Abstract
Motivation: Protein complexes integrate multiple gene products to coordinate many biological functions. Given a graph representing pairwise protein interaction data one can search for subgraphs representing protein complexes. Previous methods for performing such search relied on the assumption that complexes form a clique in that graph. While this assumption is true for some complexes, it does not hold for many others. New algorithms are required in order to recover complexes with other types of topological structure.
Results: We present an algorithm for inferring protein complexes from weighted interaction graphs. By using graph topological patterns and biological properties as features, we model each complex subgraph by a probabilistic Bayesian network (BN). We use a training set of known complexes to learn the parameters of this BN model. The log-likelihood ratio derived from the BN is then used to score subgraphs in the protein interaction graph and identify new complexes. We applied our method to protein interaction data in yeast. As we show our algorithm achieved a considerable improvement over clique based algorithms in terms of its ability to recover known complexes. We discuss some of the new complexes predicted by our algorithm and determine that they likely represent true complexes.
Availability: Matlab implementation is available on the supporting website: www.cs.cmu.edu/~qyj/SuperComplex
Contact: zivbj@cs.cmu.edu
1 INTRODUCTION
Protein–protein interactions (PPI) are fundamental to the biological processes within a cell. Correctly identifying the interaction network among proteins in an organism is useful for deciphering the molecular mechanisms underlying given biological functions. Beyond individual interactions, there is a lot more systematic information contained in protein interaction graphs. Complex formation is one of the typical patterns in this graph and many cellular functions are performed by these complexes containing multiple protein interaction partners. As the number of species for which global high throughput protein interaction data is measured becomes larger (Ito et al., 2001; Rual et al., 2003; Stelzl et al., 2005; Uetz et al., 2000), methods for accurately identifying complexes from such data become a bottleneck for further analysis of the resulting interaction graphs.
High-throughput experimental approaches aiming to specifically determine the components of protein complexes on a proteome-wide scale suffer from high false positive and false negative rates (von Mering et al., 2002). In particular, mass spectrometry methods (Gavin et al., 2002; Ho et al., 2002) may miss complexes that are not present under the given conditions; tagging may disturb complex formation and weakly associated components may dissociate and escape detections. Therefore, accurately identifying protein complexes remains a challenge.
The logical connections between proteins in complexes can be best represented as a graph where the nodes correspond to proteins and the edges correspond to the interactions. Extracting the set of protein complexes from these graphs can help obtain insights into both the topological properties and functional organization of protein networks in cells. Previous attempts at automatic complex identification have mainly involved the use of binary protein–protein interaction graphs. Most methods utilized unsupervised graph clustering for this task by trying to discover densely connected subgraphs.
Automatic complex identification approaches can be divided into five categories: (1) Graph segmentation. To identify complexes King et al. (2004) partitioned the nodes of a given graph into distinct clusters using a cost-based local search algorithm. Zotenko et al. (2006) proposed a graph-theoretical approach to identify functional groups and provided a representation of overlaps between functional groups in the form of the ‘Tree of Complexes’. (2) Overlapping clustering. Since some proteins participate in multiple complexes or functional modules, a number of approaches allow overlapping clusters. Bader et al. (2003b) detected densely connected regions in large PPI networks using vertex weights representing local neighborhood density. Pereira-Leal et al. (2004) used the line graph strategy of the network (where a node represents an interaction between two proteins and edges share interactors between interactions) to produce an overlapping graph partitioning of the original PPI network. Adamcsek et al. (2006) identified overlapping densely interconnected groups in a given undirected graph using the k-clique percolation clusters in the network. Spirin et al. (2003) discovered molecular modules that are densely connected with themselves but sparsely connected with the rest of the network by analyzing the multibody structure of the PPI network. (3) New similarity measures. Rives et al. (2003) applied standard clustering algorithms to group similar nodes on the interaction graph. The cluster similarity is calculated on vectors of nodes’ attributes, such as their shortest path distances to other nodes. (4) Conservation across species. Sharan et al. (2005) used conservation alignment to find protein complexes that are common to yeast and bacteria. They formulated a log-likelihood ratio model to represent individual edges between proteins and assumed a clique structure for a protein complex. (5) Spatial constraints analysis. By utilizing the spatial aspects of complex formation, Scholtens et al. (2005) applied a local modeling method to better estimate the protein complex membership from direct mass spectrometry complex data and Y2H binary interaction data. Chu et al. (2006) proposed an infinite latent feature model to identify protein complexes and their constituents from large-scale direct mass spectrometry sets.
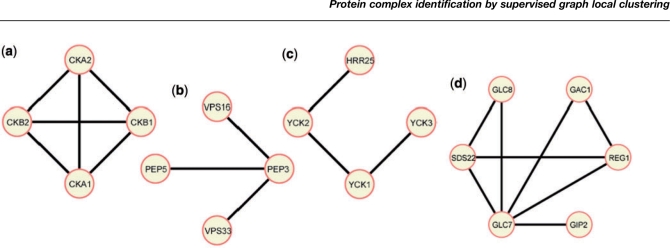
The methods presented above are based on the assumption that complexes form a clique in the interaction graph. While this is true for many complexes, there are many other topological structures that may represent a complex on a PPI graph. One example is a ‘star’ or ‘spoke’ model, in which all vertices connect to a ‘Bait’ protein (Bader et al., 2003a). Another possible topology is a structure that links several small densely connected components with loose linked edges. This topology is especially attractive for large complexes: due to spatial limitations, it is unlikely that all proteins in a large complex can interact with all others. See Figure 1 for some examples of real complexes with different topologies.
