Abstract
Meiotic lamin C2 is the only A-type lamin expressed during mammalian spermatogenesis. Typical for this short lamin is the unique hexapeptide GNAEGR, which substitutes the nonhelical amino terminus and part of the α-helical rod domain present in somatic lamins. Meiotic lamin C2 also lacks a carboxyl-terminal CaaX box, which is modified by isoprenylation and involved in nuclear envelope (NE) association of somatic isoforms. The mechanism by which lamin C2 becomes localized in the NE is totally unknown. Here we demonstrate that the hexapeptide GNAEGR is essential for this process: (i) Its deletion resulted in a diffuse distribution of lamin C2 within nuclei of transfected COS-7 cells; (ii) Mutated somatic lamin C, containing the sequence GNAEGR at its amino terminus, was located at the NE. The mass spectrometric analysis of the amino terminus of lamin C2 revealed that it is modified by myristoylation. Correspondingly, the substitution of the first glycine residue abolishes the NE association of lamin C2. We conclude that NE association of lamin C2 is achieved by a mechanism different from that of somatic lamins.
Keywords: meiosis, myristoylation, nuclear lamins, spermatogenesis
The nuclear lamina is a proteinaceous component of the nuclear envelope (NE) that is intimately associated with the inner nuclear membrane. The nuclear lamina fulfills a structural role at the nuclear periphery and has been involved in the functional organization of components of the nuclear interior (refs. 1–5; for reviews see refs. 6 and 7).
The major protein components of the nuclear lamina are the lamins, i.e., the only intranuclear members of the intermediate filament protein family (8, 9). Within the lamin family, it can be distinguished between A-type and B-type lamins (10). B-type lamins (B1 and B2) are ubiquitous proteins, whereas A-type lamins (A and C) are expressed solely in differentiated somatic cells (11). More recently, it has been demonstrated that mutations in the lamin A gene are responsible for three different types of autosomal dominant dystrophies in humans (for a review, see ref. 12).
Lamins are fibrillar molecules with a molecular mass of about 60–75 kDa. They are composed of a characteristic central α-helical domain that forms coiled coil structures flanked by nonhelical amino and carboxyl termini. The amino terminus and the first part of the helical domains have been shown to be of importance for the lamin dimerization and for the head-to-tail association of dimers (for reviews, see refs. 6, 7, and 13). The presence of a nuclear localization signal (NLS) and of the CaaX box (C, cysteine; a, aliphatic; X, any amino acid) in the carboxyl-terminal domain are further important features. Isoprenylation of the cysteine residue of the CaaX box is essential for lamin attachment to the inner nuclear membrane (14–18). An exception to this scheme is lamin C (a short splicing variant of the lamin A gene), which lacks a CaaX box and requires the presence of the other lamins for NE association (19–22).
The organization of the nucleus of meiotic cells differs substantially from that of somatic cells. A main characteristic of the meiotic prophase nucleus is its dynamics. During most of the meiotic prophase, homologous chromosomes are attached via their telomeres to the NE and move actively, which is of importance for the processes of chromosome pairing and recombination. Despite the high biological relevance of these processes, very little is known about the chromosome attachment to the nuclear periphery of meiotic prophase cells and the mechanisms responsible for chromosome movements (23, 24). When compared with somatic cells, the nuclear periphery of mammalian meiotic cells presents a series of peculiarities (25–30). During meiotic prophase, lamins A, B2, and C have not been detected (27). Instead, the lamina structure is composed of lamin B1 together with the lamins C2 and B3 (25–29). The last two are shorter splicing variants of the lamin A and B2 genes, respectively (26, 31). Lamins C2 and B3 (about 50 kDa each) appear to be specific for meiosis and show remarkable structural differences with respect to their somatic counterparts (25, 26, 28, 29). The complete amino-terminal end and part of the central rod domain of somatic lamins are replaced in lamins C2 and B3 by short sequences that are unique for them. In the case of rodent lamin C2, this short sequence is only six amino acids long (GNAEGR). The sequence of the other domains of lamin C2 is identical to that of lamin C (28, 29). More recently, we have shown that lamin C2 is distributed not as a continuous sheet at the nuclear periphery of rat pachytene spermatocytes, but rather in the form of discontinuous domains. Telomeres of meiotic chromosomes, for their part, were found to be associated to the NE only at regions containing lamin C2 (32).
