Abstract
We have addressed the search of novel genetic prognostic markers in a selected cohort of patients with stroma-poor localized resectable neuroblastoma (NB) who underwent relapse or progression (group 1) or complete remission (group 2) over a minimum follow-up of 32 months from diagnosis. Twenty-three Italian patients with localized resectable NB (stages 1 and 2) diagnosed from 1994 through 2005 were studied. All patients received surgical treatment. Chemotherapy was administered only to the three stage 2 patients who had MYCN-amplified tumors. High-resolution array-comparative genomic hybridization (CGH) DNA copy-number analysis technology was used to identify novel prognostic markers. Chromosome 1p36.22p36.32 loss and 1q22qter gain, detected almost exclusively in group 1 patients, were significantly associated with poor event-free survival (EFS) (p = 0.0024 and p = 0.024, respectively). In contrast, patients with 7p11.2p22 gain, who belonged predominantly to group 2, had a significantly better EFS (p = 0.015). The frequency of 17q gain or 3p and 11q losses did not differ significantly in group 1 versus group 2 NBs. The sensitive technique allowed us to define the smallest region of 1p deletion. In conclusion, 1q22qter gain and 7p11.2p22 gain might represent new prognostic markers in localized resectable NB, but the small study size and the retrospective nature of the findings warrant further validation of the results in larger studies.
Keywords: 1q gain, 7p gain, array-CGH, localized resectable neuroblastoma, prognostic markers
Neuroblastoma (NB) accounts for 9%–10% of pediatric tumors. More than 10,000 children a year worldwide develop NB, which represents the most frequent extracranial solid tumor and the main cause of cancer-related death in preschool-age children.1 Clinical variables associated with poor disease outcome include age greater than 1 year and metastatic disease at diagnosis, together with unfavorable histopathology according to the system of Shimada et al.2–4 MYCN proto-oncogene amplification (MNA) is strongly associated with rapid disease progression and poor outcome.5–8 In addition, DNA ploidy is an independent prognostic factor in patients younger than 1 year at diagnosis.5
Half of NB patients present with metastatic disease at diagnosis, and approximately one-third survive at 5 years.9 In contrast, patients with localized resectable NB have an excellent prognosis, with approximately 10% of them developing only local recurrences or metastatic progression.10 These latter patients may benefit from early and aggressive treatment based upon predictive risk factors, such as MNA, unfavorable histopathology, or positive lymph nodes.11–14 These factors, however, identify only a subset of patients with localized resectable NB at risk of relapse and/or progression, and the search for novel prognostic markers is warranted.
During the last decade, comparative genomic hybridization (CGH), which allows one-step screening of DNA copy number gains and losses across the entire tumor genome, has been developed.15 A further refinement of CGH is array-CGH, which makes use of the microarray technology to increase the sensitivity by 10- to 15-fold compared with chromosomal CGH.16 Array-CGH holds great promise for the discovery of subtle genetic abnormalities in localized tumors that were previously undetected with less sensitive techniques. The aim of this study was to search for novel prognostic factors able to predict the risk of local recurrence and/or progression in a small cohort of selected patients with localized resectable NB by using array-CGH.
Materials and Methods
Patients
Twenty-three Italian patients with localized resectable NB (stages 1 and 2) diagnosed from 1994 to 2005 were studied. All patients received surgical treatment. Chemotherapy was administered only to the three stage 2 patients who had MNA tumors. Age at diagnosis ranged from 2 to 116 months. Tumor staging was performed according to the International Neuroblastoma Staging System.3 Tumors were stage 1 in 11 cases and stage 2 in 12 cases. After surgical resection, nine patients suffered from local recurrence and/or metastatic progression (group 1), whereas 14 patients remained disease-free (group 2) over a minimum follow-up of 32 months. All tumors were classified according to the histology-prognostic group classification (International Neuroblastoma Pathology Classification/Shimada)4 as favorable (F; 19 patients) or unfavorable (U; four patients) (Table 1). The study was conducted following the approval of a local investigation committee. Based on the results of recent studies, we divided our patients in two age groups using a cutoff of 18 months rather than 12 months.17, 18
Table 1.