Fig. 1.
Projection of selected yeast MIPS complexes on our PPI graph (weight thresholded). (a) Example of a clique. All nodes are connected by edges. (b) Example of a star-shape, also referred to as the spoke model. (c) Example of a linear shape. (d). Example for a hybrid shape where small cliques are connected by a common node.
While some previous work was carried out to identify such structures in PPI networks [most notable by looking for network motifs (Yeger-Lotem et al., 2004)], these structure were not exploited for complex discovery. In this article we present a computational framework that can identify complexes without making strong assumptions about their topology. Instead of the ‘cliqueness’ assumption, we derive several properties from known complexes, and use these properties to search for new complexes. Since our method relies on real complexes, it does not assume any prior model for complexes. Our algorithm is probabilistic. Following training to determine the importance of different properties, it can assign a score to any subgraph in the graph. By thresholding this likelihood ratio score we can label some of the subgraphs as complexes. Our model results in a significantly improved F1-score when compared to the density-based approaches. Using a cross validation analysis we show that the graphs discovered by our method highly coincide with complexes from the hand-curated MIPS database and a recent high confidence mass spectrometry dataset (Gavin et al., 2006). The top-ranked new complexes are likely to provide novel hypotheses for the mechanism of action or definition of function of proteins within the predicted complex as we discuss in Section 3.
2 METHODS
The main feature of our method is that it considers the possibility of multiple factors defining complexes in protein interaction graphs. Instead of assuming a specific topological model, we design a general framework which learns to weigh possible subgraph patterns based on the available known complexes.
Previous analysis of known PPI graphs has already revealed multiple shapes forming subgraphs. For example, Bader et al. (2003a) proposed two topological models in the context of protein complexes. The first is the ‘matrix model’ which assumes that each of the members in the complex physically interact with all other members (leading to a clique-like structure). The second shape is the ‘spoke model’ that assumes that all proteins in a complex directly interact with one ‘bait’ protein leading to a star shape. Hybrids of these or other models are also possible, resulting in more complex topologies.
Besides graph structures, there could be other features that characterize complexes. In particular, complexes have certain biological, chemical or physical properties that distinguish them from non-complexes. For example, the physical size of a complex may be an important feature. There is a physical limitation of creating large complexes because inner proteins become inaccessible and therefore more difficult to regulate. By incorporating such additional features into our supervised learning framework, the proposed model is able to integrate multiple evidence sources to identify new complexes in the PPI graph.
The input to our algorithm is a weighted graph of interacting proteins. The network is modeled as a graph, where vertexes represent proteins and edges represent interactions. Edge weights represent the likelihoods for the interactions. Since the current data does not provide any directionality information, the PPI graph considered in this article is a weighted undirected graph. Our objective is to recover the protein complexes from this undirected PPI graph. Computationally speaking, complexes are one special kind of subgraphs on the PPI network. A subgraph represents a subset of nodes with a specific set of edges connecting them. The number of distinct subgraphs, or clusters, grows exponentially with the number of nodes.
2.1 Complex features
Extracting appropriate features for subgraphs representing complexes is related to the problem of measuring the similarity between complex subgraphs. This task has been studied for other networks, specifically social networks (Chakrabarti et al., 2005; Robins et al., 2005; Virtanen, 2003). In general, these previous approaches either (1) utilize properties of nodes or edges (indegree, outdegree, cliqueness (Borgwardt et al., 2007), or (2) rely on comparing non-trivial substructures such as triangles or rectangles (Przulj et al., 2007; Yan et al., 2002). We use both types to arrive at a list of properties for a feature vector that describes a subgraph in the PPI network. The properties include topological measurements about the subgraph structures and biological properties of the group of proteins in the subgraph.
Table 1 presents the set of features we use. We rely in part on prior work (Bader et al., 2003b; Barabasi et al., 2004; Chakrabarti et al., 2005; Stelzl et al., 2005; Zhu et al., 2005) to determine which features may be useful for this complex identification task. Each row in Table 1 represents one group of features. Totally 33 features were extracted from 10 groups.
Table 1.
Features for representing protein complex properties
| No | Group | Reference | Graph type | Num. features |
|---|---|---|---|---|
| 1 | Node size | Chakrabarti et al. (2005) | Binary | 1 |
| 2 | Graph density | Chakrabarti et al. (2005) | Binary | 1 |
| 3 | Degree statistics | Barabasi et al. (2004) | Binary | 4 |
| 4 | Edge weight statistics | Chakrabarti et al. (2005) | Weight | 4 |
| 5 | Density wrt. weight cutoffs | Chakrabarti et al. (2005) | Weight | 7 |
| 6 | Degree correlation statistics | Stelzl et al. (2005) | Binary | 3 |
| 7 | Clustering coefficient statistics | Barabasi et al. (2004) | Binary | 3 |
| 8 | Topological coefficient statistics | Stelzl et al. (2005) | Binary | 3 |
| 9 | First Eigenvalues | Chakrabarti et al. (2005) | Binary | 3 |
| 10 | Protein weight/size statistics | Cherry et al. (1997) | 4 |
Each row represents a group of similar features. We use 33 features divided into 10 groups. See supporting website for more details. The second column lists the name of the feature group and the third column provides the references. The fourth column specifies which type of graph is used to derive the property.