Taken together, the information available presently points to an essential role of the NE during meiotic process (for a review, see ref. 24). In this context, lamin C2 would play an important role because of its condition as a structural protein of the NE that is enriched at the attachment sites of meiotic chromosomes (32). Unfortunately, meiotic cells of higher eukaryotes are not well suited for manipulations under culture conditions, a limitation that has often hampered the functional analysis of meiotic proteins. As an alternative, properties of meiotic proteins may be investigated in transiently transfected somatic cells. Thus, we were able to investigate the sequence requirements for NE association of C2, i.e., a lamin lacking a CaaX box.
Materials and Methods
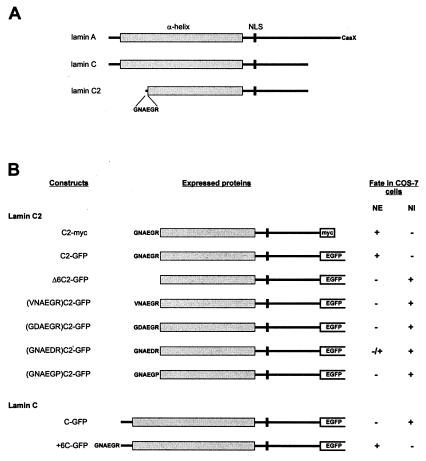
Construction of Plasmids for Expression of EGFP (Enhanced Green Fluorescent Protein) and myc-Tagged Fusion Proteins in Mammalian Cells.
To express EGFP-fusion proteins, cDNAs coding for proteins of interest were cloned in frame into the 5′ multiple cloning site of mammalian expression vector pEGFP-N2 (CLONTECH). cDNAs were derived from parental clones for either rat lamin C2 (29) or human lamin C (9). To generate fusion vectors with the complete coding sequences of these cDNAs, parental clones were used for PCR amplification. 5′ primers contained the starting codon ATG flanked by a Kozak-sequence (33). In the 3′ primers, stop codons were removed to get an ORF in the pEGFP-fusion vector. Nucleotide sequences with restriction sites suitable for cloning the cDNAs into the vector were linked to the primers. In the case of mutant cDNAs, 5′ primers were modified as follows. To create a lamin C2 mutant lacking the amino-terminal hexapeptide GNAEGR, a 5′ primer with a deletion of the corresponding nucleotides was used for amplification. To create a mutant lamin C containing hexapeptide GNAEGR, a 5′ primer with insertion of the corresponding nucleotides between the starting codon ATG and the first wild-type codon was used. Amino acid substitutions in hexapeptide GNAEGR were produced by using point mutated 5′ primers. PCR amplification was done according to standard procedures. Amplified cDNAs were directly cloned into PCR cloning vector PCRII-TOPO (Invitrogen). Before ligation into pEGFP-N2, cDNAs were isolated with the aid of appropriate restriction enzymes cutting in the primer sites. To generate an expression vector coding for a lamin C2-myc fusion protein, the C2-GFP plasmid was used as a template for PCR amplification. Appropriate primers were used to introduce a carboxyl-terminal myc-tag (LAAEQKLISEEDLNGAA) and to remove the EGFP coding sequence in the same reaction step. The resulting PCR product was purified and recirculated by ligation. In all cases, clones were verified by sequencing according to standard methods.
Cell Line and Transfection.
Cell line COS-7 (green monkey kidney) was cultured according to standard procedures. As determined by immunoblotting, this cell line expresses the lamins A, C, B1, and B2 (data not shown). For transfection, 4 × 105 cells were transferred to a 6-cm Petri dish containing cover slips, grown overnight, and preincubated with 3 ml of Optimem (GIBCO/BRL) for 30 min after washing twice with PBS. For cell transfections and culture conditions after transfection, we followed the instructions provided by the supplier of Lipofectamine (GIBCO/BRL). Briefly, cells were transfected in 1 ml of Optimem containing 12 μl of Lipofectamine and 5 μg of plasmid DNA. After 2 h of incubation (37°C, 5% CO2), transfection solution was exchanged by fresh standard culture medium. Cells were incubated overnight at 37°C and 5% CO2. The cells were fixed as described below 24 h after the beginning of the transfection.
Antibodies and Immunofluorescence Microscopy.
The following primary monoclonal or polyclonal antibodies have been used: (i) mAb L3f4 against lamins A, B1, B2, and C (27); (ii) mAb X223 specific for lamin B2 (34); (iii) a human anti-DNA antibody (Kallestad Laboratories, Chaska, MN); and (iv) the anti-c-myc mAb 9E10 (Sigma, Deisenhofen, Germany). Secondary antibodies conjugated to DTAF or Texas red were purchased by Dianova (Hamburg, Germany).