Clinical-pathologic characteristics of 23 localized neuroblastoma patients and copy number aberrations detected by array-CGH
| ID | Sex | Age (Months) | Primary Site/Recurrence | DNA Index | Stage | Histology | Survival (Months) | Outcome | Losses | Gains |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 25 | Adrenal/disseminated | 1.52 | 1 | Favorable | 146 | ANED | 1p36.22p 36.32, 3, X, Y | 8q24.3, 11p15.5, 11q24.2qter, 17q12 qter, 20p12.3, 20q11.12qter, 22 |
| 2 | F | 2 | Retroperitoneal/disseminated | 1.6 | 1 | Favorable | 130 | ANED | 1p35.2pter, 2p21qter, 3p 21.32pter, 4, 8, 10, 11, 12, 14q23.1qter, X | 1p35.1qter, 2p21pter, 3p21.32qter, 17q21.1qter, 22q13.32qter |
| 3 | M | 4 | Adrenal/disseminated | 1 | 1 | Favorable | 105 | ANED | 4, 11q13.3qter, 21q22.12qter | 11q12.1q13.3, 17q11.2qter |
| 4 | M | 6 | Adrenal/abdomen | 1 | 1 | Unfavorable | 2 | DOD | 1p34.2pter, 2p23.2, 3p25.2 pter, 16q23.2 qter | amp2p24.1p25.1, 7q34qter, 13q14.2qter, 17q11qter |
| 5 | M | 55 | Adrenal/disseminated | 1.09 | 2A | Unfavorable | 13 | DOD | 1p13.3pter | amp2p24.3p25.1, 8q24.3, 9q33.1 qter, 10q25.1qter, 11p15.5pter, 17q 21.3qter, 22q13.3 |
| 6 | F | 76 | Adrenal/disseminated | 1 | 2B | Favorable | 10 | DOD | X | 1q22qter, 6, 7, 13, 19p13.2qter, 21q21.3 |
| 7 | F | 43 | Adrenal/disseminated | 1.89 | 2B | Unfavorable | 47 | DOD | 1p31.3pter, 16p13.3pter, 17p11.2pter | 1p31.2qter, 17q21.31qter |
| 8 | M | 2 | Adrenal/abdomen | 1.67 | 2B | Favorable | 10 | DOD | 4, 21, X, Y | 6p21.2qter, 13pterq33.3, 17, 18 |
| 9 | M | 3 | Adrenal/abdomen | 1.90 | 2 | Favorable | 32 | AWED | 1p32.2pter, 14q21.1qter, 19p13 | 12q24.11qter, 17q12qter |
| 10 | M | 2 | Adrenal/N | 1.57 | 1 | Favorable | 126 | ANED | 4, X, Y | 6, 7, 9, 12, 13, 17, 21, 22 |
| 11 | M | 2 | Adrenal/N | 1.68 | 1 | Favorable | 147 | ANED | 16, 19 | 2, 5, 6, 8, 12, 13, 18 |
| 12 | F | 116 | Retroperitoneal/N | 1 | 1 | Favorable | 118 | ANED | 19p13.1qter | 7p21.1, 17q21.2qter |
| 13 | F | 2 | Adrenal/N | 2.77 | 1 | Favorable | 109 | ANED | 4, 9p24.1pter, 9q21.3qter, 11p15.4pter, 11p11.2q23, 14q11.2q24.3, 14q32.2qter, 19, X | 6, 7p21.3q21.11, 13, 17, 18 |
| 14 | M | 26 | Adrenal/N | 2.12 | 1 | Favorable | 95 | ANED | 11q14.1qter | 2p25.2, 11p15.5 pter, 11q12.1q 13.2, 17q12.1qter |
| 15 | F | 17 | Adrenal/N | 1.43 | 1 | Favorable | 76 | ANED | 3, 4, 5, 6, 9, 10, 11, 13, 16p13.3qter, 17, 19q12qter, 21q11.2q22.11, 22, Xpterq27.3 | 7, 8q24.3 |
| 16 | M | 116 | Adrenal/N | 1 | 1 | Favorable | 64 | ANED | 16q24.1qter, 22q11.22 | 8q24.3, 11p15.5, 17q25.1qter, 19p13.3pter, 20q13.3qter |
| 17 | M | 76 | Thorax/N | 1.87 | 2A | Favorable | 125 | ANED | — | 7, 13, 18 |
| 18 | M | 14 | Retroperitoneal/N | 1.55 | 2A | Favorable | 67 | ANED | 4, 9, 11, 14, 21, X | 2, 7, 12, 17 |
| 19 | F | 83 | Thorax/N | — | 2A | Unfavorable | 58 | ANED | 19p13.3, 22q 11.21-qter | 7, 9q33.3qter |
| 20 | M | 13 | Abdomen/N | 1.62 | 2B | Favorable | 100 | ANED | 4, X | 2, 6, 7, 8, 12, 13, 17, 20, 22q12.1qter |
| 21 | F | 28 | Adrenal/N | 1.63 | 2B | Favorable | 59 | ANED | 9, 21 | 13, 17 |
| 22 | M | 16 | Thorax/N | 2.32 | 2B | Favorable | 58 | ANED | 3p25pter | 6, 7, 10, 13, 17q11.2qter, 18 |
| 23 | M | 33 | Adrenal/N | 1 | 2B | Favorable | 38 | ANED | 1p21.3pter, 6p24pter, 8q12.1q13.2 | amp2p24.2p24.3, 17q21.2qter |
Abbreviations: M, male; ANED, alive no evidence of disease; F, female; DOD, dead of disease; AWED, alive with evidence of disease; N, none. The relevant copy number aberrations are shown in boldface.
Sample Preparation
Aliquots of primary tumor tissue were obtained at diagnosis and snap frozen at − 80°C. Cryosections were examined by a pathologist to identify samples with at least 90% tumor cells for subsequent DNA extraction. Each tumor specimen was tested for DNA index by flow cytometry.19
Genomic DNA was extracted from frozen tissues by using the GenElute Mammalian Genomic DNA Mini-prep Kit (Sigma Chemical Co., St. Louis, MO, USA) according to the manufacturer’s instructions.