Below we briefly discuss each of the feature types used. The numbers match the numbers in Table 1.
Given a complex subgraph G=(V,E), with |V| vertexes and |E| edges, the first property we considered is the number of nodes in the subgraph: |V|.
The density is defined as |E| divided by the theoretical maximum number of possible edges |E|max. Since we do not consider self interactions in the input weighted PPI graph, |E|max=|V|*(|V|−1)/2. As mentioned above, in the ‘matrix’ model the graph density is expected to be very high, whereas it may be lower for the ‘spoke’ shape.
Degree statistics are calculated from the degree of nodes in the candidate subgraph. Degree is defined as the number of partners for a node. This group includes mean degree, degree variance, degree median and degree maximum.
The edge weight feature includes mean and variance of edge weights considering two different cases (with and without missing edges).
Density of weight cutoffs evaluate the possibility of topological changes under different weight cutoffs.
Degree correlation property measures the neighborhood connectivity of nodes within the subgraph. For each node it is defined as the average number of links of the nearest neighbors of the protein. We use mean, variance and maximum of this property in the feature set.
Clustering coefficient (CC) measures the number of triangles that go through nodes. To compute this feature we calculate the number of neighbors (q) and the number of links (t) connecting the q neighboring nodes. We set CC=2t/q(q−1). This feature will have a small value for ‘star’ or ‘linear’ shapes while ‘matrix’ or ‘hybrid’ shapes receive a higher value.
The topological coefficient (TC) is a relative measure of the extent to which a protein shares interaction partners with other proteins. It reflects the number of rectangles that pass through a node. See supporting website for details.
The first three largest singular values (SV) of the candidate subgraph's adjacency matrix. Different shapes have distinct value distributions for these three SV. For instance when comparing subgraphs with the same size, the ‘matrix’ shape has higher value for the first SV than other shapes and the ‘star’ shape has a lower value of the third SV. See supporting website for details.
As for biological properties (No. 10), we use average and maximum protein length and average and maximum protein weight of each subgraph. This feature is based on the intuition that protein complexes are unlikely to grow indefinitely, because proteins within the center of large complexes become inaccessible to interactions with other putative partners.
Our framework described below is general and it is straightforward to add other features if they are deemed relevant.
2.2 A supervised Bayesian network (BN) to model complexes
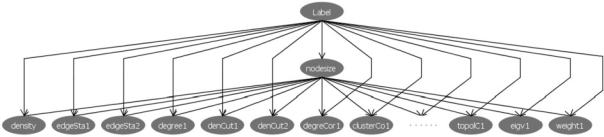
We assume a generative probabilistic model for complexes. Figure 2 presents an overview framework of our model. Our method uses a BN model. Features are generated, independently, based on two parameters, (1) whether the subgraph is a complex or not (C) and (2) the number of nodes in the subgraph (N). The main reason we pay special attention to N and do not model it as another complex property is because of the tendency of other properties to depend on N. For example, the larger the complex the more unlikely it is that all members will interact with each other (due to spatial constraints). Thus, the density property is directly related to the size. Similarly other properties such as ‘mean of edge weight’ and ‘average clustering coefficient’ also depend on N. While it would have been useful to assume more dependency among other features, the more dependencies our model has the more data we need in order to estimate its parameters. We believe that the current model strikes a good balance between the need to encode feature dependencies and the available training data. Thus, other feature descriptors, X1…Xm are assumed to be independent given the size (N) and the label (C) of the subgraph.
Fig. 2.
A Bayesian probabilistic model for scoring a subgraph in our framework. The root node ‘Label’ is the binary indicator for complexes (1 if this subgraph is a complex, 0 otherwise). The second level node ‘nodeSize’ represents the number of nodes in the subgraph. The remaining nodes are all located on the third level and each represents a feature property described in Table 1.
For a subgraph in our PPI network we can compute the conditional probability of how likely it represents a complex using the following Equation (4).
| (1) |
| (2) |
| (3) |
| (4) |
The second row uses Bayes rule. The third row utilizes the chain rule. The fourth equation uses the conditional independence encoded in our graphical model to decompose the probability to products of different features. Similarly, we can compute a posterior probability for a non complex by replacing 1 with 0 in the above equation.
Using these two posteriors we can compute a log likelihood ratio score for each candidate subgraph:
| (5) |
| (6) |
Applying Bayes’ rule and canceling common terms in the numerator and denominator, the only terms we need to compute for the likelihood ratio L are the prior probability P(Ci) and the conditional probabilities P(N|C) and P(Xk|N,Ci).
Maximum likelihood estimation is used for learning these conditional dependencies from training data. We first discretized the continuous features and then used the multinomial distribution to model their probabilities. We uniformly discretized each feature into 10 equal width bins in the experiments presented in Section 3. Due to the small sample size of the training data, we apply a Bayesian Beta Prior to smooth the multinomial parameters in extreme cases (Manning et al., 1999). As for the prior p(C=1) of complexes, we assign a default value of 0.0001 which leads to good performance in cross validation experiments.
