Abstract
Sugar transporters are central machineries to mediate cross-membrane transport of sugars into the cells, and sugar availability may serve as a signal to regulate the sugar transporters. However, the mechanisms of sugar transport regulation by signal sugar availability remain unclear in plant and animal cells. Here, we report that a sucrose transporter, MdSUT1, and a sorbitol transporter, MdSOT6, both localized to plasma membrane, were identified from apple (Malus domestica) fruit. Using a combination of the split-ubiquitin yeast two-hybrid, immunocoprecipitation, and bimolecular fluorescence complementation assays, the two distinct sugar transporters were shown to interact physically with an apple endoplasmic reticulum-anchored cytochrome b5 MdCYB5 in vitro and in vivo. In the yeast systems, the two different interaction complexes function to up-regulate the affinity of the sugar transporters, allowing cells to adapt to sugar starvation. An Arabidopsis (Arabidopsis thaliana) homolog of MdCYB5, AtCYB5-A, also interacts with the two sugar transporters and functions similarly. The point mutations leucine-73 → proline in MdSUT1 and leucine-117 → proline in MdSOT6, disrupting the bimolecular interactions but without significantly affecting the transporter activities, abolish the stimulating effects of the sugar transporter-cytochrome b5 complex on the affinity of the sugar transporters. However, the yeast (Saccharomyces cerevisiae) cytochrome b5 ScCYB5, an additional interacting partner of the two plant sugar transporters, has no function in the regulation of the sugar transporters, indicating that the observed biological functions in the yeast systems are specific to plant cytochrome b5s. These findings suggest a novel mechanism by which the plant cells tailor sugar uptake to the surrounding sugar availability.
The sugar transporters are central machineries to mediate cross-membrane transport of sugars into the cells and the distribution of sugars throughout the plant (Lalonde et al., 1999, 2004; Williams et al., 2000; Delrot et al., 2001; Kühn, 2003). Suc is the most prevalent sugar translocated in higher plants. Besides Suc, the sugar alcohols, such as mannitol and sorbitol, and raffinose can also be transported. The Suc transporters have been identified in a variety of plant species and are the best-characterized sugar transporters (for review, see Rentsch et al., 1998; Williams et al., 2000; Delrot et al., 2001; Kühn, 2003; Lalonde et al., 2004). The transporters for mannitol and sorbitol have also been identified in various plants, such as the mannitol transporters in celery (Apium graveolens; Noiraud et al., 2001) and the sorbitol transporters in sour berry (Prunus cerasus; Gao et al., 2003), apple (Malus domestica; Watari et al., 2004; Gao et al., 2005), common plantain (Plantago major; Ramsperger-Gleixner et al., 2004), and Arabidopsis (Arabidopsis thaliana; Reinders et al., 2005). Both Suc and sugar alcohol transporters belong to the major facilitator superfamily (Marger and Saier, 1993). Additional members of this superfamily, identified in many plants, are hexose transporters that are generally sink specific and that are postulated to catalyze Glc and Fru import into sink cells after phloem-unloaded Suc is hydrolyzed by extracellular invertases (Sauer et al., 1994; Lalonde et al., 1999, 2004; Büttner and Sauer, 2000; Williams et al., 2000).
It has been believed that, for maintaining sugar homeostasis in cells, the sugar transporters are tightly regulated at multiple levels of biological organization by altered sugar concentrations (Roitsch, 1999). The gene expression of Suc transporters was reported, in most cases, to be down-regulated by high levels of Suc (Weber et al., 1997; Chiou and Bush, 1998; Vaughn et al., 2002; Li et al., 2003a; Ransom-Hodgkins et al., 2003) but up-regulated by Suc starvation (Li et al., 2003b). However, in some cases, such as in the developing rice (Oryza sativa) embryo, inducing effects of the Suc transporter OsSUT1 expression by high levels of Suc and Glc occurred, whereas a sugar starvation-induced gene expression for OsSUT1 was not observed (Matsukura et al., 2000), suggesting that the sugar-induced effects on the transporter expression may be different, or otherwise the thresholds of sugar concentrations for induction or repression may change, according to organs/tissues or developmental stages. Similar to most Suc transporters, the expression of the grape (Vitis vinifera) hexose transporter VvHT1 was induced by low levels of its substrate Glc (Atanassova et al., 2003; Çakir et al., 2003; Conde et al., 2006), but both the transcripts and protein products of the VvHT1 gene as well as its transport activity were repressed by higher levels of Glc (Conde et al., 2006). The repressing effects of high concentrations of Glc on the hexose transporter activity were also observed in suspension-cultured cells of olive (Olea europaea; Oliveira et al., 2002). Additionally, the sugar transporters have also been shown to be regulated by abiotic effects such as wounding or pathogen infection (Truernit et al., 1996), salt stress (Noiraud et al., 2000), and light (Kühn et al., 1997; Stadler et al., 2003) and by other biotic factors than sugars such as the phytohormones abscisic acid (Fillion et al., 1999; Çakir et al., 2003) and cytokinins (Ehness and Roitsch, 1997). These biotic and abiotic stimuli may act either directly as physical or biochemical signals or indirectly by affecting sugar concentrations in cells.
It is known that sugar availability can exert a hormone-like signal in a variety of eukaryotic cell types. In yeast (Saccharomyces cerevisiae) that is used as an excellent model for studying sugar signaling, four signaling pathways have been identified to mediate Glc signaling (for review, see Rolland et al., 2001, 2002b, 2006). In addition to the Hexokinase2 (Hxk2)-triggered “main Glc repression pathway” involving several downstream components such as the Suc nonfermenting1 (Snf1) kinase (Moreno et al., 2005) and the cAMP-PKA-regulated pathway that relays signal from both the G protein-coupled receptor Gpr1 (Lemaire et al., 2004) and multiple hexose kinases (Hxk1/2 and Glk1 sensors) as well as trehalose signaling, the well-characterized signaling pathway concerns the hexose transporter homologs Snf3 and Rgt2, which are inactive in hexose transport but function to perceive Glc signal. The Snf3/Rgt2-mediated signal is relayed by activating the casein kinase Yck1 that inactivates the Rgt1 transcription repressor through a ubiquitin-mediated degradation pathway (Johnston and Kim, 2005). The high-affinity sensor Snf3 perceives low Glc signal, up-regulating the high-affinity Glc transporters, but the low-affinity sensor Rgt2 perceives high Glc signal, up-regulating the low-affinity Glc transporters (Boles and Hollenberg, 1997; Rolland et al., 2001, 2006).
Several sugar-signaling pathways, analogous to those in yeast, have been identified in plants. The Arabidopsis Hexokinase1 (HXK1) has been shown to be a Glc sensor (Jang et al., 1997; Rolland et al., 2002a, 2006; Moore et al., 2003; Cho et al., 2006), which controls a nuclear multiprotein complex to modulate specific target gene transcription (Cho et al., 2006). A series of the phytohormone abscisic acid-synthesizing enzymes and -signaling components (Arenas-Huertero et al., 2000; Cheng et al., 2002) and ethylene-signaling components (Zhou et al., 1998; Yanagisawa et al., 2003) have been identified as sugar-signaling regulators and may function downstream of the HXK1 sensor (Yanagisawa et al., 2003; Rolland et al., 2006). The Snf1-related protein kinases SnRK1s have also been suggested to regulate plant sugar signaling (Halford et al., 2003; Rolland et al., 2006), and SnRK1A was recently reported to be involved in the signaling pathway in rice (Lu et al., 2007). Additionally, several lines of evidence supported the regulatory roles of the Arabidopsis G protein-coupled receptor RGS1 as sensor (Chen et al., 2003; Johnston et al., 2007) and trehalose as a metabolic signal in plant Glc signaling (Avonce et al., 2004, 2005; Rolland et al., 2006).
In contrast to this progress, the mechanisms of sugar transport regulation by signal sugar availability remain largely unclear in plants as in other eukaryotic cells except for yeasts. Sugar transporters have been thought to act as a potential extracellular Glc or Suc detection system like Snf3 and Rgt2 in yeast (Lalonde et al., 1999). The homologs of Snf3 and Rgt2 in Arabidopsis and tomato (Solanum lycopersicum), AtSUT2 and LeSUT2, have no function in Suc transport and were suggested to be putative Suc sensors to modulate the activity of low- and high-affinity Suc transporters (Barker et al., 2000), but evidence against this point of view was provided as well (Barth et al., 2003). Recently, a Golgi-localized hexose transporter was reported to be a downstream player of heterotrimeric G protein-mediated sugar signaling in Arabidopsis early growth (Wang et al., 2006). Pharmaceutical experiments suggested that the hexose-regulated expression of the grape hexose transporter VvHT1 may involve the HXK-sensing pathway (Conde et al., 2006). Nevertheless, molecular and cellular insights into the regulation of the plant sugar transporters in relation to their transport functions have been lacking. Here, we report that both a Suc transporter, MdSUT1, and a sorbitol transporter, MdSOT6, from apple (Malus domestica) interact with cytochrome b5 in vitro and in vivo and that this interaction results in up-regulation of the affinity of both transporters to their substrate sugars, thus allowing cells to better adjust to sugar deficiency. These findings suggest a novel mechanism by which the plant cells tailor sugar uptake to the surrounding sugar availability.
RESULTS
Characterization of Apple Suc Transporter MdSUT1 and Sorbitol Transporter MdSOT6
We isolated from apple fruits two full-length cDNA clones: one encodes a putative Suc transporter and the other encodes a putative sorbitol transporter, designated MdSUT1 and MdSOT6, respectively. The MdSUT1 and MdSOT6 genes encode proteins of 499 amino acid residues (Supplemental Fig. S1A) and 533 amino acid residues (Supplemental Fig. S2A), respectively. The predicted amino acid sequences of both MdSUT1 and MdSOT6 exhibit the modular structures typical for the members of a major facilitator superfamily (Marger and Saier, 1993; Sauer and Tanner, 1993) comprising 12 transmembrane-spanning regions, which could be resolved into two parts of hydrophilic loops separated by a large central hydrophilic loop on the cytosolic side (Supplemental Figs. S1B and S2B). Homology searches with the deduced amino acid sequence indicated that MdSUT1 should belong to the SUT4 subfamily of low-affinity Suc transporters (Weise et al., 2000) like its closer homologs LeSUT4, StSUT4, AtSUT4, and OsSUT2 (Supplemental Fig. S3A; for review, see Kühn, 2003). MdSOT6 shares strong identity with an identified sorbitol transporter from apple MdSOT1 (Gao et al., 2005), but a fragment of 42 amino acids at the N terminus was lost for MdSOT1 compared with MdSOT6 (Supplemental Figs. S2A and S3B).
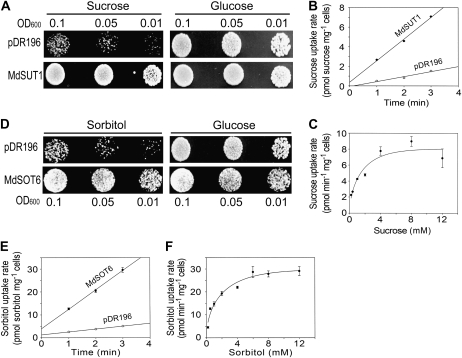
Functional expression of the MdSUT1 cDNA in the Suc uptake-deficient yeast mutant SUSY7/ura3 (Reinders et al. 2002) and the MdSOT6 cDNA in the sorbitol uptake-deficient mutant RS453 (Sauer and Stadler, 1993), rescuing the mutant yeast growth in the medium with Suc (for the MdSUT1-SUSY7/ura3 transformants) or sorbitol (for the MdSOT6-RS453 transformants) as sole carbon source, demonstrated that MdSUT1 is a functional Suc transporter (Fig. 1A) and MdSOT6 is a functional sorbitol transporter (Fig. 1D). Kinetic analysis of the [14C]Suc uptake by the SUSY7/ura3 cells expressing MdSUT1, assayed at the optimum pH 5.0 (Fig. 1, B and C; Supplemental Fig. S4A), showed a Km for Suc of 0.63 mm with a Vmax of 6.54 pmol Suc min−1 mg−1 cells (Supplemental Fig. S4E). This value of Km indicates that MdSUT1 is a high-affinity Suc transporter in spite of its sequence similarity to the low-affinity transporters (Supplemental Fig. S3A; Kühn, 2003). The kinetic assay for [3H]sorbitol uptake at the optimum pH 4.5 (Fig. 1, E and F; Supplemental Fig. S4C) in the MdSOT6-transgenic RS453 yeast cells indicated a Km for sorbitol of 1.57 mm with a Vmax of 28 pmol sorbitol min−1 mg−1 cells (Supplemental Fig. S4F). Competition assays by various sugars or sugar alcohols showed that MdSUT1 is a Suc transporter with high affinity for and specificity to Suc and MdSOT6 is a sorbitol transporter that may also transport Glc and Fru but apparently with a lower affinity than for sorbitol (Supplemental Fig. S4, B and D).
Figure 1.
Identification of MdSUT1 and MdSOT6 as functional sugar transporters. A, Expression of MdSUT1 in the yeast strain SUSY7/ura3 allows growth on Suc as the sole carbon source. SUSY7/ura3 yeast cells were transformed with the empty pDR196 vector (indicated by pDR196; top panels) or MdSUT1 in the vector pDR196 (indicated by MdSUT1; bottom panels). Yeast cells of 8 μL were spotted at three concentrations (OD600 = 0.1, 0.05, and 0.01) on synthetic minimal media containing 2% (w/v) Suc or Glc and 20 mg L−1 Trp at pH 5.0 and grown at 30°C for 4 d for observations. The assays were repeated three times with the same results. B, Time course of Suc uptake by SUSY7/ura3 expressing MdSUT1. SUSY7/ura3 yeast transformed with MdSUT1 in pDR196 and empty pDR196 vector were assayed for [14C]Suc uptake at 0.4 mm Suc and pH 5.0. C, Suc uptake kinetics of SUSY7/ura3 expressing MdSUT1. Transport assays were performed at pH 5.0, and uptake rates were plotted against Suc concentrations in the assays. D, Expression of MdSOT6 in the yeast strain RS453 allows growth on sorbitol as the sole carbon source. RS453 yeast cells were transformed with the empty pDR196 vector (indicated by pDR196; top panels) or MdSOT6 in the vector pDR196 (indicated by MdSOT6; bottom panels). Cells of 8 μL were spotted at three concentrations (OD600 = 0.1, 0.05, and 0.01) on synthetic minimal media containing 2% (w/v) sorbitol or Glc and 20 mg L−1 adenine, Leu, Trp, and His at pH 5.0 and grown at 30°C for 4 d for observations. The assays were repeated three times with the same results. E, Time course of sorbitol uptake by RS453 expressing MdSOT6. RS453 yeast transformed with MdSOT6 in pDR196 or empty pDR196 vector were assayed for [3H]sorbitol uptake at 0.25 mm sorbitol and pH 4.5. F, Sorbitol uptake kinetics of RS453 expressing MdSOT6. Transport assays were performed at pH 4.5, and uptake rates were plotted against sorbitol concentrations in the assays. Values in B, C, E, and F are means ± se (n = 3).
Both MdSUT1 and MdSOT6 are expressed in different organs/tissues such as growing shoots, young and mature leaves, developing fruits, and flowers except for drying seeds (Supplemental Fig. S5). Two different sugar transporters reside similarly in both the fruit phloem and parenchyma sink cells and apparently localize to plasma membranes of the cells (Supplemental Figs. S6 and S7; Supplemental Methods S1).