Coverslips with the attached cells were fixed with methanol (−20°C, 7 min), postfixed with acetone (−20°C, 3 min), and air dried. After incubation with the primary antibodies, the coverslips were incubated with the corresponding secondary antibody for 15 min and conjugated to Texas red. After being washed in PBS, the cells were dehydrated in ethanol and embedded in Mowiol (32). Double-label immunofluorescences, using antibodies specific for the myc-tag and DNA, were performed as already described (35). The preparations were evaluated by confocal laser scanning microscopy as previously described (32, 36). As judged by fluorescence microscopy, about 70% of the cells were successfully transfected.
Enzymatic Digestions.
Sequencing grade endoproteinase Lys-C and sequencing grade modified porcine trypsin were obtained from Roche Diagnostics and Promega, respectively. α-Cyano-4-hydroxycinnamic acid was purchased from Sigma-Aldrich.
COS-7 cells, expressing either construct C2-GFP or GFP alone, were cultured as described above. The cells were homogenized and the proteins separated by two-dimensional gel electrophoresis 24 h after transfection set in (37). The Coomassie-stained spot corresponding to the lamin C2 fusion protein (as determined in parallel by immunoblotting) was excised from the gel and cut into small pieces (≈1 mm × 1 mm). The gel pieces were washed twice with water and a 50% solution of water/acetonitrile, and finally shrunk with acetonitrile. The protein was digested in-gel with either endoproteinase Lys-C or trypsin in 40 mM ammonium bicarbonate overnight at 37°C. The reaction was stopped by freezing.
Matrix-Assisted Laser Desorption/Ionization (MALDI) Mass Spectrometry.
MALDI mass spectra were recorded in the positive ion mode with delayed extraction on a Reflex II time-of-flight instrument (Bruker-Daltonik GmbH, Bremen, Germany) equipped with a SCOUT multiprobe inlet and a 337 nm nitrogen laser. Ion acceleration voltage was set to 20.0 kV, the reflector voltage was set to 21.5 kV, and the first extraction plate was set to 15.4 kV. Mass spectra were obtained by averaging 50–200 individual laser shots. Calibration of the spectra was performed externally by two-point linear fit using angiotensin I and oxidized insulin B-chain for the Lys-C digest, and internally with two autolysis products of trypsin at m/z 842.50 and m/z 2211.10 for the tryptic digest. Sample preparation was achieved by using the thin film preparation techniques (38). Briefly, 0.3 μl aliquots of a nitrocellulose containing a saturated solution of α-cyano-4-hydroxycinnamic acid in acetone were deposited onto individual spots on the target. Subsequently, 0.8 μl of 10% formic acid and 0.4 μl of the digest sample were loaded on top of the thin film spots and allowed to dry slowly at room temperature. To remove salts from the digestion buffer, the spots were washed with 10% formic acid and H2O.
Results
To investigate the properties of meiotic lamin C2, different lamin constructs were cloned into the expression vector pEGFP-N2, which codes for EGFP, and expressed in COS-7 cells (Fig. 1). For control, the cells were transfected in a first series of experiments with a construct coding for somatic lamin C and EGFP (construct C-GFP; Fig. 1). As shown in Fig. 2, the corresponding fusion protein was found in the nuclear compartment, where it formed several circle-shaped aggregates. Most of these aggregates appear to be free in the nucleoplasm, whereas only some are in contact with the nuclear periphery. As expected, mAb L3f4, which is specific for somatic lamins, recognizes the lamin C-GFP aggregates as well as the nuclear periphery of the transfected cells (Fig. 2 A–A"). These results are consistent with those of previous studies, which demonstrated that the association with the nuclear periphery of lamin C is facilitated during interphase by the presence of similar amounts of lamin A, i.e., the A-type lamin that possesses a CaaX box. Otherwise, the surplus of lamin C remains largely in the nucleoplasm where it forms aggregates (19, 21, 22).
Figure 1.
(A) Schematic representation of A-type lamins A, C, and C2. (B) Schematic representation and fate of the fusion proteins expressed in COS-7 cells. NI, nuclear interior.
Figure 2.
Expression of somatic lamin C in cultured COS-7 cells as investigated by confocal laser scanning microscopy. (A and B) Protein C-GFP is localized within nuclei. (A′) The distribution in the same cell as in A of the transfected and endogenous lamins (L) is shown with the aid of mAb L3f4, which is specific for lamins A/C, B1, and B2. (B′) The distribution of DNA in the transfected cell shown in B is revealed by anti-DNA antibodies. Overlays are seen in (A" and B"). (Bar = 10 μm.)