The BN structure in Figure 2 was manually selected. We have also tried to learn the BN structure using tree augmented structure learning techniques (Witten et al., 2000). However, the resulting performance of the learned network is not significantly better than our proposed structure (Fig. 2). Since our structure is simpler we omit the related results here. However potential improvements may be possible with more training examples and better BN structure learning approaches.
2.3 Searching for new complexes
The above model can be used to evaluate candidate subgraphs. If the log-likelihood ratio exceeds a certain threshold the subgraph is predicted to be a complex. This reduces the problem of identifying proteins complexes to the problem of searching for high scoring subgraphs in our PPI network. However, as we prove in the following lemma this search problem is NP-hard.
LEMMA 2.1. —
Identifying the set of maximally scoring subgraph in our PPI graph is NP-hard
PROOF. —
We prove this by reducing our search problem to max-clique, a NP hard problem (Cormen et al., 2001). To reduce our model to max-clique we will assume that we are only using one property, the graph density and that all edges in our graph have a weight of 1. Furthermore, we set the probability of a complex given a subgraph to:
For this model, the only subgraphs with positive scores are the cliques in our graph. In addition, the bigger the clique the higher our score and so finding the highest scoring subgraph is equivalent to finding the maximal clique.
The NP-hardness implies that there are no efficient (polynomial time) algorithms that can find an optimal solution for the search problem defined above. Thus, heuristic algorithms are needed. There are many approaches for local graph search proposed in the literature, which include hill climbing, simulated annealing, heuristic based greedy search, or tabu-search heuristic (Virtanen, 2003). All these strategies try to find local optima for certain fitness functions.
Here we choose to employ the iterated simulated annealing (ISA) search (Ideker et al., 2002; Virtanen, 2003), using the complex ratio score as the objective function (see Equation (6)). The basic idea for ISA is: after each round of modifying the current cluster, we accept the new cluster candidate if it has a higher score L′ than the current score L, but even if the score decreases, we accept the new cluster with probability exp((L−L′)/T), where T is the temperature of the system. This allows the algorithm to avoid local minima in some cases. After each round, the temperature is decreased by a scaling factor α by setting T′=αT. The initial temperature T0, the scaling factor α, and the number of rounds are parameters of the search process. After the algorithm terminates the highest scoring subgraph is returned and the search continues. Ideker et al. (2002) pointed out that given a suitable parameter setting, ISA could identify the global optimum even though this setting is generally unknown and can be impractically hard to find.
At the beginning, we connect each seeding protein to its highest weight neighbor and then use the pair as the starting cluster. Beginning from these clusters, we pursue the cluster modification process and the simulated annealing search. A number of heuristics could be used for modifying the current cluster. The order in which we add new proteins to the cluster is based on their impact on the cluster ratio score. We also explore the option of removing nodes from the cluster and merging of two clusters. We chose to limit the rounds of iterative search to 20. This restricts the size of the complexes we search for is between 3 and 20. We use cross validation to choose best values for the temperature and scaling factor parameters. To avoid revisiting the same/similar clusters, we keep checking the overlap ratio between the current cluster to the investigated clusters so far. If the ratio is higher than a threshold, we stop searching for the current seed. See supporting website for details about the complexity of the algorithm and values for the parameters it uses.
The complete proposed algorithm for complex identification is presented in Table 2. Our input is the weighted PPI graph and a set of known complexes and non-complexes (random collections of genes) as training data. First, we learn model parameters for the probabilistic BN model from the training data. Next, we search for subgraphs to identify candidate complexes. The final output clusters are those clusters found to have a ratio score larger than a predefined threshold.
Table 2.
Protein complex identification algorithm
| Input |
| - Weighted PPI matrix; |
| - A training set of complexes and non-complexes; |
| Output |
| - Discovered list of protein complexes; |
| Complex model parameter estimation |
| - Extract property features from positive and negative training examples; |
| - Discretize the continuous features; |
| - Calculate the BN MLE parameters for different features properties on the multinomial distribution; |
| Search for complexes |
| - Starting from the seeding subgraphs, apply simulated annealing search to expand and identify candidate complexes; |
| - Output subgraphs with ratio scores exceeding a certain threshold |
2.4 Weighted undirected PPI graph
As discussed above, we assume that our model input is a weighted undirected graph representing the PPI network. The edge weight describes how likely an interaction happens between the two related proteins based on the following rationale: While high-throughput experimental data for PPI is available, it has suffered from high false positive and false negative rates (von Mering et al., 2002). In addition to direct experimental interaction data there are many indirect sources that may contain information about protein interactions. As has been shown in a number of recent papers (Jansen et al., 2003), such indirect data can be combined with the direct data to improve the accuracy of protein interaction prediction. This type of analysis usually results in an interaction probability or confidence score assigned to each protein pair. Edges in our graph are weighted using this interaction probability which is computed as follows. In previous work (Qi et al., 2006), we assembled a large set of biological features (a total of 162 features representing 17 distinct groups of biological data sources) for the task of pairwise protein interaction prediction. Considering our current goal of complex identification we remove the features derived from the two high throughput mass spectrometry data sets (Gavin et al., 2002; Ho et al., 2002). Training is based on the small scale physical PPI data in the DIP database (Xenarios et al., 2002). Based on our previous evaluation, the support vector machine (SVM) classifier (Joachims et al., 2002) performs as well or better than any of the other classifiers suggested for this physical interaction task. We have thus used the results of our SVM analysis [see details in Qi et al. (2006)] to obtain weights for edges in our graph. Weights range from minus infinity to infinity where larger values indicate a higher likelihood to be an interacting pair. To reduce the number of edges in our graph we apply a cutoff and remove all edges with weights below the cutoff. We have chosen a cutoff of 1.0 such that the number of remaining edges roughly corresponds to previous estimates of the number of protein interaction pairs in yeast (von Mering et al., 2002).