Both MdSUT1 and MdSOT6 Interact with Apple Cytochrome b5 MdCYB5
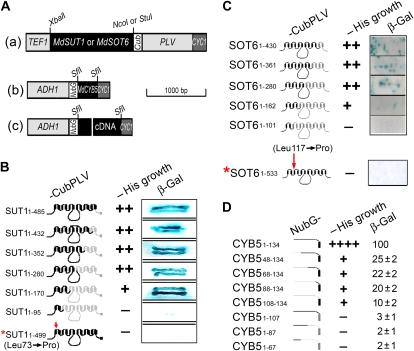
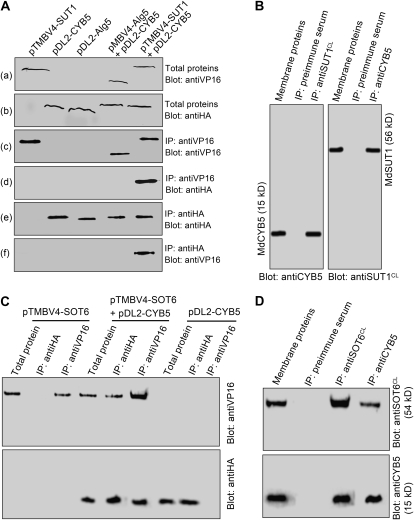
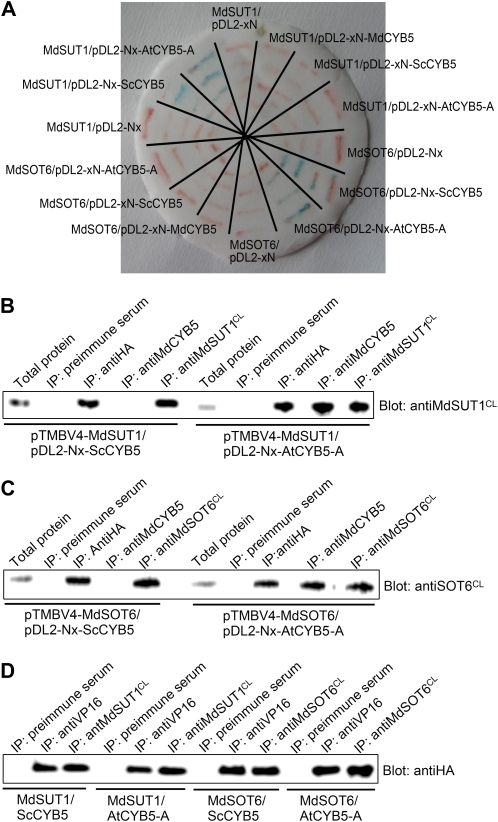
We used a yeast two-hybrid split-ubiquitin system to screen the interaction partners of the two sugar transporters MdSUT1 and MdSOT6 in a cDNA library from apple fruit. The system was tested in the preliminary experiments and shown to be efficient and reliable for the two-hybrid screening (Supplemental Fig. S8). Furthermore, the N-terminal half of the MdSUT1 was found to interact with its C-terminal half (Supplemental Fig. S9, A and B), supporting the view that Suc transporter may interact intracellularly (Reinders et al., 2002) and providing additional proof for the reliability of this experimental system. Using MdSUT1 or MdSOT6 as bait protein (Fig. 2A), 3 × 104 yeast transformants were screened. We confirmed about 500 clones by yeast growth assays and sequenced 100 clones. More than 15 putative interactors were identified in the 100 sequenced clones, including about 20 false-positive clones, in which a putative cytochrome b5 (MdCYB5, for Malus domestica cytochrome b5) was identified in 17 independent clones as putative interactor of MdSUT1 and in 20 independent clones as putative interactor of MdSOT6. The interaction between either of the two transporters and the putative cytochrome b5 MdCYB5 was further confirmed in the assays of the yeast growth and the reporter β-galactosidase (β-gal) activity (Fig. 2, B and C). Consistently, the pull-down assays showed that both MdSUT1 and MdSOT6 were immunocoprecipitated with the MdCYB5 in both the yeast (Fig. 3, A and C) and apple fruit proteins (Fig. 3, B and D), supporting the results from the yeast growth and β-gal assays.
Figure 2.
A putative cytochrome b5 MdCYB5 interacts with the MdSUT1 and MdSOT6 in the yeast split-ubiquitin system. A, Schematic presentation of the constructs for the two-hybrid analysis in the split-ubiquitin system. (a) Construct MdSUT1- or MdSOT6-CubPLV in the pTMBV4 bait vector. The restriction sites XbaI-NcoI were used for MdSUT1 and XbaI-StuI were used for MdSOT6. (b) Construct NubG-MdCYB5 in the pDL2 prey vector. (c) Construct NubG-cDNA in the pDL2 prey vector, where cDNA represents apple fruit cDNA. ADH1, Alcohol dehydrogenase1 promoter; Cub, C terminus of ubiquitin; CYC1, cytochrome c terminator; NubG, the mutated N-terminal (with mutation Ile-13→Gly) subdomain of ubiquitin; PLV, an artificial transcription factor consisting mainly of Lex A and VP16; TEF1, transcription elongation factor1 promoter. Bar = 1,000 bp for length. The restriction sites are indicated above the scheme. See “Materials and Methods” for detailed information. B, Interactions of the different MdSUT1 truncations from the C terminus of MdSUT1 with MdCYB5 (prey construct NubG-CYB5). The numbers 1-n after SUT1 indicate the amino acid numbers from the N-terminal first amino acid to the truncated site (n) in the protein (left panel). The schematic structures of the MdSUT1 (middle-left panel) are presented: the black portions indicate the sequences retained and the gray portions indicate the sequences removed. The middle-right (−His growth) panel indicates the transformant yeast growth in the His-deficient medium, with symbol ++ indicating good status of growth and − indicating no apparent growth of the yeast cells. The right panel (β-Gal) indicates the β-gal activity developed on filter papers. The asterisk indicates the MdSUT1 with a point mutation at Leu-73→Pro. All other symbols or abbreviations are as described in A, and the single + indicates a decreased growth compared with the growth expressed by ++. C, Interactions of the different MdSOT6 truncations from the C terminus of MdSOT6 with MdCYB5 (prey construct NubG-CYB5). The numbers 1-n after SOT6 indicate the amino acid numbers from the N-terminal first amino acid to the truncated site (n) in the protein (left panel). The schematic structures of the MdSOT6 (middle-left panel) are presented: the black portions indicate the sequences retained and the gray portions indicate the sequences removed. The asterisk indicates the MdSOT6 with a point mutation. All other symbols or abbreviations are as described in A and B. D, Interactions of the different truncations of MdCYB5 with MdSOT6 (bait construct SOT6-CubPLV) with MdCYB5. The numbers after CYB5 indicate the amino acid numbers of the beginning and the end of the fragments left after truncation (left panel). The schematic structures of the MdCYB5 (middle-left panel) are presented: the black portions indicate the sequences retained and the gray portions indicate the sequences removed. All other symbols or abbreviations are as described in A to C. β-Gal activity was quantified (right panel; relative units).
Figure 3.
Interaction of MdCYB5 with the MdSUT1 and MdSOT6 identified by pull-down assays. A, MdCYB5 coimmunoprecipitates with MdSUT1 in the yeast split-ubiquitin system. The total proteins were prepared from the yeast cells transformed with different constructs, including the pTMBV4 vector harboring MdSUT1 (pTMBV4-SUT1), the pDL2 vector harboring MdCYB5 (pDL2-CYB5), the pDL2-Alg5 control vector, the pMBV4-Alg5 control vector plus pDL2-CYB5 (pMBV4-Alg5+pDL2-CYB5), and pMBV4-SUT1 plus pDL2-CYB5. The total proteins were immunoblotted with the antiserum against the tag VP16 for MdSUT1 (a) or with the antiserum against the tag HA for CYB5 (b) for two positive controls. The two immunodetected bands migrating as higher molecular mass proteins should be the complex MdSUT1-VP16, and that migrating at low molecular mass should be VP16. The total proteins were precipitated with the anti-VP16 serum (IP: antiVP16), and the immunoprecipitates were blotted with the anti-VP16 serum as the third positive control [Blot: antiVP16 (c)] or anti-HA serum [Blot: antiHA (d)]. The immunodetected bands with higher molecular mass indicate the complex MdSUT1-VP16, and that with lower molecular mass indicates VP16 (c). The immunoprecipitates obtained with the anti-HA serum from the total proteins were blotted with either the anti-HA serum [as the fourth positive control (e)] or the anti-VP16 serum (f). B, MdCYB5 coimmunoprecipitates with MdSUT1 in the membrane proteins from apple. The membrane proteins were isolated from the total proteins and immunoblotted with the anti-MdCYB5 serum (Blot: antiCYB5) or anti-MdSUT1CL serum (Blot: antiSUT1CL) as two positive controls. The membrane proteins were precipitated with anti-MdSUT1CL serum (IP: antiMdSUT1CL), and the immunoprecipitates were blotted with the anti-MdCYB5 serum, which detected a 15-kD MdCYB5 signal (left panel). The immunoprecipitates obtained with the anti-MdCYB5 serum from the membrane proteins were blotted with the anti-MdSUT1CL serum, which detected a 56-kD MdSUT1 signal (right panel). In both cases, the immunoprecipitates obtained with the preimmune serum from the total proteins (IP: preimmune serum) served as negative controls. C, MdCYB5 coimmunoprecipitates with MdSOT6 in the yeast split-ubiquitin system. The total proteins were prepared from the yeast cells transformed with different constructs, including the pTMBV4 vector harboring MdSOT6 (pTMBV4-SOT6), the pDL2 vector harboring MdCYB5 (pDL2-CYB5), and pTMBV4-SOT6 plus pDL2-CYB5 (pTMBV4-SOT6+pDL2-CYB5). The total proteins and the immunoprecipitates isolated from the total proteins with either the anti-HA serum (IP: antiHA) or the anti-VP16 serum (IP: antiVP16) were immunoblotted with the anti-VP16 serum (Blot: antiVP16; top panel) or with the anti-HA serum (Blot: antiHA; bottom panel). D, MdCYB5 coimmunoprecipitates with MdSOT6 in the membrane proteins from apple. The membrane proteins and the immunoprecipitates obtained from the membrane proteins with either the anti-MdSOT6CL serum (IP: anti-MdSOT6CL) or the anti-MdCYB5 serum (IP: antiCYB5) were immunoblotted with the anti-MdSOT6CL serum (Blot: antiSOT6CL; top panel shows a 54-kD immunosignal detected) or anti-MdCYB5 serum (Blot: antiCYB5; the bottom panel shows a 15-kD immunosignal detected). The immunoprecipitates obtained with the preimmune serum from the membrane proteins (IP: preimmune serum) served as negative controls.
We further tested the sequence or key amino acid residues responsible to the transporter-MdCYB5 interactions using a series of truncations of the transporters as bait proteins in the split-ubiquitin system. The results revealed an N-terminal minimum fragment covering the sequence from the first amino acid to amino acid 170 (fragment 1–170), which is indispensable for the interaction between MdSUT1 and MdCYB5 (Fig. 2B), and an N-terminal minimum fragment from amino acid 1 to amino acid 162 (fragment 1–162), which is indispensable for the interaction between MdSOT6 and MdCYB5 (Fig. 2C). The point mutations Leu-73→Pro in the fragment 1–170 of MdSUT1 (data not shown) or in the full-length MdSUT1 (Fig. 2B) and Leu-117→Pro in the fragment 1–162 of MdSOT6 (data not shown) or in the full-length MdSOT6 (Fig. 2C) disrupted the MdSUT1-MdCYB5 and MdSOT6-MdCYB5 interactions, respectively, whereas the mutations replacing other amino acids in these fragments of MdSUT1 or MdSOT6 did not affect the bimolecular interactions (data not shown). These results showed that Leu-73 is an essential amino acid for MdSUT1 and that Leu-117 an essential amino acid for MdSOT6 to interact with MdCYB5. However, the mutation of Leu-73 to Pro in MdSUT1 and Leu-117 to Pro in MdSOT6 are both located in the transmembrane regions. How can the cytosolic domain of CYB5 interact with this site? It is a difficult question to answer at this time. We postulate that the mutation may induce significant conformational changes in the sugar transporters or alter the transmembrane status. Alternatively, the sugar transporter-CYB5 interactions may need some extracellular stimuli perceived by the mutation-covered domains of the sugar transporters. The mutations may affect the interactions of the sugar transporters with their interaction partners in such an indirect way.
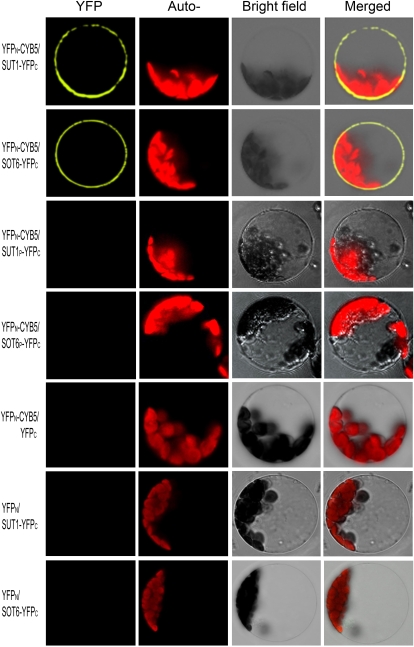
The interactions of both transporters with MdCYB5 were further tested in vivo with the bimolecular fluorescence complementation (BiFC) assay. The fluorescence from the yellow fluorescent protein (YFP) in the transgenic onion (Allium cepa) epidermis and Arabidopsis protoplasts was observed in the plasma membrane region but not in the cytoplasm (Fig. 4; Supplemental Fig. S10), where the MdCYB5 protein resides in the endoplasmic reticulum (ER; see below). The point mutations Leu-73→Pro in MdSUT1 and Leu-117→Pro in MdSOT6 disrupted the MdSUT1-MdCYB5 and MdSOT6-MdCYB5 interactions, respectively, in the BiFC assays (Fig. 4; Supplemental Fig. S10), which provides additional in vivo evidence for the essential roles of Leu-73 for MdSUT1 and Leu-117 for MdSOT6 in the bimolecular interactions. The negative controls, such as the deletion of MdSUT1 or MdSOT6 from the C-terminal half of YFP (YFPC) or deletion of MdCYB5 from the N-terminal half of YFP (YFPN), disrupted the fluorescence (Fig. 4), showing the specificity of the BiFC assays. These data indicate clearly that, on the one hand, MdCYB5 is an interaction partner of the two transporters in living cells, and on the other hand, the bimolecular interaction takes place on the MdSUT1/MdSOT6 transporter-colonized plasma membranes when the moving ER-localized MdCYB5 touches the membranes.
Figure 4.
In vivo interaction of MdCYB5 with MdSUT1 and MdSOT6 in the BiFC system. The laser-scanning confocal microscopy images show fluorescence (indicated by YFP) and merged images of the double transgenic cells with the YFPN-MdCYB5 and MdSUT1-YFPC fusions (YFPN-CYB5/SUT1-YFPC) or with the YFPN-CYB5/SOT6-YFPC construct pair. The wild-type MdSUT1 and MdSOT6 were replaced in the above-mentioned constructs by the mutated MdSUT1P (Leu-73→Pro; YFPN-CYB5/SUT1P-YFPC) and mutated MdSOT6P (Leu-117→Pro; YFPN-CYB5/SOT6P-YFPC), respectively, which were used to transform cells that served as negative controls. The construct pairs YFPN-CYB5/YFPC, YFPN/SUT1-YFPC, and YFPN/SOT6-YFPC were also used as negative controls to transform the protoplasts. The chlorophyll autofluorescence (Auto-) and the bright-field images are also presented.
To test the specificity of the interaction of the MdCYB5 with plant sugar transporters, we assayed the possible interaction of MdCYB5 with three sugar transporters of Arabidopsis. A Suc transporter, AtSUC7, and a polyol transporter, AtPLT5, interacted with MdCYB5, while a hexose transporter, AtSTP4, did not (Supplemental Fig. S11; Supplemental Methods S2), suggesting that MdCYB5 is able to interact with many members of Suc and polyol transporters but not with hexose transporters.
MdSUT1 and MdSOT6 Colocalize with MdCYB5 in Tissues and Cells
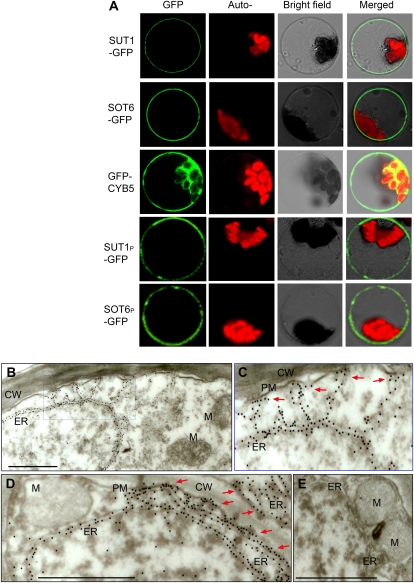
The MdCYB5 was shown to express in different organs/tissues with a similar profile to that of MdSUT1 and MdSOT6 (Supplemental Figs. S5–S7 and S12). The subcellular localization assays of the two transporters and MdCYB5 protein were performed with transient expression in both the onion epidermis and Arabidopsis protoplasts and using GFP as a reporter. The results showed clearly that MdSUT1 and MdSOT6 both localize to plasma membrane (Fig. 5A; Supplemental Fig. S13), which is consistent with the results from the immunochemical assays (Supplemental Figs. S6 and S7). The plasma membrane localization of MdSUT1 is different from that of its homolog AtSUT4 in Arabidopsis, which was previously shown to localize to vacuolar membrane (Endler et al., 2006; Reinders et al., 2008). We confirmed this different localization between MdSUT1 and AtSUT4 with the transient expression system in onion epidermis (Supplemental Fig. S13; Supplemental Methods S3). The MdCYB5 protein apparently resides in cytoplasm, except for the chloroplast (Fig. 5A; Supplemental Fig. S13), and the immunochemical assays consistently showed similar localization (Supplemental Fig. S12). It has been reported that the CYB5s localize generally to ER and/or mitochondrial outer membrane in mammalian, yeast, and plant cells (Borgese et al., 1993, 2003; Zhao et al., 2003; Hwang et al., 2004; Maggio et al., 2007) and could also distribute on chloroplast outer membranes in plant cells (Maggio et al., 2007). So we performed further assays of MdCYB5 localization with immunogold labeling, and the results revealed clearly that MdCYB5 localizes to ER (Fig. 5, B–E), which can apparently approach plasma membrane (Fig. 5, C and D). Therefore, the localization of MdCYB5 apparently in cytoplasm and partly around plasma membrane, which was visualized in the transient expression cells (Fig. 5A) and in the immunolabeled tissues (Supplemental Fig. S12), should be the ER-localized MdCYB5 molecules that approached the plasma membrane (Fig. 5, C and D). This intracellular trafficking should be important to facilitate the interactions of MdCYB5 with plasma membrane proteins such as the sugar transporters. Taken together, these results showed that, like their similar expression profiles in different tissues, the three proteins MdSUT1, MdSOT6, and MdCYB5 apparently colocalize in cells, which supports the transporter-MdCYB5 bimolecular interactions in the same compartment of cells.