Array-Based CGH Analysis
Array-based CGH analysis was performed using commercially available oligonucleotide microarrays containing about 60-mer probes (Human Genome CGH Microarray 244A Kit; Agilent Technologies, Santa Clara, CA, USA). This platform allows genomewide survey and molecular profiling of genomic aberrations with a mean resolution of approximately 6.4 kb. Labeling and hybridization were performed following the protocols provided by Agilent Technologies. Briefly, DNA purified from tumors and reference DNA from normal male or female controls (Promega Corporation, Madison, WI, USA) were double-digested with RSAI and AluI for 2 h at 37°C. After 20 min at 65°C, DNA labeling was performed according to the Agilent protocol using the random primers labeling kit for 2 h. Each DNA sample was labeled with Cy5-coupled deoxyuridine 5’-triphosphate (Cy5-dUTP) and DNA controls with Cy3-dUTP. Labeled products were column purified, and the labeling efficiency was checked with a Nanodrop ND-1000 spectrophotometer. Cy5 and Cy3 incorporations were measured at 650 and 550 nm, respectively. Test and reference DNA were pooled and mixed with 50 μg human Cot-1 DNA (Bethesda Research Laboratories, Gaithersburg, MD, USA), 50 μl blocking buffer, and 250 μl hybridization buffer (Agilent Technologies). Before hybridization to the array, the mix was denatured at 95°C for 10 min and then preassociated at 37°C for 20 min. Hybridization was carried out for 40 h at 65°C in a rotating oven (20 rpm). The microarray slides were washed according to the manufacturer’s protocol with wash buffers supplied with the Agilent Microarray 244A Kit. The slides were dried and scanned at 532 nm (Cy3) and 635 nm (Cy5) using the Agilent G2565BA DNA microarray scanner and the Feature Extraction software (version 9.1.3; Agilent Technologies). Graphical overview was obtained using Agilent CGH Analytics software (version 3.4.27).
Statistical Analysis
Descriptive statistics were performed and quantitative parameters were reported as means and SD, or as medians with minimum and maximum values in case of skewed distributions. Qualitative data were reported as frequencies and percentages. Comparison of qualitative data (gender, stage, etc.) among two groups of patients (group 1 vs. group 2) was made by the chi-square test or by Fisher’s exact test in cases of expected frequencies less than five. Comparison of quantitative data (age at diagnosis, number of structural aberrations, etc.) between group 1 and group 2 was made by means of the Mann-Whitney U-test because the normality assumption was not fulfilled.
Overall survival (OS) and event-free survival (EFS) curves were drawn by the different groups of patients (group 1 vs. group 2) or by different types of copy number aberrations (CNAs; e.g., 1p loss vs. no 1p loss); survival curves were constructed with the Kaplan-Meier method, and the log-rank test was used to compare these curves.
All tests were two sided, and p < 0.05 was considered statistically significant. Statistica, release 6 (StatSoft Corp., Tulsa, OK, USA), was used for all the analyses.
Results
Demographic Features of Patients with Localized Resectable NB
Group 1 included five patients (four stage 1, one stage 2) younger and four patients (one stage 1, three stage 2) older than 18 months (range, 2–76 months). Tumor DNA was mostly near-diploid/tetraploid (Table 1).
Group 2 included seven patients (four stage 1, three stage 2) younger and seven (three stage 1, four stage 2) older than 18 months (age range, 2–116 months). Tumor DNA ranged from diploid to pentaploid (Table 1).
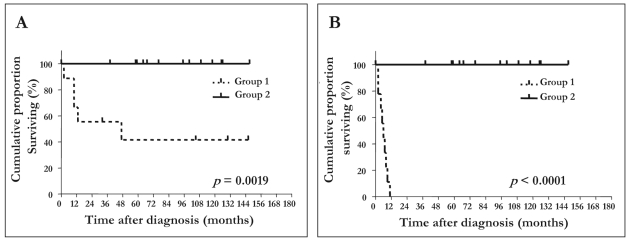
OS and EFS of group 1 and group 2 patients are shown in Fig. 1. Group 2 patients had significantly better EFS and OS than did group 1 patients (log-rank test: EFS, p < 0.0001; OS, p = 0.0019). Of the 23 patients, five died of disease, and 18 are alive (16 in complete remission, two with evidence of disease) (Table 1). The five patients who died belonged to group 1, and three of them had tumors with unfavorable histology.
Fig. 1.
Overall survival (A) and event-free survival (B) for 23 localized group 1 and group 2 resectable neuroblastoma patients.
Group 1 and 2 patients showed no significant difference in age, gender, disease stage, or DNA ploidy.
Array-CGH Analysis of Localized Resectable NB Tumors
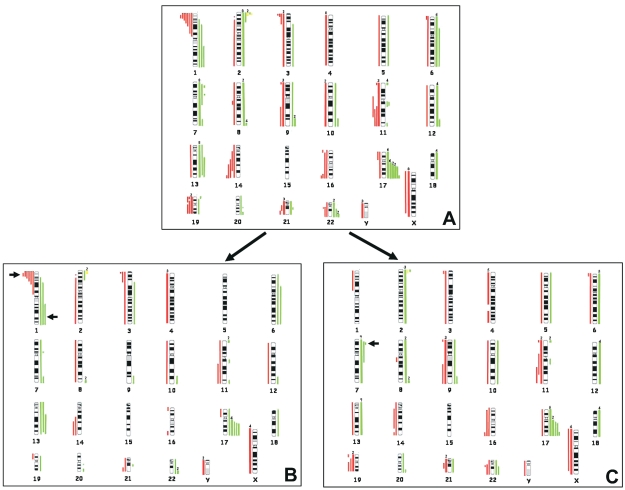
Genomic typing using array-CGH disclosed multiple CNAs in all NB samples (Table 1). The cumulative losses and gains are summarized in Fig. 2A.
Fig. 2.
(A) Summary of the patterns of total gains (right, green lines) and losses (left, red lines) detected in 23 localized resectable neuroblastoma samples using array-comparative genomic hybridization (CGH). (B) Aberration patterns present in group 1 tumors. (C) Aberration patterns present in group 2 tumors. Each line represents a copy number aberration in the individual tumors. The thick lines indicate the number of tumors, shown above the lines, with the same imbalances. The arrows indicate the major differences between the two CGH results.