To further improve the quality of the PPI graph we filter the predicted weighted graph using a newly published Yeast interaction data set from Reguly et al., (2006). For each of the remaining interactions we keep the weight learned from our integrated data analysis. This data contains a comprehensive database of genetic and protein interactions in yeast, manually curated from over 31 793 abstracts and online publications. A total of 35 244 interactions are reported, including literature curated and high throughput interactions. To allow fair comparisons we removed those interactions coming from the high-throughput mass spectrometry experiments in this data set.
3 EXPERIMENTS AND RESULTS
3.1 Reference sets
The MIPS (Mewes et al., 2004) protein complex catalog is a curated set of 260 protein complexes for yeast that was compiled from the literature and is thus more accurate than large scale mass spectrometry complex data. After filtering away those complexes composed of a single or a pair of proteins, 101 complexes in MIPS remained. The size of the complexes in MIPS is distributed as a power law, with most of the complexes having fewer than five proteins. We use the projection of the MIPS complexes on our PPI graphs as the positive training examples. See Figure 1 for four examples of such a projection.
As another independent positive set we used the core set of protein complexes from a newly published TAP-MS experiment (Gavin et al., 2006), one of the most comprehensive genome-wide screens for complexes in budding yeast. Again, we removed those complexes with only two proteins leading to 152 complexes that were used as positive examples to test our method.
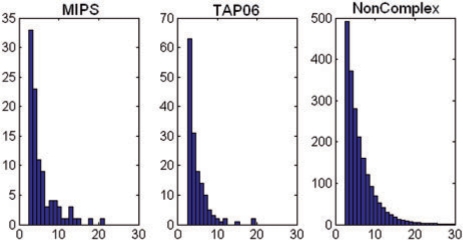
Since we are using a supervised learning method we also need negative training data, which we generated by randomly selecting nodes in the graph. The size distribution of these non-complexes follows the same power law distribution of the known complexes in MIPS. Figure 3 presents the histogram of these distributions for each of the three reference sets: ‘MIPS’, ‘TAP06’ and ‘Non-complexes’. As can be seen, all roughly follow the same ‘power law’ distributions.
Fig. 3.
Histogram of number of proteins in each of the three reference sets: ‘MIPS’, ‘TAP06’ and ‘Non-complexes’. Note that all resemble ‘power law’ distributions. Horizontal axis is the number of proteins. Vertical axis is the number of subgraphs (complexes).
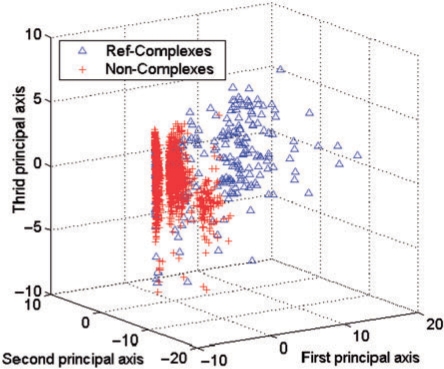
Figure 4 presents the distribution of two classes for real complexes (blue) versus negative examples (red) when projected on the first three principal coordinates after applying SVD on the features. The distribution strongly indicates that the proposed features can separate the two sets reasonably.
Fig. 4.
Reference examples’ distribution when projected with the first three principle components after applying SVD to the features.
3.2 Performance measures
In order to quantify the success of different methods in recovering the set of known complexes we define three descriptors for each pair of a known and predicted complex:
A: Number of proteins only in the predicted complex
B: Number of proteins only in the known complex
C: Number of proteins in the overlap between two
We say that a predicted complex recovers a known complex if
| (7) |
where p is an input parameter between 0 and 1 which we set to 0.5. Thus we require that the majority of the proteins in the complex be recovered and that the majority of the proteins in the predicted complex belong to that known complex.
Based on the above definition, three evaluation criteria are applied to quantify the quality of different protein complex identification methods:
Recall (r): Measures the fraction of known complexes detected by predicted complexes, divided by the total number of positive examples in the test set.
Precision (p): Measures the fraction of the predicted complexes that match the positive complexes among all predicted complexes.
F1: The F1 score combines the precision and recall scores. It is defined as 2pr/(p+r).
All three values range from 0 to 1, with 1 being the best score. Recall quantifies the extent to which a solution set captures the labeled examples. Precision measures the accuracy of the solution set. A good protein complex detector should have both high precision and high recall. The F1 measure provides a reasonable combination for both precision and recall. These three criterions are frequently used in many computational areas (Jones et al., 1981).
3.3 Performance comparison
To assess the performance in complex identification, we conducted experiments using MIPS as the positive training set and TAP06 as a test set and vice versa. There are a total of 1376 proteins in the MIPS and TAP06 complexes. Thus, we applied our train-test analysis on a PPI graph containing theses genes. The resulting graph used contains 1376 proteins and 10 918 weighted edges.