Figure 5.
Localization of MdSUT1, MdSOT6, and MdCYB5 in cells. A, Transient expression of MdSUT1-GFP (and its mutated form MdSUT1P), MdSOT6-GFP (and its mutated form MdSOT6P), and MdCYB5-GFP fusion proteins in the Arabidopsis protoplasts. The laser-scanning confocal microscopy images show the MdSUT1 (indicated by SUT1-GFP), MdSOT6 (SOT6-GFP), GFP-MdCYB5 (GFP-CYB5), MdSUT1P (SUT1P-GFP), and MdSOT6P (SOT6P-GFP) fluorescence (GFP) and the merged images. The chlorophyll autofluorescence (Auto-) and the bright-field images are also presented. Note that the mutation of MdSUT1P did not change the plasma membrane localization of MdSUT1, and the mutation of MdSOT6P did not change the plasma membrane localization of MdSOT6. B to E, Immunogold labeling of MdCYB5 in apple fruit cells. The protein reacting with anti-MdCYB5N (visualized by immunogold particles) mainly resides on the ER (B and D). A blowup (C) of the area from B shows more clearly the ER localization of MdCYB5; note that some of the ER hosting MdCYB5 immunoparticles distribute near the plasma membrane (PM), which are shown by arrows. D shows more clearly the distribution of MdCYB5-hosting ER around the plasma membrane (indicated by arrows). No substantial signal was detected in other cellular compartments (B–D) such as the cell wall (CW), mitochondrion (M), and cytoplasm. No substantial gold particles were found in the control cells without antiserum treatment (E). Bars = 0.5 μm.
MdSUT1 and MdSOT6 Interact Also with the Arabidopsis CYB5-A and Yeast CYB5
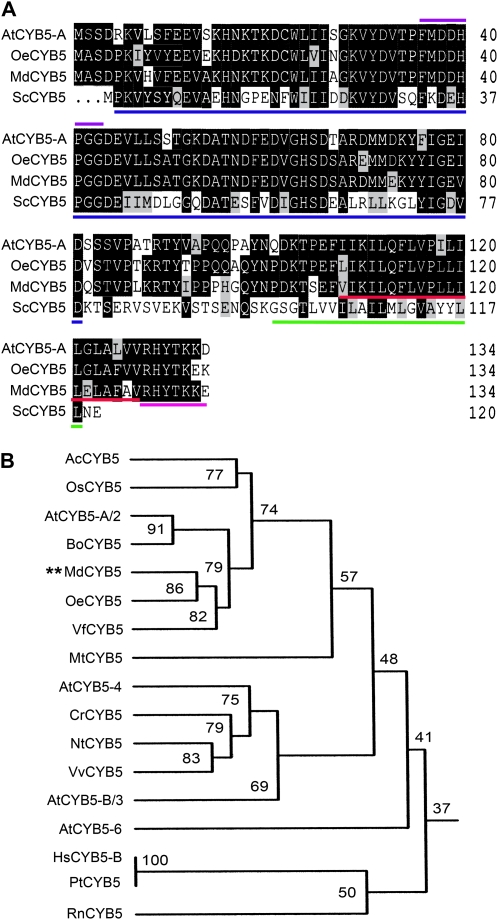
CYB5s are a class of small-molecular-mass (about 15 kD), tail-anchored proteins that are structurally characterized by an N-terminal, globular cytosolic heme-binding domain, a short connecting region, and a hydrophobic anchoring domain followed by seven polar residues at the extreme C terminus (Borgese et al., 1993, 2003). The apple MdCYB5 has all of these structural features as, for example, two of its closest homologs from Arabidopsis (AtCYB5-A) and olive (OeCYB5; Fig. 6). To assess whether other members of the CYB5s also interact with the sugar transporters, we assayed the possible interactions of the transporters with AtCYB5-A (also called AtCYB5-2), one of the closest homologs of MdCYB5 from Arabidopsis (Fig. 6), and a yeast (Saccharomyces cerevisiae) CYB5 (ScCYB5) that has low similarity of amino acid sequence with MdCYB5 (Fig. 6A). In the yeast growth (visualized by β-gal activity on filter; Fig. 7A) and pull-down (Fig. 7, B–D) assays, both sugar transporters were shown to interact with the Arabidopsis AtCYB5-A. Surprisingly, the two transporters interact also with the yeast ScCYB5 (Fig. 7), suggesting that cytochrome b5 may be a general partner of physical interaction with the sugar transporters.
Figure 6.
Alignment of MdCYB5 and its closed homologs, and homology of plant cytochrome b5s. A, Deduced amino acid sequence alignment of AtCYB5, OeCYB5, MdCYB5, and ScCYB5, showing the presence of conserved features and including the predicted heme-binding domain (underlined in blue), heme-binding signature (violet line above), C-terminal transmembrane domain (underlined in red), and C-terminal polar sequence (underlined in pink) among the three plant CYB5s (AtCYB5-A, OeCYB5, and MdCYB5). The C-terminal transmembrane domain of the yeast ScCYB5 is underlined in green. Numbers at right indicate numbers of amino acid residues in the predicted sequences. Gaps, indicated by dots, were induced to maximize alignment. Identical amino acid residues between two or more CYB5s are indicated by white letters on a black background, and similar residues are indicated by black letters on a gray background. B, Homology tree of plant cytochrome b5. The prefixes of the CYB5 names represent the following plant species: Ac, Anana comosus; At, Arabidopsis thaliana; Bo, Brassica oleracea; Cr, Cuscuta reflexa; Hs, Homo sapiens; Md, Malus domestica; Mt, Medicago truncatula; Nt, Nicotiana tabacum; Os, Oryza sativa; Pt, Pan troglodytes; Rn, Rattus norvegicus; Vf, Vernicia fordii; Vv, Vitis vinifera. Percentages of amino acid sequence identity are indicated. The corresponding accession numbers of these cytochrome b5s are given at the end of “Materials and Methods.”
Figure 7.
Both the Arabidopsis CYB5-A (AtCYB5-A) and yeast CYB5 (ScCYB5) interact with MdSUT1 and MdSOT6. A, Interactions of AtCYB5-A and ScCYB5 with MdSUT1 and MdSOT6 tested by β-gal activity through filter assays in the yeast split-ubiquitin system. The MdSUT1 and MdSOT6 were recombined into the bait vector pTMBV4, and the CYB5s, including MdCYB5, AtCYB5-A, and ScCYB5, were recombined into the prey vector pDL-Nx or pDL-xN (where x indicates the CYB5 cDNAs and N denotes the mutated N terminus of the subdomain of ubiquitin). The yeast cells were cotransformed with the construct pairs mentioned (with slashes between the names of two constructs). The vectors pDL2-Nx and pDL2-xN containing no CYB5s were used as negative controls (where x indicates the control Alg5 cDNA). The streaks stained blue indicate the interactions of MdSUT1 with AtCYB5-A (MdSUT1/pDL2-Nx-AtCYB5-A) and with ScCYB5 (MdSUT1/pDL2-Nx-ScCYB5) and those of MdSOT6 with AtCYB5-A (MdSOT6/pDL2-Nx-AtCYB5-A) and with ScCYB5 (MdSOT6/pDL2-Nx-ScCYB5). The yeast cells transformed with the other constructs showed no blue signal. B to D, AtCYB5-A and ScCYB5 coimmunoprecipitate with both MdSUT1 and MdSOT6 in the yeast split-ubiquitin system. The total proteins were prepared from the yeast cells that had been transformed with the construct pairs pTMBV4-MdSUT1/pDL2-Nx-AtCYB5-A (MdSUT1/AtCYB5-A in D), pTMBV4-MdSUT1/pDL2-Nx-ScCYB5 (MdSUT1/ScCYB5 in D), pTMBV4-MdSOT6/pDL2-Nx-AtCYB5-A (MdSOT6/AtCYB5-A in D), and pTMBV4-MdSOT6/pDL2-Nx-ScCYB5 (MdSOT6/ScCYB5 in D). The immunoprecipitates were isolated from the total proteins with the anti-VP16 serum (IP: antiVP16), anti-HA serum (IP: antiHA), anti-MdCYB5 serum (IP: antiCYB5), anti-MdSUT1CL serum (IP: antiMdSUT1CL), anti-MdSOT6CL serum (IP: antiMdSOT6CL), and the preimmune serum (IP: preimmune serum; as a negative control). The total proteins and immunoprecipitates were immunoblotted with the anti-MdSUT1CL serum (Blot: anti-MdSUT1CL; B), anti-MdSOT6CL serum (Blot: anti-MdSOT6CL; C), and anti-HA serum (Blot: antiHA; D).
Cytosolic N Terminus Leads Cytochrome b5s to Interact with the Sugar Transporters, and the C-Terminal Anchor Tail of Cytochrome b5s Is Also Critical for the Interactions
It is necessary to explore the sites in the CYB5s allowing these tail-anchored proteins to interact with the plasma membrane-localized sugar transporters. It has been known clearly that the tail-anchored proteins such as cytochrome b5 are inserted into the phospholipid bilayer membranes by a hydrophobic segment close to the C terminus and have their N-terminal functional domains exposed to the cytosolic face of membranes (Borgese et al., 2003). Using a series of truncations of the MdCYB5 protein as prey proteins to perform the two-hybrid assays in the split-ubiquitin system, we showed that the interactions (estimated by β-gal activities) of MdCYB5 with MdSOT6 (Fig. 2D) and MdSUT1 (data not shown) were largely reduced by the deletions of a fragment of 48 amino acids at the N terminus of MdCYB5, indicating that the extreme N-terminal sequence or the intact N-terminal functional domains are important for the interactions. However, an intact N-terminal portion but with the deletion of the C-terminal hydrophobic region of a minimum fragment of 27 amino acids that allows CYB5 to localize to membranes abolished the interactions (Fig. 2D; data not shown), indicating that the extreme C terminus determining a correct membrane localization of CYB5s is as important as its N terminus for its interaction with the transporters. All of these results showed that the maximum interactions between the transporters and MdCYB5 need a full-length MdCYB5 with both the intact N-terminal functional domains and C-terminal subcellular localization domain.
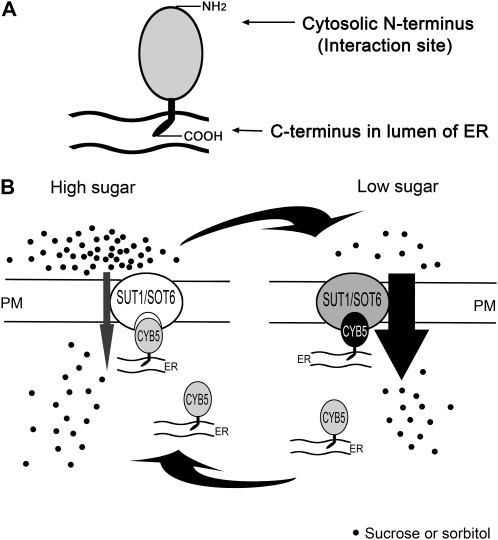
Based on these findings, we proposed a model for the transporter-CYB5 interaction where the cytosolic N terminus of CYB5s, due to their subcellular localization, has a unique opportunity to approach and interact with the plasma membrane-associated sugar transporters, whereas the C terminus, inserted into the endomembranes such as in ER, mitochondrion, or chloroplast, spatially cannot meet the sugar transporters but is indispensable in this interaction event because the features of the tail anchoring the CYB5s to the bilayer membranes are critical for the bimolecular interactions (see Fig. 11 below). We further tested this model in the split ubiquitin system. The CYB5s (MdCYB5, AtCYB5-A, and ScCYB5) were linked to the mutated N terminus of subdomain of ubiquitin (NubG) with the N terminus of CYB5s to form the constructs NubG-CYB5s or with the C terminus of CYB5s to form the constructs CYB5s-NubG (Fig. 7A; Table I). The NubG-CYB5s-transgenic yeast cells displayed the transporter-CYB5 interactions, shown by positive β-gal activities (Fig. 7A) and yeast growth in the medium deficient in Leu, Trp, and His nutrients (Table I), which is consistent with all the results for the transporter-MdCYB5 interactions where the NubG-MdCYB5 constructs were used (Figs. 2 and 3). Like the negative controls, however, the CYB5s-NubG-transgenic yeast cells showed no such bimolecular interactions (Fig. 7A; Table I). These data indeed provide further supporting evidence for the above-mentioned model, showing that the NubG-linked cytosolic N terminus of CYB5s targeted the CYB5s directly on the transporters and the NubG-linked C terminus could not allow the CYB5s to interact with the transporters, probably because the C terminus-linked NubG is hidden in the lumens of their anchoring organelles or, otherwise, that the C terminal-linked NubG changed subcellular localization of the CYB5s by disturbing the C-terminal tail for anchoring the CYB5s correctly to their host organelles.
Figure 11.
A model of sugar sensing through the MdSUT1-MdCYB5 or MdSOT6-MdCYB5 complex. A, A scheme showing the localization of MdCYB5 to ER. The C-terminal tail is anchored to ER membrane, with the extreme C terminus displayed in the lumen of the ER. The N terminus is present in cytosol and functions to interact with the plasma membrane (PM)-localized sugar transporters MdSUT1 and MdSOT6. B, MdSUT1 (SUT1) or MdSOT6 (SOT6) interacts with MdCYB5 (CYB5) to form an MdSUT1-MdCYB5 or MdSOT6-MdCYB5 bimolecular complex in response to low Suc or sorbitol supply, which enhances the affinity of MdSUT1 or MdSOT6 transporter to its substrate sugar, stimulating sugar uptake by cells to maintain a relatively stable internal level of the limiting sugar. The intensity of the transporter-MdCYB5 interactions and the affinity of the transporter return to normal levels in higher sugar supply.
Table I.
Growth status of the yeast cells expressing different constructs
Yeast cells were cotransformed with the vector pairs harboring the bait construct (pTMBV4-MdSUT1 or MdSOT6) and prey construct (pDL2-Nx- or pDL2-xN-AtCYB5-A, MdCYB5 or ScCYB5, with empty pDL2-Nx or pDL2xN as negative controls; x indicates the CYB5 cDNAs or the negative control Alg5 cDNA in the empty vector, and N denotes the mutated N terminus of the subdomain of ubiquitin). The transgenic yeast cells were grown in the common SD medium deficient in Leu and Trp nutrients (SD-Leu-Trp) and then grown further in Leu, Trp, and His (SD-Leu-Trp-His, with the addition of 5 mm 3-amino-1,2,4-triazole to inhibit growth due to self-activation). The yeast growth status was recorded 3 d after inoculation. ++ indicates good growth status, and − indicates no growth. The assays were repeated biologically three times with the same results.
| Bait Construct | Prey Construct | Yeast Growth in SD-Leu-Trp | Yeast Growth in SD-Leu-Trp-His (with 5 mm 3-Amino-1,2,4-Triazole) |
|---|---|---|---|
| pTMBV4-MdSOT6 | pDL2-Nx | ++ | − |
| pTMBV4-MdSOT6 | pDL2-Nx-MdCYB5 | ++ | ++ |
| pTMBV4-MdSOT6 | pDL2-Nx-ScCYB5 | ++ | ++ |
| pTMBV4-MdSOT6 | pDL2-Nx-AtCYB5-A | ++ | ++ |
| pTMBV4-MdSOT6 | pDL2-xN | ++ | − |
| pTMBV4-MdSOT6 | pDL2-xN-MdCYB5 | ++ | − |
| pTMBV4-MdSOT6 | pDL2-xN-ScCYB5 | ++ | − |
| pTMBV4-MdSOT6 | pDL2-xN-AtCYB5-A | ++ | − |
| pTMBV4-MdSUT1 | pDL2-Nx | ++ | − |
| pTMBV4-MdSUT1 | pDL2-Nx-MdCYB5 | ++ | ++ |
| pTMBV4-MdSUT1 | pDL2-Nx-ScCYB5 | ++ | ++ |
| pTMBV4-MdSUT1 | pDL2-Nx-AtCYB5-A | ++ | ++ |
| pTMBV4-MdSUT1 | pDL2-xN | ++ | − |
| pTMBV4-MdSUT1 | pDL2-xN-MdCYB5 | ++ | − |
| pTMBV4-MdSUT1 | pDL2-xN-ScCYB5 | ++ | − |
| pTMBV4-MdSUT1 | pDL2-xN-AtCYB5-A | ++ | − |
Interactions of MdSUT1 and MdSOT6 Transporters with Two Plant Cytochrome b5s, But Not Yeast Cytochrome b5, Are Stimulated by Low Sugar Supply
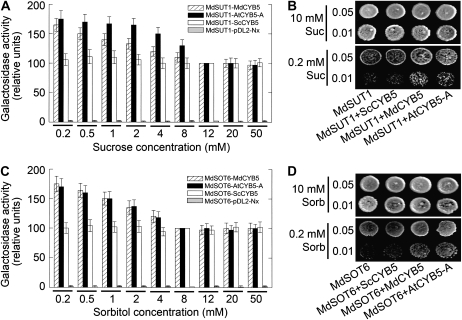
To explore the possible biological significance of the sugar transporter-CYB5 interactions, we assayed the effects of the substrate sugar concentrations on the interactions. In the split-ubiquitin two-hybrid system, we found that the low Suc concentrations (≤8 mm) stimulated the interactions (estimated by β-gal activities) of the transporter MdSUT1 with the two plant CYB5s (AtCYB5-A and MdCYB5; Fig. 8A) and the low sorbitol concentrations (≤4 mm) showed the same stimulating effects on the MdSOT6-CYB5 interactions (Fig. 8C). The stimulating effects were apparently negatively correlated with the substrate sugar concentrations in the range of the assayed low levels (Fig. 8, A and C). However, the interactions of the transporters with the yeast ScCYB5 were not found to be affected in the low sugar concentrations (Fig. 8, A and C). The stimulating effects disappeared when replacing Suc by sorbitol in the MdSUT1-CYB5 interactions (Supplemental Fig. S14A) or replacing sorbitol by Suc in the MdSOT6-CYB5 interactions (Supplemental Fig. S14B), indicating that the effects of the substrate sugars were specific. Further assays with the same transgenic yeast cells showed that the yeast cells cotransformed with either of the two transporters and either AtCYB5-A or MdCYB5 could substantially grow in the low level of Suc (0.2 mm Suc for the MdSUT1 transgenic cells; Fig. 8B) or in low level of sorbitol (0.2 mm sorbitol for the MdSOT6 transgenic cells; Fig. 8D), but the yeast cells cotransformed with any of the two transporters and the yeast ScCYB5 showed significantly much lower tolerance to such sugar starvation (Fig. 8, B and D). These results suggest a possible involvement of the transporter-CYB5 interactions in the sugar transport regulation through a mechanism responsive to low sugar availability in cells.