The total number of overrepresented regions detected in individual tumors exceeded that of underrepresented regions. Only 3 of 23 tumors had MNA. The most frequent minimal common regions (MCRs) of genomic gains were found on chromosomes 17q25.1qter (18 cases), 7p11.2p22 (10 cases), 13q14.2q33.3 (10 cases), 7q34qter (9 cases), 2p (8 cases), 6p21.2qter (7 cases), 8q24.3 (6 cases), 12q24.11qter (5 cases), and whole chromosome 18 (5 cases). The most frequent MCRs of genomic losses involved chromosomes Xpterq27.3 (9 cases), whole chromosome 4 (8 cases), 1p36.22p36.32 (7 cases), 11q14.1q23 (6 cases), 3p25.2pter (5 cases), 9p24.1pter (4 cases), 9q21.3qter (4 cases), 11p15.4pter (4 cases), 14q23.1q24.3 (4 cases), 16q24.1qter (4 cases), 19q12qter (4 cases), and 21q11.2q22.11 (4 cases) (Fig. 2A). CNAs present in less than 20% of tumor samples were excluded from the above analyses.
Comparison of Array-CGH Profiles in Group 1 and Group 2 NB Patients
Next, array-CGH profiles were compared in group 1 versus group 2 NB tumors (Fig. 2B, C). Group 1 tumors showed structural CNAs commonly detected in NB (1p, 3p, and 11q losses, 17q gain, MNA),6–8,20–23 as well as partial CNAs not frequently observed in NB (1q22qter gain, 13q14.2q33.3 gain, 22q13.32 gain, 6p21.2qter gain, 11p15.5 gain, 21q22.12qter loss), and occasional whole-chromosome numerical aberrations (X and 4 losses). The most frequent MCRs of imbalances were 17q21.31qter gain (8 of 9), 1p36.22p36.32 loss (6 of 9), X loss (4 of 9), 1q22qter gain (3 of 9), 3p21.32pter loss (3 of 9), 4 loss (3 of 9), 13q14.2q33.3 gain (3 of 9), 22q13.32 gain (3 of 9), 6p21.2qter gain (2 of 9), 6q12qter gain (2 of 9), 7q34qter gain (2 of 9), 8q24.3 gain (2 of 9), 11q13.3qter loss (2 of 9), 11p15.5 gain (2 of 9), 14q23.1qter loss (2 of 9), and 21q22.12qter loss (2 of 9) (each present in more than 20% of tumors) (Fig. 2B).
Group 2 NBs showed whole chromosomal gains or losses, but also segmental CNAs reported in NB (i.e., 11q loss, 17q gain, MNA),6–8,20–23 and partial imbalances of other chromosomes (8q12.1q13.2 loss, 21q 11.2q22.11 loss, 22q11.22 loss). The most frequent MCRs of imbalances were i) 17q25.1qter gain, detected in 10 of 14 group 2 tumors (5 of which had gain of whole chromosome 17); ii) 7p11.2p22 gain (9 of 14), 13q12qter gain (7 of 14), Xpterq27.3 loss (5 of 14), 9p24.1pter loss (4 of 14), 9q21.3qter (4 of 14), 19q12qter loss (4 of 14), 8q24.3 gain (4 of 14), 11p15.4pter loss (3 of 14), 16q24.1qter loss (3 of 14), 21q11.2q22.11 loss (3 of 14), and 22q11.22 loss (3 of 14); and iii) whole-chromosome numerical aberrations including chromosome 6 gain (5 of 14), chromosome 12 gain (4 of 14), chromosome 18 gain (4 of 14), and chromosome 4 loss (5 of 14) (Fig. 2C).
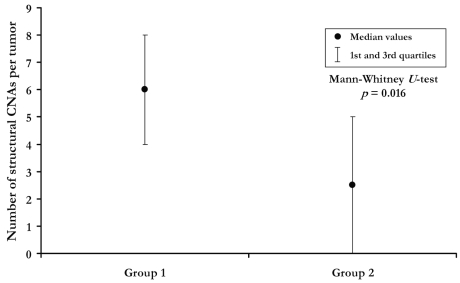
Group 1 NBs had a significantly higher frequency of 1p loss (p = 0.0049, Fisher’s exact test), 1q22qter gain (p = 0.0474; Fig. 2B), and structural changes per tumor (medians: 6 in group 1 vs. 2.5 in group 2; p = 0.016) than did group 2 NBs (Fig. 3).
Fig. 3.
Median number of structural copy number aberrations (CNAs) per tumor in the two groups of patients (group 1, relapsed tumors; group 2, nonrelapsed tumors). Mann-Whitney U-test, p = 0.016.
The latter tumors showed higher but not significant frequency of numerical changes per tumor (medians: 4.5 in group 2 vs. 1 in group 1, p = 0.17) and whole chromosome 12 gain (p = 0.13) than did group 1 NBs. 7p11.2p22 gain occurred significantly more frequently in group 2 than in group 1 (p = 0.029). Other CNAs detected in group 2 but below statistical significance were 9p24.1pter loss (p = 0.127), 9q21.3qter (p = 0.127), and 19q12qter loss (p = 0.127) (Fig. 2C).