We have compared our method, referred to as ‘SCI-BN’, with three other methods suggested for complex identification. (1) ‘Density’ uses the the same search algorithm discussed in Section 2. However, unlike our method which maximizes the BN likelihood ratio, for ‘Density’ we simply try to find the maximally dense subgraphs in the graph. (2) The ‘MCODE’ complex detection method was proposed by Bader et al. (2003b). MCODE finds clusters (highly interconnected regions) in any network loaded into Cytoscape. The method was developed for PPI in which these clusters correspond to protein complexes (Bader et al., 2003b). (3) ‘SCI-SVM’ is used to determine whether the BN structure helps in identifying complexes. It uses the same features as our method but instead of using a BN it uses a SVM (Joachims et al., 2002).
The performance comparison is presented in Table 3. For each method, we report the precision, recall and F1, separately. As can be seen our method dominates all other methods in all measures. The recall rate of our method is around 50%. This number is impressive when considering the fact that the training and testing were done on different datasets. Our precision is lower (between 20–30%). However, since many of the complexes are not included in either gold standard sets, this precision value can be the result of correct predictions that are not included in the available data. We discuss some of these complexes below. As for the other methods, surprisingly, the recall and F1 values reported by MCODE are much lower than both the ‘Density’ and ‘SCI-SVM’ methods. We investigated the clusters identified by ‘MCODE’ and determined that they were relatively large compared to clusters determined by other methods which may have hurt performance. Interestingly the performance of ‘SCI-SVM’ is not as good as ‘SCI-BN’. This is largely caused by the unique way BN can handle the ‘node size’ feature. For the ‘Density’ approach, it performs reasonably well for the Recall measure but not as good in terms of precision.
Table 3.
Performance comparison between our algorithm (‘SCI-BN’), SVM with the same set of features (‘SCI-SVM’), Clique based method using only the density feature (‘Density’) and the ‘MCODE’ methods (Bader et al., 2003b) (‘MCODE’)
| Train | Test | Method | Precision | Recall | F1 |
|---|---|---|---|---|---|
| MIPS | TAP06 | Density | 0.217 | 0.409 | 0.283 |
| MIPS | TAP06 | MCODE | 0.293 | 0.088 | 0.135 |
| MIPS | TAP06 | SCI-SVM | 0.247 | 0.377 | 0.298 |
| MIPS | TAP06 | SCI-BN | 0.312 | 0.489 | 0.381 |
| TAP06 | MIPS | Density | 0.143 | 0.515 | 0.224 |
| TAP06 | MIPS | MCODE | 0.146 | 0.063 | 0.088 |
| TAP06 | MIPS | SCI-SVM | 0.176 | 0.379 | 0.240 |
| TAP06 | MIPS | SCI-BN | 0.219 | 0.537 | 0.312 |
Evaluation is based on precision, recall and the F1 measure. Experiments carried out with either MIPS as positive training set and TAP06 as test set, or vice versa.
4 VALIDATION
Using a threshold of 1.0 for the weights of the edges, our yeast PPI network contains 5234 proteins and 19 246 interaction edges. To identify and validate new complexes within this network graph, we trained a new BN model on all of the MIPS manual complexes as positive examples and used 2000 randomly selected non-complexes subgraphs as negative examples. Within the resulting full graph, we predict 987 complexes using the ‘SCI-BN’ search method.
To identify new complexes within the predicted graph, we compared the predicted clusters with those reported in five reference datasets, the manually curated MIPS dataset (Mewes et al., 2004) and four large-scale complex datasets obtained using high-throughput experimental approaches (Gavin et al., 2002, 2006; Ho et al., 2002; Krogan et al., 2006). After filtering those clusters matching reference complexes, we are left with 570 novel predictions. These are either entirely new complexes or extensions to known complexes by adding new proteins.
Amongst the new complexes, most highly ranked were of size 3–4. The size distribution agrees with the distribution of known complexes. While many of these top scoring complexes took the shape of cliques, others displayed more diverse shapes. Examples are shown in Figure 5. Black edges in Figure 5 represent interactions with SVM score higher than 4.0 (indicating strong evidence for interactions between proteins).
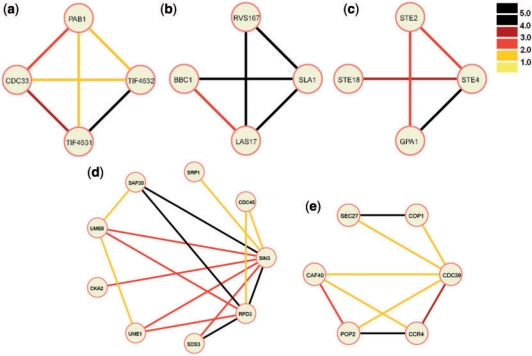
Fig. 5.
Projection of predicted complexes on our weighted PPI graph. The edge weights are thresholded and color coded. See color legend (top right corner bar) for edge weights. Descriptions for each predicted complex are provided in the ‘Validation’ section.