Figure 8.
Low sugar supply stimulates the interactions of MdSUT1 and MdSOT6 with MdCYB5 and AtCYB5-A, and the interactions promote yeast growth in sugar deficiency. A, Low concentrations of Suc stimulate the interactions of MdSUT1 with MdCYB5 and AtCYB5-A (but not ScCYB5). The DSY-1 yeast cells were cotransformed with the vector pairs pTMBV4-MdSUT1 and pDL2-Nx-AtCYB5-A (MdSUT1-AtCYB5-A), pTMBV4-MdSUT1 and pDL2-Nx-MdCYB5 (MdSUT1-MdCYB5), pTMBV4-MdSUT1 and pDL2-Nx-ScCYB5 (MdSUT1-ScCYB5), and pTMBV4-MdSUT1 and pDL2-Nx empty vector (MdSUT1-pDL2-Nx; as a control). The transgenic cells were grown in liquid dropout SD medium lacking Trp and Leu, supplemented with 10 mm Glc and 0.2, 0.5, 1, 2, 4, 8, 12, 20, and 50 mm Suc, respectively, instead of 2% Glc (about 50 mm) supplementation for the common SD medium, and β-gal activities were determined in the lysates of the cells. Relative β-gal activities, normalized relative to the corresponding values obtained from the cells grown at 12 mm Suc, were compared within the same yeast line transformed with the same construct pair. Note that the values of the controls (MdSUT1-pDL2-Nx) were scarcely detectable. The experiments were repeated biologically five times. Values are means ± se (n = 5). B, The interactions of MdSUT1 with MdCYB5 and AtCYB5-A, but not ScCYB5, enhance the yeast growth at low Suc supply. The DSY-1 yeast cells were cotransformed with the vector pairs as described in A. The symbols of the vectors are also the same as in A except for the vector pair pTMBV4-MdSUT1 and pDL2-Nx empty vector, which is called MdSUT1 here. Drop tests were used to observe the transformed yeast growth on low Suc concentration. Cells of 8 μL were spotted at two concentrations (OD600 = 0.01 and 0.05) on the media supplemented with 10 or 0.2 mm Suc as sole carbon source and grown at 30°C for 4 d for observations. The assays were repeated three times with the same results. C, Low concentrations of sorbitol stimulate the interactions of MdSOT6 with MdCYB5 and AtCYB5-A (but not ScCYB5). The DSY-1 yeast cells were cotransformed with the vector pairs pTMBV4-MdSOT6 and pDL2-Nx-AtCYB5-A (MdSOT6-AtCYB5-A), pTMBV4-MdSOT6 and pDL2-Nx-MdCYB5 (MdSOT6-MdCYB5), pTMBV4-MdSOT6 and pDL2-Nx-ScCYB5 (MdSOT6-ScCYB5), and pTMBV4-MdSOT6 and pDL2-Nx empty vector (MdSOT6-pDL2-Nx; as a control). The transgenic cells were grown as described in A but with 0.2, 0.5, 1, 2, 4, 8, 12, 20, and 50 mm sorbitol (instead of Suc) in the SD medium, and β-gal activities were determined in the lysates of the cells. Relative β-gal activities, normalized relative to the corresponding values obtained from the cells grown at 8 mm sorbitol, were compared within the same yeast line transformed with the same construct pair. Note that the values of the controls (MdSOT6-pDL2-Nx) were scarcely detectable. The experiments were repeated biologically five times. Values are means ± se (n = 5). D, The interactions of MdSOT6 with MdCYB5 and AtCYB5-A, but not ScCYB5, enhance the yeast growth at low sorbitol supply. The DSY-1 yeast cells were cotransformed with the vector pairs as described in C. The symbols of the vectors are also the same as in C except for the vector pair pTMBV4-MdSOT6 and pDL2-Nx empty vector, which is called MdSOT6 here. Drop tests were conducted as described in B at 10 or 0.2 mm sorbitol as sole carbon source in SD medium. The assays were repeated three times with the same results.
Interactions of MdSUT1 and MdSOT6 Transporters with Two Plant Cytochrome b5s Promote Both the Affinity of the Transporters to Their Substrates and Tolerance of Cells to Sugar Starvation
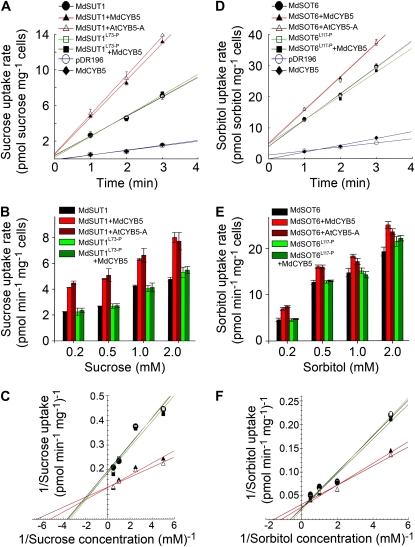
We further observed, using the yeast strains defective in Suc (the SUSY7/ura3 strain) or in sorbitol (the RS453 strain) uptake, that the cotransformation of the yeast cells with the transporter MdSUT1 and either AtCYB5-A or MdCYB5 significantly enhanced the affinity of MdSUT1 to Suc in comparison with the transformation of the yeast cells only with MdSUT1 (Fig. 9, A–C; Table II; Supplemental Fig. S15A). The cotransformation of the yeast cells with MdSOT6 and either of the plant CYB5s had the same stimulating effects on the affinity of MdSOT6 to sorbitol in comparison with the transformation of the yeast cells only with MdSOT6 (Fig. 9, D–F; Table II; Supplemental Fig. S15B). The mutations Leu-73→Pro in MdSUT1 and Leu-117→Pro in MdSOT6, which disrupt, respectively, the MdSUT1-CYB5 and MdSOT6-CYB5 interactions (Figs. 2, B and C, and 4) without changing the subcellular localization of the sugar transporters (Fig. 5A), did not significantly affect the transport activities of the MdSUT1 and MdSOT6 transporters (Fig. 9; Table II; Supplemental Fig. S15). However, these point mutations in both transporters abolished the stimulating effects of combining the transporters with the plant CYB5s on the affinity of the transporters to their substrate sugars (Fig. 9; Table II; Supplemental Fig. S15). This reveals clearly that the interactions of the transporters with the CYB5s are responsible for the stimulating effects on the affinity of the transporters.
Figure 9.
Expression of MdCYB5 and AtCYB5-A enhances the affinity of MdSUT1 and MdSOT6 to their substrate sugars. A to C, Expression of MdCYB5 and AtCYB5-A enhances the affinity of MdSUT1 to Suc. SUSY7/ura3 yeast cells were transformed with the pDR196 vector harboring MdSUT1 (indicated by MdSUT1), mutated MdSUT1 at Leu-73→Pro (MdSUT1L73-P), and MdCYB5 (as a negative control) or with the empty pDR196 vector (as another negative control). The yeast cells were also cotransformed with the pDR196 vector pairs pDR196-MdSUT1 plus pDR196-MdCYB5 (MdSUT1+MdCYB5), pDR196-MdSUT1 plus pDR196-AtCYB5-A (MdSUT1+AtCYB5-A), and pDR196-MdSUT1L73-P plus pDR196-MdCYB5 (MdSUT1L73-P+MdCYB5). The time course of Suc uptake by the transgenic yeast cells was assayed for [14C]Suc uptake at 0.4 mm Suc and pH 5.0 (A). Suc uptake kinetics of the transgenic yeast cells were assayed at pH 5.0 at low Suc concentrations ranging from 0.2 to 2 mm (B). The Suc uptake rates were plotted against the Suc concentrations (C) according to the values presented in B. A linear regression fit of the Michaelis-Menten equation was obtained for each yeast line expressing the mutated MdSUT1L73-P or wild-type MdSUT1 and CYB5s. The Suc uptake kinetics parameters for these transgenic yeast lines are presented in Table II. Symbols in C are as described in A. D to F, Expression of MdCYB5 and AtCYB5-A enhances the affinity of MdSOT6 to sorbitol. RS453 yeast cells were transformed with the pDR196 vector harboring MdSOT6 (indicated by MdSOT6), mutated MdSOT6 at Leu-117→Pro (MdSOT6L117-P), and MdCYB5 (as a negative control) or with the empty pDR196 vector (as another negative control). The yeast cells were also cotransformed with the pDR196 vector pairs pDR196-MdSOT6 plus pDR196-MdCYB5 (MdSOT6+MdCYB5), pDR196-MdSOT6 plus pDR196-AtCYB5-A (MdSOT6+AtCYB5-A), and pDR196-MdSOT6L117-P plus pDR196-MdCYB5 (MdSOT6L117-P+MdCYB5). The time course of sorbitol uptake by the transgenic yeast cells was assayed for [3H]sorbitol uptake at 0.25 mm sorbitol and pH 4.5 (D). Sorbitol uptake kinetics of the transgenic yeast cells were assayed at pH 4.5 at low sorbitol concentrations ranging from 0.2 to 2 mm (E). The sorbitol uptake rates were plotted against the sorbitol concentrations (F) according to the values presented in E. A linear regression fit of the Michaelis-Menten equation was obtained for each yeast line expressing the mutated MdSOT6L117-P or wild-type MdSOT6 and CYB5s. The sorbitol uptake kinetics parameters for these transgenic yeast lines are presented in Table II. Symbols in F are as described in D.
Table II.
Comparison of affinity (Km) to substrate sugars of the yeast cells expressing different constructs
The experiments were repeated biologically five times with substantially the same results. The listed equations are obtained with the means of the five groups of data. The values of Km are calculated according to the corresponding listed equations, which are substantially the same as the means from the five different equations corresponding, respectively, to the five repetitions. The values of se were calculated according to the five different equations obtained from each of the five repetitions. Values marked with the same letter (a, b, or c) indicate statistically no significant difference between/among them. The relative units (%) are also calculated relative to the corresponding values obtained from the cells expressing MdSUT1alone (for all of the yeast lines expressing the mutated MdSUT1L73-P at Leu-73→Pro or wild-type MdSUT1) or MdSOT6 alone (for all of the yeast lines coexpressing the mutated MdSOT6L117-P at Leu-117→Pro or wild-type MdSOT6).
| cDNA Expressed | Equation (r2) | Km | |
|---|---|---|---|
| mm (means ± se) | % (calculated from the means) | ||
| MdSUT1 | y = 0.0535x + 0.1955 (0.93) | 0.27 ± 0.03 a | 100 |
| MdSUT1-MdCYB5 | y = 0.0247x + 0.1297 (0.92) | 0.19 ± 0.02 b | 70 |
| MdSUT1-AtCYB5-A | y = 0.0202x + 0.1294 (0.92) | 0.16 ± 0.02 b | 59 |
| MdSUT1L73-P | y = 0.0556x + 0.1883 (0.92) | 0.30 ± 0.04 a | 111 |
| MdSUT1L73-P-MdCYB5 | y = 0.0522x + 0.1864 (0.90) | 0.28 ± 0.03 a | 104 |
| MdSOT6 | y = 0.0379x + 0.0242 (0.97) | 1.57 ± 0.20 a | 100 |
| MdSOT6-MdCYB5 | y = 0.0229x + 0.0264 (0.98) | 0.87 ± 0.08 b | 55 |
| MdSOT6-AtCYB5-A | y = 0.0201x + 0.032 (0.97) | 0.63 ± 0.06 c | 40 |
| MdSOT6L117-P | y = 0.0393x + 0.0201 (0.97) | 1.95 ± 0.24 a | 124 |
| MdSOT6L117-P-MdCYB5 | y = 0.0365x + 0.0233 (0.97) | 1.56 ± 0.17 a | 99 |
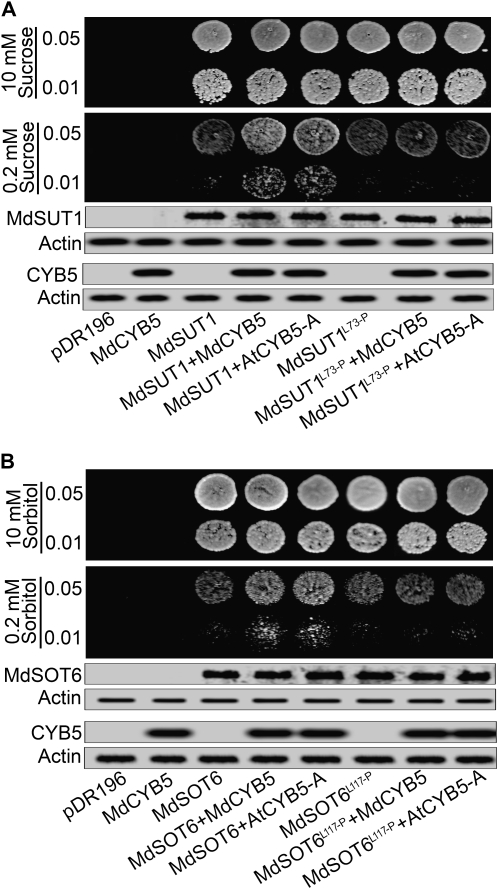
Using the same transgenic SUSY7/ura3 and RS453 yeast cells as used above, we performed the yeast growth assays and found that the yeast cells cotransformed with the transporter MdSUT1 and either AtCYB5-A or MdCYB5 could substantially grow in the low Suc supply (0.2 mm) and that the yeast cells cotransformed with MdSOT6 and either AtCYB5-A or MdCYB5 showed the same phenotypes tolerating low sorbitol level (0.2 mm; Fig. 10). However, the yeast cells transformed only with MdSUT1 or MdSOT6 showed a much lower ability to tolerate such Suc or sorbitol deficiencies (Fig. 10). The mutations of Leu-73→Pro in MdSUT1 and Leu-117→Pro in MdSOT6 did not change the status of the yeast growth in the low sugar supply in comparison with the wild-type MdSUT1 and MdSOT6, respectively (Fig. 10). However, the cotransformation of the mutated MdSUT1 or MdSOT6 with either of the plant CYB5s annulled the tolerating effects for the yeast growth to sugar deficiency (0.2 mm Suc or sorbitol; Fig. 10) in comparison with the MdSUT1- or MdSOT6-AtCYB5/MdCYB5 yeast cotransformations (Fig. 10). These results, consistent with those mentioned above in assaying transporter affinities (Fig. 9; Supplemental Fig. S15), reveal the roles of regulation of the transporters in their transport activities by the bimolecular interactions especially under sugar starvation. It is noteworthy that the protein amounts of MdSUT1 and MdSOT6 were not changed by expressing the various CYB5-containing constructs (Fig. 10), suggesting that the stimulating effects of the bimolecular interactions on the transporters may not be attributed to an increase in the transporter numbers but rather to a posttranslational regulation of these transporters.