In the next series of experiments, the fate of lamin C2 was investigated after transfection of construct C2-GFP into COS-7 cells (Fig. 1). When lamin C2-GFP was expressed in COS-7 cells, most of the fusion protein was located at the nuclear periphery where it was distributed in the form of discrete plaques (Figs. 3 A and B and 5A). This finding is remarkable because it recalls the discontinuous distribution of lamin C2 recently described in rat spermatocytes (32). Virtually the same distribution in the form of plaques was obtained in COS-7 cells expressing a lamin C2 fusion protein tagged with myc instead of GFP (Fig. 3C). This experiment was performed to rule out the possibility that plaque formation at the nuclear periphery resulted from the aggregation of the GFP moiety of the fusion protein.
Figure 3.
Expression of meiotic lamin C2 in cultured COS-7 cells as investigated by confocal laser scanning microscopy. (A and B) In these cells, most lamin C2 (construct C2-GFP) is located at the nuclear periphery where it forms plaques. The expression level appears to be higher in the cell shown in B. (C) The fate of myc-tagged lamin C2 (construct C2-myc) is indistinguishable from that of lamin C2-GFP. The distribution of DNA in the same cells is revealed with the aid of an anti-DNA antibody in A′–C′. Overlays are seen in A"–C". (Bar = 10 μm.)
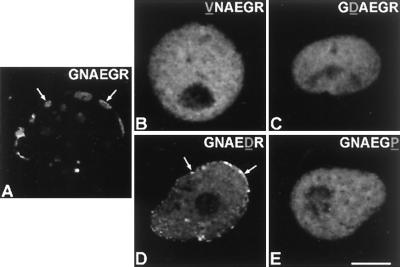
Figure 5.
Fate of lamin C2 as well as of lamin C2 carrying a mutated amino-terminal hexapeptide as investigated by confocal laser scanning microscopy. (A) Lamin C2 (construct C2-GFP) carrying the wild-type amino-terminal hexapeptide. Mutant lamins (VNAEGR)C2-GFP (B), (GDAEGR)C2-GFP (C), (GNAEDR)C2-GFP (D), and (GNAEGP)C2-GFP (E) are shown. Arrows denote some of the accumulations of expressed lamins that are located at the nuclear preriphery. (Bar = 10 μm.)
The results of Fig. 3 show that meiotic lamin C2 is localized in the NE of the transfected cells. This finding contrasts with the situation of lamin C under the same experimental conditions (Fig. 2; see also refs. 15, 19, 21, and 22). Therefore, the next step was to determine the sequence requirements for NE association of lamin C2. Because the amino-terminal hexapeptide GNAEGR is unique to lamin C2, we investigated the possible relevance of this sequence. For this purpose, we expressed a construct coding for a lamin C2 in COS-7 cells that lacked the first six amino acids (construct Δ6C2-GFP; Fig. 1). As shown in Fig. 4, the corresponding protein was diffusely distributed in the nuclear interior. Remarkably, no colocalization of the chimeric protein and the nuclear lamina structure was observed (Fig. 4 A–A").
Figure 4.
Role of hexapeptide GNAEGR in NE association as shown by confocal laser scanning microscopy. (A and B) Intranuclear distribution of mutant lamin C2 lacking the amino-terminal hexapeptide (construct Δ6C2-GFP). (C and D) Mutant lamin C carrying hexapeptide GNAEGR (construct + 6C-GFP) is located at the nuclear periphery. (A′) Endogenous lamins (L) are evidenced by using the mAb L3f4, an antibody that does not recognize lamin C2. (B′ and D′) The distribution of DNA is shown after incubation of the cells with a human anti-DNA antibody. (C′) The endogenous nuclear lamina structure is shown with the aid of the lamin B2-specific mAb X223. Overlays are seen in A"–D". (Bar = 10 μm.)
The role of hexapeptide GNAEGR was tested more directly by fusing it to the amino terminus of lamin C (construct +6C-GFP; Fig. 1). Under these conditions, virtually all lamin C was located at the NE, where it formed a continuous sheet (Fig. 4 C and D), i.e., a pattern indistinguishable from that of endogenous lamins (Fig. 4 C–C"). There was no indication of plaque formation at the nuclear periphery as in the case of lamin C2. In contrast to the experiment with the C-GFP construct (Fig. 2), almost no intranuclear protein aggregates were observed (Fig. 4 C and D).