The clique complex shown in Figure 5a represents a protein complex involved in translation. CDC33, also known as eIF4E, is a translation initiation factor. PAB1 is a Poly(A)-binding protein. TIF4632 is the 130-kD subunit of translation initiation factor eIF4F/G. TIF4631 is the 150-kD subunit of the same translation initiation factor, eIF4F/G. Being two subunits of the same protein, we expect the evidence for this binary interaction to be very strong, represented by the black edge connecting these two proteins. eIF4F/G needs to interact with eIF4E to mediate cap-dependent mRNA translation. eIF4F/G can also interact with p20, but p20 competes with eIF4F/G for binding to eIF4E. Thus, in a complex involving eIF4E (CDC33), we expect to find CDC33 or p20 but not all three proteins together. This is what is indeed observed in this complex.
Figure 5b shows a high scoring cluster that is not a clique. This cluster contains four proteins with known or presumed roles in actin cytoskeleton structure, and a complex formation between them is quite likely.
Figure 5c shows a cluster that is not listed in any of the databases used but is actually a known complex: the heterotrimeric G-protein [with alpha(GPA1)-, beta(STE4)- and gamma(STE18)-subunits] binds to activated pheromone alpha-factor receptor(STE2) (Whiteway et al., 1989). This is a transient complex and would not be identified by high-throughput screening methods, although the formation of this complex is a requirement for G protein coupled signal transduction (not only in yeast, but in all G protein coupled receptor signaling). The identification of this cluster by our methodology is particularly encouraging, as such transient complexes can have crucial cellular roles. The G protein coupled receptors are the most abundant cell surface receptors in human, and some 60% of currently marketed drugs are targeted at them (Muller, 2000).
The shape shown in Figure 5d constitutes several small cliques connected via common edges or nodes. This predicted cluster therefore potentially gives a higher-level view of the local functionalities for related proteins. Most proteins in this complex have defined roles in transcription regulation, and a subset of these was already known to form a complex earlier (SIN3, RPD3, SDS3, UME6, SAP30 are part of the histone deacetylase complex). The function of SRP1 (karyopherin-alpha) is somewhat engimatic with diverse roles in nuclear import on the one hand and protein degradation on the other hand. The prediction of SRP1 being part of this complex would be interesting to verify experimentally because it would potentially link multiple processes.
Although the detected cluster shown in Figure 5e is a subcluster of a very large cluster previously detected by high-throughput methodology (Gavin et al., 2002), we present it here because of its interesting shape of two clusters (triangle SEC27, COP1, CDC39) and (rectangle CAF40, POP2, CCR4, CDC39) being connected by a common binding partner (CDC39). The first cluster contains proteins that are part of secretory pathway vesicles (SEC27, COP1), while the second cluster contains proteins mostly with roles in transcription. CDC39 linking these two groups is itself a protein also involved in transcription. Its linking role to secretory pathway proteins is unsuspected and should be investigated experimentally.
5 CONCLUSIONS AND DISCUSSIONS
In this article we presented a probabilistic algorithm for discovering complexes in a supervised manner. Specifically we extract features that can be used to distinguish complex versus non-complexes and train a classifier using these features to identify new complexes in the PPI graph. Unlike previous methods that relied on the ‘dense’ assumption of complex subgraphs, our algorithm integrates subgraph topologies and biological evidence, and learns the importance of each of the features from known complexes. This allows our algorithm to identify complexes with topologies that are missed by previous methods. We have shown that our algorithm can achieve better precision and recall rates for previously identified complexes. Finally, we discussed examples of new complexes determined by our algorithm and their possible function.
Our framework of feature representation is general. It is straightforward to add other topological properties that are found to be relevant for this problem. It is also possible to add other types of features. For example, information about the function of proteins can be encoded in our framework as well.
We hope to extend this work and improve both feature representation and search so that we can detect other types of interaction groups. Besides complexes, pathways of logically connected proteins also play a major role in both cellular metabolism and signaling. How to detect interesting pathways on PPI graph in our framework is an interesting direction to pursue. Another interesting direction is to apply this method to other species for which protein interaction data became available recently, including humans.
ACKNOWLEDGEMENTS
This work was supported in part by National Science Foundation grants CAREER 0448453, EIA0225656, EIA0225636, CAREER CC044917, and National Institutes of Health grant LM07994-01. The authors want to express sincere thanks to Oznur Tastan of CMU for suggestions regarding one validation.
Conflict of Interest: none declared.