Figure 10.
Coexpression of MdSUT1 or MdSOT6 with MdCYB5 or AtCYB5-A promotes yeast growth in sugar deficiency. A, Coexpression of MdSUT1 with MdCYB5 or AtCYB5-A in the SUSY7/ura3 yeast cells. Yeast cells were transformed with the pDR196 vector harboring MdSUT1 (indicated by MdSUT1), mutated MdSUT1 at Leu-73→Pro (MdSUT1L73-P), and MdCYB5 (as a negative control) or with the empty pDR196 vector (pDR196, as another negative control). The yeast cells were also cotransformed with the pDR196 vector pairs pDR196-MdSUT1 plus pDR196-MdCYB5 (MdSUT1+MdCYB5), pDR196-MdSUT1 plus pDR196-AtCYB5-A (MdSUT1+AtCYB5-A), pDR196-MdSUT1L73-P plus pDR196-MdCYB5 (MdSUT1L73-P+MdCYB5), and pDR196-MdSUT1L73-P plus pDR196-AtCYB5-A (MdSUT1L73-P+AtCYB5-A). The expression of MdSUT1 was tested with immunoblotting using anti-MdSUT1CL serum, and the expression of MdCYB5 and AtCYB5-A was tested using anti-MdCYB5N serum in the yeast lysates. Actin was used as a loading control. Drop tests were used to observe the transformed yeast growth on low Suc concentration. Cells of 8 μL were spotted at two concentrations (OD600 = 0.01 and 0.05) on the SD media supplemented with 10 or 0.2 mm Suc as sole carbon source and grown at 30°C for 4 d for observations. The assays were repeated three times with the same results. B, Coexpression of MdSOT6 with MdCYB5 or AtCYB5-A in the RS453 yeast cells. Yeast cells were transformed with the pDR196 vector harboring MdSOT6 (indicated by MdSOT6), mutated MdSOT6 at Leu-117→Pro (MdSOT6L117-P), and MdCYB5 (as a negative control) or with the empty pDR196 vector (pDR196, as another negative control). The yeast cells were also cotransformed with the pDR196 vector pairs pDR196-MdSOT6 plus pDR196-MdCYB5 (MdSOT6+MdCYB5), pDR196-MdSOT6 plus pDR196-AtCYB5-A (MdSOT6+AtCYB5-A), pDR196-MdSOT6L117-P plus pDR196-MdCYB5 (MdSOT6L117-P+MdCYB5), and pDR196-MdSOT6L117-P plus pDR196-AtCYB5-A (MdSOT6L117-P+AtCYB5-A). The expression of MdSOT6 was tested with immunoblotting using anti-MdSOT6CL serum, and the expression of MdCYB5 and AtCYB5-A was tested using anti-MdCYB5N serum in the yeast lysates. Actin was used as a loading control. Drop tests were used to observe the transformed yeast growth on low sorbitol concentration as described in A, but on the SD media supplemented with 10 or 0.2 mm sorbitol as sole carbon source. The assays were repeated three times with the same results.
DISCUSSION
The Sugar Transporter-Cytochrome b5 Complex Functions to Up-Regulate the Affinity of the Sugar Transporters in Response to Low Sugar Availability
The fleshy fruits are heterotrophic, strong sink organs whose phloem-unloading pathway is predominantly apoplasmic at least during their important storage processes (Patrick, 1997; Wu et al., 2004; Zhang et al., 2004, 2006). The fruit cells thus depend highly on the sugar transporters to capture the photoassimilates from the apoplasmic space. In this report, we identified from apple fruit a Suc and a sorbitol transporter that are distinct in their structure and substrate specificity (Fig. 1; Supplemental Figs. S1–S4), although they belong to the same major facilitator superfamily. Interestingly, these two distinct sugar transporters both interact physically with an apple ER-anchored cytochrome b5 MdCYB5 (Figs. 2–7; Supplemental Figs. S9 and S10). In the yeast systems, the two different interaction complexes function as sensors to perceive sugar starvation signal in order to up-regulate the affinity of the transporters, allowing cells to adapt to sugar starvation (Figs. 8–10). Consistently, an Arabidopsis homolog of MdCYB5, AtCYB5-A, interacts also with the two sugar transporters and functions similarly (Figs. 5–10), further supporting the function of the sugar transporter-MdCYB5 complex and suggesting that the functions of the sugar transporter-cytochrome b5 interaction may be a general mechanism in plant cells. However, the yeast cytochrome b5 ScCYB5, an additional interacting partner of the two plant sugar transporters, has no function in the regulation of the sugar transporters (Figs. 6–8), indicating that the observed biological functions in the yeast systems are specific to plant cytochrome b5s. We also performed additional assays to test the possible function of the transporter-ScCYB5 interaction by disrupting the unique copy of the yeast cytochrome b5 from the yeast genome through the homologous recombination technique, and we observed that this mutation did not affect the activities of any of the transporters in the yeast systems (data not shown), showing consistently no role of the yeast cytochrome b5 in the plant transporter regulation. Because it is currently difficult to assay the transport functions of the sugar transporters in vivo in an appropriate system using the plant cells with their sugar transporters and cytochrome b5s both encoded by the multigene families conferring functional redundancy, we had to perform experiments to assess the biological functions of the bimolecular interactions in the widely used model yeast systems. In the yeast cells, however, both the MdSUT1 Suc transporter and the MdSOT6 sorbitol transporter, representing two different classes of the plant sugar transporters, share consistently the same mechanism when interacting separately with the two plant cytochrome b5s, homologous in their structure but of different plant origins (Figs. 2–10), strongly suggesting that the mechanism observed in the yeast systems operates in plant cells.
The previous reports showed that the expression of the sugar transporter genes was regulated by sugar concentrations (Weber et al., 1997; Chiou and Bush, 1998; Oliveira et al., 2002; Vaughn et al., 2002; Atanassova et al., 2003; Çakir et al., 2003; Li et al., 2003a, 2003b; Ransom-Hodgkins et al., 2003; Conde et al., 2006), which raised the hypothesis that, as in yeast cells, uptake of sugars by plant cells may be transcriptionally regulated by altering the expression of low- or high-affinity sugar transporters through a sugar-sensing process in response to sugar availability (Lalonde et al., 1999; Conde et al., 2006). Based on our findings presented in this report, we propose a model different from the transcription regulation hypothesis, in which the sugar transporters are regulated at the posttranslational level by the sugar transporter-CYB5 complexes (Fig. 11). This mechanism seems not to involve transcriptional machinery, because the amounts of the transporter proteins did not change when the mechanism driven by the bimolecular complexes was operating (Fig. 10). Therefore, the sugar transporter-CYB5 bimolecular complexes, promoting the affinity of the sugar transporters in response to low sugar availability that strengthens the sugar transporter-CYB5 interactions, stimulates sugar uptake to maintain a relatively stable internal level of the limiting sugar (Fig. 11). Taken together, these findings suggest a novel mechanism by which plant cells adapt to sugar starvation.
How Does the Sugar Transporter-Cytochrome b5 Complex Work to Allow Cells to Adapt to Sugar Deficiency?
Whereas the plant sugar transporters have been postulated to have dual functions as sugar carriers and sensors (Lalonde et al., 1999; Barker et al., 2000), our findings indicate that the complex combining a functional sugar transporter with a cytosolic factor, but not the sugar transporter alone, plays a role in sensing sugar availability to regulate the sugar transporter activity. The bimolecular complex may perceive directly the low sugar signal as a sugar sensor complex or relay the low sugar signal as a key component downstream of a sensing system to elicit the final function of sugar transporter stimulation. Given the plasma membrane localization of the sugar transporter-CYB5 interaction complex (Figs. 4, 5, and 8; Supplemental Fig. S10) and the extracellular site of the low sugar availability that acts as an initial signal to trigger this sensing process, it is most likely that the bimolecular complex functions as both a direct sensor and a final functional player to modulate the sugar uptake ability of cells. However, we cannot exclude that, unlike in yeast cells, this sensing process may also result in an alteration of gene expression of sugar transporters in plant cells, which may lead to a more complex regulation at the multiple levels of transcription, posttranscription, translation, and posttranslation for adaptation of cells to sugar starvation. Nevertheless, without necessarily calling more sugar transporters, as shown in the yeast cells (Fig. 10), this bimolecular complex-mediated low sugar sensing, using the proteins already in place, is an efficient regulatory mechanism by which the activity of the sugar transporters may be directly modulated in response to sugar availability. While this mechanism may function in parallel with the transcriptional regulation mechanism, it acts more rapidly than inducing gene expression to maintain sugar homeostasis and cell reproduction and growth under sugar deficiency (Figs. 8–10).
The plasma membrane-associated sugar transporters were shown to depend on intracellular movement of ER to interact with the ER-anchored cytochrome b5 (Figs. 4 and 5; Supplemental Fig. S10), indicating that the regulation of the sugar transporter activities is linked to both the cytochrome b5 and ER trafficking. It has been well known that the ER is not only a major component of a cell's secretory pathway but also a signaling checkpoint involved in various developmental and pathological processes, which is, like the Golgi apparatus, in constant motion within the cortical cytoplasm of cells (Hawes et al., 1999; Lippincott-Schwartz et al., 2000; Herman and Schmidt, 2004; Xu et al., 2005; Marciniak and Ron, 2006). In the lumens of ER, a series of important metabolic processes, such as glycosylation, sulfation, and phosphorylation of secretory and membrane proteins, proteoglycans, and lipids, occur (Hirschberg et al., 1998). These metabolisms in ER indicate a strong demand for energy in this compartment (Hirschberg et al., 1998; Brandizzi et al., 2002; Leroch et al., 2008), which depends primarily on sugar supply in cells. Cytochrome b5 is a heme-binding protein and functions as an electron transfer component involved in a number of oxidative reactions, such as the anabolic metabolism of lipids and the catabolism of xenobiotics and compounds of endogenous metabolism (Vetten et al., 1999; Schenkman and Jansson, 2003; Kumar et al., 2006). The oxidative reactions mediated by cytochrome b5 are also associated with sugar supply in cells. Accordingly, we postulate that an impact of the sugar deficiency on the metabolic status in the ER may probably induce ER to approach the plasma membrane and to strengthen the sugar transporter-CYB5 interaction, and in this way, a downstream feedback signal raised from the intracellular low sugar availability, besides the extracellular low sugar signal, may be an additional player to drive a sensing pathway to enhance upstream sugar transport flow. In this context, we do not exclude a more complex signaling network to be involved in this CYB5-anchored ER-moving event. The expected double signaling pathways, with the extracellular sensing by the sugar transporter-CYB5 complex and intracellular sensing via the CYB5-ER structure, may ensure an efficient response and adaptation of the cells to the environmental sugar starvation. Elucidating this mechanism will shed new light on sugar transport regulation in eukaryotic cells.
MATERIALS AND METHODS
Plant Materials, Yeast Strain, and Media
Apple (Malus domestica ‘Golden Delicious’) fruits were sampled from 8- to 9-year-old trees growing in a commercial orchard in the western suburbs of Beijing. The Suc transport function of MdSUT1 was assayed by yeast transformation using the yeast (Saccharomyces cerevisiae) strain SUSY7/ura3 (MATa, ura3-52, leu2-3, 112, trp1, mal0, suc2::URA3, ura3, Leu-2::128A2-SuSy) and the pDR196 vector (Riesmeier et al., 1992; Barker et al., 2000; Reinders et al., 2002), which were a generous gift from Dr. W.B. Frommer (Universität Tübingen). The sorbitol transporter MdSOT6 was assayed with the yeast strain RS453 (MAT alpha, ade2-1, trp1-1, leu2-3, his3-11, ura 3–52, lys +can1-100; Sauer and Stadler, 1993) and the pDR196 vector. The RS453 strain was kindly provided by Dr. N. Sauer's laboratory (Universität Erlangen-Nürnberg) through Dr. U. Stochaj (McGill University). Yeast and Escherichia coli media, including the rich, Glc-containing medium (yeast peptone dextrose) and the defined media (synthesis dextrose [SD] for MdSUT1 and synthesis complete [SC] for MdSOT6, both with appropriate supplements) were prepared using standard recipes (Sambrook et al., 1989; Rose et al., 1990).
Isolation of MdSUT1 and MdSOT6 cDNAs
The apple fruits were sampled around 80 d after full bloom for preparing total RNA. Total RNA was isolated from fruit flesh as described previously by Hu et al. (2002). Single-stranded cDNA was synthesized from the total RNA using PowerScript reverse transcriptase and SMART III Oligonucleotide/CDSIII3′ as the primer (Clontech). Using the cDNA as the template, the main part of a Suc (SUT) and a sorbitol (SOT) transporter cDNA was amplified by PCR with two degenerate primers [for SUT, forward primer 5′-A(G/A)TTCGG(G/A)TGGGCICTICAGC-3′ and reverse primer 5′-TICCCATCCAATCAGT(A/G)TCGAAIA-3′; for SOT, forward primer 5′-GAGTCGCC(T/C)CG(C/T)TGGCT(T/C)GT-3′ and reverse primer 5′-GGTTCTACCCTGTGTTTC(C/T)GG-3′] that correspond to the conserved domain of most SUTs (QFGWALQL and FDTDWMGR) and polyol transporters (ESPERWL and PETQGRT), respectively. PCR products were cloned into pMD-18T plasmid (Takara, Dalian Division) and were confirmed by sequence analysis. 5′-RACE PCR was performed using nested PCR with the gene-specific primers derived from the cloned SUT or SOT 5′ end (for SUT, reverse primer 1, 5′-GCCCACAGAGCCATATGACGC-3′, and reverse primer 2, 5′-AGCTCTTGAACGTACGGGGTC-3′; for SOT, reverse primer 1, 5′-CAGGACTCGCTTGGCTTCGC-3′, and reverse primer 2, 5′-GCAGCTTCTTTGATGTCGGC-3′) and SMART III Oligonucleotide primer as forward primer. 3′-RACE PCR was done with the gene-specific primers derived from the cloned SUT or SOT 3′ end (for SUT, forward primer 1, 5′-CCCACGGGTCTGTATGGTTAATC-3′, and forward primer 2, 5′-TGCTCTAAACTGGATTGGGTGGT-3′; for SOT, forward primer 1, 5′-GGGTGGTACCTTCTTTCTTTATGC-3′, and forward primer 2, 5′-GAGTGGCCGTGAATAGGTTGATG-3′) and CDSIII3′ as reverse primer. 5′-RACE and 3′-RACE products were cloned into pMD-18T plasmid and were sequenced. Splicing three fragments, and thus designing a pair of specific primer (for SUT, forward primer 5′-GCCATTATGGCCGGGGGGAGAGAGAGTAGTC-3′ and reverse primer 5′-TTGAACTTGTAGAAGCCTCATGTGACAGCTCTGG-3′; for SOT, forward primer 5′-GCCATTATGGCCGGGGAGGAAGATGACTGACCGGAC-3′ and reverse primer 5′-TTTCAAGAGGAAATGTTTTATTACAA-3′), two full-length cDNAs, one encoding the putative Suc transporter MdSUT1 and the other encoding the putative sorbitol transporter MdSOT6, were obtained by PCR amplification. All of the clones were sequenced to ensure that the gene sequences were correct.
Yeast Complementation
To test the functionality of MdSUT1 and MdSOT6, the open reading frames (ORFs) of MdSUT1 and MdSOT6, respectively, were cloned into the yeast expression vector pDR196 (Rentsch et al. 1995). Appropriate restriction sites and a stop codon were introduced with the designed primers (for MdSUT1, forward primer 5′-CTCGACTAGTATGCCAGCTCCAGACACAGACCGCCA-3′ and reverse primer 5′-CAGGCTGCAGTGTGACAGCTCTGGGCTTAGGTGCAGCA-3′, with the PstI and SpeI sites being underlined; for MdSOT6, forward primer 5′-GCGAATTCATGACTGACCGGACAACTGACG-3′ and reverse primer 5′-GCCTCGAGAGCTAAGAGGTGCGCCCTGTTGTCCGG-3′, with the EcoRI and XhoI sites being underlined). The PCR products were digested either with PstI and SpeI to yield MdSUT1 or with EcoRI and XhoI to yield MdSOT6. The fragments were subcloned into the corresponding restriction sites of pDR196, yielding pDR196-MdSUT1 and pDR196-MdSOT6, respectively. All of the constructs were confirmed by sequencing. Yeast strain SUSY7/ura3 was transformed with pDR196-MdSUT1 and RS453 was transformed with pDR196-MdSOT6, and transformation with the empty vector pDR196 was used as a control in both cases. For assaying yeast growth, we used the SD medium supplemented with either Glc (2%) or Suc (2%) as a carbon source for testing the MdSUT1 transformants and the SC medium supplemented with ether Glc (2%) or sorbitol (2%) as a carbon source for testing the MdSOT6 transformants. The medium pH for yeast growth assays was adjusted to 5.0 with HCl. A drop test was performed as described (“Drop Test of Yeast Growth” below).