The frequency of 17q gain, MNA, 3p, and 11q losses did not differ significantly between groups 1 and 2 (Fig. 2B, C). Finally, no significant difference in the frequency or type of CNAs was detected between patients younger and older than 18 months.17,18
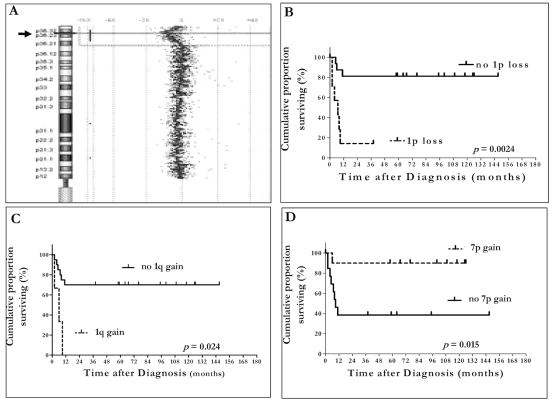
The MCR of 1p loss is shown in Fig. 4A. The break point of 1p loss falls onto 1p36 band from 1p36.22 to 1p36.32.
Fig. 4.
(A) Minimal common region of 1p loss. The arrow points to the break point position on chromosome 1p. (B–D) Event-free survival according to chromosome 1p loss (B), chromosome 1q gain (C), and chromosome 7p gain (D).
Impact of 1p Loss, 1q Gain, and 7p Gain on EFS of Localized Resectable NB Patients
Chromosome 1p36.22p36.32 loss and 1q22qter gain, detected almost exclusively in group 1 patients, were associated with significantly worse EFS (p = 0.0024 and p = 0.024, respectively) (Fig. 4B, C). In contrast, patients with 7p11.2p22 gain, who belonged predominantly to group 2, had a significantly better EFS than did those without the same CNA (p = 0.015; Fig. 4D). Chromosome 1p36.22p36.32 loss, 1q22qter gain, and 7p11.2p22 gain had no impact on OS of either patient group (p = 0.099, p = 0.064, and p = 0.23, respectively).
Discussion
Patients with localized resectable NBs usually have excellent survival rates, but a small percentage of them subsequently relapse and/or die of disease. Several genetic features have been associated with poor outcome in localized resectable NBs. MNA, a powerful indicator of poor prognosis in approximately one-third of NB patients with metastatic disease at diagnosis (stage 4), predicts tumor recurrence and/or progression only in a subset of patients with localized disease.6 Recently, deletions of 3p and 11q, thought to harbor yet unidentified tumor suppressor genes, have been associated with poor outcome,20,21 and deletion of 1p was correlated with a higher event and death rate in localized unresectable NBs (stage 2 and 3).22,24–26 The role of these genetic factors in predicting outcome in localized resectable NB patients is not established.
In this study, we have addressed the search of novel genetic prognostic markers in a selected cohort of patients with localized resectable NB and different clinical outcomes over a 3-year follow-up: local recurrence or metastatic progression (group 1) versus complete remission (group 2).
We have identified two distinct regions of gain that have not been associated previously with localized resectable NB: 1q22qter gain, detected only in group 1 patients and associated with risk of local relapse or metastatic relapse, and 7p11.2p22 gain, detected only in group 2 patients and associated with favorable prognosis. These CNAs likely harbor genes that are biologically and clinically relevant. Several genes related to oncogenesis are localized in 1q22qter region. The NDSP (neuroblastoma-derived secretory protein) gene, whose expression has been found to be upregulated and to be correlated with tumor aggressiveness and metastasis in NB,27 maps to 1q243. The NBPF15 gene, which maps to 1q21.1, was originally identified by positional cloning of a translocation break point from an NB patient.28 Expression of this gene is upregulated in many tumor types.29 The NTRK1 gene maps to 1q21-q22 and is involved with the TPM3 (tropomyosin 3) gene in a somatic rearrangement that creates the chimeric oncogene TRK.30 TRK is a gene coding for a putative receptor molecule with an associated tyrosine kinase activity that was found to be activated in 25% of patients with papillary thyroid carcinoma. In situ hybridization to human metaphase chromosomes localized the TRK gene to 1q32-q41.31 The GAC1 gene, mapping to 1q32.1, was found to be amplified and overexpressed in malignant gliomas.32 The HDGF (hepatoma-derived growth factor) gene, mapping to 1q21, is expressed ubiquitously in normal tissues and tumor cell lines.33
Different insulin-like growth factor–binding protein (IGBP) genes have been found to be involved in the control of NB growth.34 Notably, the IGBP3 gene maps on 7p11.2p22 gain.35 However, a relationship between this gene and the favorable prognosis of patients with 7p11.2p22 gain cannot be established.
In addition, numerical CNAs were more represented in localized resectable NB with favorable outcome,36 as opposed to structural CNAs that were detected more frequently in group 1 patients. These findings are at variance with a recent report showing that tumors from all stage 1 and 2 NB patients have numerical CNAs.37 Thus, it is tempting to speculate that structural CNAs due to unbalanced chromosome translocations are related to relapse/progression in patients with localized resectable NB. Further studies with larger cohorts of patients will help to confirm our findings and hypotheses.