REFERENCES
- Adamcsek B, et al. Cfinder: locating cliques and overlapping modules in biological networks. Bioinformatics. 2006;22:1021–1023. doi: 10.1093/bioinformatics/btl039. [DOI] [PubMed] [Google Scholar]
- Bader GD, Hogue CW. Analyzing yeast protein-protein interaction data obtained from different sources. Nat. Biotechnol. 2003a;20:991–997. doi: 10.1038/nbt1002-991. [DOI] [PubMed] [Google Scholar]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks, BMC Bioinformatics. 2003b;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization, Nat Rev Genet. 2004;5:101–103. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Borgwardt KM, et al. Graph kernels for disease outcome prediction from protein-protein interaction networks, Pacific Symposium on Biocomputing. 2007;12:4–15. [PubMed] [Google Scholar]
- Chakrabarti D. Ph.d. thesis. School of Computer Science, Carnegie Mellon University; 2005. Tools for Large Graph Mining. [Google Scholar]
- Chu W, et al. Identifying protein complexes in high-throughput protein interaction screens using an infinite latent feature model, Pacific Symposium on Biocomputing. 2006;11:231–242. [PubMed] [Google Scholar]
- Cherry JM, et al. Genetic and physical maps of Saccharomyces cerevisiae. Nature. 1997;387:67–73. [PMC free article] [PubMed] [Google Scholar]
- Cormen, et al. McGraw-Hill. 2001. Introduction to algorithms (Second Edition) [Google Scholar]
- Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Gavin AC, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Ho Y, et al. Systematic identification of protein complexes inSaccharomyces cerevisiaeby mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Ideker T, et al. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics. 2002;18(Suppl1):S233–S240. doi: 10.1093/bioinformatics/18.suppl_1.s233. [DOI] [PubMed] [Google Scholar]
- Ito T, et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. 2001;10:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, et al. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science. 2003;302:449–453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- Joachims T. PhD Thesis. Cornell University, Department of Computer Science; 2001. Learning to classify text using support vector machines. [Google Scholar]
- Jones KS, editor. Information Retrieval Experimental. London: Butterworths; 1981. [Google Scholar]
- Kim PM, et al. Relating three-dimensional structures to protein networks provides evolutionary insights. 2006;314:1938–1941. doi: 10.1126/science.1136174. [DOI] [PubMed] [Google Scholar]
- King AD, et al. Protein complex prediction via cost-based clustering. Bioinformatics. 2004;20:3013–3020. doi: 10.1093/bioinformatics/bth351. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, et al. Global landscape of protein complexes in yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Manning, Schutze . MIT press; 1999. Foundations of Statistical Natural Language Processing. [Google Scholar]
- Mewes HW, et al. MIPS: analysis and annotation of proteins from whole genomes. Nucleic Acids Res. 2004;32:D41–D44. doi: 10.1093/nar/gkh092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller G. Towards 3D structures of G protein-coupled receptors: a multidisciplinary approach. Curr. Med. Chem. 2000;7:861–888. doi: 10.2174/0929867003374534. [DOI] [PubMed] [Google Scholar]
- Pereira-Leal JB, et al. Detection of functional modules from protein interaction networks. Proteins. 2004;54:49–57. doi: 10.1002/prot.10505. [DOI] [PubMed] [Google Scholar]
- Przulj N. Biological network comparison using graphlet degree distribution. Bioinformatics. 2007;23:e177–e183. doi: 10.1093/bioinformatics/btl301. [DOI] [PubMed] [Google Scholar]
- Qi Y, et al. Evaluation of different biological data and computational classification methods for use in protein interaction prediction. Proteins. 2006;63:490–500. doi: 10.1002/prot.20865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguly T, et al. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J. Biol. 2006;5:11. doi: 10.1186/jbiol36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rives AW, Galitski T. Modular organization of cellular networks. Proc. Natl Acad. Sci. USA. 2003;100:1128–1133. doi: 10.1073/pnas.0237338100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins G, et al. Psychology Department, University of Melbourne; 2005. A workshop on exponential random graph (p*) models for social networks. [Google Scholar]
- Rual JF, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Scholtens D, et al. Local modeling of global interactome networks. Bioinformatics. 2005;21:3548–3557. doi: 10.1093/bioinformatics/bti567. [DOI] [PubMed] [Google Scholar]
- Sharan R, et al. Identification of protein complexes by comparative analysis of yeast and bacterial protein interaction data. J. Comput. Biol. 2005;12:835–846. doi: 10.1089/cmb.2005.12.835. [DOI] [PubMed] [Google Scholar]
- Spirin V, Mirny LA. Protein complexes and functional modules in molecular networks. Proc. Natl Acad. Sci. USA. 2003;100:12123–1218. doi: 10.1073/pnas.2032324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzl U, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:830–832. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Uetz P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Virtanen SE. Research Report. Helsinki University of Technology, Laboratory for Theoretical Computer Science. 2003. Properties of nonuniform random graph models. [Google Scholar]
- von Mering C, et al. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- Whiteway M, et al. The STE4 and STE18 genes of yeast encode potential beta and gamma subunits of the mating factor receptor-coupled G protein. Cell. 1989;56:467–477. doi: 10.1016/0092-8674(89)90249-3. [DOI] [PubMed] [Google Scholar]
- Witten IH, Frank E. San Francisco: Morgan Kaufmann; 2000. Data Mining: Practical machine learning tools with Java implementations. [Google Scholar]
- Xenarios I, et al. DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30:303, 305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Han J. Technical Report UIUCDCS-R-2002-2296. Dept. of Computer Science, UIUC; 2002. gSpan: Graph-based substructure pattern mining. [Google Scholar]
- Yeger-Lotem E, et al. Network motifs in integrated cellular networks of transcription-regulation and protein-protein interaction. Proc. Natl Acad. Sci. USA. 2004;101:5934–5939. doi: 10.1073/pnas.0306752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Qin ZS. Structural comparison of metabolic networks in selected single cell organisms. BMC Bioinformatics. 2005;6:8. doi: 10.1186/1471-2105-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotenko E, et al. Decomposition of overlapping protein complexes: A graph theoretical method for analyzing static and dynamic protein associations. Algorithms Mol. Biol. 2006;1:7. doi: 10.1186/1748-7188-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]