After showing that the amino-terminal hexapeptide of lamin C2 is essential for NE localization, we became interested in the question of the mechanisms involved. A computer search of the databases suggested that hexapeptide GNAEGR could be a target for myristoylation. Myristate is a rare 14-carbon saturated fatty acid that facilitates the association of the modified protein with membranes of a variety of cell systems (for reviews, see refs. 39 and 40). Sequence requirements for myristoylation follow several established rules, although some divergences are possible depending on the sequence context. A typical motif is at least six amino acids long. The presence of a glycine residue at position 1 is critical and myristoylation of this residue is only possible after cleavage of the starting methionine. At position 2, proline and charged or bulk hydrophobic amino acids are not allowed. Positions 3 and 4 tolerate a broad spectrum of amino acids. Proteins containing proline, large hydrophobic or charged amino acids at position 5, are poor substrates. Position 6 shows few restrictions, but proline is not tolerated (for details, see ref. 39). Taking into account these sequence restrictions, we expressed different fusion proteins in COS-7 cells composed of lamin C2 and EGFP in which the wild-type amino-terminal hexapeptide (GNAEGR) was substituted by one of the following sequences: VNAEGR, GDAEGR, GNAEDR, or GNAEGP (see Fig. 1). The results of these experiments are shown in Fig. 5. In contrast to protein C2-GFP (Figs. 3 and 5A), fusion proteins containing one of these mutated hexapeptides remained diffusely distributed in the nucleus (Fig. 5 B and E). The most dramatic effects were produced by the substitution of either the first glycine through a valine (Fig. 5B), the asparagine at position 2 through aspartic acid (Fig. 5C), and the arginine (position 6) through a proline (Fig. 5E). In addition to the diffuse nuclear distribution, most of the cells expressing the fusion protein with the mutated hexapeptide GNAEDR showed a small proportion of the protein to be located at the nuclear periphery (Fig. 5D). This result is consistent with the notion that a charged amino acid at position 5 converts the hexapeptide in a poor substrate for myristoylation (39). In summary, these results provide strong evidence that myristoylation of the amino-terminal glycine residue of lamin C2 is required for NE association of this protein.
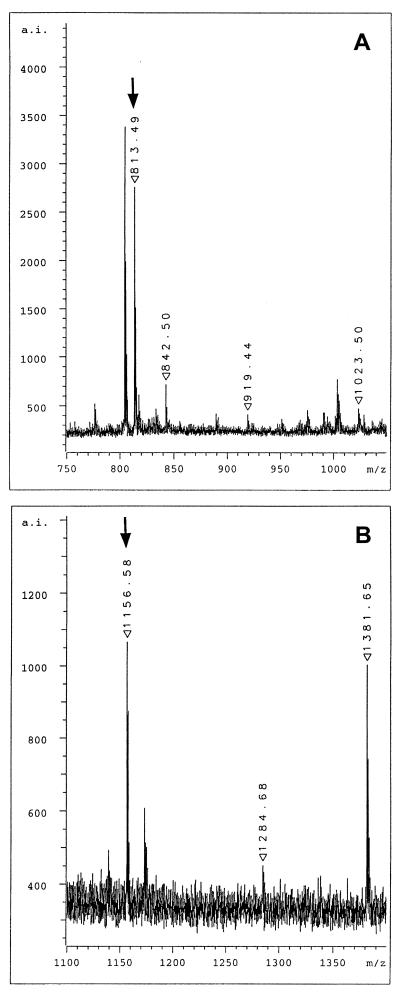
To obtain additional proof as to whether the amino terminus of lamin C2 is myristoylated, COS-7 cells expressing lamin C2 were prepared for two-dimensional gel electrophoresis (37). The spot corresponding to the lamin C2 fusion protein was cut out and the protein was digested in-gel by trypsin. The total tryptic digest was analyzed by MALDI mass spectrometry. Tryptic digestion of lamin C2 resulted in a series of different peptides. The expected sequence for the amino-terminal peptide bearing the myristoyl moiety is myrGNAEGR with a calculated monoisotopic mass for the singly protonated molecular ion of 813.48. Mass spectrometric analysis of the tryptic digest clearly showed a major ion signal at m/z 813.49 (Fig. 6A) indicating the presence of the myristoylated peptide. Ion signals at m/z 919.44 and m/z 1023.50 in the zoom view spectrum correspond to the tryptic fragments SSFSQHAR and NIYSEELR of lamin C2, respectively (for the complete sequence of rat lamin C2, see ref. 29). On the other hand, the ion signal at m/z 842.50 represents the autolysis product T8 of trypsin itself and was used for internal calibration. The ion signal at m/z 804.44 is frequently observed in mass spectra of proteins which are digested in-gel with trypsin. To support the presence of the myristoyl group at the amino terminus further, a second digestion of lamin C2 was performed by using the endoproteinase Lys-C. Because this enzyme cleaves carboxyl-terminally of lysine residues, the expected sequence for the amino-terminal peptide is myrGNAEGRNTK, i.e., three amino acids longer than the myristoylated tryptic peptide. The calculated mass for the singly protonated molecular ion is m/z 1156.67. An ion signal at m/z 1156.58 was observed in the mass spectrum of the Lys-C digest, indicating the presence of the myristoylated peptide (Fig. 6B). Additional signals in this mass range of the spectrum correspond to two further Lys-C peptides of lamin C2, namely EGDLLAAQARLK at m/z 1284.68 and NIYSEELRETK at m/z 1381.65. Nonmyristoylated forms of peptides GNAEGR and GNAEGRNTK were not detectable in the whole spectra (data not shown).