Assays of Suc and Sorbitol Uptake
Suc uptake by yeast cells was assayed as described by Weise et al. (2000). Yeast transformants were grown to an optical density at 600 nm (OD600) of 0.8 in liquid SD medium containing Glc. Cells were washed in 25 mm sodium phosphate buffer (pH 5.0) and suspended in the same buffer to OD600 = 20. Uptake assays were initiated by adding Glc to a final concentration of 10 mm into yeast cells 1 min before addition of [14C]Suc (0.5 μCi, 22.8 GBq mmol−1; Amersham Pharmacia Biotech). The final concentration of Suc was 0.4 mm (except when assaying saturation curves in different Suc concentrations), and the final volume of the reaction solution was 200 μL. After incubation at 30°C with shaking for 1 min (except when assaying the time course of the uptake rate; Fig. 1), Suc uptake was stopped by the addition of 4 mL of ice-cold Suc (10 mm) solution and filtered on glass fiber filters (GF/C; Whatman) in a vacuum filtration apparatus. Cells were rapidly washed three times with 4 mL of ice-cold Suc (10 mm) solution, transferred to liquid scintillation vials, and counted. The assays of competition of radioactive Suc uptake by different sugars are described in the figure legends (Supplemental Fig. S4).
Sorbitol uptake into transformed yeast cells was as described by Gao et al. (2003). Yeast cells were initially grown overnight in SC medium supplemented with 2% (w/v) Glc and then transferred to SC medium supplemented with 3% (w/v) glycerol and 0.05% (w/v) Glc. The cells were grown for 16 h to the early logarithmic phase, washed with distilled water, and resuspended to 1% (w/v) in SC medium containing 25 mm MES (pH 4.5). For each sorbitol uptake assay, the cell suspension (100 μL) was mixed with 100 μL of suspension buffer containing [3H]sorbitol (0.5 μCi, 40.5 GBq mmol−1; American Radiolabeled Chemicals). The final concentration of sorbitol was 0.25 mm (except when assaying saturation curves in different sorbitol concentrations; Fig. 1), and the final volume of the reaction solution was 200 μL. After incubation at 28°C with shaking for 1 min (except when assaying the time course of the uptake rate; Fig. 1), sorbitol uptake was assayed as described above for Suc uptake except for the solution used to stop the uptake and for washing, which was 4 mL of ice-cold distilled water instead of Suc solution. The assays of competition of radioactive sorbitol uptake by different sugars are described in the figure legends (Supplemental Fig. S4). All of the uptake experiments for both Suc and sorbitol were repeated at least three times with the pDR196 empty vector-transformed cells as controls, and sugar uptake by the vector controls was subtracted from the sugar uptake by yeast expressing MdSUT1 or MdSOT6.
Preparation of Antiserum against the Recombinant MdSUT1CL, MdSOT6CL, and MdCYB5N
To generate the antiserum that recognizes MdSUT1, MdSOT6, or MdCYB5, we produced MdSUT1CL, MdSOT6CL, and MdCYB5N, respectively, in E. coli as glutathione S-transferase (GST) fusion protein essentially as described by Zhu et al. (2007). The MdSUT1CL represents the variable central loop fragment of 40 amino acids (amino acids 240–280; Supplemental Fig. S1), and the MdSOT6CL represents also the central loop of 66 amino acids (amino acids 218–282; Supplemental Fig. S2). Therefore, the antisera against MdSUT1CL and MdSOT6CL should recognize specifically MdSUT1 and MdSOT6, respectively. The MdCYB5N represents the conserved region of the plant CYB5s around the N terminus of MdCYB5 (amino acids 28–107; Fig. 6A). The MdSUT1CL-, MdSOT6CL-, and MdCYB5N-corresponding regions in the ORFs of MdSUT1, MdSOT6, and MdCYB5 were amplified by PCR using Pfu DNA polymerase (Takara, Dalian Division) and synthetic oligonucleotide primers (for MdSUT1CL, forward primer 5′-TCGCGGATCCACACCTCTGGGTTCAAGTAA-3′ [BamHI] and reverse primer 5′-ACCGCTCGAGAGACCCTGGGAAATATCTAAAA-3′ [XhoI]; for MdSOT6CL, forward primer 5′-GAGGGATCCCCTCGGTGGCTCGTCATGCA−3′ [BamHI] and reverse primer 5′-GGCCTCGAGAACAAGCAACTCCTTCCACACGCC-3′ [XhoI]; and for MdCYB5N, forward primer 5′-GAGGGATCCGGGAAGGTGTATGATGTGAC-3′ [BamHI] and reserve primer 5′-GGCCTCGAGGAATTCGGATGTCTTGTCAG-3′ [XhoI]). The PCR products were cloned into the BamHI (5′ end)/XhoI (3′ end) sites of pGEX-4T-1 (Amersham Pharmacia Biotech) for the expression of N-terminal GST fusion proteins under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible tac promoter. To ensure that no errors were introduced by PCR, each construct was checked by sequencing. To produce GST tag fusion proteins, DH5α cells transformed with pGEX-4T-1/MdSUT1CL, pGEX-4T-1/MdSOT6CL, or pGEX-4T-1/MdCYB5N were induced with 0.5 mm IPTG for 4 h. The fusion proteins were purified from IPTG-induced cell pellets by a Glutathione Sepharose 4B column (Amersham Pharmacia Biotech) and analyzed by SDS-PAGE. The purified fusion proteins were used for standard immunization protocols in rabbits. Polyclonal antisera were affinity purified first by HiTrap Protein-G HP (Amersham Pharmacia Biotech) and further by an immunosorbent column coupled with GST-MdSUT1CL, GST-MdSOT6CL, or GST-MdCYB5N fusion protein. The affinity-purified antisera were evaluated by immunoblotting and shown to be highly specific.
Preparation of Membrane Fractions from Apple Fruits
The membrane fractions were prepared as described previously by Duan et al. (2003) with modifications. All of the steps were performed at 0°C to 4°C. For preparation of membrane protein, the freshly harvested fruits were frozen with liquid N2 and then crushed. The crushed flesh was homogenized with cold medium in a volume ratio of 1:3 (flesh:medium). This medium contained 250 mm Suc, 5 mm EDTA, 0.5% (w/v) casein, 0.2% β-mercaptoethanol, 2 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride (PMSF), 5 mm ascorbic acid, 10% (v/v) glycerol, 10 mm MgCl2, 3.5% polyvinylpolypyrrolidone, and 100 mm Tris-HCl (pH 7.5). The homogenate was filtered through four layer of 600-μm nylon cloth, the obtained filtrate was centrifuged at 5,000g for 15 min, and the supernatant was further centrifuged at 100,000g for 1 h. The supernatant gave the cytosolic fraction. The pellet gave the membrane fraction that was resuspended in a suspension buffer containing 250 mm mannitol, 1 mm EDTA, 1 mm dithiothreitol, 2 mm MgCl2, 5 mm ascorbic acid, 0.5 mm PMSF, and 5 mm Tris-HCl (pH 7.0). Protein concentrations were determined by the method of Bradford (1976) with bovine serum albumin (BSA) as a standard.
Immunoblotting
Proteins were separated by SDS-PAGE on 12% polyacrylamide gels, and the polypeptides were transferred to nitrocellulose membranes (0.45 μm; Amersham Life Science) in a medium consisting of 25 mm Tris-HCl (pH 8.3), 192 mm Gly, and 20% (v/v) methanol. After rinsing in the Tris-buffered saline (TBS) containing 10 mm Tris-HCl (pH 7.5) and 150 mm NaCl, the blotted membranes were preincubated for 2 h in a blocking buffer containing 3% (w/v) BSA dissolved in TBS supplemented by 0.05% (v/v) Tween 20 (TBST1) and then incubated with gentle shaking for 3 h at room temperature in the appropriate antibodies (generally diluted 1:3,000 in the blocking buffer). Following extensive washes by TBST1, the membranes were incubated with goat anti-rabbit IgG conjugated with alkaline phosphatase (1:1,000 diluted in TBST1) at room temperature for 2 h and then washed with TBST2 (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 0.1% [v/v] Tween 20) and TBS. The locations of antigenic proteins were visualized by incubating the membranes with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate.
Analysis of Protein Interaction by the Yeast Split-Ubiquitin System
Interaction between proteins was assayed by the split-ubiquitin system (Johnsson and Varshavsky, 1994; Reinders et al., 2002) using yeast strain DSY-1 [MATa, his3Δ200, trp1-901, leu2-3,112 ade2, LYS2::(lexAop)4-HIS3 URA3::(lexAop)8-lacZ GAL4] in the Split Ubiquitin System kit (Dualsystems Biotech). The assays were performed according to the protocols of the kit. In this system, a version of the modified membrane-based system developed by Stagljar et al. (1998) and Stagljar and Heesen (2000) was used, in which interaction of two membrane-bound fusion proteins can be monitored by the release of an artificial transcription factor consisting mainly of Lex A and VP16 (for virus protein 16; PLV). cDNAs of interest were cloned in frame with either the C terminus (Cub) followed by PLV (Cub-LexA-VP16) or the mutated N-terminal (NubG with mutation Ile-13→Gly; Johnsson and Varshavsky, 1994) subdomain of ubiquitin (Fig. 2). Interaction of the fusion proteins causes an association of NubG and CubPLV, leading to cleavage of the PLV transcription factor by ubiquitin-specific proteases and, in turn, expression of the lacZ and HIS3 reporters integrated in the yeast genome.
The “bait” and “prey” vectors for the split-ubiquitin system were constructed for expression of heterologous proteins in yeast. The integrative bait vector pTMBV4 contains the transcription elongation factor1 promoter, which allows high constitutive expression of the bait fusion protein, ORF of the transporter genes, and Cub in frame with PLV (Cub-LexA-VP16) as well as the LEU2 marker (Fig. 2). Two types of prey vectors, pDL2-Nx and pDL2-xN (where x indicates cDNAs of interest and N indicates NubG), contain the strong constitutive alcohol dehydrogenase1 promoter, the NubG domain, a multicloning site followed by the cytochrome c terminator, as well as the Trp1 marker (Fig. 2). To detect interaction between two halves of MdSUT1, the N-terminal half of MdSUT1, encoding amino acids 1 to 250 (MdSUT1-I), was fused to the Cub-coding sequence of vector pTMBV4 (Supplemental Fig. S9), while the C-terminal half of MdSUT1, encoding amino acids 251 to 499 (MdSUT1-II), was fused to the NubG-coding sequence of vector pDL2-xN (where x indicates MdSTU1-II; Supplemental Fig. S9). To screen the interacting partner of the two transporters, the ORFs of MdSUT1 and MdSOT6 were fused to the Cub-coding sequence of vector pTMBV4 (Fig. 2), while the cDNA, reverse transcribed from apple fruit mRNA and digested by SfiI, was fused to the NubG-coding sequence of vector pDL2-Nx (where x indicates the cDNA; Fig. 2; see the detailed description in “Apple cDNA Library Construction” below). pMBV4-Alg5 (for Asn-linked glycosylation5), provided by the supplier of the kit, was used as a control bait plasmid, and pAlg5-NubI and pAlg5-NubG were used as control prey plasmids.
For cloning, the partial or full-length sequences were amplified by PCR using Pfu DNA polymerase (Takara, Dalian Division) with cloned genes as templates. Appropriate restriction sites were introduced with the following specially designed primers: for pTMBV4-MdSUT1 construction, forward primer 5′-CGATCTAGACAAAAATGCCAGCTCCAGACACAGACC-3′ and reserve primer 5′-CACTCCATGGGTGACAGCTCTGGGCTTAGGTGC-3′; for pTMBV4-MdSUT1-I construction, forward primer 5′-CGATCTAGACAAAAATGCCAGCTCCAGACACAGACC-3′ and reserve primer 5′-CACTCCATGGTCATCAGCAAAGGGTGTAGTTCT-3′; for pDL2-xN-MdSUT1-II construction, forward primer 5′-ATGTGGATCCACATGGGGCCAGGACAGTCAAGT-3′ and reserve primer 5′-TCGACCCGGGGTGACAGCTCTGGGCTTAGGTGC-3′; for pTMBV4-MdSOT6 construction, forward primer 5′-GCTCCAGACAAAAATGACTGACCGGACAACTGACG-3′ and reserve primer 5′-GAAGGCCTGCTAAGAGGTGCGCCCTGTTGTCCGG-3′. The PCR products were subcloned at the XbaI-NcoI (MdSUT1 and MdSUT1-I) or XbaI-StuI (MdSOT6) sites of pTMBV4 or the SmaI-BamHI (MdSUT1-II) sites of pDL2-xN to yield plasmids pMdSUT1-CubPLV, pMdSOT6-CubPLV, pMdSUT1-I-CubPLV, and pNubG-MdSUT1-II, respectively. All of the constructs were confirmed by sequencing. The yeast strain DSY-1 was transformed with these plasmids. For yeast two-hybrid screening in apple fruit cDNA, the DSY-1 yeast cells transformed with the pTMBV4 vector harboring MdSUT1 or MdSOT6 were further transformed with plasmid DNA derived from an apple cDNA library. The screening and analysis of interacting proteins were performed according to the supplier's instructions with the kit.
To test the interactions of MdSUT1 and MdSOT6 with MdCYB5, AtCYB5-A, or ScCYB5, the ORFs of MdCYB5, AtCYB5-A, and ScCYB5 were subcloned at the BamHI-SmaI sites of pDL2-xN or pDL2-Nx vector (where x indicates the CYB5s). The following primers were used: for pDL2-xN-MdCYB5 construction, forward primer 5′-CGCGGATCCAAATGGCTTCAGATCCGAAAGT-3′ and reserve primer 5′-TCCCCCGGGCCCTCTTTCTTGGTGTAGTGAC-3′; for pDL2-Nx-MdCYB5 construction, forward primer 5′-GAGGGATCCATGGCTTCAGATCCGAAAGT-3′ and reverse primer 5′-TCCCCCGGGCTCTTTCTTGGTGTAGTGAC-3′; for pDL2-xN-AtCYB5-A construction, forward primer 5′-CGCGGATCCAAATGTCTTCAGATCGGAAGGT-3′ and reserve primer 5′-TCCCCCGGGGGGTCTTTCTTGGTATAGTGAC-3′; for pDL2-Nx-AtCYB5-A construction, forward primer 5′-CGCGGATCCATGTCTTCAGATCGGAAGGT-3′ and reserve primer 5′-TCCCCCGGGGGTCTTTCTTGGTATAGTGAC-3′; for pDL2-xN-ScCYB5 construction, forward primer 5′-CGCGGATCCAAATGCCTAAAGTTTACAGTTA-3′ and reserve primer 5′-TCCCCCGGGCCTTCGTTCAACAAATAATAAG-3′; for pDL2-Nx-ScCYB5 construction, forward primer 5′-CGCGGATCCATGCCTAAAGTTTACAGTTA-3′ and reserve primer 5′-TCCCCCGGGGTTATTCGTTCAACAAATAATAAG-3′. All of the constructs were confirmed by sequencing. The yeast strain DSY-1 was cotransformed with these plasmids and pTMBV4-MdSUT1 or pTMBV4-MdSOT6.
To search for key amino acid sequences responsible for the transporter-MdCYB5 interactions (Fig. 2), the fragments of MdSUT1, MdSOT6, or MdCYB5 cDNA were PCR amplified using the primers described in Supplemental Table S1. The PCR products for the fragments were then fused to CubPLV (as for MdSUT1 and MdSOT6) or NubG (as for MdCYB5, in the form of pDL2-Nx, where x indicates the fragments of MdCYB5) as described above.
To determine the precise sites responsible for the MdSUT1-MdCYB5 and MdSOT6-MdCYB5 interactions (Figs. 2, 4, 9, and 10), we introduced point mutations in MdSUT1 and MdSOT6. First, we cloned the 5′ ORF sequences with the following primers: forward primer 5′-ATGCCAGCTCCAGACACAGA-3′ and reverse primer 5′-AAGGGGCTGGACCACAGGGCCCGATAA-3′ for MdSUT1P and forward primer 5′-ATGACTGACCGGACAACTGACG-3′ and reverse primer 5′-GTTGGTGGAGAACCCCATGGGAATCGCTCCGAT-3′ for MdSOT6P. The underlined nucleotides are the mutated sites. Then, using the PCR products as forward primers for MdSUT1P and MdSOT6P, together with reverse primer 5′-TGTGACAGCTCTGGGCTTAG-3′ for MdSUT1P and reverse primer 5′-AGCTAAGAGGTGCGCCCTGT-3′ for MdSOT6P, we obtained the ORFs of the mutated MdSUT1 (at Leu-73→Pro; MdSUT1P) and MdSOT6 (at Leu-117→Pro; MdSOT6P). The PCR products for the mutated MdSUT1 or MdSOT6 were fused to CubPLV (as for wild-type MdSUT1 and MdSOT6) as described above. All of the clones were sequenced to ensure that the sequences were correct.
cDNA Isolation of ScCYB5 and AtCYB5-A
Single-stranded cDNA was synthesized from the total RNA of the yeast strain DSY-1 cells and Arabidopsis (Arabidopsis thaliana) leaves using PowerScript reverse transcriptase and SMART III Oligonucleotide/CDSIII3′ as the primer (Clontech). Using the cDNA as the template, the ORFs of cDNA were amplified by PCR with two primers (for ScCYB5, forward primer 5′-ATGCCTAAAGTTTACAGTTA-3′ and reverse primer 5′-TTCGTTCAACAAATAATAAG-3′; for AtCYB5-A, forward primer 5′-ATGTCTTCAGATCGGAAGGT-3′ and reverse primer 5′-GTCTTTCTTGGTATAGTGAC-3′). PCR products were cloned into pMD18-T plasmid (Takara, Dalian Division) and confirmed by sequence analysis.
