smooth muscle in lymphangioleiomyomatosis. Am J Respir Cell Mol Biol 2001;24:414–418. [DOI] [PubMed] [Google Scholar]
- 29.Galijasevic S, Saed GM, Diamond MP, Abu-Soud HM. Myeloperoxidase up-regulates the catalytic activity of inducible nitric oxide synthase by preventing nitric oxide feedback inhibition. Proc Natl Acad Sci USA 2003;100:14766–14771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eiserich JP, Hristova M, Cross CE, Jones AD, Freeman BA, Halliwell B, van der Vliet A. Formation of nitric oxide-derived inflammatory oxidants by myeloperoxidase in neutrophils. Nature 1998;391:393–397. [DOI] [PubMed] [Google Scholar]
- 31.Ohshima H, Celan I, Chazotte L, Pignatelli B, Mower HF. Analysis of 3-nitrotyrosine in biological fluids and protein hydrolyzates by high-performance liquid chromatography using a postseparation, on-line reduction column and electrochemical detection: results with various nitrating agents. Nitric Oxide 1999;3:132–141. [DOI] [PubMed] [Google Scholar]
- 32.Wu W, Chen Y, Hazen SL. Eosinophil peroxidase nitrates protein tyrosyl residues; implications for oxidative damage by nitrating intermediates in eosinophilic inflammatory disorders. J Biol Chem 1999;274:25933–25944. [DOI] [PubMed] [Google Scholar]
- 33.Moncada S, Higgs A. The L-arginine-nitric oxide pathway. N Engl J Med 1993;329:2002–2012. [DOI] [PubMed] [Google Scholar]
- 34.Weitzberg E, Lundberg JO. Nonenzymatic nitric oxide production in humans. Nitric Oxide 1998;2:1–7. [DOI] [PubMed] [Google Scholar]
- 35.Reutov VP, Sorokina EG. NO-synthase and nitrite-reductase components of nitric oxide cycle. Biochemistry (Mosc) 1998;63:874–884. [PubMed] [Google Scholar]
- 36.Sethi S. Bacterial infection and the pathogenesis of COPD. Chest 2000;117:286S–291S. [DOI] [PubMed] [Google Scholar]
- 37.Zweier JL, Wang P, Samouilov A, Kuppusamy P. Enzyme-independent formation of nitric oxide in biological tissues. Nat Med 1995;1:804–809. [DOI] [PubMed] [Google Scholar]
- 38.Duncan C, Dougall H, Johnston P, Green S, Brogan R, Leifert C, Smith L, Golden M, Benjamin N. Chemical generation of nitric oxide in the mouth from the enterosalivary circulation of dietary nitrate. Nat Med 1995;1:546–551. [DOI] [PubMed] [Google Scholar]
- 39.Tannock IF, Rotin D. Acid pH in tumors and its potential for therapeutic exploitation. Cancer Res 1989;49:4373–4384. [PubMed] [Google Scholar]
- 40.Jahde E, Rajewsky MF. Tumor-selective modification of cellular microenvironment in vivo: effect of glucose infusion on the pH in normal and malignant rat tissues. Cancer Res 1982;42:1505–1512. [PubMed] [Google Scholar]
- 41.Wike-Hooley JL, Haveman J, Reinhold HS. The relevance of tumour pH to the treatment of malignant disease. Radiother Oncol 1984;2:343–366. [DOI] [PubMed] [Google Scholar]
- 42.Dejam A, Hunter CJ, Schechter AN, Gladwin MT. Emerging role of nitrite in human biology. Blood Cells Mol Dis 2004;32:423–429. [DOI] [PubMed] [Google Scholar]
- 43.Beckman JS. Oxidative damage and tyrosine nitration from peroxynitrite. Chem Res Toxicol 1996;9:836–844. [DOI] [PubMed] [Google Scholar]
- 44.Goodman JE, Hofseth LJ, Hussain SP, Harris CC. Nitric oxide and p53 in cancer-prone chronic inflammation and oxyradical overload disease. Environ Mol Mutagen 2004;44:3–9. [DOI] [PubMed] [Google Scholar]
- 45.van der Vliet A, O'Neill CA, Halliwell B, Cross CE, Kaur H. Aromatic hydroxylation and nitration of phenylalanine and tyrosine by peroxy-nitrite: evidence for hydroxyl radical production from peroxynitrite. FEBS Lett 1994;339:89–92. [DOI] [PubMed] [Google Scholar]
- 46.Ischiropoulos H. Biological tyrosine nitration: a pathophysiological function of nitric oxide and reactive oxygen species. Arch Biochem Biophys 1998;356:1–11. [DOI] [PubMed] [Google Scholar]
- 47.Ohshima H, Friesen M, Brouet I, Bartsch H. Nitrotyrosine as a new marker for endogenous nitrosation and nitration of proteins. Food Chem Toxicol 1990;28:647–652. [DOI] [PubMed] [Google Scholar]