Localized NBs in group 1 patients were characterized by multiple structural CNAs likely related to unbalanced translocations dominated by 1p loss. Loss of heterozygosity at the distal short arm of chromosome 1 (1p36) was detected in six of nine group 1 patients, three of whom died of disease; two are alive without evidence of disease, and one is alive with disease. In previous studies, allelic loss of 1p36 was detected in about 35% of all NB patients and found to correlate with high-risk tumors characterized by unfavorable prognosis.22,25,36,38,39 Loss of 1p36 is detected in stage 4 NB mostly in association with MNA.36,38–40 In this study, only two of six tumors bearing 1p36 loss had concomitant MNA, pointing to 1p36 loss as an independent prognostic marker of relapse/progression in localized resectable NB patients. This finding is in line with the conclusions of other studies22,24–26 and consistent with recent reports indicating that structural abnormalities of chromosome 1p are observed more frequently in relapsing tumors.36,40–43 Recently, the CHD5 gene, a new member of a chromatin remodeling gene family mapping to 1p36.22p36.32, was shown to function as a tumor suppressor in mice genetically engineered to have a germ line hemizygous deletion,44 supporting evidence that an NB tumor suppressor gene resides in the 1p chromosomal region.45–47 The 1p deleted region, which was previously found to be usually quite large (1p32-pter, 1–7.03 Mb) in primary NB tumors,48 has been recently restricted to 2 Mb in NB cell lines.46 Accordingly, we have here defined a minimal region of 1p deletion in the specific band 1p36.22p36.32 (5.4-Mb MCR deletion) in primary tumors from group 1 NB patients. Finally, 17q gain was detected in the majority of patients and was devoid of prognostic relevance because it was equally distributed in all groups. In contrast, few tumors displayed 3p or 11q losses.20,21
In conclusion, 1q22qter gain and 7p11.2p22 gain might represent new prognostic markers in localized resectable NB, but the small study size and the retrospective nature of the findings warrant further validation of the results obtained in larger studies.
Acknowledgments
We thank pathologists, surgeons, and pediatric oncologists of participating institutions, Dr. Anna Cremonini for collecting patient data, and Dr. Gianpaolo Tonini (Translational Pediatric Oncology, National Institute for Cancer Research, Genoa, Italy) for providing some tumor specimens. Dr. Federica Parodi and Dr. Francesca Negri are fellows of the Italian Neuroblastoma Foundation. This study has been presented in part at the Advances in Neuroblastoma Research 2008 meeting (Chiba, Japan, May 21–24, 2008).
Appendix
Participating Institutions Belonging to the Associazione Italiana di Ematologia e Oncologia Pediatrica (AIEOP)
Oncologia Pediatrica, Istituto Nazionale Tumori, Milano; Ematoncologia Pediatrica, Azienda Ospedaliera Ciaccio, Catanzaro; Ematoncologia Pediatrica, Policlinico Universitario, Catania; Oncoematologia Pediatrica, Ospedale Civico Benefratelli, Palermo; Pediatria Oncologica, Ospedale Infantile Regina Margherita, Torino; Centro di Emato-Oncologia, Ospedale Burlo-Garofalo, Trieste; Clinica Pediatrica, Ospedali Civili di Brescia, Brescia; Clinica Pediatrica, Policlinico di Bari, Bari; Oncologia-Ematologia Pediatrica, Ospedale S. Orsola, Bologna; Servizio di Oncologia, Dipartimento di Pediatria, II Ateneo di Napoli; Italy.
References
- 1.Maris JM. The biologic basis for neuroblastoma heterogeneity and risk stratification. Curr Opin Pediatr. 2005;17:7–13. doi: 10.1097/01.mop.0000150631.60571.89. [DOI] [PubMed] [Google Scholar]
- 2.Breslow N, McCann B. Statistical estimation of prognosis for children with neuroblastoma. Cancer Res. 1971;31:2098–2103. [PubMed] [Google Scholar]
- 3.Brodeur GM, Pritchard J, Berthold F, et al. Revisions of the international criteria for neuroblastoma diagnosis, staging, and response to treatment. J Clin Oncol. 1993;11:1466–1477. doi: 10.1200/JCO.1993.11.8.1466. [DOI] [PubMed] [Google Scholar]
- 4.Shimada H, Ambros IM, Dehner LP, et al. The International Neuroblastoma Pathology Classification (the Shimada system) Cancer. 1999;86:364–372. [PubMed] [Google Scholar]
- 5.Look AT, Hayes FA, Shuster JJ, et al. Clinical relevance of tumor cell ploidy and MYCN gene amplification in childhood neuroblastoma: a pediatric oncology group study. J Clin Oncol. 1991;9:581–591. doi: 10.1200/JCO.1991.9.4.581. [DOI] [PubMed] [Google Scholar]
- 6.Seeger RC, Brodeur GM, Sather H, et al. Association of multiple copies of the MYCN oncogene with rapid progression of neuroblastomas. N Engl J Med. 1985;313:1111–1116. doi: 10.1056/NEJM198510313131802. [DOI] [PubMed] [Google Scholar]
- 7.Schwab M, Alitalo K, Klempnauer KH, et al. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumor. Nature. 1983;305:245–248. doi: 10.1038/305245a0. [DOI] [PubMed] [Google Scholar]