Apple cDNA Library Construction
The construction of an expression cDNA library was done essentially according to the protocols of the BD Matchmaker library construction kit (Clontech). Single-stranded cDNA was synthesized from mRNA of apple fruit as described above (see “Cloning of MdSUT1 and MdSOT6”). Double-stranded cDNA was obtained by long-distance PCR. The double-stranded cDNA was ligated into the SfiI-SfiI sites of the yeast prey vector pDL2-Nx (Fig. 2). The cDNA expression library was constructed and amplified through transducing E. coli XL1-Blue. The titer of the amplified library was 2.37 × 107 plaque-forming units μg−1 DNA. Thus, the cDNA expression library fused downstream of the split ubiquitin NubG was ready to be introduced into yeast.
β-Gal Assay
To assess protein-protein interactions in the yeast split-ubiquitin system, the β-gal activity was tested using filter assays as described previously (Ramer et al., 1992). Cells were streaked out on filter papers (Whatman), placed on top of SD agar plates containing appropriate supplements but lacking Trp and Leu, and grown for 2 d at 30°C. Filters were then removed from the agar, frozen briefly in liquid nitrogen, defrosted, and placed in petri dishes onto filter paper saturated with buffer 1 (60 mm Na2HPO4, 40 mm NaH2PO4, 10 mm KCl, and 1 mm MgSO4) containing 35 mm β-mercaptoethanol and 1.5 mg mL−1 5-bromo-4-chloro-3-indolyl-3-d-galactoside (Sigma). For development, filters were incubated at 37°C for 15 min.
The β-gal activity was also quantitatively determined as described by Stagljar et al. (1998). Cells were grown in liquid dropout common SD medium lacking Trp and Leu to an OD546 of 0.5 to 0.7. Cells (1.5 mL) were pelleted, washed once in buffer 2 (113 mm Na2HPO4, 40 mm NaH2PO4, 10 mm KCl, and 1 mm MgCl2, pH 7.0), and suspended in 300 μL of buffer 2. One hundred microliters was taken and lysed by three freeze/thaw cycles. Buffer 2 (700 μL) containing 0.27% (v/v) 2-mercaptoethanol and 160 μL of 2-nitrophenyl-β-d-galactopyranoside (4 mg mL−1 in Z buffer) was added and incubated for 5 to 15 min at 30°C. Four hundred microliters of 1 m NaCO3 was added, the sample was centrifuged, and the OD420 was measured. β-Gal units were calculated as 1,000 × OD420/OD546 × min.
Coimmunoprecipitation
Yeast cell lysates were prepared as described previously (Nam and Li, 2002). Yeast strains were grown on SD-Leu-Trp-His medium to OD546 = 1.0 at 28°C. Total proteins were prepared from yeast cells with an extraction buffer (2 mL g−1 cells) containing 50 mm HEPES (pH 7.4), 10% (v/v) glycerol, 10 mm EDTA, 0.1% (v/v) Triton X-100, 200 μm PMSF, and 2 μg mL−1 each aprotinin, leupeptin, and pepstatin A. The antibodies used were mouse antibody (Clontech) specific to VP16-tagged MdSUT1 or MdSOT6 protein and rabbit antibody specific to hemagglutinin peptide epitope (HA; Santa Cruz)-tagged MdCYB5 protein. Immunoprecipitation experiments were performed with protein A/G Plus-agarose beads (Santa Cruz), following the manufacturer's protocol. In brief, cell lysates were precleared and incubated with the antibodies for 6 h at 4°C in the extraction buffer. The beads were washed twice extensively with buffer A (50 mm Tris, pH 8.0, 150 mm NaCl, and 0.1% [v/v] Triton X-100) and buffer B (50 mm Tris, pH 8.0, and 0.1% [v/v] Triton X-100) and then resuspended in SDS-PAGE sample buffer. The immunoprecipitates were separated by 12% SDS-PAGE and analyzed by immunoblotting with anti-VP16 or anti-HA serum.
For immunoprecipitation in apple fruit extracts, the membrane proteins (100 mg) were resuspended in 1.0 mL of membrane solubilization buffer containing 10 mm Tris-HCl, pH 7.3, 150 mm NaCl, 1 mm EDTA, 1% glycerol, 1 mm PMSF, 1 μg mL−1 leupeptin, 10 μg mL−1 aprotinin, and 10 μg mL−1 chymostatin. The immunoprecipitation was done with the same procedures as described above but using the anti-MdSUT1CL, anti-MdSOT6CL, or anti-MdCYB5N serum as appropriate.
Immunogold Labeling
Subcellular immunogold labeling of MdCYB5 was done essentially as described previously (Zhang et al., 2004). Briefly, the ultrathin sections were prepared from Lowicryl K4M-embedded specimen. They were first blocked for 30 min at room temperature by floating the grids on droplets of phosphate-buffered saline containing 8 mm Na2HPO4, 1.5 mm KH2PO4, 3 mm KCl, and 500 mm NaCl (pH 7.4) supplemented with 50 mm Gly and continuously blocked with phosphate-buffered saline supplemented with 0.1% (w/v) gelatin, 0.5% (w/v) BSA, and 0.1% (v/v) Tween 20 (pH 7.4; PBGT). Without rinsing, the sections were incubated with the rabbit anti-MdCYB5N polyclonal antibody (diluted 1:100 in PBGT buffer) overnight at 4°C. Following extensive washes with PBGT buffer, the sections were incubated with secondary antibody (goat anti-rabbit IgG antibody conjugated with 10-nm gold) at 1:100 dilution in PBGT buffer for 2 h at room temperature. The sections were rinsed consecutively with PBGT and double-distilled water. The sections were then stained with 2% uranyl acetate in 50% ethanol for 5 min at 25°C. After extensively washing with double distilled water, the ultrathin sections were examined with an electron microscope.
The specificity and reliability of the immunogold labeling were tested by two negative controls. In the first one, the antiserum was omitted to test possible unspecific labeling of the goat anti-rabbit IgG antibody-gold conjugate. In the second one, rabbit preimmune serum was used instead of the rabbit antiserum before immunogold labeling to test the specificity of the antiserum. More than three repetitions of the control experiments were conducted for each sample.
Transient Expression in Arabidopsis Protoplasts
For observation of the subcellular localization of MdSUT1, MdSOT6, and MdCYB5, the ORFs of their cDNAs were PCR amplified. The primers used were as follows: for MdSUT1, forward primer 5′-GGGGTACCATGCCAGCTCCAGACACAGACC-3′ and reverse primer 5′-CGGGATCCGTGACAGCTCTGGGCTTAG-3′, with the KpnI (5′ end) and BamHI (3′ end) sites; for MdSOT6, forward primer 5′-CCCAAGCTTATGACTGACCGGACAACTGACG-3′ and reverse primer 5′-CGCGGATCCGCTAAGAGGTGCGCCCTGT-3′, with the HindIII (5′ end) and BamHI (3′ end) sites; for MdCYB5, forward primer 5′-CCCAAGCTTATGGCTTCAGATCCGAAAGT-3′ and reverse primer 5′-CGGGATCCTCTTTCTTGGTGTAGTGAC-3′, with the HindIII (5′ end) and BamHI (3′ end) sites. Each PCR product was then fused upstream of the enhanced GFP in the cauliflower mosaic virus 35S-EGFP-Ocs 3′ vector (p-EZS-NL vector; http://deepgreen.stanford.edu). This vector does not express GFP well without adding coding sequence to the 5′ end of the ORF of GFP; thus, the control cells do not show fluorescence of GFP. Protoplasts were isolated from the leaves of 3- to 4-week old plants of Arabidopsis (ecotype Columbia) and transiently transformed using polyethylene glycol essentially according to Ueda et al. (2001). Fluorescence of GFP was observed with a confocal laser-scanning microscope (Zeiss; LSM 510 META) after incubation at 23°C for 16 h.
BiFC Assay
We performed BiFC assays in vivo as described (Walter et al., 2004). The ORFs of MdSUT1, MdSOT6, and MdCYB5 were amplified by PCR with the following primers: for MdSUT1, forward primer 5′-GCTCTAGAATGCCAGCTCCAGACACAGA-3′ and reverse primer 5′-CGCGGATCCTGTGACAGCTCTGGGCTTAG-3′, with the XbaI (5′ end) and BamHI (3′ end) sites; for MdSOT6, forward primer 5′-GCTCTAGAATGACTGACCGGACAACTGACG-3′ and reverse primer 5′-CGCCTCGAGAGCTAAGAGGTGCGCCCTGT-3′, with the XbaI (5′ end) and XhoI (3′ end) sites. The ORFs of the mutated MdSUT1 (at Leu-73→Pro; MdSUT1P) and MdSO61 (at Leu-117→Pro; MdSOT6P) were obtained as mentioned above. The ORFs were cloned into the plasmid pSPYCE-35S/pUC-SPYCE; thus, the plasmids pUC-SPYCE-MdSUT1 and pUC-SPYCE-MdSOT6 were expressed as the MdSUT1-YFPC and MdSOT6-YFPC fusion proteins. Coding sequences of MdCYB5 were obtained by PCR with forward primer 5′-GCTCTAGAATGGGGAAGGTGTATGATGTGAC-3′ and reverse primer 5′-CGCCTCGAGCTCTTTCTTGGTGTAGTGAC-3′. The coding sequence was cloned into plasmid pSPYNE-35S/pUC-SPYNE at the XbaI and XhoI sites to give rise to plasmid pUC-SPYNE-MdCYB5 expressed as the MdCYB5-YFPN fusion protein. The Arabidopsis protoplasts were transiently transformed, the strips of onion (Allium cepa) bulb epidermis were bombarded, and the fluorescence of YFP was observed as described above.
Drop Test of Yeast Growth
Yeast cells expressing various constructs were grown on the indicated media overnight and then transferred to fresh liquid media to OD600 = 0.2. After a further incubation of 2 h, the OD values were measured. The cells were diluted in sterile water, and 8 μL was spotted at the indicated concentrations (OD600 = 0.1, 0.01, and 0.05) on the media supplemented with the indicated sugars as carbon source. The yeast cells were further grown at 30°C for 4 d for observations.
Sequence data from this article can be found in the GenBank data libraries under following accession numbers (in parentheses): for sugar transporters, AtPLT5 (Arabidopsis genomic locus tag At3G18830), AtSUC7 (At1G66570), AtSTP4 (At3G19930), AtSUT4 (NM100870), LeSUT4 (AF176950), MdSOT1 (AAO88964), MdSOT6 (AY592976), MdSUT1 (AY445915), OsSUT2 (DQ072592), StSUT4 (AF237780), and yeast ScHXT1 (M82963); for cytochrome b5, AcCYB5 (AAM28288), AtCYB5-A/2 (At5g53560), AtCYB5-4 (At2g32720), AtCYB5-B/3 (At5g48810), AtCYB5-6 (At1g26340), BoCYB5 (P40934), CrCYB5 (P49097), HsCYB5-B (O43169), MdCYB5 (EF614285), MtCYB5 (ABE93515), NtCYB5 (S46306), OeCYB5 (CAA04703), OsCYB5 (ABB47889), PtCYB5 (XP_511067), RnCYB5 (AAB67609), VfCYB5 (AAT84458), VvCYB5 (CAN78289), and yeast ScCYB5 (L22494).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Structure of MdSUT1.
Supplemental Figure S2. Structure of MdSOT6.
Supplemental Figure S3. Homology of plant Suc transporters (SUTs) and polyol transporters (PLTs).
Supplemental Figure S4. Characterization of the MdSUT1- and MdSOT6-mediated sugar uptake by yeast cells.
Supplemental Figure S5. Temporal and spatial expression of MdSUT1, MdSOT6, and MdCYB5.
Supplemental Figure S6. Expression of MdSUT1 in different tissues or cells.
Supplemental Figure S7. Expression of MdSOT6 in different tissues or cells.
Supplemental Figure S8. Identification of the recombinant bait vectors for the yeast two-hybrid split-ubiquitin system.
Supplemental Figure S9. Interactions between two halves of MdSUT1 and between MdSUT1 and MdCYB5.
Supplemental Figure S10. In vivo interaction of MdCYB5 with MdSUT1 and MdSOT6 in the BiFC system in the onion epidermis cells.
Supplemental Figure S11. MdCYB5 interacts with a Suc and a polyol transporter but not with a hexose transporter.
Supplemental Figure S12. Expression of MdCYB5 in different tissues or cells.
Supplemental Figure S13. Transient expression of MdSUT1-, MdSOT6-, and MdCYB5-GFP fusion proteins in the onion epidermis cells.
Supplemental Figure S14. Stimulation of the sugar transporter-CYB5 interactions by low sugar supply is specific to the sugar substrates.
Supplemental Figure S15. Expression of MdCYB5 and AtCYB5-A enhances the affinity of MdSUT1 and MdSOT6 to their substrate sugars.
Supplemental Table S1. Primers used for generating the fragments of MdSUT1 (SUT1-n), MdSOT6 (SOT1-n), and MdCYB5 (CYB1-n or CYBn-134).
Supplemental Methods S1. Immunohistochemical technique.
Supplemental Methods S2. cDNA isolation of several sugar transporters from Arabidopsis.
Supplemental Methods S3. Transient expression in onion epidermis.
Supplementary Material
Acknowledgments
We thank Dr. W.B. Frommer for his generous gift of the yeast strain SUSY7/ura3 and the vector pDR196. We also thank Dr. N. Sauer and Dr. U. Stochaj for giving us the RS453 yeast strain.