Figure 6.
MALDI mass spectrometric analysis of total enzymatic digests of lamin C2. (A) Selected view of the mass spectrum of the tryptic digest. The ion signal at m/z 813.49 represents the myristoylated amino-terminal hexapeptide. (B) Selected view of the mass spectrum of the Lys-C digest. The ion signal at m/z 1156.58 represents the myristoylated amino-terminal peptide myrGNAEGRNTK. Arrows denote the ion signal of the myristoylated peptides.
Discussion
It has been demonstrated in previous studies that isoprenylation of the carboxyl-terminal CaaX box is necessary for efficient integration of lamins in the NE (14–18, 41). However, these studies do not provide an answer to the question of how lamin C2 becomes associated with the NE because, as mentioned before, this lamin isoform lacks a CaaX box (28, 29) and is expressed in meiotic cells in the absence of other A-type lamins (27).
The results of the present study clearly demonstrate that hexapeptide GNAEGR is essential for NE association of lamin C2: (i) After deletion of hexapeptide GNAEGR, lamin C2 remained diffusely distributed in the nucleoplasm (Fig. 4 A and B). (ii) However, the fusion of the hexapeptide to the amino terminus of lamin C (i.e., a somatic lamin isoform lacking a CaaX box) resulted in the NE localization of this protein (Fig. 4 C and D). We also provide compelling evidence that the hexapeptide GNAEGR is myristoylated: (i) Mass spectra corresponding to peptides myrGNAEGR and myrGNAEGRNTK were detected after proteolytic cleavage of lamin C2 by the endoproteases trypsin and Lys-C, respectively (Fig. 6). (ii) The substitution of the amino acids of the amino-terminal hexapeptide that are essential for the action of the myristoyltransferase (39) abolished the NE association of lamin C2 (Fig. 5). These observations are remarkable, because myristoylation confers affinity for cellular membranes to the modified proteins, as already demonstrated for other classes of proteins (for reviews see refs. 39 and 40). Taken together, the results of the present study provide strong evidence that NE association of lamin C2 is mediated by the amino-terminal myristoylglycine. The involvement of myristoylation in NE association of a member of the lamin family is without precedent in the literature.
The mammalian germ cell lamin B3 is a short splicing variant of the lamin B2 gene which lacks, like lamin C2, the amino terminus as well as a significant part of the α-helical rod domain, which are present in the somatic isoform. These domains are substituted in lamin B3 by a unique 84-amino acid sequence (26). Furukawa and Hotta (26) showed that, in transfection experiments using COP5 cells, lamin B3 was localized largely at the nuclear periphery and that it induced nuclear deformations. In contrast, lamin B3 mutants lacking the first 59 or 81 amino acids, remained in the nucleoplasm and had no effects on the nuclear shape of transfected cells. A plausible interpretation for these results is that the unique amino-terminal domain of lamin B3 is responsible for the observed nuclear deformations (26). Based on our results with lamin C2, an alternative interpretation would be possible: that the amino-terminal domain of lamin B3 contains information necessary for NE association.
Myristoylation and isoprenylation are not sufficient per se to stably anchor the modified protein to a cellular membrane. Membrane binding can be greatly enhanced by palmitoylation or by the presence of a protein basic domain (for a review, see ref. 40). However, lamins, including lamin C2, lack both a palmitoylation site and a polybasic domain. In the case of isoprenylated lamins, permanent attachment to the nuclear envelope is achieved by their polymerization into the preexisting lamina, a step that requires the integrity of the α-helical rod domain (refs. 14, 19, 21, 42, and 43; for a review, see ref. 13). In which way the newly synthesized lamin C2 molecules become permanently attached to the nuclear periphery cannot be answered at present. This question is particularly relevant for understanding how the nuclear periphery of meiotic cells is organized. Because of the peculiarities of meiotic lamins (e.g., shorter rod domain, unique amino terminus), mechanisms of permanent attachment different from those of somatic lamins cannot be excluded a priori.