- 48.Szabo C, Ohshima H. DNA damage induced by peroxynitrite: subsequent biological effects. Nitric Oxide 1997;1:373–385. [DOI] [PubMed] [Google Scholar]
- 49.Gow AJ, Farkouh CR, Munson DA, Posencheg MA, Ischiropoulos H. Biological significance of nitric oxide-mediated protein modifications. Am J Physiol Lung Cell Mol Physiol 2004;287:L262–L268. [DOI] [PubMed] [Google Scholar]
- 50.Gow A, Duran D, Thom SR, Ischiropoulos H. Carbon dioxide enhancement of peroxynitrite-mediated protein tyrosine nitration. Arch Biochem Biophys 1996;333:42–48. [DOI] [PubMed] [Google Scholar]
- 51.Sampson JB, Ye Y, Rosen H, Beckman JS. Myeloperoxidase and horseradish peroxidase catalyze tyrosine nitration in proteins from nitrite and hydrogen peroxide. Arch Biochem Biophys 1998;356:207–213. [DOI] [PubMed] [Google Scholar]
- 52.Thomas DD, Espey MG, Vitek MP, Miranda KM, Wink DA. Protein nitration is mediated by heme and free metals through Fenton-type chemistry: an alternative to the NO/O2- reaction. Proc Natl Acad Sci USA 2002;99:12691–12696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wattanapitayakul SK, Weinstein DM, Holycross BJ, Bauer JA. Endothelial dysfunction and peroxynitrite formation are early events in angiotensin-induced cardiovascular disorders. FASEB J 2000;14:271–278. [DOI] [PubMed] [Google Scholar]
- 54.Ferdinandy P, Danial H, Ambrus I, Rothery RA, Schulz R. Peroxynitrite is a major contributor to cytokine-induced myocardial contractile failure. Circ Res 2000;87:241–247. [DOI] [PubMed] [Google Scholar]
- 55.Oyama J, Shimokawa H, Momii H, Cheng X, Fukuyama N, Arai Y, Egashira K, Nakazawa H, Takeshita A. Role of nitric oxide and peroxynitrite in the cytokine-induced sustained myocardial dysfunction in dogs in vivo. J Clin Invest 1998;101:2207–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kondo S, Toyokuni S, Tsuruyama T, Ozeki M, Tachibana T, Echizenya M, Hiai H, Onodera H, Imamura M. Peroxynitrite-mediated stress is associated with proliferation of human metastatic colorectal carcinoma in the liver. Cancer Lett 2002;179:87–93. [DOI] [PubMed] [Google Scholar]
- 57.Chung-man Ho J, Zheng S, Comhair SA, Farver C, Erzurum SC. Differential expression of manganese superoxide dismutase and catalase in lung cancer. Cancer Res 2001;61:8578–8585. [PubMed] [Google Scholar]
- 58.Xu W, Kaneko FT, Zheng S, Comhair SA, Janocha AJ, Goggans T, Thunnissen FB, Farver C, Hazen SL, Jennings C, et al. Increased arginase II and decreased NO synthesis in endothelial cells of patients with pulmonary arterial hypertension. FASEB J 2004;18:1746–1748. [DOI] [PubMed] [Google Scholar]
- 59.Souza JM, Radi R. Glyceraldehyde-3-phosphate dehydrogenase inactivation by peroxynitrite. Arch Biochem Biophys 1998;360:187–194. [DOI] [PubMed] [Google Scholar]
- 60.Unwin RD, Craven RA, Harnden P, Hanrahan S, Totty N, Knowles M, Eardley I, Selby PJ, Banks RE. Proteomic changes in renal cancer and co-ordinate demonstration of both the glycolytic and mitochondrial aspects of the Warburg effect. Proteomics 2003;3:1620–1632. [DOI] [PubMed] [Google Scholar]
- 61.Cobbs CS, Whisenhunt TR, Wesemann DR, Harkins LE, Van Meir EG, Samanta M. Inactivation of wild-type p53 protein function by reactive oxygen and nitrogen species in malignant glioma cells. Cancer Res 2003;63:8670–8673. [PubMed] [Google Scholar]
- 62.Chazotte-Aubert L, Hainaut P, Ohshima H. Nitric oxide nitrates tyrosine residues of tumor-suppressor p53 protein in MCF-7 cells. Biochem Biophys Res Commun 2000;267:609–613. [DOI] [PubMed] [Google Scholar]
- 63.Pino I, Pio R, Toledo G, Zabalegui N, Vicent S, Rey N, Lozano MD, Torre W, Garcia-Foncillas J, Montuenga LM. Altered patterns of expression of members of the heterogeneous nuclear ribonucleoprotein (hnRNP) family in lung cancer. Lung Cancer 2003;41:131–143. [DOI] [PubMed] [Google Scholar]
- 64.Bastian BC. Annexins in cancer and autoimmune diseases. Cell Mol Life Sci 1997;53:554–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.