- 8.Brodeur GM, Seeger RC, Schwab M, et al. Amplification of N-myc in untreated human neuroblastoma correlates with advanced disease stage. Science. 1984;224:1121–1124. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- 9.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 10.Navarro S, Amann G, Beiske K, et al. Prognostic value of international neuroblastoma pathology classification in localized resectable peripheral neuroblastic tumors: a histopathologic study of localized neuroblastoma European Study Group 94.01 trial and protocol. J Clin Oncol. 2006;24:695–699. doi: 10.1200/JCO.2004.00.8631. [DOI] [PubMed] [Google Scholar]
- 11.Berthold F, Hero B, Breu H, et al. The recurrence patterns of stage I, II and III neuroblastoma: experience with 77 relapsing patients. Ann Oncol. 1996;7:183–187. doi: 10.1093/oxfordjournals.annonc.a010547. [DOI] [PubMed] [Google Scholar]
- 12.Nitschke R, Smith EI, Shochat S, et al. Localized neuroblastoma treated by surgery. A Pediatric Oncology Group study. J Clin Oncol. 1988;6:1271–1279. doi: 10.1200/JCO.1988.6.8.1271. [DOI] [PubMed] [Google Scholar]
- 13.Matthay KK, Sather HN, Seeger RC, et al. Excellent outcome of stage II neuroblastoma is independent of residual disease and radiation therapy. J Clin Oncol. 1989;7:236–244. doi: 10.1200/JCO.1989.7.2.236. [DOI] [PubMed] [Google Scholar]
- 14.Perez CA, Matthay KK, Atkinson JB, et al. Biologic variables in the outcome of stage I and II neuroblastoma treated with surgery as primary therapy: a children’s cancer group study. J Clin Oncol. 2000;18:18–26. doi: 10.1200/JCO.2000.18.1.18. [DOI] [PubMed] [Google Scholar]
- 15.Kallioniemi A, Kallioniemi OP, Sudar D, et al. Comparative genomic hybridization for molecular cytogenetics analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 16.Lucito R, Healy J, Alexander J, et al. Representational oligonucleotide microarray analysis: a high-resolution method to detect genome copy number variation. Genome Res. 2003;13:2291–2205. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.London WB, Boni L, Simon T, et al. The role of age in neuroblastoma risk stratification: the German, Italian, and Children’s Oncology Group perspectives. Cancer Lett. 2005;228:257–266. doi: 10.1016/j.canlet.2004.12.054. [DOI] [PubMed] [Google Scholar]
- 18.London WB, Castleberry RP, Matthay KK, et al. Evidence for an age cutoff greater than 365 days for neuroblastoma risk group stratification in the Children’s Oncology Group. J Clin Oncol. 2005;23:6459–6465. doi: 10.1200/JCO.2005.05.571. [DOI] [PubMed] [Google Scholar]
- 19.Ladenstein R, Ambros IM, Pötschger U, et al. Prognostic significance of DNA ditetraploidy in neuroblastoma. Med Pediatr Oncol. 2001;36:83–92. doi: 10.1002/1096-911X(20010101)36:1<83::AID-MPO1020>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 20.Spitz R, Hero B, Ernestus K, et al. Deletions in chromosome arms 3p and 11q are new prognostic markers in localized and 4s neuroblastoma. Clin Cancer Res. 2003;9:52–58. [PubMed] [Google Scholar]
- 21.Spitz R, Hero B, Simon T, Berthold F. Loss in chromosome 11q identifies tumors with increased risk for metastatic relapses in localized and 4S neuroblastoma. Clin Cancer Res. 2006;12:3368–3373. doi: 10.1158/1078-0432.CCR-05-2495. [DOI] [PubMed] [Google Scholar]
- 22.Simon T, Spitz R, Hero B, et al. Risk estimation in localized unresectable single copy MYCN neuroblastoma by the status of chromosomes 1p and 11q. Cancer Lett. 2006;237:215–222. doi: 10.1016/j.canlet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 23.Bown N, Cotterill S, Lastowska M, et al. Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. N Engl J Med. 1999;340:1954–1961. doi: 10.1056/NEJM199906243402504. [DOI] [PubMed] [Google Scholar]
- 24.Caron H, van Sluis P, de Kraker J, et al. Allelic loss of chromosome 1p as a predictor of unfavorable outcome in patients with neuroblastoma. N Engl J Med. 1996;334:225–230. doi: 10.1056/NEJM199601253340404. [DOI] [PubMed] [Google Scholar]
- 25.Maris JM, Weiss MJ, Guo C, et al. Loss of heterozygosity at 1p36 independently predicts for disease progression but not decreased overall survival probability in neuroblastoma patients: a children’s cancer group study. J Clin Oncol. 2000;18:1888–1899. doi: 10.1200/JCO.2000.18.9.1888. [DOI] [PubMed] [Google Scholar]
- 26.Spitz R, Hero B, Westermann F, et al. Fluorescence in situ hybridization analyses of chromosome band 1p36 in neuroblastoma detect two classes of alterations. Genes Chrom Cancer. 2002;34:299–305. doi: 10.1002/gcc.10070. [DOI] [PubMed] [Google Scholar]
- 27.Vasudevan SA, Russell HV, Okcu MA, et al. Neuroblastoma-derived secretory protein messenger RNA levels correlate with high-risk neuroblastoma. J Pediat Surg. 2007;42:148–152. doi: 10.1016/j.jpedsurg.2006.09.064. [DOI] [PubMed] [Google Scholar]
- 28.Laureys G, Speleman F, Opdenakker G, et al. Constitutional translocation t(1;17)(p36;q12–21) in a patient with neuroblastoma. Genes Chrom Cancer. 1990;2:252–254. doi: 10.1002/gcc.2870020315. [DOI] [PubMed] [Google Scholar]