We have shown in previous studies that lamin C2 is located at the nuclear envelope of mammalian spermatocytes, where it is distributed in the form of discontinuous domains, to which the ends of the synaptonemal complexes are attached. These lamin C2-containing domains have been interpreted as representing local reinforcements of the NE involved in proper attachment and movements of meiotic chromosomes (32). It was interesting to observe that, in transfected COS-7 cells, lamin C2 forms discontinuous plaques at the nuclear periphery (Figs. 3 and 5A). Virtually no transfected cells were found in which lamin C2 was distributed as a continuous sheet. This remarkable result suggests that the formation of discontinuous domains at the nuclear periphery may represent a property of lamin C2. To clarify this point, further studies on the polymerization properties of lamin C2 are required. Finally, it is tempting to speculate that the differences in the mechanisms of association of lamin subtypes with the NE (i.e., myristoylation vs. isoprenylation) could promote different patterns of assembly that, in turn, might yield significant effects on the gross organization of the nuclear periphery.
Acknowledgments
This article is dedicated to Professor Werner W. Franke (Heidelberg) on the occasion of his 60th birthday. We thank Georg Krohne and Robert Hock for many stimulating discussions and suggestions; Georg Krohne for human lamin C clone and mAb X223; Corinna Zünkler for excellent technical assistance; and Rosie Rudd and Carmen Ramos for correcting the manuscript. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (Be 1168/4–2 and 4–3) to R.B.
Abbreviations
- EGFP
enhanced green fluorescent protein
- NE
nuclear envelope
- MALDI
matrix-assisted laser desorption ionization
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.240466597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.240466597
References
- 1.Burke B, Gerace L. Cell. 1986;44:639–652. doi: 10.1016/0092-8674(86)90273-4. [DOI] [PubMed] [Google Scholar]
- 2.Benavente R, Krohne G. J Cell Biol. 1986;103:1847–1854. doi: 10.1083/jcb.103.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newport J W, Wilson K L, Dunphy W G. J Cell Biol. 1990;111:2247–2259. doi: 10.1083/jcb.111.6.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lenz-Böhme B, Wisnar J, Fuchs S, Reifegerste R, Buchner E, Betz H, Schmitt B. J Cell Biol. 1997;137:1001–1016. doi: 10.1083/jcb.137.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spann T P, Moir R D, Goldman A E, Stick R, Goldman R D. J Cell Biol. 1997;136:1201–1212. doi: 10.1083/jcb.136.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moir R D, Spann T P, Goldman R D. Int Rev Cytol. 1995;162B:141–182. doi: 10.1016/s0074-7696(08)62616-9. [DOI] [PubMed] [Google Scholar]
- 7.Stuurman N, Hein S, Aebi U. J Struct Biol. 1998;122:42–66. doi: 10.1006/jsbi.1998.3987. [DOI] [PubMed] [Google Scholar]
- 8.McKeon F D, Kirschner M W, Caput D. Nature (London) 1986;319:463–468. doi: 10.1038/319463a0. [DOI] [PubMed] [Google Scholar]
- 9.Fisher D Z, Chaudhary N, Blobel G. Proc Natl Acad Sci USA. 1986;83:6450–6454. doi: 10.1073/pnas.83.17.6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krohne G, Benavente R. Exp Cell Res. 1986;162:1–10. doi: 10.1016/0014-4827(86)90421-0. [DOI] [PubMed] [Google Scholar]
- 11.Röber R A, Weber K, Osborn M. Development (Cambridge, UK) 1989;105:365–378. doi: 10.1242/dev.105.2.365. [DOI] [PubMed] [Google Scholar]
- 12.Wilson K L. Trends Cell Biol. 2000;10:125–129. doi: 10.1016/s0962-8924(99)01708-0. [DOI] [PubMed] [Google Scholar]
- 13.Krohne G. In: Subcellular Biochemistry: Intermediate Filaments. Herrmann H, Harris J R, editors. New York: Plenum; 1998. pp. 563–586. [Google Scholar]
- 14.Holtz D, Tanaka R A, Hartwig J, McKeon F. Cell. 1989;59:969–977. doi: 10.1016/0092-8674(89)90753-8. [DOI] [PubMed] [Google Scholar]