This work was supported by the National Natural Science Foundation of China (grant nos. 30671444 to D.-P.Z., 30771759 to C.-C.P., and 30700546 to X.-Y.Z.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Da-Peng Zhang (zhangdp@tsinghua.edu.cn).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Arenas-Huertero F, Arroyo A, Zhou L, Sheen J, Leon P (2000) Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev 14 2085–2096 [PMC free article] [PubMed] [Google Scholar]
- Atanassova R, Leterrier M, Gaillard C, Agasse A, Sagot E, Coutos-Thevenot P, Delrot S (2003) Sugar-regulated expression of a putative hexose transport gene in grape. Plant Physiol 131 326–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avonce N, Leyman B, Mascorro-Gallardo JO, Van Dijck P, Thevelein JM, Iturriaga G (2004) The Arabidopsis trehalose-6-P synthase AtTPS1 gene is a regulator of glucose, abscisic acid, and stress signaling. Plant Physiol 136 3649–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avonce N, Leyman B, Thevelein JM, Iturriaga G (2005) Trehalose metabolism and glucose sensing in plants. Biochem Soc Trans 33 276–279 [DOI] [PubMed] [Google Scholar]
- Barker L, Kuhn C, Weise A, Schulz A, Gebhardt C, Hirner B, Hellmann H, Schulze W, Ward JM, Frommer WB (2000) SUT2, a putative sucrose sensor in sieve elements. Plant Cell 12 1153–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth I, Meyer S, Sauer N (2003) PmSUC3: characterization of a SUT2/SUC3-type sucrose transporter from Plantago major. Plant Cell 15 1375–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles E, Hollenberg CP (1997) The molecular genetics of hexose transport in yeasts. FEMS Microbiol Rev 21 85–111 [DOI] [PubMed] [Google Scholar]
- Borgese N, Colombo S, Pedrazzini E (2003) The tale of tail-anchored proteins: coming from the cytosol and looking for a membrane. J Cell Biol 161 1013–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese N, D'Arrigo A, DeSilvestris M, Pietrini G (1993) NADH-cytochrome b5 reductase and cytochrome b5: the problem of posttranslational targeting to the endoplasmic reticulum. Subcell Biochem 21 313–341 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Snapp EL, Roberts AG, Lippincott-Schwartz J, Hawes C (2002) Membrane protein transport between the endoplasmic reticulum and the Golgi in tobacco leaves is energy dependent but cytoskeleton independent: evidence from selective photobleaching. Plant Cell 14 1293–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner M, Sauer N (2000) Monosaccharide transporters in plants: structure, function and physiology. Biochim Biophys Acta 1465 263–274 [DOI] [PubMed] [Google Scholar]
- Çakir B, Agasse A, Gaillard C, Saumonneau A, Delrot S, Atanassova R (2003) A grape ASR protein involved in sugar and abscisic acid signaling. Plant Cell 15 2165–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Willard FS, Huang J, Liang J, Chasse SA, Jones AM, Siderovski DP (2003) A seven-transmembrane RGS protein that modulates plant cell proliferation. Science 301 1728–1731 [DOI] [PubMed] [Google Scholar]
- Cheng WH, Endo E, Zhou L, Penney J, Chen HC, Arroyo A, Leon P, Nambara E, Asami T, Seo M, et al (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14 2723–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Bush DR (1998) Sucrose is a signal molecule in assimilate partitioning. Proc Natl Acad Sci USA 95 4784–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, Yoo SD, Sheen J (2006) Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127 579–589 [DOI] [PubMed] [Google Scholar]
- Conde C, Agasse A, Glissant D, Tavares R, Geros H, Delrot S (2006) Pathways of glucose regulation of monosaccharide transport in grape cells. Plant Physiol 141 1563–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delrot S, Atanassova R, Gomès E, Coutos-Thévenot P (2001) Plasma membrane transporters: a machinery for uptake of organic solutes and stress resistance. Plant Sci 161 391–404 [Google Scholar]
- Duan CQ, Shen YY, Liang XE, Zhang DP (2003) Membrane-associated protein kinase activities in developing apple fruit. Physiol Plant 118 105–113 [DOI] [PubMed] [Google Scholar]
- Ehness R, Roitsch T (1997) Coordinated induction of mRNA for extracellular invertase and a glucose transporter in Chenopodium rubrum by cytokinins. Plant J 11 539–548 [DOI] [PubMed] [Google Scholar]
- Endler A, Meyer S, Schelbert S, Schneider T, Weschke W, Peters SW, Keller F, Baginsky S, Martinoia E, Schmidt UG (2006) Identification of a vacuolar sucrose transporter in barley and Arabidopsis mesophyll cells by a tonoplast proteomic approach. Plant Physiol 141 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillion L, Ageorges A, Picaud S, Coutos-Thevenot P, Lemoine R, Romieu C, Delrot S (1999) Cloning and expression of a hexose transporter gene expressed during the ripening of grape berry. Plant Physiol 120 1083–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Jayanty S, Beaudry R, Loescher W (2005) Sorbitol transporter expression in apple sink tissues: implication for fruit sugar accumulation and watercore development. J Am Soc Hortic Sci 130 261–268 [Google Scholar]
- Gao Z, Maurousset L, Lemoine R, Yoo SD, van Nocker S, Loescher W (2003) Cloning, expression, and characterization of sorbitol transporters from developing sour cherry fruit and leaf sink tissues. Plant Physiol 131 1566–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford NG, Hey S, Jhurreea D, Laurie S, McKibbin RS, Paul M, Zhang Y (2003) Metabolic signaling and carbon partitioning: role of Snf1-related (SnRK1) protein kinase. J Exp Bot 54 467–475 [DOI] [PubMed] [Google Scholar]
- Hawes CR, Brandizzi F, Andreeva AV (1999) Endomembranes and vesicle trafficking. Curr Opin Plant Biol 2 454–461 [DOI] [PubMed] [Google Scholar]
- Herman E, Schmidt M (2004) Endoplasmic reticulum to vacuole trafficking of endoplasmic reticulum bodies provides an alternate pathway for protein transfer to the vacuole. Plant Physiol 136 3440–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberg CB, Robbins PW, Abeijon C (1998) Transporters of nucleotide sugars, ATP, and nucleotide sulfate in the endoplasmic reticulum and Golgi apparatus. Annu Rev Biochem 67 49–69 [DOI] [PubMed] [Google Scholar]
- Hu CG, Honda C, Kita M, Zhang ZL, Tsuda T, Moriguchi T (2002) A simple protocol for RNA isolation from fruit trees containing high levels of polysaccharides and polyphenol compounds. Plant Mol Biol Rep 20 67–69 [Google Scholar]
- Hwang YT, Pelitire SM, Henderson MPA, Andrews DW, Dyer JM, Mullen RT (2004) Novel targeting signals mediate the sorting of different isoforms of the tail-anchored membrane protein cytochrome b5 to either endoplasmic reticulum or mitochondria. Plant Cell 16 3002–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JC, Leon P, Zhou L, Sheen J (1997) Hexokinase as a sugar sensor in higher plants. Plant Cell 9 5–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson N, Varshavsky A (1994) Split ubiquitin as a sensor of protein interactions in vivo. Proc Natl Acad Sci USA 91 10340–10344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CA, Taylor JP, Gao Y, Kimple AJ, Grigston JC, Chen JG, Siderovski DP, Jones AM, Willard FS (2007) GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. Proc Natl Acad Sci USA 104 17317–17322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M, Kim JH (2005) Glucose as a hormone: receptor-mediated glucose sensing in the yeast Saccharomyces cerevisiae. Biochem Soc Trans 33 247–252 [DOI] [PubMed] [Google Scholar]
- Kühn C (2003) A comparison of the sucrose transporter systems of different plant species. Plant Biol 5 215–232 [Google Scholar]
- Kühn C, Franceschi VR, Schulz A, Lemoine R, Frommer WB (1997) Macromolecular trafficking indicated by localization and turnover of sucrose transporters in enucleate sieve elements. Science 275 1298–1300 [DOI] [PubMed] [Google Scholar]
- Kumar R, Wallis JG, Skidmore C, Browse J (2006) A mutation in Arabidopsis cytochrome b5 reductase identified by high-throughput screening differentially affects hydroxylation and desaturation. Plant J 48 920–932 [DOI] [PubMed] [Google Scholar]
- Lalonde S, Boles E, Hellmann H, Barker L, Patrick JW, Frommer WB, Ward JM (1999) The dual function of sugar carriers: transport and sugar sensing. Plant Cell 11 707–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde S, Wipf D, Frommer WB (2004) Transport mechanisms for organic forms of carbon and nitrogen between source and sink. Annu Rev Plant Biol 55 341–372 [DOI] [PubMed] [Google Scholar]
- Lemaire K, Van de Velde S, Van Dijck P, Thevelein JM (2004) Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol Cell 16 293–299 [DOI] [PubMed] [Google Scholar]
- Leroch M, Neuhans HE, Kirchberger S, Zimmermann S, Melzer M, Gerhold J, Tjaden J (2008) Identification of a novel adenine nucleotide transporter in the endoplasmic reticulum of Arabidopsis. Plant Cell 20 438–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CY, Shi JX, Weiss D, Goldschmidt EE (2003. a) Sugars regulate sucrose transporter gene expression in citrus. Biochem Biophys Res Commun 306 402–407 [DOI] [PubMed] [Google Scholar]
- Li CY, Weiss D, Goldschmidt EE (2003. b) Effects of carbohydrate starvation on gene expression in citrus root. Planta 217 11–20 [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Roberts TH, Hirschberg K (2000) Secretory protein trafficking and organelle dynamics in living cells. Annu Rev Cell Dev Biol 16 557–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CA, Lin CC, Lee KW, Chen JL, Huang LF, Ho SL, Liu HJ, Hsing YL, Yu SM (2007) The SnRK1A protein kinase plays a key role in sugar signaling during germination and seedling growth of rice. Plant Cell 19 2484–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggio C, Barbante A, Ferro F, Frigerio L, Pedrazzini E (2007) Intracellular sorting of the tail-anchored protein cytochrome b5 in plants: a comparative study using different isoforms from rabbit and Arabidopsis. J Exp Bot 58 1365–1379 [DOI] [PubMed] [Google Scholar]
- Marciniak SJ, Ron D (2006) Endoplasmic reticulum stress signaling in disease. Physiol Rev 86 1133–1149 [DOI] [PubMed] [Google Scholar]
- Marger MD, Saier JMH (1993) A major super-family of transmembrane facilitators that catalyze uniport, symport and antiport. Trends Biochem Sci 18 13–20 [DOI] [PubMed] [Google Scholar]
- Matsukura C, Saitoh T, Hirose T, Ohsugi R, Perata P, Yamaguchi J (2000) Sugar uptake and transport in rice embryo: expression of companion cell-specific sucrose transporters (OsSUT1) induced by sugar and light. Plant Physiol 124 85–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J (2003) Roles of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300 332–336 [DOI] [PubMed] [Google Scholar]
- Moreno F, Ahuatzi D, Riera A, Palomino CA, Herrero P (2005) Glucose sensing through the Hxk2-dependent signaling pathway. Biochem Soc Trans 33 265–268 [DOI] [PubMed] [Google Scholar]
- Nam KH, Li J (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110 203–212 [DOI] [PubMed] [Google Scholar]
- Noiraud N, Delrot S, Lemoine R (2000) The sucrose transporter of celery: identification and expression during salt stress. Plant Physiol 122 1447–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noiraud N, Maurousset L, Lemoine R (2001) Identification of a mannitol transporter, AgMat1, in celery phloem. Plant Cell 13 695–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira J, Tavares RM, Geros H (2002) Utilization and transport of glucose in Olea europaea cell suspensions. Plant Cell Physiol 43 1510–1517 [DOI] [PubMed] [Google Scholar]
- Patrick JW (1997) Phloem unloading: sieve element unloading and post-sieve element transport. Annu Rev Plant Physiol Plant Mol Biol 48 191–222 [DOI] [PubMed] [Google Scholar]
- Ramer SW, Elledge SJ, Davis RW (1992) Dominant genetics using a yeast genomic library under the control of a strong inducible promoter. Proc Natl Acad Sci USA 89 11589–11593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsperger-Gleixner M, Geiger D, Hedrich R, Sauer N (2004) Differential expression of sucrose transporter and polyol transporter genes during maturation of common plantain companion cells. Plant Physiol 134 147–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransom-Hodgkins R, Vaughn MW, Bush DR (2003) Protein phosphorylation plays a key role in sucrose-mediated transcriptional regulation of a phloem-specific proton-sucrose symporter. Planta 217 483–489 [DOI] [PubMed] [Google Scholar]
- Reinders A, Panshyshyn JA, Ward JM (2005) Analysis of transport activity of Arabidopsis sugar alcohol permease homolog AtPLT5. J Biol Chem 280 1594–1602 [DOI] [PubMed] [Google Scholar]
- Reinders A, Silvitz AB, Starker CG, Gantt JS, Ward JM (2008) Functional analysis of LjSUT4, a vacuolar sucrose transporter from Lotus japonicus. Plant Mol Biol 68 289–299 [DOI] [PubMed] [Google Scholar]
- Reinders A, Thaminy S, Stagljar I, Frommer WB, Ward JM (2002) Intra- and intermolecular interactions in sucrose transporters at the plasma membrane detected by the split-ubiquitin system and functional assays. Structure 10 763–772 [DOI] [PubMed] [Google Scholar]
- Rentsch D, Boorer KJ, Frommer WB (1998) Structure and function of plasma membrane amino acid, oligopeptide and sucrose transporters from higher plants. J Membr Biol 162 177–190 [DOI] [PubMed] [Google Scholar]
- Rentsch D, Laloi M, Rouhara I, Schmelzer E, Delrot S, Frommer WB (1995) NTR1 encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Lett 370 264–268 [DOI] [PubMed] [Google Scholar]
- Riesmeier JW, Willmitzer L, Frommer WB (1992) Isolation and characterization of a sucrose carrier cDNA from spinach by functional expression in yeast. EMBO J 11 4705–4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitsch T (1999) Source-sink regulation by sugar and stress. Curr Opin Plant Biol 2 198–206 [DOI] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J (2006) Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol 57 675–709 [DOI] [PubMed] [Google Scholar]
- Rolland F, Moore B, Sheen J (2002. a) Sugar sensing and signaling in plants. Plant Cell (Suppl) 14: S185–S205 [DOI] [PMC free article] [PubMed]
- Rolland F, Winderickx J, Thevelein JM (2001) Glucose-sensing mechanisms in eukaryotic cells. Trends Biochem Sci 26 310–317 [DOI] [PubMed] [Google Scholar]
- Rolland F, Winderickx J, Thevelein JM (2002. b) Glucose-sensing and -signaling mechanisms in yeast. FEMS Yeast Res 2 183–201 [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P (1990) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 119–187
- Sambrook J, Fritsch EF, Maniatis T (1989). Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sauer N, Baier K, Gahrtz M, Stadler R, Stolz J, Truernit E (1994) Sugar transport across the plasma membranes of higher plants. Plant Mol Biol 26 1671–1679 [DOI] [PubMed] [Google Scholar]
- Sauer N, Stadler R (1993) A sink-specific H+/monosaccharide co-transporter from Nicotiana tabacum: cloning and heterologous expression in baker's yeast. Plant J 4 601–610 [DOI] [PubMed] [Google Scholar]
- Sauer N, Tanner W (1993) Molecular biology of transporters in plants. Bot Acta 106 277–286 [Google Scholar]
- Schenkman JB, Jansson I (2003) The many roles of cytochrome b5. Pharmacol Ther 97 139–152 [DOI] [PubMed] [Google Scholar]
- Stadler R, Buttner M, Ache P, Hedrich R, Ivashikina N, Melzer M, Shearson SM, Smith SM, Sauer N (2003) Diurnal and light-regulated expression of AtSTP1 in guard cells of Arabidopsis. Plant Physiol 133 528–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagljar I, Heesen S (2000) Detecting interactions between membrane proteins in vivo using chimeras. Methods Enzymol 327 190–198 [DOI] [PubMed] [Google Scholar]
- Stagljar I, Korostensky C, Johnsson N, Heesen S (1998) A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc Natl Acad Sci USA 95 5187–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E, Schmid J, Epple P, Illig J, Sauer N (1996) The sink-specific and stress-regulated Arabidopsis STP4 gene: enhanced expression of a gene encoding a monosaccharide transporter by wounding, elicitors, and pathogen challenge. Plant Cell 8 2169–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Yamaguchi M, Uchimiya H, Nakano A (2001) Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis. EMBO J 20 4730–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn MW, Harrington GN, Bush DR (2002) Sucrose-mediated transcriptional regulation of sucrose symporter activity in the phloem. Proc Natl Acad Sci USA 99 10876–10880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetten ND, Horst JT, Van Schaik HP, Boer AD, Mol J, Koes R (1999) A cytochrome b5 is required for full activity of flavonoid 3′,5′-hydroxylase, a cytochrome P450 involved in the formation of blue flower colors. Proc Natl Acad Sci USA 96 778–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40 428–438 [DOI] [PubMed] [Google Scholar]
- Wang HX, Weerasinghe RR, Perdue TD, Cakmakci NG, Taylor JP, Marzluff WF, Jones AM (2006) A Golgi-localized hexose transporter is involved in heterotrimeric G protein-mediated early development in Arabidopsis. Mol Biol Cell 17 4257–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watari J, Kobae Y, Yamaki S, Yamada K, Toyofuku K, Tabuchi T, Shiratake K (2004) Identification of sorbitol transporters expressed in the phloem of apple source leaves. Plant Cell Physiol 45 1032–1041 [DOI] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Heim U, Sauer N, Wobus U (1997) A role for sugar transporters during seed development: molecular characterization of a hexose and a sucrose carrier in faba bean seeds. Plant Cell 9 895–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise A, Barker L, Kühn C, Lalonde S, Buschmann H, Frommer WB, Ward JM (2000) A new subfamily of sucrose transporters, SUT4, with low affinity/high capacity localized in enucleate sieve elements of plants. Plant Cell 12 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LE, Lemoine R, Sauer N (2000) Sugar transporters in higher plants: a diversity of roles and complex regulation. Trends Plant Sci 5 283–290 [DOI] [PubMed] [Google Scholar]
- Wu GL, Zhang XY, Zhang LY, Pan QH, Shen YY, Zhang DP (2004) Phloem unloading in developing walnut fruit is symplasmic in the seed pericarp and apoplasmic in the fleshy pericarp. Plant Cell Physiol 45 1461–1470 [DOI] [PubMed] [Google Scholar]
- Xu C, Bailly-Maitre B, Reed JC (2005) Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest 115 2656–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S, Yoo SD, Sheen J (2003) Differential regulation of EIN3 stability by glucose and ethylene signaling in plants. Nature 425 521–525 [DOI] [PubMed] [Google Scholar]
- Zhang LY, Peng YB, Pelleschi ST, Fan Y, Lu YF, Lu YM, Gao XP, Shen YY, Delrot S, Zhang DP (2004) Evidence for apoplasmic phloem unloading in developing apple fruit. Plant Physiol 135 574–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XY, Wang XL, Wang XF, Xia GH, Pan QH, Fan RC, Wu FQ, Yu XC, Zhang DP (2006) A shift of phloem unloading from symplasmic to apoplasmic pathway is involved in developmental onset of ripening in grape berry. Plant Physiol 142 220–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Onduka T, Kinoshita J, Honsho M, Kinoshita T, Shimazaki K, Ito A (2003) Dual subcellular distribution of cytochrome b5 in plant, cauliflower, cells. J Biochem 133 115–121 [DOI] [PubMed] [Google Scholar]
- Zhou L, Jang JC, Jones TL, Sheen J (1998) Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc Natl Acad Sci USA 95 10294–10299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu SY, Yu XC, Wang XJ, Zhao R, Li Y, Fan RC, Shang Y, Du SY, Wang XF, Wu FQ, et al (2007) Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19 3019–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.