- 29.Vandepoele K, Van Roy N, Staes K, et al. A novel gene family NBPF: intricate structure generated by gene duplications during primate evolution. Mol Biol Evol. 2005;22(11):2265–2274. doi: 10.1093/molbev/msi222. [DOI] [PubMed] [Google Scholar]
- 30.Greco A, Mariani C, Miranda C, et al. Characterization of the NTRK1 genomic region involved in chromosomal rearrangements generating TRK oncogenes. Genomics. 1993;18:397–400. doi: 10.1006/geno.1993.1482. [DOI] [PubMed] [Google Scholar]
- 31.Miozzo M, Pierotti M, Sozzi G, et al. Human TRK proto-oncogene maps to chromosome 1q32-q41. Oncogene. 1990;5:1411–1414. [PubMed] [Google Scholar]
- 32.Almeida A, Zhu XX, Vogt N, et al. GAC1, a new member of the leucine- rich repeat superfamily on chromosome band 1q32.1, is amplified and overexpressed in malignant gliomas. Oncogene. 1998;16:2997–3002. doi: 10.1038/sj.onc.1201828. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura H, Izumoto Y, Kambe H, et al. Molecular cloning of complementary DNA for a novel human hepatoma-derived growth factor: its homology with high mobility group-1 protein. J Biol Chem. 1994;269:25143–25149. [PubMed] [Google Scholar]
- 34.Russo VC, Schütt BS, Andaloro E, et al. Insulin-like growth factor binding protein-2 binding to extracellular matrix plays a critical role in neuroblastoma cell proliferation, migration, and invasion. Endocrinology. 2005;146:4445–4455. doi: 10.1210/en.2005-0467. [DOI] [PubMed] [Google Scholar]
- 35.Ehrenborg E, Larsson C, Stern I, et al. Contiguous localization of the genes encoding human insulin-like growth factor-binding proteins 1 (IGBP1) and 3 (IGBP3) on chromosome 7. Genomics. 1992;12:497–502. doi: 10.1016/0888-7543(92)90440-4. [DOI] [PubMed] [Google Scholar]
- 36.Spitz R, Oberthuer A, Zapatka M, et al. Oligonucleotide array-based comparative genomic hybridization (aCGH) of 90 neuroblastomas reveals aberration patterns closely associated with relapse pattern and outcome. Genes Chrom Cancer. 2006;45:1130–1142. doi: 10.1002/gcc.20376. [DOI] [PubMed] [Google Scholar]
- 37.Vandesompele J, Bacolod M, Selvanayagam Z, et al. Unequivocal delineation of clinicogenetic subgroups and development of a new model for improved outcome prediction in neuroblastoma. J Clin Oncol. 2005;10:22280–22299. doi: 10.1200/JCO.2005.06.104. [DOI] [PubMed] [Google Scholar]
- 38.White PS, Maris JM, Beltinger C, et al. A region of consistent deletion in neuroblastoma maps within human chromosome 1p36.2–36.3. Proc Natl Acad Sci USA. 1995;92:5520–5524. doi: 10.1073/pnas.92.12.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Attiyeh EF, London WB, Mosse YP, et al. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med. 2005;353:2243–2253. doi: 10.1056/NEJMoa052399. [DOI] [PubMed] [Google Scholar]
- 40.Luttikhuis ME, Powell JE, Rees SA, et al. Neuroblastomas with chromosome 11q loss and single copy MYCN comprise a biologically distinct group of tumors with adverse prognosis. Br J Cancer. 2001;85:531–537. doi: 10.1054/bjoc.2001.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caron H, van Sluis P, de Kraker J, et al. Allelic loss of chromosome 1p as a predictor of unfavorable outcome in patients with neuroblastoma. N Engl J Med. 1996;334:225–230. doi: 10.1056/NEJM199601253340404. [DOI] [PubMed] [Google Scholar]
- 42.Chen QR, Bilke S, Wei JS, et al. cDNA array-CGH profiling identifies genomic alterations specific to stage and MYCN amplification in neuroblastoma. BMC Genomics. 2004;5:70. doi: 10.1186/1471-2164-5-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schleiermacher G, Michon J, Huon I, et al. Chromosomal CGH identifies patients with a higher risk of relapse in neuroblastoma without MYCN amplification. Br J Cancer. 2007;97:238–246. doi: 10.1038/sj.bjc.6603820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bagchi A, Papazoglu C, Wu Y, et al. CHD5 is a tumor suppressor at human 1p36. Cell. 2007;128:459–475. doi: 10.1016/j.cell.2006.11.052. [DOI] [PubMed] [Google Scholar]
- 45.White PS, Thompson PM, Gotoh T, et al. Definition and characterization of a region of 1p36.3 consistently deleted in neuroblastoma. Oncogene. 2005;24:2684–2694. doi: 10.1038/sj.onc.1208306. [DOI] [PubMed] [Google Scholar]
- 46.Okawa ER, Gotoh T, Manne J, et al. Expression and sequence analysis of candidates for the 1p36.31 tumor suppression gene deleted in neuroblastomas. Oncogene. 2007;14:1–8. doi: 10.1038/sj.onc.1210675. [DOI] [PubMed] [Google Scholar]
- 47.Wang Q, Diskin S, Rappaport E, et al. Integrative genomics identifies distinct molecular classes of neuroblastoma and shows that multiple genes are targeted by regional alterations in DNA copy number. Cancer Res. 2006;66:6050–6062. doi: 10.1158/0008-5472.CAN-05-4618. [DOI] [PubMed] [Google Scholar]
- 48.Mosse YP, Diskin SJ, Wasserman N, et al. Neuroblastomas have distinct genomic DNA profiles that predict clinical phenotype and regional gene expression. Genes Chrom Cancer. 2007;46:936–949. doi: 10.1002/gcc.20477. [DOI] [PubMed] [Google Scholar]