- 15.Krohne G, Waizenegger I, Höger T H. J Cell Biol. 1989;109:2003–2011. doi: 10.1083/jcb.109.5.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitten G, Nigg E A. J Cell Biol. 1991;113:13–23. doi: 10.1083/jcb.113.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hennekes H, Nigg E A. J Cell Sci. 1994;107:1019–1029. doi: 10.1242/jcs.107.4.1019. [DOI] [PubMed] [Google Scholar]
- 18.Frimbach-Kraft I, Stick R. J Cell Biol. 1995;129:17–24. doi: 10.1083/jcb.129.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horton H, McMorrow I, Burke B. Eur J Cell Biol. 1992;57:172–183. [PubMed] [Google Scholar]
- 20.Ye Q, Worman H J. Exp Cell Res. 1995;219:292–298. doi: 10.1006/excr.1995.1230. [DOI] [PubMed] [Google Scholar]
- 21.Pugh G E, Coates P J, Lane B, Raymond Y, Quinlan R A. J Cell Sci. 1997;110:2483–2493. doi: 10.1242/jcs.110.19.2483. [DOI] [PubMed] [Google Scholar]
- 22.Broers J L V, Machiels B M, van Eys G J J M, Kuijpers H J H, Manders E M M, van Driel R, Ramaekers F C S. J Cell Sci. 1999;112:3463–3475. doi: 10.1242/jcs.112.20.3463. [DOI] [PubMed] [Google Scholar]
- 23.Wilson E B. The Cell in Development and Heredity. New York: Macmillan; 1925. [Google Scholar]
- 24.Zickler D, Kleckner N. Annu Rev Genet. 1998;32:619–697. doi: 10.1146/annurev.genet.32.1.619. [DOI] [PubMed] [Google Scholar]
- 25.Smith A, Benavente R. Differentiation. 1992;52:55–60. doi: 10.1111/j.1432-0436.1992.tb00499.x. [DOI] [PubMed] [Google Scholar]
- 26.Furukawa K, Hotta Y. EMBO J. 1993;12:97–106. doi: 10.1002/j.1460-2075.1993.tb05635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vester B, Smith A, Krohne G, Benavente R. J Cell Sci. 1993;104:557–563. doi: 10.1242/jcs.104.2.557. [DOI] [PubMed] [Google Scholar]
- 28.Furukawa K, Inagaki H, Hotta Y. Exp Cell Res. 1994;212:426–430. doi: 10.1006/excr.1994.1164. [DOI] [PubMed] [Google Scholar]
- 29.Alsheimer M, Benavente R. Exp Cell Res. 1996;228:181–188. doi: 10.1006/excr.1996.0315. [DOI] [PubMed] [Google Scholar]
- 30.Glasenapp E, von & Benavente R. Chromosoma. 2000;109:117–122. doi: 10.1007/s004120050419. [DOI] [PubMed] [Google Scholar]
- 31.Nakajima N, Abe K. FEBS Lett. 1995;365:108–114. doi: 10.1016/0014-5793(95)00453-g. [DOI] [PubMed] [Google Scholar]
- 32.Alsheimer M, von Glasenapp E, Hock R, Benavente R. Mol Biol Cell. 1999;10:1235–1245. doi: 10.1091/mbc.10.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozak M. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Höger T H, Zatloukal K, Waizenegger I, Krohne G. Chromosoma. 1990;99:379–390. doi: 10.1007/BF01726689. [DOI] [PubMed] [Google Scholar]
- 35.Kralewski M, Benavente R. Chromosoma. 1997;106:304–307. doi: 10.1007/s004120050251. [DOI] [PubMed] [Google Scholar]
- 36.Hock R, Wilde F, Scheer U, Bustin M. EMBO J. 1998;17:6992–7001. doi: 10.1093/emboj/17.23.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Farrell P H. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 38.Jensen O N, Podtelejnikov A, Mann M. Rapid Commun Mass Spectrom. 1996;10:1371–1378. doi: 10.1002/(SICI)1097-0231(199608)10:11<1371::AID-RCM682>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 39.Towler D A, Gordon J I, Adams S P, Glaser L. Annu Rev Biochem. 1988;57:69–99. doi: 10.1146/annurev.bi.57.070188.000441. [DOI] [PubMed] [Google Scholar]
- 40.Resh M D. Biochim Biophys Acta. 1999;1451:1–16. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- 41.Mical T I, Monteiro M J. J Cell Sci. 1998;111:3471–3485. doi: 10.1242/jcs.111.23.3471. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt-Zachmann M S, Dargemont C, Kühn L C, Nigg E A. Cell. 1993;74:439–504. doi: 10.1016/0092-8674(93)80051-f. [DOI] [PubMed] [Google Scholar]
- 43.Monteiro M J, Hicks C, Gu L, Janicki S. J Cell Biol. 1994;127:1327–1343. doi: 10.1083/jcb.127.5.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]